Abstract
Adaptive mutations are mutations that occur in nondividing or very slowly dividing microbial cells during prolonged nonlethal selection and that are specific to the challenge of the selection in the sense that the only mutations that can be detected are those that provide a growth advantage to the cell. The phoPQ genes encode a two-component positively acting regulatory system that controls expression of at least 25 to 30 genes in Escherichia coli and Salmonella typhimurium. PhoPQ responds to a variety of environmental stress signals including Mg2+ starvation and nutritional deprivation. Here I show that disruption of phoP or phoQ by Tn10dCam significantly reduces the adaptive mutation rate to ebgR, indicating that the adaptive mutagenesis machinery is regulated, directly or indirectly, by phoPQ. The finding that it is regulated implies that adaptive mutagenesis does not simply result from a failure of various error correction mechanisms during prolonged starvation.
Adaptive mutations differ from growth-dependent mutations in two key respects. First, adaptive mutations occur in nondividing or slowly dividing cells which are under selection for a particular phenotype, whereas growth-dependent mutations occur in dividing cells that are not under strong selection. Second, adaptive mutations produce only those phenotypes which allow the cells to grow (13), whereas growth-dependent mutations occur randomly with respect to their effects on fitness. The mechanisms for producing adaptive mutations remain unknown although numerous speculative models have been proposed.
Adaptive mutations are usually observed by spreading a population of bacteria or yeast onto medium upon which growth cannot occur unless a known mutation reverts. The first revertants to appear are presumed to be the result of mutations that were present in the population prior to plating. Typically, additional revertant colonies continue appear for periods of several days up to a month, and it is those late-appearing colonies that are said to result from adaptive mutations.
A variety of experiments have shown that adaptive mutations are specifically beneficial. The clearest demonstration of the specificity of adaptive mutations showed that when selection was applied to a specific nucleotide in the ebgA gene to permit lactulose utilization, mutations at that nucleotide accumulated over the course of several days, but in that same population no mutations accumulated at another, equally mutable nucleotide in the same gene where mutations permitted utilization of lactose but not of lactulose (13).
The process or processes that produce adaptive mutations are not well understood. The most widely used system, reversion of the F′-borne lacI33 frameshift mutation during selection for growth on lactose, led to the conclusion that genes whose products are involved in recombination are necessary for adaptive mutagenesis (1, 15). That conclusion has been cast into doubt by the finding that when the lacI33 frameshift allele is located on the chromosome, rather than on the F′ episome, adaptive reversion is not affected by lesions in recombination genes (4).
In an effort to identify the genes that are involved in adaptive mutagenesis, I have conducted a large-scale mutant hunt. Here I report that disruption of phoP or phoQ, which together encode a global two-component regulatory system, significantly reduces adaptive mutagenesis.
MATERIALS AND METHODS
Organisms.
Escherichia coli K-12 strain SJ134 is F− ΔlacZ4680 lacY+ ebgR+ ebgA51 rpsL. E. coli K-12 strain SJ2 is F− ΔlacZ4680 lacY+ ebgR+ ebgA+ rpsL metC. E. coli K-12 strain FCY2 is F− trpA46 (10). Bacteriophage λ strain NK1324 is Tn10dCam cI857 Pam80 nin5 b522 att− (18).
Media.
The phosphate-buffered minimal medium has been previously described (13). Glucose medium contained 2 g of glucose per liter. Lactulose minimal medium contained 1 g of lactulose (4-O-β-d-galactopyranosyl-d-fructofuranose) per liter, 20 mg of X-Gal (5-bromo-4-chloro-3-indolyl β-galactoside) per liter, and 2 × 10−4 M IPTG (isopropyl-β-d-thiogalactopyranoside). TAD medium (11) contained 5 μM tryptophan, a concentration that limits trpA46 strains to grow to about 109 cells per plate. Media were solidified with 15 g of purified agar (Sigma) per liter. L agar consisted of LB agar (20) plus 1 g of glucose per liter. MacConkey lactose medium was prepared from MacConkey agar base (Difco) according to the manufacturer’s instructions and contained 2 × 10−4 M IPTG.
Identification of Tn10dCam insertion mutants.
Strain SJ134 was infected with λNK1324 as described by Kleckner (18), and chloramphenicol-resistant colonies were selected on glucose minimal medium containing 15 μg of chloramphenicol per ml. Individual colonies were grown as 0.5-ml cultures in minimal medium containing 0.1 g of glucose per liter to limit cell density to about 108/ml, dimethyl sulfoxide was added to 10% (vol/vol), and the cultures were stored at −80°C; 100 μl of each culture was spread onto a lactulose minimal medium plate, and the plates were incubated at 30°C. Four days and seven days after inoculation, any ebgR− (lactulose-positive) mutant colonies were marked. Typical cultures produced 5 to 10 colonies on day 4 and by day 7 had accumulated 60 to 80 colonies. Cultures that exhibited unusually high or unusually low (including zero) numbers of colonies were retested. Those that exhibited the same unusual numbers were purified by restreaking, and fresh cultures were again tested. Those that again reproduced the unusual numbers were retained for further study. Isolates that failed to generate any ebgR mutants within 7 days were streaked onto MacConkey lactose-IPTG medium. Lactose, although a weak inducer of ebgR+, permits sufficient synthesis of Ebg β-galactosidase to produce light red colonies in MacConkey lactose, while those in which ebgAC, lacY, or any other gene required for transport and hydrolysis of β-galactoside sugars has been disrupted produce white colonies. Cultures producing white colonies were discarded.
Reconstruction tests.
Reconstruction tests were used to determine the time required for an ebgR mutant cell to form a visible colony on lactulose minimal medium. Typically three independent ebgR mutants that had arisen on different days from a particular Tn10dCam mutant were tested. About 100 cells of the ebgR mutant were plated onto lactulose minimal medium in the presence and in the absence of about 108 strain SJ2 scavenger cells. Because strain SJ2 is both ebgR+ and ebgA+, two mutations are required in order for that strain to utilize lactulose, and such double mutants do not arise prior to 2 weeks of incubation (12). The population of SJ2 cells mimics the situation when rare ebgR mutants arise within a population of the parent cells. The time required for approximately the same number of colonies to appear on the plates with SJ2 cells and without SJ2 cells is taken as the typical time required for that strain to form visible ebgR mutant colonies.
RESULTS
A library of 19,616 independent mini-Tn10 (chloramphenicol) (Tn10dCam) insertions into random sites in the chromosome of E. coli K-12 strain SJ134 was screened to determine the effect of the Tn10dCam insertion on adaptive mutagenesis (Materials and Methods).
Instead of monitoring adaptive mutagenesis by selecting for reversion of a known mutation, the forward (loss-of-function) mutation rate at ebgR, a gene that specifies a repressor which controls expression of the ebgAC-encoded Ebg β-galactosidase (8), was monitored. The advantages of using ebgR as a reporter locus are that (i) adaptive mutation at ebgR has already been demonstrated to occur (12) and (ii) selecting for loss of function makes it more likely that any genes so identified will affect adaptive mutations that occur by a variety of events (base substitutions, frameshifts, insertions, etc.), rather than affecting just a single class of mutations as is common in reversion studies.
Strain SJ134 and its derivatives carry the ebgA51 allele. ebgA encodes the α subunit of the Ebg β-galactosidase, but the wild-type Ebg enzyme does not hydrolyze the β-galactoside sugar lactulose at a rate that permits utilization of lactulose as a sole carbon and energy source. The ebgA51 allele alters the kinetic properties of Ebg enzyme to permit effective lactulose hydrolysis (9). ebgA expression is under control of the ebgR-encoded repressor that does not respond to lactulose as an inducer (8); thus, ebgR− mutants of strain SJ134 can utilize lactulose as a sole carbon and energy source.
The mutant hunt has led to the identification of four genes in which Tn10dCam insertions increase the adaptive mutation rate and seven genes in which the insertions decrease the adaptive mutation rate. The identities and characterizations of the genes other than those discussed below will be reported elsewhere.
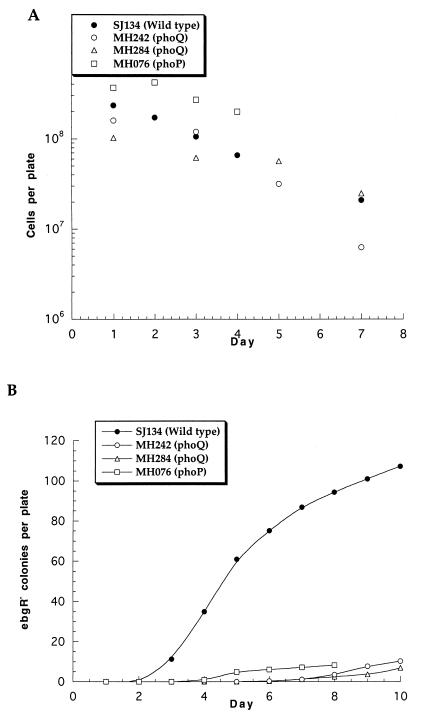
To measure the adaptive mutation rate at ebgR approximately 107 cells are spread onto lactulose minimal medium at 30°C (12). The populations grow at the expense of trace contaminants to a density of about 108 cells per plate, after which the population declines slowly (Fig. 1A). ebgR colonies appear beginning 3 days after inoculation, and they continue to accumulate for the next several days (Fig. 1B). Each day for 4 days, cells were washed off of two plates, suitably diluted, and plated onto L agar to determine the number of viable cells per plate. Once ebgR colonies began to appear, on a subset of plates those colonies were immediately eliminated, with little disturbance of surrounding cells, through the use of a diathermy probe (Hyfrecator Plus model 7-796; Birtcher Medical Supplies), an electrosurgery device that delivers an intense spark which kills the cells in the colony. Those treated plates were used to estimate the number of viable cells.
FIG. 1.
(A) Population densities on lactulose minimal medium as a function of time at 30°C; (B) appearance of ebgR mutant colonies on lactulose minimal medium during incubation at 30°C.
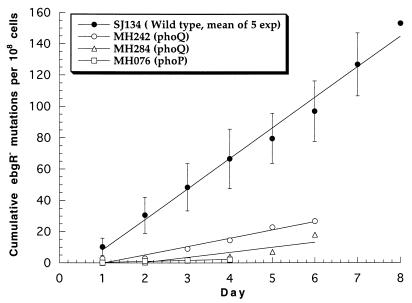
Reconstruction tests showed that ebgR mutants of the wild-type strain SJ134 require 3 days to form detectable colonies on lactulose medium. The plating efficiencies of the ebgR mutants in the presence and absence of the scavenger strain SJ2 are indistinguishable. Thus, colonies that appear on day 3 are the result of mutations that were present in the initial population, those that appear on day 4 are the result of mutations that occurred in the population that was present on day 1, etc. The mutation rate each day is the average number of new mutant colonies that appeared 3 days later divided by the number of cells on the plate on that day. Figure 2 shows the cumulative mutations per 108 cells, and the slope of the best-fit line is the average mutation rate over the course of the experiment. For the wild-type strain SJ134, the average mutation rate to ebgR, over five independent experiments, was (1.9 ± 0.2) × 10−7 per cell per day (Fig. 2).
FIG. 2.
Cumulative ebgR mutations per viable cell during selection on lactulose minimal medium at 30°C. Symbols indicate the days on which the mutations occurred. Mutant colonies appeared either 3 days (SJ134) or 4 days (other strains) after mutations occurred (see text). Lines are least-squares fit to the data points. For strain SJ134, symbols indicate mean values for five independent experiments and error bars indicate the standard errors of those values.
Three Tn10dCam mutants, MH076, MH242, and MH284, yielded mutants at much lower rates than did the wild-type strain, although the population densities were comparable to those of the wild-type strain (Fig. 1). Reconstruction tests showed that ebgR mutants of each of the three Tn10dCam mutants grew slightly more slowly than ebgR mutants of the wild-type strain, requiring 4 days to form visible colonies. The mutation rates to ebgR, based on 4 days for visible colonies to appear, were 6.3 × 10−9, 4.8 × 10−8, and 3.2 × 10−8 per cell per day for MH076, MH242, and MH284, respectively.
Identities of the genes that were disrupted by Tn10dCam were determined by cloning Tn10dCam and its flanking regions into plasmid pBluescript KS+ (Stratagene) and sequencing. Genomic DNA was digested with one of several restriction enzymes that do not cut within Tn10dCam, ligated into similarly cut plasmid, and transformed into strain JM109 by electroporation, and transformants were selected on L agar containing 25 mg of chloramphenicol per liter. Plasmid DNA was sequenced on an ABI model 377 automated sequencing apparatus, using primers corresponding to bases 78 to 102 or 1359 to 1383 of Tn10dCam. Those primers allowed sequencing of about 90 bp of Tn10dCam outward into the region flanking the insertion. In strain MH076, Tn10dCam was inserted into phoP at bp 676 of the ECOPHOP (Genbank accession no. M81433) sequence. In strains MH242 and MH284, Tn10dCam was inserted into phoQ at bp 2176 and 3029, respectively, of ECOPHOP.
Although the effects of phoP and phoQ cannot be accounted for on the basis of slow growth, it is possible that the disruptions affect the probability that a newly arisen mutant will survive long enough to begin to grow. The Tn10dCam::phoP and Tn10dCam::phoQ alleles of strains MH076 and MH284 were transduced into strain SJ2 (metC ebgR+ ebgA51). To mimic the effect of a newly arisen ebgR mutant allele in a carbon-starved population, a known ebgR ebgA51 allele was transduced into cultures of SJ2 and the phoP and phoQ derivatives that had been carbon starved for 3 days. The transduced populations were plated onto lactulose-methionine medium to select ebgR transductants and onto methionine-free glucose medium to select metC+ transductants. The recovery of newly arisen ebgR mutants was expressed as the transduction efficiency of ebgR selected on lactulose medium relative to efficiency of metC+ selected on methionine-free medium. For strain SJ2, the relative transduction efficiency was 0.19. For the phoP and phoQ derivatives, the relative efficiencies were 0.23 and 0.19, respectively; i.e., newly arisen ebgR alleles were recovered as efficiently in phoPQ as in wild-type backgrounds. Thus, the observed effects on adaptive mutagenesis cannot be attributed either to slow growth or inefficient recovery of mutants, and disruption of phoP or phoQ must affect adaptive mutagenesis itself.
The observation that three independent Tn10dCam disruptions of the phoP and phoQ genes produce the same adaptive mutation phenotype suggests that it is those insertions themselves, not some possible accompanying random unidentified mutation, that is responsible for the phenotype. The Tn10dCam::phoQ allele of strain MH284 was transduced back into the parent strain, SJ134, and the adaptive mutation rate at ebgR was measured. That rate was 4.2 × 10−8 per cell per day, not very different from the rate in MH284 itself, confirming that it is disruption of phoPQ that is responsible for the reduced adaptive mutation rate at ebgR.
Fluctuation tests (19) were used to determine whether disruption of phoPQ affects growth-dependent, as well as adaptive, mutagenesis at ebgR. Independent 200-μl cultures of the wild-type strain SJ134 and of MH284 (Tn10dCam::phoQ) were grown in 0.01% (vol/vol) glycerol to yield populations of about 2 × 107 cells per culture. Five cultures were suitably diluted and plated onto L agar to determine the number of viable cells per culture, and each remaining culture was spread onto a lactulose selection plate. SJ134 colonies were counted 3 days and MH284 colonies were counted after 4 days incubation at 30°C. The Stewart et al. method for analysis of fluctuation tests (23), as implemented by Stewart’s DataFit program, permits one to estimate both the average number of mutations that occurred prior to plating during the growth of each culture and the average number of mutations that occurred after plating. For strain SJ134, 127 cultures at 2.4 × 107 cells per culture produced an average of 1.34 mutations per culture prior to plating to give a growth-dependent mutation rate of 5.6 × 10−8 per cell division. For strain MH284, 40 cultures at an average of 1.5 × 107 cells per culture produced an average of 0.7 mutation per culture prior to plating, to give a growth-dependent mutation rate of 4.7 × 10−8 mutations per cell division. Thus, disruption of phoQ affects adaptive, but not growth-dependent, mutation at ebgR.
To determine whether disruption of phoPQ would affect adaptive mutagenesis at other loci, adaptive reversion of a trpA46 missense mutation was examined, and adaptive activation of the bgl operon was examined. The adaptive mutation spectra at both ebgR (14a) and bgl (14) are dominated by insertion element mediated mutations. The Tn10dCam::phoQ allele of strain MH284 was transduced into strain FCY2 (trpA46), and the adaptive reversion rates of trpA46 in strain FCY2 and in FCY2MH284 were compared on TAD medium by monitoring the rate at which Trp+ colonies appeared on the trpA46 lawns in a manner comparable to that used to measure adaptive mutation rates at ebgR. In strain FCY2 the adaptive reversion rate of trpA46 was 6 × 10−10 per cell per day, whereas it was 6.6 × 10−10 per cell per day in the Tn10dCam::phoQ derivative, strain FCY2MH284.
The adaptive mutation rate at bgl was measured in strain MH284 on arbutin selective medium as previously described (14). The bgl adaptive mutation rate in strain SJ134 is 3.4 × 10−7 mutations per cell per day (14). In strain MH284 that rate was 2.9 × 10−7 mutations per cell per day, not very different from the wild-type rate.
DISCUSSION
Disruption of phoPQ reduces the adaptive mutation rate at ebgR by about a factor of 6 but has no effect on the growth-dependent mutation rate at the same locus. That reduction cannot be accounted for either by slow growth or by reduced efficiency of recovery of ebgR mutants under the selective conditions used. At the same time, disruption of phoQ has no effect on the adaptive mutation rate at either trpA or bgl. The different responses of the two adaptive mutagenesis reporter systems is consistent with the idea that there are multiple systems for adaptive mutagenesis and that adaptive mutagenesis at ebgR is affected by a system different from those affecting trpA and bgl. The difference is surprising because adaptive mutagenesis at both ebgR and bgl is primarily mediated by transposition of insertions elements.
The phoQ gene encodes the sensor kinase component, and phoP encodes the positive-regulatory component of a two-component regulatory system PhoP/PhoQ that controls expression of at least 30 loci in Salmonella typhimurium (21). PhoP, initially described by virtue of its involvement in expression of phoN-encoded acid phosphatase (17), is necessary for resistance to defensins, antimicrobial peptides found in macrophages (2, 6). PhoP/PhoQ-activated genes are regulated in response to Mg2+ levels (21), and the PhoQ protein includes a Mg2+ binding site (24). The system responds to a wide variety of environmental stress signals; indeed, Groisman et al. (5) point out the “involvement of PhoP in response to stress situations such as those which might be present during stationary phase, when a microorganism may face nutritional deprivation, exposure to toxic by-products of metabolism, or both.” PhoP/PhoQ regulation is both direct and indirect in that PhoP/PhoQ activates transcription of pmrAB (7), which in turn regulates expression of at least seven of the Salmonella genes that are under PhoP/PhoQ control (22).
In E. coli at least 50 genes are under control of PhoP/PhoQ (16), including genes for resistance to the antimicrobial peptide Magainin 2 (5). Several genes that are induced by starvation for carbon, nitrogen, or phosphorus are thought to be under PhoP/PhoQ control, as are the pex genes which are involved in development of the resistant state during entry into stationary phase (5).
Because PhoP/PhoQ is a global regulator that responds to a wide variety of environmental signals, particularly including starvation, and because disruption of phoP or phoQ significantly reduces the adaptive mutation rate to ebgR, it seems likely that adaptive mutagenesis is a regulated process that is subject to positive control by PhoPQ.
Adaptive mutations at ebgR appear to require the expression of some gene or genes under control of PhoPQ. This finding is consistent with the view that adaptive mutagenesis is a programmed response to prolonged environmental stress in which a portion of the stressed population transiently enters a hypermutable state as a last desperate effort to find a genetic solution to the current environmental problem (3, 10).
ACKNOWLEDGMENTS
I am grateful to the American Cancer Society (grant NP-932) for supporting this work.
I thank Jacqueline Toner for expert technical assistance.
REFERENCES
- 1.Cairns J, Foster P L. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fields P I, Groisman E A, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 3.Foster P L. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster P L, Trimarchi J M. Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc Natl Acad Sci USA. 1995;92:5487–5490. doi: 10.1073/pnas.92.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groisman E A, Heffron F, Soloman F. Molecular genetic analysis of the Escherichia coli phoP locus. J Bacteriol. 1992;174:486–491. doi: 10.1128/jb.174.2.486-491.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groisman E A, Parra-Lopez C, Salcedo M, Lipps C J, Hefron F. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11939–11943. doi: 10.1073/pnas.89.24.11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunn J S, Miller S I. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall B G. Regulation of newly evolved enzymes. IV. Directed evolution of the ebg repressor. Genetics. 1978;90:673–691. doi: 10.1093/genetics/90.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall B G. Changes in the substrate specificities of an enzyme during directed evolution of new functions. Biochemistry. 1981;20:4042–4049. doi: 10.1021/bi00517a015. [DOI] [PubMed] [Google Scholar]
- 10.Hall B G. Spontaneous point mutations that occur more often when they are advantageous than when they are neutral. Genetics. 1990;126:5–16. doi: 10.1093/genetics/126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall B G. Adaptive evolution that requires multiple spontaneous mutations: mutations involving base substitutions. Proc Natl Acad Sci USA. 1991;88:5882–5886. doi: 10.1073/pnas.88.13.5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall B G. Adaptive mutations in E. coli as a model for the multiple-mutational origins of tumors. Proc Natl Acad Sci USA. 1995;92:5669–5673. doi: 10.1073/pnas.92.12.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall B G. On the specificity of adaptive mutations. Genetics. 1997;145:39–44. doi: 10.1093/genetics/145.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall B G. Activation of the bgl operon by adaptive mutation. Mol Biol Evol. 1998;15:1–5. doi: 10.1093/oxfordjournals.molbev.a025842. [DOI] [PubMed] [Google Scholar]
- 14a.Hall, B. G. Unpublished data.
- 15.Harris R H, Longerich S, Rosenberg S M. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 16.Kasahara M, Nakata A, Shinagawa H. Molecular analysis of the Escherichia coli phoP-phoQ operon. J Bacteriol. 1992;174:492–498. doi: 10.1128/jb.174.2.492-498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kier L D, Weppleman R M, Ames B N. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J Bacteriol. 1979;138:155–161. doi: 10.1128/jb.138.1.155-161.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 19.Luria S E, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 21.Soncini F C, Véscovi E G, Solomon F, Groisman E. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soncini F C, Groisman E A. Two-component regulatory signals can interact to process multiple environmental signals. J Bacteriol. 1996;178:6796–6801. doi: 10.1128/jb.178.23.6796-6801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart F M, Gordon D M, Levin B R. Fluctuation analysis: the probability distribution of the number of mutants under different conditions. Genetics. 1990;124:175–185. doi: 10.1093/genetics/124.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Véscovi E G, Ayala Y M, Di Cera E, Groisman E A. Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+ J Biol Chem. 1997;272:1440–1443. doi: 10.1074/jbc.272.3.1440. [DOI] [PubMed] [Google Scholar]