Abstract
Nociception and pain sensation are important neural processes in humans to avoid injury. Many proteins are involved in nociception and pain sensation in humans; however, the evolution of these proteins in animals is unknown. Here, we chose nociception- and pain-related proteins, including G protein-coupled receptors (GPCRs), ion channels (ICs), and neuropeptides (NPs), which are reportedly associated with nociception and pain in humans, and identified their homologs in various animals by BLAST, phylogenetic analysis and protein architecture comparison to reveal their evolution from protozoans to humans. We found that the homologs of transient receptor potential channel A 1 (TRPA1), TRAPM, acid-sensing IC (ASIC), and voltage-dependent calcium channel (VDCC) first appear in Porifera. Substance-P receptor 1 (TACR1) emerged from Coelenterata. Somatostatin receptor type 2 (SSTR2), TRPV1 and voltage-dependent sodium channels (VDSC) appear in Platyhelminthes. Calcitonin gene-related peptide receptor (CGRPR) was first identified in Nematoda. However, opioid receptors (OPRs) and most NPs were discovered only in vertebrates and exist from agnatha to humans. The results demonstrated that homologs of nociception and pain-related ICs exist from lower animal phyla to high animal phyla, and that most of the GPCRs originate from low to high phyla sequentially, whereas OPRs and NPs are newly evolved in vertebrates, which provides hints of the evolution of nociception and pain-related proteins in animals and humans.
Keywords: Nociception, pain, GPCRs, transient receptor potential, ion channels, neuropeptides
Introduction
Nociception is defined as the ability to detect stimuli that elicit damage to the body or the potential for such damage, 1 and pain is defined as “an unpleasant sensory and emotional experience associated with actual or potential tissue damage, or described in terms of such damage” by the International Association for the Study of Pain (IASP).2,3 The definition of pain emphasizes subjectivity and a conscious experience. Nociception and pain are closely related processes. Activation of nociceptors and nociceptive pathways by noxious and partially harmful stimuli can give rise to pain, and the pain signal can be sent to the central nervous system, where perception of pain could occur. Nociception and pain in humans are expressed via language and various behaviors, such as defense, protection, avoidance, or aversion. Nonverbal animals express nociception via behavior in a species-specific manner.4,5 For example, the large marine snail Aplysia californica withdraws its siphon in response to tactile stimuli, and this reflex can be facilitated (sensitized) by training the animal and can last for up to 3 weeks. 6 In Drosophila larvae, menthol stimulation induces nocifensive rolling behavior, 7 and amputating the leg of an adult Drosophila was observed to cause allodynia-like mechanical hypersensitivity, lasting for at least 3 weeks. 7 The lepidopteran insect Manduca sexta reduced its threshold for defensive responses to punctate mechanical stimuli 2 h after the ingestion or injection of heat-killed bacteria. 8 The molecular evolution of nociception and pain in vertebrates and invertebrates is unclear.
The neural substrate for nociception and pain perception in the vertebrate begins in the activation of peripheral sensory neurons with their cell bodies located in the dorsal root ganglion (DRG), which transmit the nociceptive signal to the spinal cord dorsal horn and further downstream to various brain regions. Many molecules and their receptors are involved in nociception and pain. For example, hundreds of mammalian genes are reported to be involved in the mediation of nociception and pain,9 -11 and some of the key proteins were chosen here, such as G protein-coupled receptors (GPCRs), neuropeptides (NPs) and ion channels (ICs). GPCRs and their ligands, mostly neuropeptides, play important roles in transducing nociceptive and pain signals. One example is preprotachykinin receptor 1 (TACR1) and its ligand preprotachykinin 1 (TAC1). TACR1 is also named Neurokinin-1 (NK-1) receptor or substance-P receptor, which binds Substance P (SP)/neurokinin 1 (NK1). 12 A range of somatotopically directed pain-related behaviors are observed upon activation of spinal cord neurons that express TACR1 in the absence of noxious input. 13 TAC1+ spinal cord neurons are required for pain-related coping responses in mice. 14 Another is somatostatin receptor type 2 (SSTR2), one of the receptors of somatostatin (SST). SST-expressing interneurons in the dorsal horn of the spinal cord are specifically implicated in mediating mechanical pain. 15 SST potently inhibits neuropathic pain by activating SSTR2 in mouse dorsal root ganglia (DRG). 16 In addition, calcitonin receptor-like receptor (CGRPR, or CALCRL, CRLR, LMPHM8 in the genome) is the receptor of the NP calcitonin gene-related peptide (CGRP, also known as CALCA, CT, KC, PCT, CALC1, CGRP1, CGRP-I, CGRP-alpha). CGRP is found in sensory fibers arising from the dorsal root and trigeminal ganglia, as well as in the CNS. Growing evidence indicates that CGRP plays a key role in peripheral sensitization and that it is associated with enhanced pain. 17 Most well-known opioid receptors (OPRs), including OPR-μ (OPRM), OPR-κ (OPRK), and OPR-δ (OPRD), bind endogenous opioid peptides, namely, pro-opiomelanocortin (POMC), proenkephalin-A (PENK), and proenkephalin-B (PDYN), respectively. 18 Opioid peptides and receptors reduce responses to painful stimuli and stress, and they also influence reward processing and mood. 19
Other nociception and pain-related NPs have been recently identified. NPs mediate pain sensation in the spinal cord and brain. 7 Neuromedin U (NMU) is the ligand of the GPCR NMUR1. NMU is closely related to stress, nociception, mast cell degranulation, and energy homeostasis.20,21 C-type natriuretic peptide (NPPC, or CNP) is the major natriuretic peptide in the CNS. It potentiates TRPV1 activation and causes thermal hyperalgesia. 22 Cholecystokinin (CCK) antagonizes the antinociceptive function of opioids. 23 Peptide YY (PYY) is a gut hormone that suppresses food intake 24 and reportedly has a hypoalgesic effect on somatic thermal and visceral chemical pain as a pain inhibitor. 25
Ion channels (ICs) include members of multiple gene families, such as transient receptor potential (TRP) channels, acid sensing ion channels (ASICs) involved in the detection of nociceptive signals,26 -28 and voltage gated Na+ channels (VDSC) and voltage gated Ca2+ channels (VDCC) for the propagation and transmission of the pain signal, which are critical for detecting noxious stimuli in primary sensory neurons. 29 TRP channels are involved in pain sensations generated by chemical or thermal stimulation. The TRP channel superfamily includes TRP vanilloid (TRPV), TRP canonical (TRPC), TRP mucolipin (TRPML), TRP polycystin (TRPP), TRP ankyrin (TRPA), and TRP melastatin (TRPM).26,30 -32 Several TRP channels were found to be activated by specific temperatures ranging from noxious hot to cold.33,34 TRPV1 is activated by heat and capsaicin.35 -37 TRPM3 also participates in heat sensation in mice, 38 and TRPM8, known as the cold receptor, is a key target for treating pathological cold hypersensitivity in humans.39,40 TRPA1 serves as a sensor of tissue injury.41,42 TRP channels are also involved in migraine, 43 pain caused by fungal infections, 43 and osteoinflammation. 44
Acid-sensing ICs (ASICs) are important acid sensors; they are involved in neural modulation in the CNS and pain-associated tissue acidosis in the peripheral system. 45 Six isoforms have been identified in mammals: ASIC1a, ASIC1b, ASIC2a, ASIC2b, ASIC3, and ASIC4,46 -48 in which the most interesting protein is ASIC3 in the context of pain, which is predominantly expressed in dorsal root ganglion (DRG) neurons and involved in modulating moderate- to high-intensity pain sensation. 29 ASIC3 senses acid and mechanical stimuli in somatosensory nerves. 49 ASIC1b is selectively expressed in peripheral sensory neurons, 50 which may contribute to peripheral sensitization and pain hypersensitivity. 51
Voltage-dependent sodium channels (VDSCs, or voltage-gated Na+ channels, VGSCs, NaV) are responsible for the initiation and propagation of action potentials in neurons.52,53 The human genome includes 10 VGSC α-subunit genes encoding sodium channel proteins (SCNs). 54 NaV1.7 and NaV1.8 are encoded by SCN9A and SCN10A, 55 respectively, and are involved in trigeminal pain.56,57 Multiple VGSCs are expressed in the DRG. Sensory DRG neurons detect noxious stimuli and transmit this sensory information to regions within the CNS where activity is perceived as pain. The expression and distribution of NaV1.7 and NaV 1.8 in the DRG are altered after peripheral neuropathy.58,59
Voltage-dependent calcium channels (VDCCs) or voltage-gated calcium channels (VGCCs) are responsible for the transmission of pain signals to the central nervous system (CNS). 60 VDCCs comprise 5 subtypes, L-, N-, P/Q-, R-, and T-type. 61 To date, 10 genes in VDCCs have been identified and grouped into 3 families: CaV1, CaV2 and CaV3. 60 Several studies have found that N-type Ca2+ channels (CaV2.2) are critical for the transmission of pain signals at the spinal cord level. 62 VDCC subunit alpha-2/delta-3 isoform (α2δ3) is expressed in primary nociceptors. 63 Sodium and calcium channels in eukaryotes share a common molecular structure in which the major protein subunit is a large polypeptide of approximately 2000 amino acids, consisting of 4 homologous domains, which fold to form the bulk of the channel structure, including the ion conducting pore and the voltage-activated gating machinery. 64
Although these nociception- and pain-related proteins have been identified in humans and other mammals, little is known about the proteins and the evolution of the proteins in lower vertebrates and invertebrates, 65 which limits our understanding of nociception and pain. In the present study, we analyzed various key proteins, including GPCRs, ICs, and NPs, related to the detection, propagation and transmission of nociceptive and pain signals using a diverse range of animal taxa from invertebrates to vertebrates. This study yields interesting insights into the evolution of nociception- and pain-related genes in animals and paves the way for comparative studies of the evolutionary relationship of the proteins and the biology of nociception and pain. Comparative/evolutionary studies can also help identify fundamentally conserved processes of nociception and provide insights into the mechanisms of pathological pain conditions, which promote the development of novel therapeutic strategies to treat pain.
Materials and Methods
Selection of nociception and pain-related proteins
GPCRs, ICs, and NPs related to nociception and pain perception were chosen in our analysis. Human protein sequences, including those of GPCRs (such as TACR1, SSTR2, OPRs, and CGRPRs), TRPs, ASICs, VDSCs (VGSCs), VDCCs (VGCCs), and NPs, which have been reported to be related to nociception and pain (see introduction), were used as query sequences to perform Basic Local Alignment Search Tool (BLAST) searches in NCBI GenBank databases (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Animal taxa used in the study
Fourteen species in different phyla were used for the BLAST search, including vertebrates [eg, Homo sapiens (humans), Osteichthyes Cyprinus carpio (fish), Agnatha Petromyzon marinus (lamprey, Cyclostomata), and Protochordata Branchiostoma belcheri (amphioxus)] and invertebrates [eg, Echinodermata Strongylocentrotus purpuratus (purple sea urchin); Arthropoda-Diptera Drosophila melanogaster (fruit fly) and Arthropoda-Lepidoptera Helicoverpa armigera (cotton bollworm); Molluska Crassostrea gigas (oyster), Octopus vulgaris (octopus); Nematoda (Pseudocoelomata) Caenorhabditis elegans (worm); Platyhelminthes Schmidtea mediterranea Protopolystoma xenopodis and Opisthorchis viverrini; Coelenterata Hydra vulgaris; Porifera Amphimedon queenslandica (sponges); and Protozoa Tetrahymena thermophila (a single cell animal)]. To better support the analysis, we partially included some genomic sequences from other animals for BLAST searches, such as Mus musculus (Mammalia), Clonorchis sinensis, Schistosoma japonicum and Macrostomum lignano (Platyhelminthes). The homologs of nociception- and pain-related proteins were searched in the genome sequences of invertebrates (from unicellular to deuterostome organisms) as well as vertebrates (from Agnatha, Cyclostomata to Gnathostomata, Osteichthyes) in NCBI.
BLAST search
The human protein sequences of nociception and pain-related proteins were used as query sequences to perform Basic Local Alignment Search Tool (BLAST) searches in NCBI GenBank databases (https://blast.ncbi.nlm.nih.gov/Blast.cgi). For most protein sequences in the BLAST search, we obtained several sequences with different scores, query coverage, E-values and identities, and we usually selected the top 1 to 3 candidate sequences for further analysis. For some protein BLAST searches, we could not obtain results by using human sequences in the lower invertebrates, and we used homologous sequences from other invertebrates, such as Drosophila and C. elegans, to query genome sequence data for confirmation.
Criteria used for data selection
The sequences obtained by BLAST were further filtered by the following methods. The domain architectures were analyzed using the online SMART (Simple Modular Architecture Research Tool, http://smart.embl.de/) for the proteins in different animals and compared with the corresponding human sequences. Proteins with similar domain architectures were used for further study. We also performed sequence alignment and phylogenetic analysis for the obtained sequences. The sequences were deleted if they were not grouped with their homologs in the phylogenetic analysis and/or had different domain architectures.
Multiple sequence alignments of nociception and pain-related proteins
Multiple amino acid sequence alignments were performed using sequences of nociception and pain-related proteins from over 14 species. Alignments were performed using ClustalW. 62 The MSA (multiple sequence alignment) parameters for ClustalW during alignment were as follows: for pairwise alignment, the gap opening penalty was 10, and the gap extension penalty was 0.1; for multiple alignment, the gap opening penalty was 10, and the gap extension penalty was 0.2.
Phylogenetic analysis
The phylogenetic trees were constructed with MEGA7 (https://www.megasoftware.net/) software using the neighbor-joining method: bootstrap = 1000; substitution model, Poisson model; rate among sites, uniform rates; gaps/missing data treatments, complete deletion. The bootstrap test 66 is provided for the neighbor-joining methods in the MEGA7 software. In the test, the same number of sites are randomly sampled with replacement from the original sequences, and a phylogenetic tree is constructed from the resampled data. This process is repeated many times, and the reliability of a sequence cluster is evaluated by its relative frequency of the appearance in bootstrap replications. 67 Because of the low sequence similarity of NPs, we also used the NP sequences reported in invertebrates for the BLAST search, and we used the number of differences (Model/Method) in neighbor-joining tree construction.
Domain architecture of proteins
The domain architectures of proteins were analyzed using a web-based tool (Simple Modular Architecture Research Tool, http://smart.embl.de/). The proteins searched from the genome of different species were examined by comparison of the domain architecture with that of proteins from H. sapiens to identify the homologs of nociception and pain-related proteins in animals.
Results
Nociception and pain-related GPCRs
Nociception and pain-related GPCR homologs in different animals were searched by human nociception and pain-related GPCRs, including 3 OPRs, SSTR2, TACR1, and CGRPR. The domain architecture of nociception- and pain-related GPCRs was analyzed from low to high animals to determine their classification. However, they were found to possess a similar domain architecture with the presence of the 7tm_GPCR_Srsx domain, such as OPR and SSTR (Figure S1). These findings suggested that the architecture of nociception and pain-related GPCRs is similar in organisms, and the GPCRs could not be distinguished by their domain architectures. Therefore, the animal GPCRs that shared high identity with human GPCRs were combined for phylogenetic analysis.
The GPCRs were gathered into 4 clusters, including OPRs and SSTR2s, TACR1s, GPCR sequences from A. queenslandica and CGRPRs (Figure 1).
Figure 1.
Molecular phylogenetic analysis of 6 types of GPCRs by the neighbor-joining method. Evolutionary analyses were performed in MEGA7. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Evolutionary distances were computed using the Poisson correction method. The star shows the sequence from humans.
By domain architecture analysis, OPRs and SSTR2s possess similar 7TM_GPCR_Srsx domains (Fig. S1), and these 2 family proteins (OPRs and SSTR2s) formed a large cluster, which contained 4 subclusters in the phylogenetic tree (different colors in Figure 1): an OPR subcluster, which was only identified in vertebrates; a deuterostome SSTR2 subcluster, identified in S. purpuratus, B. belcheri, P. marinus, C. carpio, and H. sapiens; and 2 invertebrate SSTR2-like subclusters, one comprising S. mediterranea, C. elegans, C. gigas, O. vulgaris, D. melanogaster, and H. armigera, while the second comprised S. purpuratus, S. mediterranea, O. vulgaris, and C. gigas. SSTR2 was first found in Platyhelminthes (S. mediterranea). No SSTR2 sequence was discovered in H. vulgaris using human SSTR2 as a query sequence. Surprisingly, OPRs, including OPRM, OPRK, and OPRD, were identified only in vertebrates (P. marinus, C. carpio, and H. sapiens). OPRD (XP_032817657.1) identified and named in the P. marinus genome was grouped with human OPRM (Figure 1). The results of the amino acid sequence alignment of OPRs support the phylogenetic analysis, and OPRD (XP_032817657.1) of P. marinus has the same sequence motif (YWE/DNLL) as human OPRM (Figure S2).
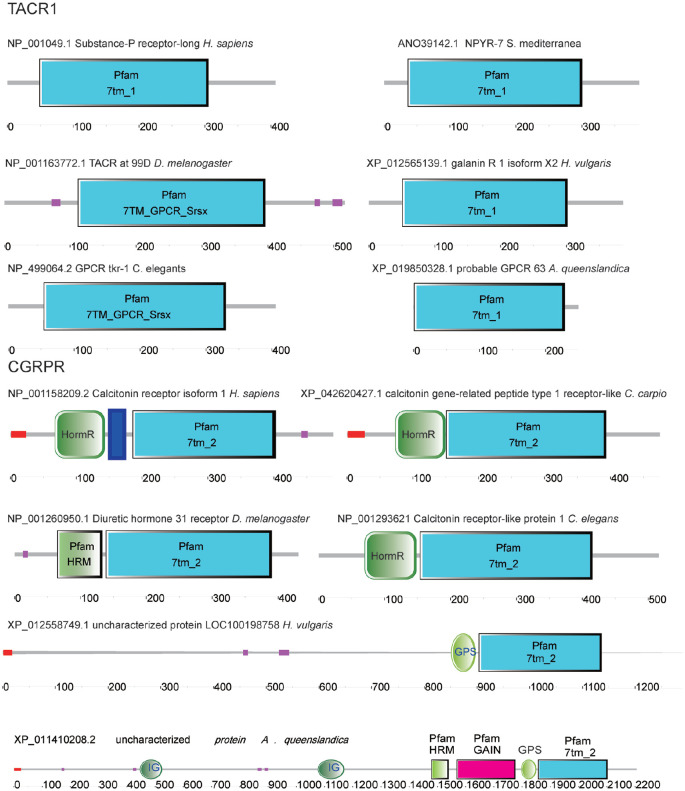
TACR1s formed the second cluster in the phylogenetic tree. Although the sequences of the family are not conserved (Figure S3), they have the conserved domain 7tm-1 or 7TM-GPCR-Srsx domain (Figure 2). All sequences of the TACR1s from different animals formed 2 subclusters: TACR1-like sequences from H. vulgaris and other sequences from the remaining animals. TACR1s were found from H. vulgaris (Coelenterata) to H. sapiens (Mammalia) (Figure 1).
Figure 2.
Domain architecture of TACR1s and CGRPRs analyzed using SMART. Pfam: protein families; 7tm-1, 7tm-2, 7TM-GPCR-Srsx: seven-transmembrane G protein-coupled receptor class of proteins; HomR: domain in hormone receptors; HRM: hormone receptor domain. 7tm_2: secretin-like family 2 GPCRs. GPS: GPCR proteolytic site. IG: immunoglobulin. GAIN: GPCR autoproteolysis-inducing domain. The deep blue block: transmembrane region.
The GPCR sequences from A. queenslandica formed the third cluster in the phylogenetic tree. Although 2 SSTR2-like sequences were named and obtained in A. queenslandica (sponges) using human SSTR2 as a query sequence, they formed a separate cluster distinct from SSTRs and other GPCR sequences. Similarly, the other 2 GPCRs obtained in A. queenslandica using human TACR1 were grouped separately; thus, they were not considered homologs of humans (Figure 1).
CGRPRs formed the fourth cluster, which contained 2 subclusters. The sequences from A. queenslandica and H. vulgaris formed one subcluster, and the rest formed another subcluster (Figure 1). The sequence identity of CGRPRs is quite low (Fig. S3); however, they have conserved domain architecture (HomR domain and 7tm-2 domain) (Figure 2). The protein from H. vulgaris had no HomR domain, and the protein from A. queenslandica is a large protein with multiple domains, including immunoglobulin (Ig), gain, and HomR domains and 7 transmembrane regions. Thus, they were not considered authentic CGRPRs. No homologous sequence was identified in Platyhelminthes by BLAST analysis using human and C. elegans CGRPRs. CGRPR was first found in C. elegans.
TRP channels
Human TRPM3, 7 and 8, TRPA1, and TRPV1 sequences were used as query sequences for BLAST analyses to obtain corresponding sequences from invertebrates and vertebrates. The sequences obtained that shared high identity to humans were used for phylogenetic analysis. Three clusters, TRPM (including TRPM3 TRPM7, and TRPM8), TRPA, and TRPV, were obtained in the phylogenetic tree (Figure 3). TRPM domain architecture shows fewer ankyrin repeat domains, an ion transport domain and other domains, such as a short antiparallel coiled-coil tetramerization domain (TRPM_tetra), SCOP d1gw5a_domain, and α-kinase domain (Figure S4). The homologs of TRPM3, 7 and 8 were clearly identified in vertebrates, but it is difficult to identify the 3 types of TRPMs in invertebrates. In general, the members of the TRPM family were identified from Porifera to humans. TRPM was also identified in other Platyhelminthes animals, including Clonorchis sinensis, Schistosoma japonicum, and Macrostomum lignano (Figure 3).
Figure 3.

Molecular phylogenetic analysis of TRPs by the neighbor-joining method. Evolutionary analyses were performed in MEGA7. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Evolutionary distances were computed using the Poisson correction method. Different colors indicate different clusters. The sequences from humans are highlighted in bold.
TRPA1 homologs were identified from A. queenslandica to humans. All TRPA1 proteins formed a cluster in the phylogenetic tree (Figure 3). Four TRPA1 homologs were identified in A. queenslandica (GenBank accession nos. XP_019857678.1, XP_019859938.1, XP_019860065.1, and XP_011402710.1) (Figure 3). TRPA1 architecture shows many ankyrin repeat domains and an ion transport domain, including TRPA1 from A. queenslandica (Figure S5).
The TRPV1 cluster was divided into 2 subgroups: vertebrate and invertebrate. The latter included proteins from various organisms, ranging from M. lignano (Platyhelminthes) to B. belcheri (Protochordata) (Figure 3). TRPV1 showed more ankyrin repeat domains than TRP-M but fewer than TRPA1. The protein from T. thermophila had no Ion_trans domain and was not identified as TRPV (Figure 4).
Figure 4.
Domain architectures of TRP-V from humans and different animals. Abbreviations: ANK, ankyrin repeats; Ion_trans, ion transport protein domain found in sodium, potassium, and calcium IC proteins; PARP_reg: poly (ADP-ribose) polymerases. 3SUIIB: unknown function. The blue block: transmembrane region. WGR: a nucleic acid binding domain.
ASICs
The ASICs comprise several isoforms, including ASIC1 to ASIC4. The ASIC1 and ASIC3 isoforms that we chose for evolution analysis are particularly important in sensory neurons. ASIC-like proteins were found from Porifera (A. queenslandica) to Mammalia (H. sapiens). In the phylogenetic analysis, ASIC-like proteins clustered into 2 groups. The first group included all ASICs from Protochordata and vertebrates. The second group included all invertebrates from Spongia (A. queenslandica) to Echinodermata (S. purpuratus) animals, which were clustered into 4 subgroups. The first ASIC protein was found in A. queenslandica (Spongia) (Figure 5A). Domain architecture analyses revealed that all ASICs have an ASC domain (Figure 5B). The results suggested that ASIC was present in all of the animal phyla analyzed in the study.
Figure 5.
Molecular phylogenetic analysis of the ASICs by the neighbor-joining method and their domain architectures. (A) The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The evolutionary distances were computed using the Poisson correction method. Evolutionary analyses were conducted in MEGA7. (B) Domain architectures of ASICs from H. sapiens to A. queenslandica. ASC: The apical membrane of many tight epithelia contains sodium channels that are primarily characterized by their high affinity to the diuretic blocker amiloride.
VDSCs
The sequences of NaV 1.7 and NaV 1.8 of VDSCs in mammals were used to search for homologous sequences from other animals. In the phylogenetic analysis, all VDSCs were divided into 2 clusters: deuterostome and protostome VDSCs. VDSCs were not found in Nematoda. The VDSC protein was first found in Platyhelminthes animals (Echinococcus multilocularis and Hymenolepis diminuta) (Figure 6A).
Figure 6.
Evolutionary relationships of VDSCs and their domain architectures. (A) The evolutionary history was inferred using the neighbor-joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Evolutionary distances were computed using the Poisson correction method, with units representing the number of amino acid substitutions per site. Evolutionary analyses were performed in MEGA7. (B) Domain architectures of VDSCs from humans and other animals. Ion_trans: domain found in sodium, potassium, and calcium IC proteins. The proteins have 6 transmembrane helices in which the last 2 helices flank a loop, which determines ion selectivity. Na_trans_cytopl: cytoplasmic domain of voltage-gated Na+ ion channel. PKD_channel: polycystic kidney disease channel domain; GPHH: domain found in voltage-dependent l-type calcium channel proteins in eukaryotes. The domain is closely associated with the isoleucine–glutamine (IQ) domain. IQ: voltage-gated calcium channels control cellular calcium entry in response to changes in membrane potential.
VDSCs showed similar domain architectures in animals: in H. sapiens, Nav 1.8 protein contains 3 Ion_trans domains, 1 Na_trans_domain, and a PKD channel domain. Nav 1.7 contains 3 Ion_trans domains, 2 Na_trans_domains, a PKD channel domain, and an IQ domain (calmodulin-binding motif). The VDSCs from Platyhelminthes H. diminuta and Molluska C. gigas are similar to human Nav 1.8, and the VDSC from D. melanogaster is similar to Nav 1.7 (Figure 6B).
VDCCs
We chose the N-type (CaV2.2) and the α2/δ-1 auxiliary subunits for evolutionary analysis according to the known function of these proteins in nociception and pain. In the phylogenetic analysis, VDCCs were divided into 2 clusters, including the α2/δ-1 subunit and N-type Ca2+ channel. α2/δ-1 clusters were found from A. queenslandica to humans. The N-type was identified from all selected animals from A. queenslandica to humans, except H. vulgaris (Coelenterata). The VDCC α2/δ-1 and CaV2.2 proteins were all first identified in A. queenslandica (Figure 7).
Figure 7.
Evolutionary relationships of VDCCs. The evolutionary history was inferred using the neighbor-joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of evolutionary distances used to infer the phylogenetic tree. Evolutionary distances were computed using the Poisson correction method, with units representing the number of amino acid substitutions per site. Evolutionary analyses were performed in MEGA7.
VDCCs belong to a domain-conserved protein family. In α2/δ-1 VDCC, the VWA_N, VWA, and VGCC domains were found in most proteins, and VGCC domains were absent in C. elegans, M. lignano, and A. queenslandica. In N-type VDCCs, the Ion_trans domain is conserved from A. queenslandica to humans (Figure 8).
Figure 8.
Domain architectures of VDCC proteins from humans and other animals. VWA_N: domain found at the N-terminus of proteins containing von Willebrand factor (vWF) type A domains. VWA: vWF type A domain. vWF is a multimeric glycoprotein essential for the normal arrest of bleeding after tissue injury (hemostasis). VGCC: Voltage-dependent Ca2+ channels. dCache: extracellular domain predicted to play a role in small-molecule recognition in many proteins, including the dihydropyridine-sensitive voltage-gated Ca2+ channel α-2δ subunit. The term “Cache” is derived from CAlcium channels and CHEmotaxis receptors. The Cache domain, also known as the extracellular PAS domain, consists of an N-terminal part with 3 predicted strands and an alpha-helix and a C-terminal part with a strand dyad followed by a relatively unstructured region. RPT 1: an internal repeat. PKD_channel: polycystic kidney disease channel domain. GPHH: domain found in voltage-dependent L-type calcium channel proteins in eukaryotes. Cacan-IQ: voltage-gated calcium channels control cellular calcium entry in response to changes in membrane potential. The IQ motif in the voltage-gated calcium channel IQ domain interacts with hydrophobic pockets of Ca2+/calmodulin. SCOP d1lsha3: unknown function.
NPs
We assessed 10 NPs, NPPC, CGRP, SST, TAC1, NMU, CCK, PYY, POMC, PENK, and PDYN, that are known to be related to nociception and pain in humans and searched for their homologs in other animals. Because of the low sequence identity of NPs, we also used the reported NP sequences of invertebrates as query sequences to search different animal genomes. The obtained sequences were used for phylogenetic analysis. The results showed that the NPs were grouped into 8 clusters, named TAC, NMU, CGRP, unknown, SST and NPPC, CCK, PYY, POMC and PENK and PDYN (Figure 9); however, the real homologs of human NPs could not be identified by phylogenetic analysis.
Figure 9.
Evolutionary relationships of NPs. The evolutionary history was inferred using the neighbor-joining method. The tree was drawn to scale, with branch lengths in the same units as those of evolutionary distances used to infer the phylogenetic tree. Evolutionary distances were computed using the p-distance method, with units representing the number of amino acid differences per site. Evolutionary analyses were performed in MEGA7.
Therefore, the architectures of NPs were further analyzed to identify the homologs of human nociception and pain-NPs using human NPs as models. The TAC1 homologs were identified from humans, C. carpio and D. melanogaster according to their TK or RPT1 domains in the peptides (Figure S6A). NMU1 was identified from fish and humans by an NMU (neuromedin-U) domain (Figure S6B). The peptide in Ciona intestinalis (XP_018669232.1) in the unknown cluster was likely a TAC (Figure S6C). CGRPs were identified in B. belcheri, P. marinus, C. carpio and humans according to the CALCI TONIN domain (Figure S7). SSTs, NPPCs, and CCKs were only identified from vertebrates. SST had a somatostatin domain, NPPC had a NAT PEP domain, and CCK had a GASTRIN domain (Figure S8). PYYs, POMC, PENK and PDYN were also found in vertebrates. PYY peptides contain a PAH (pancreatic hormones/neuropeptide F/peptide YY family) domain. Although a peptide from B. belcheri (XP_019630162.1) was named peptide YY-like, it has no PAH domain and thus was not identified as PYY. POMC, PENK, and PDYN contained an opioid neuropeptide domain; however, POMC had NPP and ACTH domains, and PENK and PDYN contained RPT domains (Figure 10).
Figure 10.
Domain architecture of PYY, PENK, PDYN, and POMC peptides from humans and other animals. PAH: pancreatic hormones/neuropeptide F/peptide YY family domain. Opioids-neuropep: opioidsneuropeptide. Red block: The precursors of opioid neuropeptides. RPT 1: internal repeat 1. CYCc: Adenylyl-/guanylyl cyclase, catalytic domain. NPP: Pro-opiomelanocortin, N-terminal region. ACTH: corticotropin ACTH domain.
Discussion
In the present study, we assessed the evolution of several key nociception- and pain-related proteins. We found that the homologs of nociception- and pain-related proteins originated in different invertebrates and that ICs and most GPCRs exist from lower animal phyla to higher animal phyla, whereas OPRs and NPs are newly evolved in vertebrates, which has not been reported previously. Our findings promote the understanding of the origin and evolution of these proteins in various organisms, ranging from sponges to humans. Although various nociception and pain related proteins have been identified in different animal species, relatively little is known about their evolution and biological roles, especially in low animal phyla. Despite the distant evolutionary relationship between mammals and invertebrates, the identification of nociception and pain related proteins in invertebrates might provide insights into the essential molecular modules that are conserved during the evolution of detecting nociceptive stimuli. Studies using the proteins from a variety of animal species offer us the opportunity to explore the evolution of nociception and pain, and will provide insights into the fundamental mechanisms of pathological pain and explore approach to treat the pain.
There are at least 40 GPCRs that are considered to be potential therapeutic targets for the regulation of pain. 68 Previous work shows that the vertebrate opioid system was established in the first jawed vertebrates. 69 In the present study, we found that OPRM was identified in a jawless (agnatha) vertebrate, P. marinus, and that OPRK and OPRD were exclusively found in jawed vertebrates from fish to humans.
Activation of SSTR2 in the mouse DRG by SST potently inhibits neuropathic pain. 16 SSTR2 has been reported to regulate proinflammatory, microbial and obesity-related signals in humans. 70 The first 2 members of the SSTR family, SSTR1 and SSTR2, were identified in humans and mice, 71 and five receptor subtypes of SSTRs (termed SSTR1-5) were identified in mammals and other vertebrates. 72 In our study, we found that SSTR2 homologs were identified in invertebrates from Platyhelminthes. The function of SSTR2 in nociception in invertebrates is unclear.
CGRPRs are widely distributed in different animals, including vertebrates and invertebrate Molluska.73 -75 In our study, we found that the CGRPR homolog appeared in Nematoda; however, the nociception- and pain-related functions of CGRPR in invertebrates need to be clarified.
NPs serve as neurotransmitters, neuromodulators or neurohormones by activating different types of GPCRs. Molecular evolution of peptides and their receptors in bilaterians has shown that NPs and their receptors coevolved. 76 CGRP is expressed mainly in the thyroid gland and nervous system in mammals. 77 Diuretic hormone 31 (DH31) is considered the ortholog of the chordate calcitonin peptides. Calcitonin-type NPs have been reported to be present in lophotrochozoan animals, such as C. gigas, and the diuretic hormone 31 (DH31) type in ecdysozoan animals, such as insects. 74 In the present study, although DH31 was found in insects, it had no CALCITONIN domain as the CGRPs in vertebrates; thus, real CGRP was first identified in Protochordata. By architecture identification, the homologs of human nociception and pain-related NPs, including NPPC, SST, NMU, CCK, PYY, POMC, PENK, and PDYN, were not found in invertebrates, except TAC1, which appeared in insects, indicating their delayed appearance during evolution.
Ion channels contain members of multiple gene families, which are activated by various nociceptive stimuli. In mammals, 9 thermoTRPs have been identified and classified into 3 subfamilies, TRPA, TRPM, and TRPV, 78 after the first TRP-V1 was identified. 79 Phylogenetic analysis of thermoTRP homologs from vertebrates revealed that TRPV1-4, TRPM2, TRPM4, TRPM5, and TRPM8 are unique to vertebrates. 80 The chordate TRPM family is typically divided into 8 distinct paralogs (TRPM1–TRPM8). 80 TRPs share common structural features, containing 6 putative transmembrane domains and all TRPs are cation channels. In our study, we found that a homolog of TRPA1 is first present in A. queenslandica, and TRPM3 and 7 in H. vulgaris (Coelenterata), TRPV1 in M. lignano (Platyhelminthes), and TRPM8 in C. gigas (Molluska).
ASICs are involved in the perception of different types of stimuli, especially extracellular H+ (low pH values), and they were previously identified in protochordates, including Tunicata and cephalochordates. 81 Here, we found that ASIC1-like is first found in A. queenslandica.
VDSCs are key players in electrical conduction along axons, including those of primary sensory neurons. Nav 1.7 and Nav 1.8 are thought to have evolved relatively recently because their sequences have not been reported from nonmammalian species thus far. 82 In our study, VDSC-like proteins were first present in the platyhelminthes S. mediterranea.
VDCCs regulate the entry of extracellular calcium into excitable cells and serve as signal transduction centers, playing a role in diverse neuronal physiological processes. VDCCs are composed of several subunits, α1, β, α2δ, and γ. Studies on Ca2+ channel regulation in nociception and pain have mainly focused on the β and α2δ auxiliary subunits in humans. 83 In the present study, α2/δ-1 and N-type VDCCs were found in the ancient species A. queenslandica.
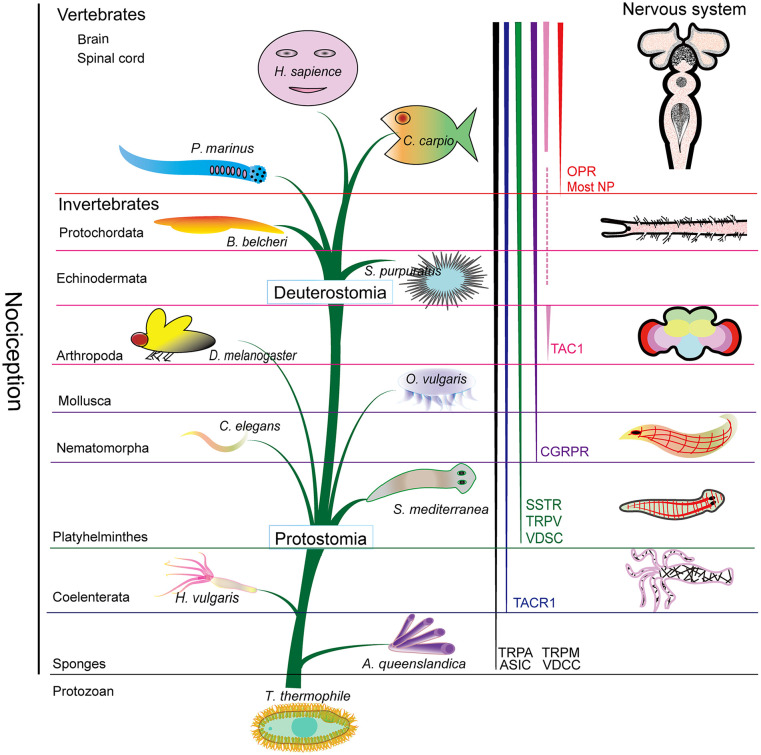
Vertebrates and invertebrates have quite different nervous systems. It has been suggested that only animals with a discernible nervous system can centrally process nociceptive information. 84 Invertebrate nervous systems are smaller and contain fewer neurons than those of vertebrates. Unicellular protozoans and lower multicellular animals, such as A. queenslandica (Porifera), do not have true nervous systems. However, genes associated with neuronal development have been identified in A. queenslandica. 85 Therefore, electrical signaling in some sponges by syncytial tissues involves calcium–potassium action potentials, regulating the feeding current and preventing the uptake of unwanted particles. 86 Accordingly, some calcium–potassium transport-related IC proteins, such as TRPA, TRPM, ASICs, and VDSCs, were identified in A. queenslandica (Figure 11 and Table 1).
Figure 11.
Evolution of nociception and pain-related proteins from invertebrates to vertebrates with different nervous systems (see the text in the discussion for details).
Table 1.
Appearance of nociception and pain-related proteins in animals.
| Human nociception and pain-related proteins and GenBank accession number | NP_000905.3 mu-type opioid receptor MOR-1 | NP_000903.2 kappa-type opioid receptor | NP_000902.3 delta-type opioid receptor | NP_001041.1 Somatostatin receptor 2 | NP_001049.1 Substance-P receptor long | NP_001158209.2 Calcitonin receptor isoform 1 | XP_011510112.1 TRP-M8 isoform X1XP_005254543.1TRP-M7 isoform 3, NP_001007472.2 TRP-M3 | NP_061197.4 TRP-V member 1 | XP_011515926.1 member 1 X1 | NP_001086.2 ASIC 1 isoform bNP_004760.1 acid-sensing ion channel 3 isoform a | Q15858.3 Voltage-gated sodium channel alpha Nav 1.7Q9Y5Y9.2 Voltage-gated sodium channel alpha Nav 1.8 | NP_000713.2 VDCC alpha-2/delta-1 isoform 1NP_000709.1 VDCC N-type alpha-1B isoform 1 | NP_077720.1 C-type natriuretic peptide | NP_001029124.1 Calcitonin preproprotein (calcitonin related polypeptide alpha) | NP_001039.1 somatostatin preproprotein | NP_003173.1 protachykinin-1 | NP_001278974.1 neuromedin-U | NP_000720.1 cholecystokinin preproprotein | NP_004151.4 peptide YY preproprotein | NP_000930.1 pro-opiomelanocortin | NP_001129162.1 proenkephalin-A | NP_001177821.1 proenkephalin-B | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Abbreviation of the proteins | OPRM | OPRK | OPRD | SSTR2 | TACR1 | CGRPR | TRPM8 and M7TRPM3 | TRPV1 | TRPA1 | ASIC | VDSC | VDCC | NPPC | CGRP | SST | TAC1 | NMU | CCK | PYY | POMC | PENK | PDYN | |
| Phylum | Organism | GPCR | IC | NP | |||||||||||||||||||
| Chordata | H. sapiens | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| C. carpio | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| P. marinus | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | − | + | − | |
| Protochordata | B. belcheri | − | − | − | + | + | + | + | + | + | + | + | + | − | + | − | − | − | − | − | − | − | − |
| Echinodermata | S. purpuratus | − | − | − | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − |
| Arthropoda | D. melanogaster | − | − | − | + | + | + | + | + | + | + | + | + | − | − | − | + | − | − | − | − | − | − |
| H. armigera | − | − | − | + | + | + | + | + | + | + | + | + | − | − | − | + | − | − | − | − | − | − | |
| Mollusca | O. vulgaris | − | − | − | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − |
| C. gigas | − | − | − | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | |
| Nematoda | C. elegans | − | − | − | + | + | + | + | + | + | + | − | + | − | − | − | − | − | − | − | − | − | − |
| Platyhelminthes | S. mediterranea | − | − | − | + | + | − | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − |
| Coelenterata | H. vulgaris | − | − | − | − | + | − | + | − | + | + | − | + | − | − | − | − | − | − | − | − | − | − |
| Porifera | A. queenslandica | − | − | − | − | − | − | + | − | + | + | − | + | − | − | − | − | − | − | − | − | − | − |
| Protozoan | T. thermophile | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Abbreviations: ASIC, acid-sensing ion channel; CCK, cholecystokinin; CGRPR, calcitonin gene-related peptide receptor; C-type natriuretic peptide; GPCR, G protein-coupled receptor; IC, ion channel; NMU, neuromedin-U; NP, neuropeptide; NPPC, CGRP (calcitonin related polypeptide alpha, CALCA, also known as CT; KC; PCT; CGRP; CALC1; CGRP1; CGRP-I; CGRP-alpha); OPRD, opioid receptor-δ; OPRM, opioid receptor-μ; OPRK opioid receptor-κ; PDYN, proenkephalin-B; PENK, proenkephalin-A; POMC, pro-opiomelanocortin; PYY, peptide YY; SST, somatostatin; SSTR2, somatostatin receptor type 2; TAC1, protachykinin; TACR, preprotachykinin receptor (substance-P receptor isoform long of Homo sapiens); TRPA, transient receptor potential channel ankyrin; TRPM, transient receptor potential channel canonical; TRPV, transient receptor potential channel vanilloid; VDCC, voltage-dependent calcium channel; VDSC (VGSC, SCN), sodium channel protein.
+, present of the protein homolog and −, absent of the protein homolog according to the phylogenetic analysis and domain architecture.
Coelenterates (one of the basal groups within metazoans), such as H. vulgaris, show dispersed, diffused neural networks, and the central nervous system (CNS) is still not formed. 87 Neurons, cells that transmit nerve stimuli, are found in Coelenterata. The coelenterate nervous systems only show one or several “diffuse” nerve nets, while the CNS is absent. 87 Herein, most nociception and pain-related proteins, such as TACR1 and some IC proteins, were identified in Coelenterata.
The CNS is present in Platyhelminthes, such as S. mediterranea, showing ventral, lateral, and dorsal pairs of nerve cords. Typically, the ventral cord—the main cord—is more strongly developed. All flatworms possess a relatively compact anterior brain resembling a typical invertebrate ganglion. 88 SSTR2 and TRPV1 were first found in Platyhelminthes.
In Nematoda, The CNS form a ring around the anterior intestinal system. Furthermore, longitudinal neurites or neurite bundles, as well as entire or partial circular neurites connecting the longitudinal neurites, are present. In the anterior part, neurons associate with each other to form nerve threads attached to the nerve ganglia. 89 Most nociception and pain-related proteins, except OPRs and NPs, were found in Nematoda.
A relatively simple nervous system is present in Bivalvia (Molluska), and 3 pairs of ganglia are found: the cerebropleural ganglia (fusion product of the cerebral and pleural ganglia), pedal ganglia, and visceral ganglia. The pedal ganglia are connected to the cerebropleural ganglia by 2 connectives. Bivalves have a remarkably high number of sensory organs, particularly at the mantle folds and in the oral region. Cephalopods (Molluska) show a highly centralized and complex CNS and have a large cerebroid ganglion that resembles the brain of vertebrates. 90 Most nociception and pain-related proteins, except OPRs and NPs, were identified in Bivalvia and Cephalopods.
In Arthropoda, the nervous systems are well developed and show greater cephalization and centralization in comparison with other lower invertebrates. They exhibit the basic pattern of the CNS, which includes a brain and one or more nerve cords. 91 Most nociception- and pain-related proteins, except OPRs and several NPs, were identified in the phylum.
Echinoderms are deuterostomes, while all other invertebrates are protostomes. Two neural units of the nervous system exist in echinoderms: one is the basiepidermal nerve plexus, which processes sensory stimuli locally, and one is the radial nerve cords and the peripheral nerves constituting a centralized control system. 92 Although the CNS is missing, nociception and pain-related proteins were found in echinoderms.
In the phylum Chordata, Protochordata (B. belcheri) are the most closely related to vertebrates, and they possess a CNS with a tubular nerve cord. The nervous system is highly centralized and cephalized in vertebrates. The CNS is composed of a prominent brain and spinal cord. The spinal cord is composed of numerous neuronal populations that sense different stimuli. Projection neurons transfer information to the brainstem and thalamus and then to several brain regions. In this study, we chose 3 classes, Cyclostomata (P. marinus), Osteichthyes (C. carpio), and Mammalia (H. sapiens), to identify nociception- and pain-related proteins, and most nociception- and pain-related proteins were identified in vertebrates. In humans, nociception and pain are perceived by peripheral nociceptors and transmitted to the spinal cord and then to the brain. 93 Nociception and pain are perceived and modulated by proteins in both the periphery and central nervous system.
Conclusion and Further Directions
In conclusion, different nociception and pain-related proteins originate from different animal phyla. ICs originate from lower animal phyla, for example, TRPA, TRPM, ASIC1, and VDCC originate from the primitive phylum Porifera (Sponges), and TRPV and VDSC originated lately from Platyhelminthes. GPCRs originate from different phyla: TACR1 appeared from Coelenterata, SSTR originated from Platyhelminthes, CGRPR appeared from Nematoda, and OPRs appeared from vertebrates. For neuropeptides, TAC1 was identified from Arthropoda, CGRP from Protochordata, and NPPC, SST, NMU, CCK, PYY, POMC, PEPCK, and PDYN were specifically found in vertebrates (Table 1 and Figure 11). Although the nociception and pain-related protein homologs existing in animals cannot fully explain the nociception and pain perception in invertebrates and vertebrates, the data show the evolution of the proteins in animals from invertebrates to vertebrates, which provides hints of the evolution of nociception and pain-related proteins in animals and humans. Future functional studies are required to demonstrate the contribution and functions of each protein in nociception and pain sensation in animals.
Nociceptors are transducers of pain, and activation of nociceptors could result in pain. It is known that subjective experience associated with physical or potential harm to the body refers to pain. Nociception has been reported in various species; however, it is contentious whether pain could occur in lower vertebrates and invertebrates. In our study, we provide evidence that nociception- and pain-related proteins are broadly distributed in both invertebrates and vertebrates, and the phylogenetic relationships of nociception- and pain-related proteins are closely related to the evolution of nervous systems (Figure 11) and could be used to address whether a fundamental conserved process of nociception and pain is present in the animal kingdom.
Limitations of the study
To obtain a better understanding of the evolutionary relationship of the proteins and the biology of nociception and pain, we performed this bioinformatic analysis of the nociception and pain-related proteins in different animal phyla. However, the results presented here are only a rough sketch of the evolution of nociception and pain-related proteins, as the sequence database is limited in some representative species. More animal taxa and sequences should be chosen to examine the evolution of the proteins. In addition, the molecular identification of the target used in this paper is only based on amino acid sequences, and the gene structure of the molecules needs to be analyzed if the data sources are available. Moreover, sequence similarities of some selected proteins are quite low between mammals and invertebrates, therefore we could not find the proteins in invertebrates using the protein sequences from humans. For example, NPs are reported in several invertebrates, such as insects and crustaceans, but we did not identify the orthologous NPs in most of invertebrates (Table 1) in our study. Whole genome sequencing data is required to identify the relevant neuropeptides in invertebrates. 75 Functional studies of the proteins involved in nociception and pain, especially in invertebrates, are also needed.
Supplemental Material
Supplemental material, sj-docx-1-evb-10.1177_11769343231216914 for Phylogenetic Analysis Provides Insight Into the Molecular Evolution of Nociception and Pain-Related Proteins by Rujun Zhai and Qian Wang in Evolutionary Bioinformatics
Footnotes
Author Contributions: RZ and QW contributed to the conception and design of the study and analyzed the data. QW wrote the manuscript. All authors contributed to manuscript revision and approved the submitted version.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article and its supplementary information files.
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Sneddon LU. Comparative physiology of nociception and pain. Physiology. 2018;33:63-73. [DOI] [PubMed] [Google Scholar]
- 2. Apkarian AV. Definitions of nociception, pain, and chronic pain with implications regarding science and society. Neurosci Lett. 2019;702:1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gonçalves Dos Santos G, Delay L, Yaksh TL, Corr M. Neuraxial cytokines in pain states. Front Immunol. 2019;10:3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elwood RW. Discrimination between nociceptive reflexes and more complex responses consistent with pain in crustaceans. Philos Trans R Soc Lond B Biol Sci. 2019;374:20190368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sneddon LU, Elwood RW, Adamo SA, Leach MC. Defining and assessing animal pain. Anim Behav. 2014;97:201-212. [Google Scholar]
- 6. Pinsker HM, Hening WA, Carew TJ, Kandel ER. Long-term sensitization of a defensive withdrawal reflex in Aplysia. Science. 1973;182:1039-1042. [DOI] [PubMed] [Google Scholar]
- 7. Himmel NJ, Letcher JM, Sakurai A, et al. Drosophila menthol sensitivity and the Precambrian origins of transient receptor potential-dependent chemosensation. Philos Trans R Soc Lond B Biol Sci. 2019;374:20190369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adamo SA, McMillan LE. Listening to your gut: immune challenge to the gut sensitizes body wall nociception in the caterpillar Manduca sexta. Philos Trans R Soc Lond B Biol Sci. 2019;374:20190278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mogil JS, Yu L, Basbaum AI. Pain genes? natural variation and transgenic mutants. Annu Rev Neurosci. 2000;23:777-811. [DOI] [PubMed] [Google Scholar]
- 10. Costigan M, Befort K, Karchewski L, et al. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng Y, Liu P, Bai L, et al. Deep sequencing of somatosensory neurons reveals molecular determinants of intrinsic physiological properties. Neuron. 2019;103: 598-616.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gadais C, Piekielna-Ciesielska J, De Neve J, et al. Harnessing the anti-nociceptive potential of NK2 and NK3 ligands in the design of new multifunctional μ/δ-opioid agonist-neurokinin antagonist peptidomimetics. Molecules. 2021; 26:5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barik A, Sathyamurthy A, Thompson J, et al. A spinoparabrachial circuit defined by tacr1 expression drives pain. eLife. 2021;10:e61135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang T, Lin SH, Malewicz NM, et al. Identifying the pathways required for coping behaviours associated with sustained pain. Nature. 2019;565:86-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duan B, Cheng L, Bourane S, et al. Identification of spinal circuits transmitting and gating mechanical pain. Cell. 2014;159:1417-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiang Q, Li JJ, Li CY, Tian RB, Li XH. Somatostatin type 2 receptor antibody enhances mechanical hyperalgesia in the dorsal root ganglion neurons after sciatic nerve-pinch injury: evidence of behavioral studies and bax protein expression. CNS Neurol Disord Drug Targets. 2019;18:791-797. [DOI] [PubMed] [Google Scholar]
- 17. Iyengar S, Ossipov MH, Johnson KW. The role of calcitonin gene-related peptide in peripheral and central pain mechanisms including migraine. Pain. 2017;158:543-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kieffer BL, Gavériaux-Ruff C. Exploring the opioid system by gene knockout. Prog Neurobiol. 2002;66:285-306. [DOI] [PubMed] [Google Scholar]
- 19. Darcq E, Kieffer BL. Opioid receptors: drivers to addiction? Nat Rev Neurosci. 2018;19:499-514. [DOI] [PubMed] [Google Scholar]
- 20. Brighton PJ, Szekeres PG, Willars GB. Neuromedin U and its receptors: structure, function, and physiological roles. Pharmacol Rev. 2004;56:231-248. [DOI] [PubMed] [Google Scholar]
- 21. Torres R, Croll SD, Vercollone J, et al. Mice genetically deficient in neuromedin U receptor 2, but not neuromedin U receptor 1, have impaired nociceptive responses. Pain. 2007;130:267-278. [DOI] [PubMed] [Google Scholar]
- 22. Loo L, Shepherd AJ, Mickle AD, et al. The C-type natriuretic peptide induces thermal hyperalgesia through a noncanonical ggβγ-dependent modulation of TRPV1 channel-dependent modulation of TRPV1 channel. J Neurosci. 2012;32:11942-11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiesenfeld-Hallin Z, Xu XJ, Hökfelt T. The role of spinal cholecystokinin in chronic pain states. Pharmacol Toxicol. 2002;91:398-403. [DOI] [PubMed] [Google Scholar]
- 24. Khoo B, Tan TM. Combination gut hormones: prospects and questions for the future of obesity and diabetes therapy. J Endocrinol. 2020;246:R65-R74. [DOI] [PubMed] [Google Scholar]
- 25. Hassan AM, Jain P, Mayerhofer R, et al. Visceral hyperalgesia caused by peptide YY deletion and Y2 receptor antagonism. Sci Rep. 2017;7:40968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jardín I, López JJ, Diez R, et al. TRPs in pain sensation. Front Physiol. 2017; 8:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Numazaki M, Tominaga M. Nociception and TRP channels. Curr Drug Targets CNS Neurol Disord. 2004;3:479-485. [DOI] [PubMed] [Google Scholar]
- 28. Wemmie JA, Taugher RJ, Kreple CJ. Acid-sensing ion channels in pain and disease. Nat Rev Neurosci. 2013;14:461-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen CC, Zimmer A, Sun WH, et al. A role for ASIC3 in the modulation of high-intensity pain stimuli. Proc Natl Acad Sci USA. 2002;99:8992-8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Young RF, Vermeulen SS, Grimm P, et al. Gamma knife thalamotomy for the treatment of persistent pain. Stereotact Funct Neurosurg. 1995;64 Suppl 1:172-181. [DOI] [PubMed] [Google Scholar]
- 31. Kashio M. Thermosensation involving thermo-TRPs. Mol Cell Endocrinol. 2021;520:111089. [DOI] [PubMed] [Google Scholar]
- 32. Julius D. TRP channels and pain. Annu Rev Cell Dev Biol. 2013;29:355-384. [DOI] [PubMed] [Google Scholar]
- 33. Vay L, Gu C, McNaughton PA. The thermo-TRP ion channel family: properties and therapeutic implications. Br J Pharmacol. 2012;165:787-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patapoutian A, Peier AM, Story GM, Viswanath V. ThermoTRP channels and beyond: mechanisms of temperature sensation. Nat Rev Neurosci. 2003;4:529-539. [DOI] [PubMed] [Google Scholar]
- 35. Caterina MJ, Schumacher MA, Tominaga M, et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816-824. [DOI] [PubMed] [Google Scholar]
- 36. Caterina MJ, Leffler A, Malmberg AB, et al. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science. 2000;288:306-313. [DOI] [PubMed] [Google Scholar]
- 37. Davis JB, Gray J, Gunthorpe MJ, et al. Vanilloid receptor-1 is essential for inflammatory thermal hyperalgesia. Nature. 2000;405:183-187. [DOI] [PubMed] [Google Scholar]
- 38. Vriens J, Owsianik G, Hofmann T, et al. TRPM3 is a nociceptor channel involved in the detection of noxious heat. Neuron. 2011;70:482-494. [DOI] [PubMed] [Google Scholar]
- 39. Bautista DM, Siemens J, Glazer JM, et al. The menthol receptor TRPM8 is the principal detector of environmental cold. Nature. 2007;448:204-208. [DOI] [PubMed] [Google Scholar]
- 40. Dhaka A, Murray AN, Mathur J, et al. TRPM8 is required for cold sensation in mice. Neuron. 2007;54:371-378. [DOI] [PubMed] [Google Scholar]
- 41. Bautista DM, Jordt SE, Nikai T, et al. TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell. 2006;124:1269-1282. [DOI] [PubMed] [Google Scholar]
- 42. Kwan KY, Allchorne AJ, Vollrath MA, et al. TRPA1 contributes to cold, mechanical, and chemical nociception but is not essential for hair-cell transduction. Neuron. 2006;50:277-289. [DOI] [PubMed] [Google Scholar]
- 43. Benemei S, Dussor G. TRP channels and migraine: recent developments and new therapeutic opportunities. Pharmaceuticals. 2019;12:1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takayama Y, Derouiche S, Maruyama K, Tominaga M. Emerging perspectives on pain management by modulation of TRP channels and ANO1. Int J Mol Sci. 2019;20:3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sluka KA, Winter OC, Wemmie JA. Acid-sensing ion channels: a new target for pain and CNS diseases. Curr Opin Drug Discov Devel. 2009;12:693-704. [PMC free article] [PubMed] [Google Scholar]
- 46. Baron A, Diochot S, Salinas M, et al. Venom toxins in the exploration of molecular, physiological and pathophysiological functions of acid-sensing ion channels. Toxicon. 2013;75:187-204. [DOI] [PubMed] [Google Scholar]
- 47. Baron A, Lingueglia E. Pharmacology of acid-sensing ion channels - physiological and therapeutical perspectives. Neuropharmacol. 2015;94:19-35. [DOI] [PubMed] [Google Scholar]
- 48. Cheng YR, Jiang BY, Chen CC. Acid-sensing ion channels: dual function proteins for chemo-sensing and mechano-sensing. J Biomed Sci. 2018;25:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee CH, Chen CC. Roles of ASICs in nociception and proprioception. Adv Exp Med Biol. 2018;1099:37-47. [DOI] [PubMed] [Google Scholar]
- 50. Chang CT, Fong SW, Lee CH, et al. Involvement of acid-sensing ion channel 1b in the development of acid-induced chronic muscle pain. Front Neurosci. 2019;13:1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Verkest C, Diochot S, Lingueglia E, Baron A. C-Jun N-terminal kinase post-translational regulation of pain-related acid-sensing ion channels 1b and 3. J Neurosci. 2021;41:8673-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wood JN, Boorman JP, Okuse K, Baker MD. Voltage-gated sodium channels and pain pathways. J Neurobiol. 2004;61:55-71. [DOI] [PubMed] [Google Scholar]
- 53. Bennett DL, Clark AJ, Huang J, Waxman SG, Dib-Hajj SD. The role of voltage-gated sodium channels in pain signaling. Physiol Rev. 2019;99:1079-1151. [DOI] [PubMed] [Google Scholar]
- 54. Denomme N, Lukowski AL, Hull JM, et al. The voltage-gated sodium channel inhibitor, 4,9-anhydrotetrodotoxin, blocks human nav1.1 in addition to nav1.6. Neurosci Lett. 2020;724:134853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanabe Y, Shiraishi S, Hashimoto K, et al. Taxane-induced sensory peripheral neuropathy is associated with an SCN9A single nucleotide polymorphism in Japanese patients. BMC Cancer. 2020;20:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mannerak MA, Lashkarivand A, Eide PK. Trigeminal neuralgia and genetics: a systematic review. Mol Pain. 2021;17:17448069211016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun JF, Xu YJ, Kong XH, Su Y, Wang ZY. Fenamates inhibit human sodium channel Nav1.7 and Nav1.8. Neurosci Lett. 2019;696:67-73. [DOI] [PubMed] [Google Scholar]
- 58. Kim CH, Oh Y, Chung JM, Chung K. Changes in three subtypes of tetrodotoxin sensitive sodium channel expression in the axotomized dorsal root ganglion in the rat. Neurosci Lett. 2002;323:125-128. [DOI] [PubMed] [Google Scholar]
- 59. Gold MS, Weinreich D, Kim CS, et al. Redistribution of Na(V)1.8 in uninjured axons enables neuropathic pain. J Neurosci. 2003;23:158-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Catterall WA, Striessnig J, Snutch TP, Perez-Reyes E. International Union of Pharmacology. XL. Compendium of voltage-gated ion channels: calcium channels. Pharmacol Rev. 2003;55:579-581. [DOI] [PubMed] [Google Scholar]
- 61. Phan NN, Wang CY, Chen CF, et al. Voltage-gated calcium channels: novel targets for cancer therapy. Oncol Lett. 2017;14:2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Altier C, Zamponi GW. Targeting Ca2+ channels to treat pain: T-type versus N-type. Trends Pharmacol Sci. 2004;25:465-470. [DOI] [PubMed] [Google Scholar]
- 63. Khuong TM, Hamoudi Z, Manion J, et al. Peripheral straightjacket (α2δ Ca2+channel subunit) expression is required for neuropathic sensitization in Drosophila. Philos Trans R Soc Lond B Biol Sci. 2019;374:20190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. French RJ, Zamponi GW. Voltage-gated sodium and calcium channels in nerve, muscle, and heart. IEEE Trans Nanobioscience. 2005;4:58-69. [DOI] [PubMed] [Google Scholar]
- 65. Burrell BD. Comparative biology of pain: what invertebrates can tell us about how nociception works. J Neurophysiol. 2017;117:1461-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783-791. [DOI] [PubMed] [Google Scholar]
- 67. Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci. 1994;10:189-191. [DOI] [PubMed] [Google Scholar]
- 68. Stone LS, Molliver DC. In search of analgesia: emerging roles of GPCRs in pain. Mol Interv. 2009;9:234-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dreborg S, Sundström G, Larsson TA, Larhammar D. Evolution of vertebrate opioid receptors. Proc Natl Acad Sci USA. 2008;105:15487-15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Memmert S, Damanaki A, Nokhbehsaim M, et al. Regulation of somatostatin receptor 2 by proinflammatory, microbial and obesity-related signals in periodontal cells and tissues. Head Face Med. 2019;15:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yamada Y, Post SR, Wang K, et al. Cloning and functional characterization of a family of human and mouse somatostatin receptors expressed in brain, gastrointestinal tract, and kidney. Proc Natl Acad Sci USA. 1992;89:251-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Patel YC. Somatostatin and its receptor family. Front Neuroendocrinol. 1999;20: 157-198. [DOI] [PubMed] [Google Scholar]
- 73. Dubos MP, Badariotti F, Rodet F, Lelong C, Favrel P. Molecular and physiological characterization of an invertebrate homologue of a calcitonin-related receptor. Biochem Biophys Res Commun. 2003;310:972-978. [DOI] [PubMed] [Google Scholar]
- 74. Schwartz J, Réalis-Doyelle E, Dubos MP, et al. Characterization of an evolutionarily conserved calcitonin signaling system in a lophotrochozoan, the Pacific oyster (Crassostrea gigas). J Exp Biol. 2019;222:jeb201319. [DOI] [PubMed] [Google Scholar]
- 75. Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11:1126-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mirabeau O, Joly JS. Molecular evolution of peptidergic signaling systems in bilaterians. Proc Natl Acad Sci USA. 2013;110:E2028-E2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Russell FA, King R, Smillie SJ, Kodji X, Brain SD. Calcitonin gene-related peptide: physiology and pathophysiology. Physiol Rev. 2014;94:1099-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Saito S, Fukuta N, Shingai R, Tominaga M. Evolution of vertebrate transient receptor potential vanilloid 3 channels: opposite temperature sensitivity between mammals and western clawed frogs. PLoS Genet. 2011;7:e1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tominaga M, Caterina MJ, Malmberg AB, et al. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron. 1998;21:531-543. [DOI] [PubMed] [Google Scholar]
- 80. Saito S, Shingai R. Evolution of thermoTRP ion channel homologs in vertebrates. Physiol Genomics. 2006;27:219-230. [DOI] [PubMed] [Google Scholar]
- 81. Pattison LA, Callejo G, St John Smith E. Evolution of acid nociception: ion channels and receptors for detecting acid. Philos Trans R Soc Lond B Biol Sci. 2019;374:20190291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dib-Hajj SD, Cummins TR, Black JA, Waxman SG. Sodium channels in normal and pathological pain. Annu Rev Neurosci. 2010;33:325-347. [DOI] [PubMed] [Google Scholar]
- 83. Jones SW. Overview of voltage-dependent calcium channels. J Bioenerg Biomembr. 1998;30:299-312. [DOI] [PubMed] [Google Scholar]
- 84. Smith ESJ, Lewin GR. Nociceptors: a phylogenetic view. J Comp Physiol A. 2009;195:1089-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Richards GS, Simionato E, Perron M, et al. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr Biol. 2008;18:1156-1161. [DOI] [PubMed] [Google Scholar]
- 86. Leys SP. Elements of a ‘nervous system’ in sponges. J Exp Biol. 2015;218:581-591. [DOI] [PubMed] [Google Scholar]
- 87. Shimizu H. Feeding and wounding responses in Hydra suggest functional and structural polarization of the tentacle nervous system. Comp Biochem Phys A. 2002;131:669-674. [DOI] [PubMed] [Google Scholar]
- 88. Ross KG, Currie KW, Pearson BJ, Zayas RM. Nervous system development and regeneration in freshwater planarians. Wiley Interdiscip Rev Dev Biol. 2017. Published online March 22, 2017. doi: 10.1002/wdev.266. [DOI] [PubMed] [Google Scholar]
- 89. Schafer W. Nematode nervous systems. Curr Biol. 2016;26:R955-R959. [DOI] [PubMed] [Google Scholar]
- 90. Hochner B. How nervous systems evolve in relation to their embodiment: wWhat we can learn from octopuses and other molluscs. Brain Behav Evol. 2013;82:19-30. [DOI] [PubMed] [Google Scholar]
- 91. Smarandache-Wellmann CR. Arthropod neurons and nervous system. Curr Biol. 2016;26:R960-R965. [DOI] [PubMed] [Google Scholar]
- 92. Formery L, Orange F, Formery A, et al. Neural anatomy of echinoid early juveniles and comparison of nervous system organization in echinoderms. J Comp Neurol. 2021;529:1135-1156. [DOI] [PubMed] [Google Scholar]
- 93. Woolf CJ. What is this thing called pain? J Clin Investig. 2010;120:3742-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-evb-10.1177_11769343231216914 for Phylogenetic Analysis Provides Insight Into the Molecular Evolution of Nociception and Pain-Related Proteins by Rujun Zhai and Qian Wang in Evolutionary Bioinformatics