Summary
The molecular mechanisms explaining why relapses of inflammatory arthritis recur at previously affected sites are unknown. In this issue of Immunity, Friščić1 et al. propose that local fibroblasts perpetuate inflammation after priming through cell-intrinsic complement C3, which reprograms their bioenergetics and activates the inflammasome.
When Nietzsche mused that ‘the advantage of a bad memory is that one can enjoy the same good things for the first time repeatedly’, he surely had no idea that the reverse was very apropos of inflammatory arthritis: the disadvantage of a good immunological memory is that one suffers the indignity of disease flares over and over again. In this issue of Immunity, Friščić et al. (Friščić1 et al., 2021) explore the robust observation that tissue-specific chronic (auto)inflammatory diseases are often characterized by relapses that affect the same tissue sites. This seemingly simple clinical observation suggests the existence of immunological memory that is site-specific, because why else would an arthritic flare show preference for previously affected sites? Although antigen-independent “training” of resident innate immune cells can be induced by danger sensing, these phenomena are not tissue-specific but rather systemic in nature and protect against recurrent infections (Netea et al., 2011). In fact, to date no adequate explanation has been provided for the site-specific immunological memory that is observed in inflammatory arthritides including rheumatoid and gouty arthritis.
The authors of the current paper hypothesized that structural tissue cells may encode prior danger sensing as epigenetic changes, thus “retaining” the ability to respond more aggressively to re-challenge at local sites – a rather heretical thought as memory is currently defined as a feature of innate and adaptive immune cells. The authors tackled this hypothesis using first rodent model systems and subsequently human patient material. In rodents, irrespective of the DAMP or PAMP used (monosodium urate crystals (MSU) or Zymosan injections into hind paws), iterative injection at the same site caused prolonged arthritis, whereas repeat injection in the previous non-injected hind paw did not, indicating local tissue priming by initial injections. Initial injection of hind paws with MSU also induced successful priming against subsequent injection with Zymosan, indicating that initial DAMP exposure sensitizes the local environment for subsequent DAMPs of a broad range. These observations remained intact in T and B cell-deficient (SCID and Rag1–/–) strains of mice, demonstrating independence from adaptive immunity. Synovial tissue macrophages were, unexpectedly, also not required, as their deletion (Cx3cr1cre:iDTR) did not prevent MSU-mediated tissue priming. However, adoptive transfer of synovial fibroblasts (SFs) from paws injected twice with MSU to non-injected (naïve) mice promoted more aggressive and prolonged arthritis on re-challenge compared to transfer of SF from non-injected mice. Fibroblast-like synoviocytes are recognized as key supporters of inflammation in rheumatoid arthritis through the production of inflammatory cytokines and other bone-destructive mediators (Bartok and Firestein, 2010). However, the data presented here suggest a much more sinister role for SFs/stromal cells in arthritis, namely that of remembering previous insults and remaining in a heightened state of alert that is detrimental to the host upon repeat DAMP sensing.
To delineate the mechanisms by which SFs mediate tissue priming, the investigators carried out (bulk and single cell) RNA- and ATAC-sequencing. In comparison to control cells, the transcriptomes of SFs from primed joints were enriched in expression of genes encoding proteins mediating inflammation, bone remodeling, cellular metabolism and components of the complement system. As expected, the epigenomes of these cells demonstrated enhanced accessibility at these loci, indicating that tissue priming had imprinted genome-wide changes in chromatin architecture, which in theory could impart prolonged gene expression. The complement system is an evolutionarily conserved component of immunity responsible for pathogen recognition and removal, as well as recruitment and activation of innate and adaptive immune cells (Merle et al., 2015). The key rate-limiting and highly regulated components are complement components (C)3 and C5. In addition to its well-described role as a hepatocyte-derived and serum-effective surveillance system, complement is also expressed and biologically active within immune cells (Liszewski et al., 2013) where it acts as a vital regulator of cellular metabolism (Kolev et al., 2015). When further probing the complement signature in SFs, the investigators found that C3 was especially induced in primed SFs and that its expression in non-hematopoietic cells was critically important to tissue priming, as C3–/– recipients of C3+/+ bone marrow showed significantly impaired tissue priming. Although the authors did not formally exclude the contribution of tissue-resident macrophages or neutrophils in tissue priming, a major contribution of this paper was further delineation of the role of SFs in this process. They carried out a key experiment by transferring C3+/+ or C3–/– CD45–CD31– SFs to non-injected paws of C3+/+ or C3–/– recipient mice. Transfer of C3–/– SFs from mice previously injected with MSU failed to imprint tissue priming in C3+/+ mice. Conversely, transfer of primed C3+/+ SF to C3–/– recipient mice did mediate inflammatory tissue priming. This indicates that SFs can act independently of other C3-expressing cells and that migrating, trans-differentiating myeloid cells are not masquerading as SFs in these models.
In human T cells, cell-intrinsic C3 and C5 orchestrate the bioenergetic pathways and the crosstalk with the NLRP3 inflammasome required for Th1 effector function (Arbore et al., 2016; Kolev et al., 2015, 2020). Similarly, Friščić et al. found that C3 replete SFs from primed sites had a metabolic profile biased towards glycolysis and mTOR activation and that C3-deficient SFs had significantly down-regulated metabolic adaptation, demonstrating features of senescence. Furthermore, C3 also supported IL1B gene transcription via HIF1α induction, maturation of IL-1β by assembly of the NLRP3 inflammasome and fostered local TNF and IL-6 secretion. Together, these potent inflammatory mediators induced osteoclast formation in the joint and perpetuated bone destruction.
Next, the authors addressed the question as to how C3 leads to modulation of cell metabolism, inflammasome activation and cytokine production in SFs of mice. C3 requires proteolytic activation into bioactive C3a and C3b fragments. C3a is a potent anaphylatoxin and mediates its cell-activating function through engagement of the G protein-coupled receptor C3aR (Verschoor et al., 2016). Indeed, iterative MSU-induced arthritis in mice with genetic ablation of C3ar1 showed significant blunting of priming and C3ar1-deficient SFs displayed reduced metabolic activity in vitro. The investigators did not prove unequivocally that processed C3a mediated priming by engaging C3aR in or on SFs because the mice were global C3ar1 knockout animals, which means that other cells in the environment could also not respond to C3a produced by SFs. A tissue-specific deletion of C3ar1 in fibroblasts is required to definitively answer whether C3a from SFs drives local priming by engaging C3aR in an autocrine/paracrine manner. It is worth considering this experiment, because if C3a engages C3aR on the surface of neighboring cells, administering an anti-C3aR monoclonal antibody could theoretically interrupt this pathogenic signaling, whereas if C3a binds C3aR on the inside of SFs, a cell-permeable inhibitor will be required instead. Similarly, the usage of mice in which C3 expression is specifically deleted in fibroblasts would have addressed remaining concerns about missing contributions of resident innate immune cells to tissue priming.
To address the relevance of their findings to human disease, the authors confirmed the presence of complement components, including C3, in SFs from patients with rheumatoid arthritis (RA) by both transcriptomic and proteomic approaches, showing a tendency to higher expression in leukocyte-rich RA. In this setting, repeated stimulation of in vitro cultured SFs from patients with TNF increased C3 expression and its activation to C3a (which agrees with our recent observation that C3 expressed by synovial T cells from RA patients is a biomarker of disease activity (Kolev et al., 2020)). Of note, cell-intrinsic C3a engaging C3aR in human T cells only supports homeostatic mTOR activity and their survival (Liszewski et al., 2013). The metabolic reprogramming necessary for effector function (IFN-γ and IL-1β secretion) requires autocrine/paracrine engagement of the C3b receptor, CD46, by T cell-intrinsic C3b (Kolev et al., 2015). CD46 is a human-specific molecule without a similar functional murine homologue on somatic tissue. Friščić et al. did not explore which C3 activation fragment contributes to SF priming in humans. Moreover, whilst mouse SFs upregulated C3aR strongly with each priming cycle, C3aR expression on human SFs did not change from baseline levels. It is therefore likely that priming in human tissues involves species-specific complement-dependent pathways, a notion that needs to be further explored.
Overall, the observations by Friščić et al. identify a cell-intrinsic role for C3 in SFs during tissue priming and relapses of arthritic disease. There are now three “million-dollar” questions – how long does C3-imprinting last, what are the transcriptional regulators of C3 in primed SFs, and what are the mechanisms for C3 protein processing? Several cytokines and integrins can initiate C3 transcription in a cell-specific manner, however, the C3 driver(s) in SFs in vivo were unexplored in this current study. The ATAC-seq data and changes in open chromatin regions noted in the paper, however, could be utilized to propose upstream transcriptional regulators based on transcription factor motif enrichment analysis. With regard to intracellular C3 protein activation, cathepsin L (CTSL) and a cell-intrinsic alternative complement pathway C3 convertase (C3bBb) can generate intracellular C3a and C3b (Yan et al., 2021). Although Friščić et al. noted increased gene expression of CTSL in primed human SFs, they did not address its functional significance, so the C3 activation pathway(s) in primed SFs remains to be identified.
The exciting identification of intrinsic C3 as driver of fibroblast priming by Friščić et al. lends further support to our growing understanding that intracellular complement (the “complosome”) is as a central player in basic physiological processes across cell populations. Since augmented complosome activity clearly contributes to human (arthritic) disease – not so nice to C you again! - it may be primetime to consider it as a worthy pharmacological target.
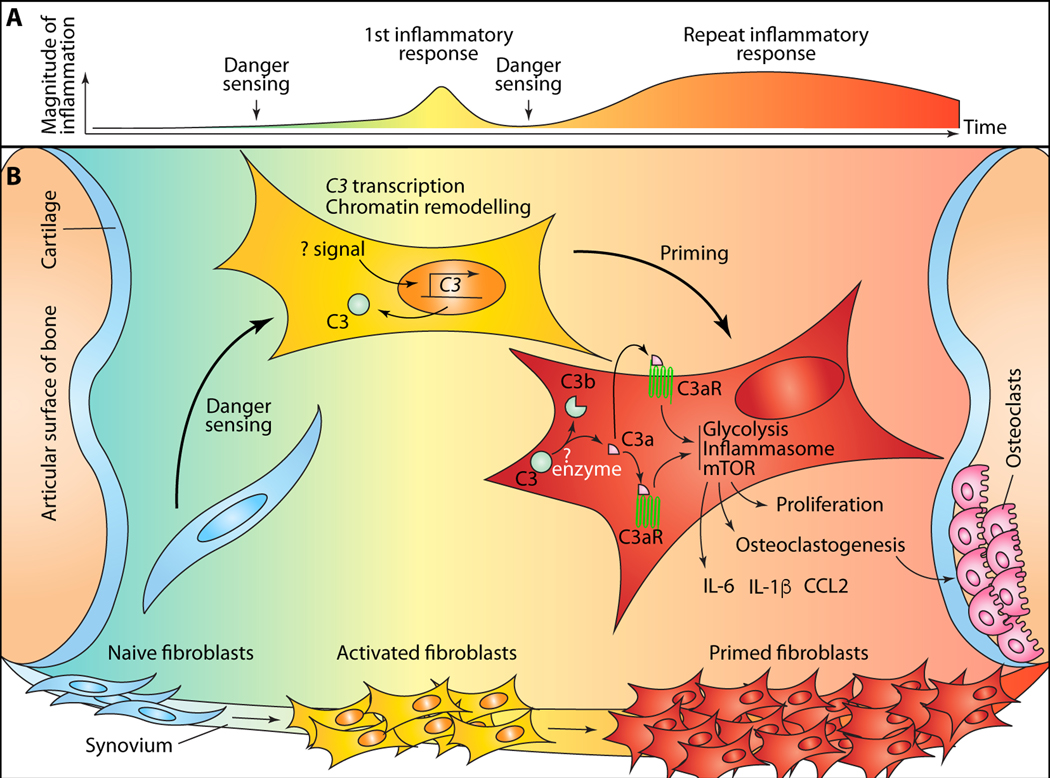
Figure 1. C3-dependent priming of fibroblasts (SFs) by danger sensing in synovial joints.
(A) naïve synovial fibroblasts (SFs) are activated and “primed” by repeated danger sensing. Phenotypically, this results in heightened and prolonged inflammation on repeat sensing of danger signals. (B) fibroblast priming events occurring within synovial tissue. Activation and priming of SFs induces chromatin remodeling and epigenetic changes that enhance accessibility at a number of loci encoding genes proteins mediating inflammation, bone remodeling, cellular metabolism and components of the complement system, including C3. The upstream signals required for transcription of C3 in SFs is currently unknown. C3 protein is enzymatically processed to activated fragments, including C3a and C3b. The enzymes mediating this process in SFs are not established. C3a ligands its cognate receptor, C3aR, either intracellularly and/or on the cell surface after secretion. Signaling mediated through the liganded receptor amplifies glycolysis and mTOR activation, and leads to inflammasome assembly. These result in secretion of inflammatory mediators, higher proliferative and invasive capacity of SFs and osteoclastogenesis.
Acknowledgements
This work was supported in part by the Intramural Research Program of the NIH, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (project number ZIA/DK075149 to B.A.) and the National Heart, Lung, and Blood Institute (NHLBI) (project number ZIA/Hl006223 to C.K.).
Footnotes
Declaration of interests
The authors declare no competing interests.
References
- Arbore G, West EE, Spolski R, Robertson AAB, Klos A, Rheinheimer C, Dutow P, Woodruff TM, Yu ZX, O’Neill LA, et al. (2016). T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4+ T cells. Science 352, aad1210–aad1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartok B, and Firestein GS (2010). Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev 233, 233–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friščić1 J, Böttcher2 M, Reinwald1 C, Bruns2 H, Wirth B, Popp S-J, Walker KI, Ackermann JA, Chen X, Turner J, et al. (2021). The complement system drives local inflammatory tissue priming by metabolic reprogramming of synovial fibroblasts. Immunity. [DOI] [PubMed] [Google Scholar]
- Kolev M, Dimeloe S, Friec GL, Navarini A, Arbore G, Povoleri GA, Fischer M, Belle R, Loeliger J, Develioglu L, et al. (2015). Complement Regulates Nutrient Influx and Metabolic Reprogramming during Th1 Cell Responses. Immunity 42, 1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev M, West EE, Kunz N, Chauss D, Moseman EA, Rahman J, Freiwald T, Balmer ML, Lötscher J, Dimeloe S, et al. (2020). Diapedesis-Induced Integrin Signaling via LFA-1 Facilitates Tissue Immunity by Inducing Intrinsic Complement C3 Expression in Immune Cells. Immunity 52, 513–527.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski MK, Kolev M, Friec GL, Leung M, Bertram PG, Fara AF, Subias M, Pickering MC, Drouet C, Meri S, et al. (2013). Intracellular complement activation sustains T cell homeostasis and mediates effector differentiation. Immunity 39, 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle NS, Church SE, Fremeaux-Bacchi V, and Roumenina LT (2015). Complement System Part I – Molecular Mechanisms of Activation and Regulation. Front Immunol 6, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Quintin J, and van der Meer JWM (2011). Trained Immunity: A Memory for Innate Host Defense. Cell Host Microbe 9, 355–361. [DOI] [PubMed] [Google Scholar]
- Verschoor A, Karsten CM, Broadley SP, Laumonnier Y, and Köhl J. (2016). Old dogs—new tricks: immunoregulatory properties of C3 and C5 cleavage fragments. Immunol Rev 274, 112–126. [DOI] [PubMed] [Google Scholar]
- Yan B, Freiwald T, Chauss D, Wang L, West E, Bibby J, Olson M, Kordasti S, Portilla D, Laurence A, et al. (2021). SARS-CoV2 drives JAK1/2-dependent local and systemic complement hyper-activation. In Press Science Immunol. [Google Scholar]