Abstract
Pyruvate, a precursor for several amino acids, can be synthesized from phosphoenolpyruvate by pyruvate kinase. Nevertheless, pyk1 pyk2 mutants of Saccharomyces cerevisiae devoid of pyruvate kinase activity grew normally on ethanol in defined media, indicating the presence of an alternative route for pyruvate synthesis. A candidate for this role is malic enzyme, which catalyzes the oxidative decarboxylation of malate to pyruvate. Disruption of open reading frame YKL029c, which is homologous to malic enzyme genes from other organisms, abolished malic enzyme activity in extracts of glucose-grown cells. Conversely, overexpression of YKL029c/MAE1 from the MET25 promoter resulted in an up to 33-fold increase of malic enzyme activity. Growth studies with mutants demonstrated that presence of either Pyk1p or Mae1p is required for growth on ethanol. Mutants lacking both enzymes could be rescued by addition of alanine or pyruvate to ethanol cultures. Disruption of MAE1 alone did not result in a clear phenotype. Regulation of MAE1 was studied by determining enzyme activities and MAE1 mRNA levels in wild-type cultures and by measuring β-galactosidase activities in a strain carrying a MAE1::lacZ fusion. Both in shake flask cultures and in carbon-limited chemostat cultures, MAE1 was constitutively expressed. A three- to fourfold induction was observed during anaerobic growth on glucose. Subcellular fractionation experiments indicated that malic enzyme in S. cerevisiae is a mitochondrial enzyme. Its regulation and localization suggest a role in the provision of intramitochondrial NADPH or pyruvate under anaerobic growth conditions. However, since null mutants could still grow anaerobically, this function is apparently not essential.
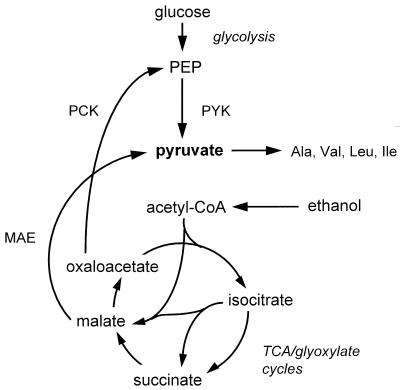
In the yeast Saccharomyces cerevisiae, pyruvate is a key intermediate in sugar dissimilation as well as a precursor for synthesis of the amino acids alanine, leucine, isoleucine, and valine (21, 28). During growth on sugars, pyruvate can be provided by pyruvate kinase (EC 2.7.1.40), the enzyme catalyzing the last reaction of the glycolytic pathway (Fig. 1).
FIG. 1.
Possible role of pyruvate kinase and malic enzyme in the formation of pyruvate as a biosynthetic precursor in S. cerevisiae. Abbreviations: PYK, pyruvate kinase; PCK, phosphoenolpyruvate carboxykinase; MAE, malic enzyme; acetyl-CoA, acetyl coenzyme A; TCA, tricarboxylic acid.
In S. cerevisiae, two structural genes (PYK1 and PYK2) that each encode an active pyruvate kinase isoenzyme have been identified (2, 3, 7, 16). PYK1 is essential for growth on sugars, and transcription of PYK1 is induced by them. The Pyk1p enzyme is strongly activated by fructose-1,6-bisphosphate. The second pyruvate kinase isoenzyme, Pyk2p, is insensitive to fructose-1,6-bisphosphate. Surprisingly, transcription of PYK2 is subject to glucose repression and is induced on ethanol. Unless it is overproduced, Pyk2p cannot sustain growth of pyk1 null mutants on sugars (3).
When S. cerevisiae is grown on ethanol or acetate, pyruvate kinase has no role in dissimilation. Nevertheless, pyruvate still has to be generated for amino acid biosynthesis. In theory, this may occur via at least two mechanisms (Fig. 1): (i) synthesis of phosphoenolpyruvate from acetyl coenzyme A via the glyoxylate cycle and the gluconeogenic enzyme phosphoenolpyruvate carboxykinase, thus providing the substrate for pyruvate kinase, and (ii) decarboxylation of malate, an intermediate of the glyoxylate cycle, to pyruvate by malic enzyme.
Malic enzyme [(S)-malate:NAD(P)+ oxidoreductase (decarboxylating); EC 1.1.1.38-40] catalyzes the oxidative decarboxylation of malate to pyruvate and carbon dioxide, using NAD+ or NADP+ as the electron acceptor [malate + NAD(P)+→pyruvate + CO2 + NAD(P)H]. The enzyme, which can also catalyze the reductive carboxylation of pyruvate to malate, occurs in a wide variety of organisms, including prokaryotes, fungi, plants, and animals. In eukaryotes, malic enzyme may reside in the cytosol, in mitochondria, or, in the case of plants, in the chloroplasts (24, 31).
Malic enzyme has been demonstrated in the yeasts Schizosaccharomyces pombe (27, 31), Rhodotorula glutinis (12), and Zygosaccharomyces bailii (15). Its status in S. cerevisiae is somewhat enigmatic. Polakis and Bartley (19) reported that this yeast did not contain NADP+-dependent malate-decarboxylating activity. Furthermore, the inability of pyruvate carboxylase-negative S. cerevisiae mutants to grow on glucose as the sole carbon source (26) indicates that reductive carboxylation of pyruvate by malic enzyme cannot replace the anaplerotic function of pyruvate carboxylase. This notwithstanding, Fuck et al. (13) reported the occurrence of very low activities of malic enzyme in cell extracts of S. cerevisiae. The partially purified S. cerevisiae malic enzyme was reported to have a high Km for malate (ca. 50 mM) and used either NAD+ or NADP+ as an electron acceptor. Occurrence of malic enzyme in S. cerevisiae is of applied significance, as conversion of malate to pyruvate (and subsequently to ethanol) can be used to decrease the acidity of wines. With this specific aim, the S. pombe MAE2 gene encoding malic enzyme has been introduced into S. cerevisiae (32).
So far, it is unknown whether and to what extent pyruvate kinase and malic enzyme contribute to the provision of pyruvate in S. cerevisiae cells growing on ethanol or acetate. Growth on ethanol of mutants lacking the two PYK genes has not been investigated in detail (3), and since no malic enzyme structural gene has been identified in S. cerevisiae, defined mutants lacking the enzyme are not available.
In the course of our work on pyruvate metabolism, evidence was obtained that in addition to the pyruvate kinase reaction, S. cerevisiae contains an alternative pathway for pyruvate synthesis. After systematic sequencing of the yeast genome, a new open reading frame (ORF) of S. cerevisiae which exhibits a high degree of similarity to malic enzyme structural genes from other organisms was identified. In this study, we demonstrate that this ORF indeed encodes an active malic enzyme. The aim of this work was to investigate the role, regulation, and subcellular localization of the S. cerevisiae malic enzyme.
MATERIALS AND METHODS
Yeast strains and maintenance.
All yeast strains used in this work were derived from isogenic strains of the CEN.PK series kindly provided by K.-D. Entian and P. Kötter (Frankfurt, Germany) and are described in Table 1. CEN.PK113-7D (MATa MAL2-8c SUC2) was used as a prototrophic wild-type strain. Chromosomal DNA from strain CEN.PK2-1C (MATa leu2-3, 112 ura3-52 trp1-289 his3-Δ1 MAL2-8c SUC2) was used to PCR amplify the MAE1 gene. Construction of strain EBY121B (MATα leu2-3, 112 ura3-52 trp1-289 his3-Δ1 MAL2-8c SUC2 pyk1Δ::HIS3 pyk2Δ::URA3) was described previously (3). For long-term maintenance, yeast strains were pregrown overnight in shake flasks at 30°C in YPE medium (10 g of Difco yeast extract, 20 g of Difco peptone, and 10 g of ethanol liter−1). After addition of glycerol to a concentration of 20% (vol/vol), 2 ml aliquots were stored at −80°C. Working stocks were maintained on YPE agar slants.
TABLE 1.
S. cerevisiae strains used
| Straina | Genotype | Source or reference |
|---|---|---|
| CEN.PK113-7D | MATa MAL2-8c SUC2 | Provided by K.-D. Entian |
| CEN.PK2-1C | MATa leu2-3, 112 ura3-52 trp1-289 his3-Δ1 MAL2-8c SUC2 | Provided by K.-D. Entian |
| EBY121B | MATα leu2-3, 112 ura3-52 trp1-289 his3-Δ1 MAL2-8c SUC2::HIS3 pyk2Δ::URA3 | 3 |
| EBY.D138A | MATa MAL2-8c SUC2 mae1Δ::lacZ-kanMX | This study |
| EBY.D149 | MAL2-8c SUC2 | This study |
| EBY.D150 | MAL2-8c SUC2 pyk1Δ | This study |
| EBY.D151 | MAL2-8c SUC2 pyk2Δ | This study |
| EBY.D153 | MAL2-8c SUC2 pyk1Δ mae1Δ::lacZ-kanMX | This study |
| EBY.D154 | MAL2-8c SUC2 pyk1Δpyk2Δ | This study |
| EBY.D155 | MAL2-8c SUC2 pyk2Δ mae1Δ::lacZ-kanMX | This study |
| EBY.D156 | MAL2-8c SUC2 pyk1Δ pyk2Δ mae1Δ::lacZ-kanMX | This study |
| CEN.PK113-13D | MATα ura3-52 MAL2-8c SUC2 | Provided by K.-D. Entian |
| EBY.D157 | MATα ura3-52 MAL2-8c SUC2 p426MET25 | This study |
| EBY.D160 | MATα ura3-52 MAL2-8c SUC2 YEpMET25-MAE1 | This study |
All strains were derived from isogenic strains of the CEN.PK family. EBY.D138A was derived from CEN.PK113-7D, EBY.D157 is CEN.PK113-13D with empty vector, and EBY.D160 is CEN.PK113-13D with MAE1 overexpression plasmid. Strains EBY.D149 to -156 are prototrophic segregants from crosses between EBY.D138A and EBY121B. Their mating types have not been determined, nor has it been analyzed whether pyk1::HIS3 or pyk2::URA3 segregants contain defective or wild-type native HIS3 and URA3 alleles, respectively.
Molecular biology techniques.
DNA and RNA were prepared and manipulated according to published procedures (22, 23, 25). Transformation of yeast cells was carried out by the freeze method (10). Escherichia coli JM101, DH5αF′, and SURE (Stratagene GmbH) were transformed by electroporation. p426MET25 (18) served as a vector.
Construction of deletion strains.
A mae1 deletion strain was constructed by using a modification of the PCR targeting technique (14, 35). pUG6 (14) was cleaved with SalI and BglII, removing the first of its two loxP sites. In their place, a 3.1-kb SalI/BglII fragment from plasmid pJJH397 (kindly provided by J. J. Heinisch, Düsseldorf, Germany) containing the lacZ gene of E. coli was inserted, resulting in plasmid pUG6lacZ. This plasmid was then used as a template to generate by PCR a DNA molecule consisting of a lacZ-kanMX marker cassette flanked by short homology regions to the MAE1 locus. For this purpose two oligonucleotides were constructed; one (S1-MAE1) [5′-ATGCTTAGAACCAGACTATCCGTTTCCGTTGCTGCTAGATCGCAACTATTCGTACGCTGCAGGTCGAC-3′]) contains the sequence from +1 (A of start codon) to +48 of the MAE1 ORF as its short flanking homology region, and the other (S2-MAE1 [5′-ACATCATGTAAAGAGCGGGTGTTGATACCAGCCCATTCCGCATCAGCATAGGCCACTAGTGGATCTG-3′]) contains the complementary sequence from +1329 to +1373 of the MAE1 ORF as its short flanking homology region. The corresponding 3′ ends of the two oligonucleotides bind at the beginning of the lacZ gene and, on the complementary strand, behind the loxP site in plasmid pUG6lacZ, respectively. PCR with the Expand High Fidelity PCR system (Boehringer, Mannheim, Germany), with S1-MAE1 and S2-MAE1 as primers and pUG6lacZ (50 ng) as the template (40 s at 95°C, 30 s at 50°C, 5 min at 68°C; 25 cycles) yielded a 4.6-kb DNA fragment. This PCR product could be used to replace the DNA sequence encoding amino acids 17 to 443 of Mae1p by the lacZ-kanMX module. Simultaneously, the β-galactosidase enzyme is fused in frame with the first 16 amino acids of Mae1p, allowing to use β-galactosidase activity as a reporter system for expression of MAE1. Five micrograms of the PCR product was transformed into strain CEN.PK113-7D, selecting for resistance to G418 (200 mg liter−1) on YPD agar plates. Whole yeast cell PCR was used to confirm the correct replacements in the G418-resistant transformants. One correct transformant strain, EBY.D138A (mae1Δ::lacZ-kanMX), was crossed with strain EBY121B (pyk1Δ pyk2Δ), the diploid strain was sporulated, and tetrads were dissected, resulting in all different combinations of MAE1, PYK1, and PYK2 wild-type and deletion alleles, all in an otherwise prototrophic background (Table 1).
Construction of plasmids.
The MAE1 gene was cloned by PCR with a pair of primers designed to amplify a DNA fragment enclosing the complete MAE1 ORF with 90 bp of its 5′ regulatory region and 23 bp of its 3′ region. Additionally, they added a BamHI and an EcoRI restriction site at the 5′ and 3′ ends of the PCR product, respectively. PCR with primers A12-MAE1 (5′-CGGGATCCAGCTTCGGATATTTGT-3′) and A42-MAE1 (5′-GGAATTCAGGCGTTGGTTATGCTT-3′), with the Expand High Fidelity PCR system and whole yeast cells of strain CEN.PK2-1C as the template (45 s at 95°C, 30 s at 50°C, 3 min at 68°C; 30 cycles), yielded a 2.1-kb DNA fragment. The fragment was cleaved with BamHI and EcoRI and cloned into the high-copy-number vector p426MET25 (18), resulting in plasmid YEpMET25-MAE1. In this plasmid, the MAE1 gene is under control of the methionine-repressible MET25 promoter and the CYC1 terminator.
Shake flask cultivation.
Specific growth rates were determined in 500-ml shake flasks containing 100 ml of a mineral medium supplemented with vitamins (30). Glucose (10 g liter−1) or ethanol (10 g liter−1) was added as the carbon source. Where indicated, l-alanine was added to a concentration of 10 mmol liter−1. Cultures were incubated at 30°C in a rotary shaker at 150 rpm and inoculated with 1 ml of an exponentially growing shake flask culture on the same medium. Optical densities at 660 nm were determined after dilution of culture samples with water to an optical density at 660 nm below 0.35. Exponential growth rates were determined in independent duplicate experiments.
Chemostat cultivation.
Aerobic and anaerobic, carbon- and energy-limited chemostat cultivation was performed at 30°C in 2-liter laboratory fermentors (Applikon, Schiedam, The Netherlands). Chemostats were operated at a dilution rate of (D) 0.10 h−1 and at a stirrer speed of 800 rpm. The culture pH was maintained at 5.0 by automatic addition of 2.0 M KOH via an Applikon ADI 1030 biocontroller. The working volume was kept at 1.0 liter by removal of effluent from the middle of the culture via an electrical level controller. This ensured that the biomass concentrations in the effluent differed by less than 2% from those taken directly from the culture. In aerobic cultures, an airflow of 0.5 liter min−1 was maintained by using a Brooks 5876 gas flow controller (Brooks B.V., Veenendaal, The Netherlands). During anaerobic cultivation, the fermenter and the medium reservoir were flushed with nitrogen gas at a flow rate of 0.5 liter min−1. To minimize oxygen diffusion, anaerobic fermentors and medium reservoirs were fitted with Norprene tubing. The dissolved oxygen tension, which was measured with an Ingold polarographic O2 electrode, remained above 30% of air saturation in aerobic cultures and below 0.1% of air saturation in anaerobic cultures. A defined mineral medium supplemented with vitamins (30) was used. Carbon sources (glucose, ethanol, and acetate) were added at a concentration of 250 mM carbon liter−1. For anaerobic cultivation, the medium was supplemented with the anaerobic growth factors Tween 80 (420 mg liter−1) and ergosterol (10 mg liter−1) (29). When asparate replaced ammonium sulfate as the nitrogen source, its concentration in the medium was 50 mmol liter−1.
Preparation of cell extracts.
Cells from exponentially growing shake flask cultures or chemostat cultures (ca. 80 mg [dry weight]) were harvested by centrifugation at 1,950 × g for 10 min, washed once, and resuspended in 5 ml of 10 mM potassium phosphate buffer (pH 7.5) containing 2 mM EDTA. After storage at −37°C, cells were thawed on ice and disrupted by sonication. When cell extracts were prepared for malic enzyme assays, cells were washed once and resuspended in 4 ml of Tris buffer (50 mM Tris-HCl [pH 7.5] containing 1 mM dithiothreitol and 2 mM MgCl2). When cell extracts were prepared for β-galactosidase assays, cells were washed once and resuspended in 50 mM phosphate buffer (pH 7.0). After resuspension in the appropriate buffer, cells were disrupted by sonication with 0.7-mm-diameter acid-washed glass beads (Sigma) at 0°C for 3 min (30-s intervals), using an MSE sonicator (150-W output, 8-μm peak-to-peak amplitude). Whole cells and debris were removed by centrifugation at 31,000 × g (20 min, 4°C). The clear supernatant was used as the cell extract. Cell extracts for malic enzyme assays were dialyzed for 4 h at 0°C against 50 mM Tris buffer by using 0.5- to 3.0-ml Slide-a-lyzer cassettes (10,000-molecular-weight cutoff; Pierce).
Isolation of mitochondria.
Mitochondria were isolated from glucose-limited, aerobic chemostat cultures (D = 0.10 h−1). The procedure involved differential centrifugation of cell homogenates obtained by controlled lysis of spheroplasts essentially as described by Bruinenberg et al. (6).
Enzyme assays.
Malic enzyme (EC 1.1.1.40) was assayed at 30°C and at 340 nm in a Hitachi model 100-60 spectrophotometer. The assay mixture (1 ml) contained 100 μmol of Tris-HCl (pH 7.5), 10 μmol of MgCl2, 0.4 μmol of NADP+, and cell extract. The reaction was initiated by addition of 100 μmol of potassium l-malate (pH 7.5). Specific activities were calculated based on an extinction coefficient of NADPH of 6.3 mM−1 cm−1. Latency of malic enzyme in mitochondrial fractions was investigated by measuring its activity in the presence and absence of 0.65 mol of sorbitol liter−1. β-Galactosidase (EC 3.2.1.23) activity in cell extracts was measured at 30°C with a discontinuous assay measuring the hydrolysis of o-nitrophenylgalactoside (17). Glucose-6-phosphate dehydrogenase (EC 1.1.1.49) was assayed as described by Postma et al. (20); cytochrome c oxidase (EC 1.9.3.1) was assayed by the method described by Douma et al. (11).
Protein determination.
Protein in cell extracts and mitochondrial fractions was assayed by the Lowry method. Bovine serum albumin (fatty acid free; Sigma) was used as a standard.
RESULTS
Growth characteristics of pyruvate kinase-deficient mutants.
To investigate whether the presence of either of the pyruvate kinase isoenzymes of S. cerevisiae is required for growth on ethanol as the sole carbon source, prototrophic mutants in which either PYK1 or PYK2 or both were deleted were constructed. As reported previously, a functional copy of PYK1 was required for growth on glucose as the sole carbon source, whereas deletion of PYK2 had no effect on the specific growth rate on glucose (Table 2). However, mutants lacking either one or both PYK genes still exhibited normal growth rates on a defined medium containing ethanol as the sole carbon source (Table 2). This finding indicated that during growth on ethanol, S. cerevisiae is able to synthesize pyruvate via an alternative pathway.
TABLE 2.
Maximum specific growth rates of the prototrophic wild-type strain S. cerevisiae EBY.D149 and of isogenic strains carrying various combinations of null mutations in PYK1, PYK2, and MAE1 (Table 1)a
| Strain | Relevant genotype | Specific growth rate (h−1)
|
||
|---|---|---|---|---|
| Glucose | Ethanol | Ethanol + alanine | ||
| EBY.D149 | PYK1 PYK2 MAE1 | 0.46 | 0.16 | 0.16 |
| EBY.D150 | pyk1Δ | No growth | 0.15 | 0.14 |
| EBY.D151 | pyk2Δ | 0.46 | 0.15 | 0.15 |
| EBY.D152 | mae1Δ | 0.45 | 0.14b | 0.18 |
| EBY.D153 | pyk1Δ mae1Δ | No growth | No growth | 0.16 |
| EBY.D154 | pyk1Δ pyk2Δ | No growth | 0.14 | 0.14 |
| EBY.D155 | pyk2Δ mae1Δ | 0.46 | 0.06 | 0.15 |
| EBY.D156 | pyk1Δ pyk2Δ mae1Δ | No growth | No growth | 0.16 |
Shake flask cultures were grown on a defined medium containing either 10 g of glucose, 10 g of ethanol, or 10 g of ethanol liter−1 plus 10 mM alanine. Data are presented as the average of at least two independent shake flask cultures; variation between duplicates was <8%.
After inoculation from an exponentially growing preculture, cultures of the mae1 null mutant on ethanol grew exponentially at a rate of 0.08 h−1 for 15 to 20 h. Then, the specific growth rate increased to ca. 0.14 h−1.
YKL029c, a possible structural gene encoding malic enzyme.
A possible explanation for the observed growth of pyk mutant strains on ethanol would be the existence of a malic enzyme, catalyzing the oxidative decarboxylation of malate to pyruvate (Fig. 1). Therefore, we compared the complete genome sequence of S. cerevisiae with the deduced amino acid sequence of the S. pombe malic enzyme, Mae2p (31). We found an ORF designated YKL029c which codes for a putative protein with 669 amino acids showing 47% amino acid identity to the S. pombe Mae2p. Moreover, amino acid residues which have been proposed to be involved in substrate or cofactor binding or are conserved between different malic enzymes (summarized in reference 31) are also conserved in S. cerevisiae Mae1p.
YKL029c/MAE1 encodes a functional malic enzyme.
To investigate whether YKL029c/MAE1 is indeed a structural gene encoding malic enzyme, malic enzyme activities were determined in cell extracts of the prototrophic wild-type strain EBY.D149 and strain EBY.D152. In the latter strain, most of the MAE1 ORF was deleted and replaced by a lacZ-kanMX cassette. In glucose-grown shake flask cultures, the wild-type strain exhibited a malic enzyme activity of 32 nmol min−1 mg of protein−1. Enzyme activities were assayed in dialyzed extracts, since high endogenous rates of NADP+ reduction were found in nondialyzed extracts. Malic enzyme activity was completely abolished in the deletion mutant (Table 3).
TABLE 3.
Specific activities of malic enzyme in dialyzed cell extracts of wild-type S. cerevisiae EBY.D149 and three isogenic mutantsa
| Strain | Relevant genotype | Malic enzyme activity (nmol min−1 mg of protein−1)
|
|
|---|---|---|---|
| Batch | Chemostat | ||
| EBY.D149 | MAE1 | 32 | 12 |
| EBY.D152 | mae1Δ | <1 | <1 |
| EBY.D157 | MAE1 p426MET25 | 26 | ND |
| EBY.D160 | MAE1 YEpMET25-MAE1 | 310 | 410 |
Cells were harvested from exponentially growing shake flask cultures on defined medium containing 10 g of glucose liter−1 or from steady-state, aerobic, and glucose-limited chemostat cultures (D = 0.10 h−1). Data represent the average from two independent experiments; variation between duplicates was less than 15%. ND, not determined.
To overexpress MAE1, the gene was placed under control of the strong methionine-repressible promoter MET25 in the multicopy vector p426MET25, which carries a URA3 selectable marker. Introduction of this construct into the wild-type strain CEN.PK113-13D in the absence of methionine resulted in an over 10-fold increase of the malic enzyme activity relative to an empty-vector reference strain and the prototrophic wild-type strain EBY.D149 (Table 2). These results are fully consistent with MAE1 being a structural gene encoding a protein with malic enzyme activity.
Relationship to malic enzymes from other organisms.
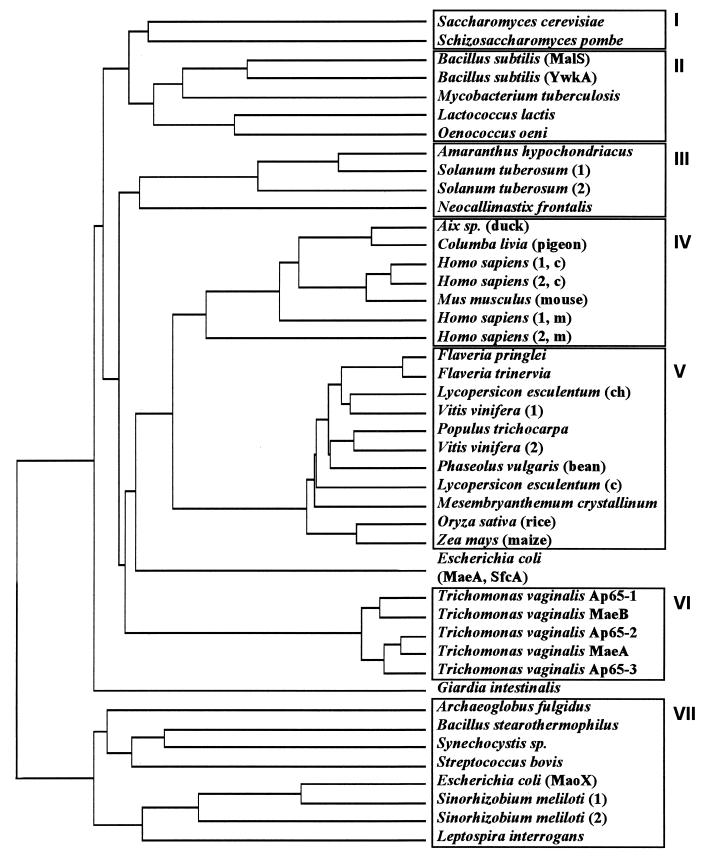
To gain some insight into the evolutionary relationships between S. cerevisiae Mae1p and malic enzymes from various organisms, we used the deduced amino acid sequences to calculate a dendrogram. Seven blocks of related malic enzymes from different organisms were found (Fig. 2). The first cluster comprises the malic enzymes of the yeasts S. pombe and S. cerevisiae. In contrast to the malic enzyme from S. pombe and most other organisms, the S. cerevisiae enzyme has a long amino-terminal extension of about 85 amino acids. The second group of sequences which are closely related to the yeast malic enzymes (about 40% identity) comprises isoenzymes from eubacteria, indicating that the yeast enzymes are more related to these bacterial isoenzymes than to those of higher eukaryotes, which comprise clusters III to VI. Moreover, there is an additional group of bacterial isoenzymes (cluster VII) which is only very distantly related to the other malic enzymes.
FIG. 2.
Sequence identity dendrogram of malic enzymes and related proteins from different organisms. The dendrogram was obtained by comparison of the deduced amino acid sequence of S. cerevisiae Mae1p against all nucleotide sequence databases (December 1997) dynamically translated in all reading frames; partial malic enzyme sequences were omitted. The sequences were clustered by the PC/GENE program (IntelliGenetics Inc., release 6.70, 1992) at standard settings. Blocks of homology are boxed and numbered I to VII. For simplicity, not all sequences of cluster VII are shown. If different malic enzyme-related sequences have been identified in an organism, this is indicated by the numbers or names in parentheses. In this case, if known, also the corresponding intracellular localization is indicated: c, cytoplasm; m, mitochondria; ch, chloroplasts.
Role of pyruvate kinase and malic enzyme during growth on ethanol.
To investigate whether malic enzyme is responsible for the synthesis of pyruvate in pyruvate kinase-negative mutants of S. cerevisiae, the mae1 deletion mutant EBY.D138A was crossed with strain EBY121B (pyk1Δ pyk2Δ), and after sporulation of the diploid strain, spores with all different combinations of the MAE1, PYK1, and PYK2 wild-type and deletion alleles in an otherwise prototrophic background were selected (Table 1).
The growth properties of the mae1 deletion strains EBY.D138A and EBY.D152 on yeast extract-peptone medium with glucose or ethanol as the carbon source were indistinguishable from the corresponding wild-type strain (data not shown). Deletion of MAE1 did not significantly affect the specific growth rate on glucose mineral medium of strains containing a wild-type PYK1 allele (Table 2). Deletion of MAE1 caused a reduction of the specific growth rate on ethanol. However, upon prolonged incubation, the specific growth rate on ethanol of mae1 null cultures increased to near wild-type levels (Table 2). The reason for this phenomenon, which was reproducibly observed in four independent growth experiments with the mae1 null strain, but not with the other strains, was not further investigated.
When both PYK1 and MAE1 were deleted, the cells were no longer able to grow on ethanol. In contrast, cells in which MAE1 and PYK2 were deleted still grew on ethanol, albeit more slowly than cells in which only MAE1 was deleted (Table 2). This finding indicates that during growth of S. cerevisiae on ethanol, pyruvate for biosynthetic purposes can be efficiently provided either by Pyk1p in combination with Pyk2p or by Mae1p. Indeed, growth of pyk1 mae1 double mutants or pyk1 pyk2 mae1 triple mutants on ethanol could be completely restored by the addition of low concentrations of alanine (Table 2) or pyruvate (data not shown). Growth of these mutants was also restored after transformation with plasmid YEpMET25-MAE1, demonstrating that the mae1 deletion is indeed the cause of the observed growth defect.
Regulation of malic enzyme synthesis.
Regulation of the MAE1 gene as a function of growth conditions was investigated by measuring malic enzyme activities in cell extracts and by Northern analysis of MAE1 transcription. Furthermore, the mae1 deletion was constructed in such a way that simultaneously the first 16 amino acids of the truncated MAE1 ORF were fused to the E. coli lacZ gene. This enabled us to study regulation of MAE1 by measuring β-galactosidase activities in cell extracts of the mae1 deletion mutant EBY.D152.
Experiments in shake flask cultures indicated that MAE1 was expressed at low levels during growth on glucose, ethanol, and acetate. Malic enzyme assays in the shake flask cultures suggested a twofold-higher activity in glucose-grown cultures than in cultures growing exponentially on ethanol or acetate (Table 4). However, experiments with the strain carrying the MAE1-lacZ fusion yielded β-galactosidase activities that were not substantially different for the three substrates tested. Also in the Northern experiments, the levels of the MAE1 transcripts did not differ more than twofold between the different conditions in shake flask cultures (Fig. 3). Addition of alanine, which could complement the growth defect of mae1Δ pyk1Δ double mutants (Table 2), had no effect on MAE1 expression (Table 4; Fig. 3).
TABLE 4.
Regulation of MAE1 expression in shake flask cultures and carbon-limited chemostat cultures (D = 0.10 h−1)a
| Cultivation method | Carbon source | Malic enzyme sp act (nmol min−1 mg of protein−1) | β-Galactosidase sp act (μmol min−1 mg of protein−1) |
|---|---|---|---|
| Shake flask | Glucose | 32 | 0.10 |
| Ethanol | 17 | 0.12 | |
| Ethanol + alanine | 19 | 0.08 | |
| Chemostat | Glucose, aerobic | 12 | 0.08 |
| Glucose, aerobic (N source, aspartate) | 13 | 0.10 | |
| Glucose, anaerobic | 49 | 0.18 | |
| Ethanol, aerobic | 12 | 0.08 | |
| Acetate, aerobic | 11 | 0.07 |
Unless indicated otherwise, ammonium sulfate was added as the nitrogen source. Specific activities of malic enzyme were measured in dialyzed cell extracts of the wild-type strain EBY.D149. In addition, activities of β-galactosidase were assayed in strain EBY.D152, in which the promoter region of the MAE1 gene had been fused with the E. coli lacZ gene. In the latter strain, malic enzyme activities were below the detection limit (1 nmol min−1 mg of protein−1) under all conditions tested. Data are the average of two independent assays; variation between duplicates was <15% for the malic enzyme assays and <20% for the β-galactosidase assays.
FIG. 3.
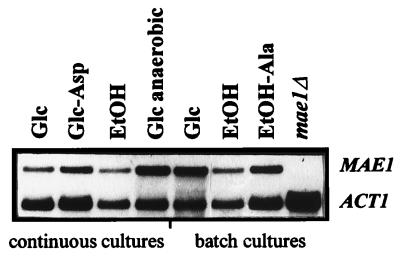
Northern analysis of MAE1 transcription. Cells of the wild-type strain EBY.D149 (lanes 1 to 7) and the mae1 deletion strain EBY.D152 (lane 8) were grown in carbon-limited chemostat cultures (continuous cultures) or in shake flask cultures (batch cultures) with different carbon sources (glucose or ethanol [EtOH]), with different nitrogen sources (ammonia or, in lane 2, aspartate), or supplemented with alanine (lane 7), either aerobically or anaerobically (lane 4). The mae1Δ strain was grown on YPD medium. Total RNA was prepared from these cultures. The filter was probed with a mixture of a 2.1-kb PCR fragment of the MAE1 coding region and a 3.2-kb DNA fragment containing the complete actin sequence (loading control), both labeled with 32P, in a 1:1 count ratio. The autoradiogram was exposed to Fuji RX.
In contrast to shake flask cultivation, chemostat cultivation offers the possibility to control specific growth rate via the supply rate of the growth-limiting nutrient. Thus, gene expression on different carbon sources or under different cultivation conditions can be compared at a fixed specific growth rate. Moreover, other important growth parameters such as dissolved oxygen concentration and culture pH can be kept constant, thus allowing studies on the impact of a single growth parameter on gene expression.
In aerobic, carbon-limited chemostat cultures of S. cerevisiae grown at a fixed specific growth rate of 0.10 h−1, MAE1 expression did not differ substantially when glucose, ethanol, or acetate was used as the carbon source (Fig. 3; Table 4). Also, provision of aspartate as a nitrogen source, which leads to an increased production of four-carbon metabolites by the cells, did not lead to a substantial increase of MAE1 expression (Fig. 3; Table 4). A clear induction of MAE1 expression was observed only during anaerobic cultivation on glucose, with a ca. threefold increase at the transcriptional level (Fig. 3; Table 4) and a ca. fourfold increase of the enzyme activity in cell extracts (Table 4) relative to the aerobic, glucose-limited reference cultures.
Subcellular localization of malic enzyme.
The subcellular localization of the S. cerevisiae malic enzyme was investigated by differential centrifugation after controlled lysis of spheroplasts, prepared from aerobic, glucose-limited chemostat cultures (D = 0.10 h−1). Initial experiments with wild-type cultures revealed enzyme activity in the particulate fraction. However, enzyme activities in the homogenates (in which the protein content was lower than in the particulate fractions) were below the detection limit, which precluded calculation of malic enzyme recoveries in the various fractions (data not shown). Since this result might have been due to the low activities of malic enzyme in wild-type cultures (Tables 3 and 4), fractionation experiments were repeated with the MAE1-overexpressing strain EBY.D160.
Similar to the mitochondrial marker enzyme cytochrome c oxidase, malic enzyme was completely recovered in the particulate fraction (Table 5). Conversely, the cytosolic enzyme glucose-6-phosphate dehydrogenase was recovered in the supernatant obtained after high-speed centrifugation of cell homogenates (Table 5).
TABLE 5.
Subcellular localization of malic enzymea
| Enzyme | Homogenate
|
Mitochondrial pellet
|
Supernatant
|
|||
|---|---|---|---|---|---|---|
| Sp act (μmol min−1 mg of protein−1) | Total activity (%) | Sp act (μmol−1 min−1 mg of protein−1) | Total activity (%) | Sp act (μmol−1 min−1 mg of protein−1) | Total activity (%) | |
| Glucose-6-phosphate dehydrogenase | 0.72 | 100 | 0.17 | 2 | 0.88 | 107 |
| Cytochrome c oxidase | 0.28 | 100 | 3.05 | 93 | 0.025 | 9 |
| Malic enzyme | 0.022b | 100 | 0.30 | 116 | <0.002 | 0 |
Recovery of malic enzyme and of the marker enzymes cytochrome c oxidase and glucose-6-phosphate dehydrogenase after differential centrifugation of a homogenate obtained after controlled lysis of spheroplasts of the Mae1p-overproducing strain S. cerevisiae EBY.D160. Cells were pregrown in an aerobic, glucose-limited chemostat culture (D = 0.10 h−1). Specific enzyme activities were determined without addition of sorbitol to the assays. Total amounts of protein in fractions: homogenate, 135.7 mg; mitochondria, 11.6 mg; supernatant, 121.8 mg.
The specific activity of malic enzyme in the homogenate obtained after Zymolase treatment was about 15-fold lower than that in dialyzed cell extracts obtained by sonication. This phenomenon was also observed in fractionation experiments with wild-type cultures (data not shown).
In the enzyme assays performed to calculate recoveries in the various fractions, no precautions were taken to osmotically stabilize the mitochondria. To investigate whether malic enzyme activity in the particulate fraction exhibited latency and consequently was of intraorganellar origin, are carried out control experiments in which malic enzyme assays were performed in the presence of 0.6 M sorbitol. Under these conditions, only a very low activity (22 nmol min−1 mg of protein−1) was measured in the particulate fraction. Malic enzyme activity increased to 300 nmol min−1 mg of protein−1 either after sonication (30 s) of the mitochondrial pellet or after addition of 0.05% Triton X-100 to the sorbitol-containing assay mixture prior to the enzyme assay.
DISCUSSION
Characteristics and compartmentation of malic enzyme in S. cerevisiae.
Our work supports the original report of Fuck et al. (13) that S. cerevisiae contains a genuine malic enzyme activity. We have identified and analyzed the corresponding structural gene and determined characteristics of its regulation.
Malic enzymes from different organisms differ in subcellular localization, being found in the cytosol, in mitochondria, or in chloroplasts. Moreover, they differ in cofactor requirement, being able to use either NAD+ or NADP+ or both as electron acceptors (24, 31). In subcellular fractionation experiments, malic enzyme activity was recovered in a particulate fraction (Table 5) from which it could be liberated after destruction of the organellar structures. The technique used in this study does not discriminate between mitochondria and other organelles. Although present at very low numbers in glucose-limited chemostat cultures, especially peroxisomes might contaminate the particulate fractions. However, as yeast peroxisomes are rather fragile (9, 37), the complete recovery and high degree of latency in the particulate fraction (Table 5) argues against a peroxisomal location of the S. cerevisiae malic enzyme. A further indication that Mae1p is a mitochondrial protein was obtained from its predicted amino acid sequence, which shows typical features of mitochondrial targeting sequences. Those targeting signals are N-terminal sequences of about 15 to 30 amino acids. They show a preponderance of basic and hydroxyl-carrying residues (Arg, Ser, and Leu) and a corresponding paucity of acidic residues (1, 34). The function of mitochondrial targeting sequences is largely determined by the overall balance between these basic, hydrophobic, and hydroxylated amino acids (1), and they may form amphiphilic helices (34). The 30 N-terminal amino acids of Mae1p include seven Arg, five Leu, and five Ser residues but no acidic amino acid. This clearly fulfills the demands of mitochondrial targeting sequences and supports our experimental results.
So far, the subcellular localization of malic enzymes in other yeasts has not been investigated. Especially interesting in this respect is the S. pombe malic enzyme, Mae2p (31), the predicted amino acid sequence of which does not contain a mitochondrial targeting sequence (our observation). Introduction of the S. pombe MAE2 gene together with a malate permease-encoding gene into S. cerevisiae has been successfully applied to reduce the malic acid content of wines (32). The rather inefficient degradation of malate by the recombinant strains was attributed to regulatory constraints on the S. pombe malic enzyme in S. cerevisiae. It will be interesting to see whether overexpression of S. cerevisiae MAE1 or of a truncated version of this enzyme lacking the mitochondrial targeting sequence can circumvent some of these difficulties.
In contrast to the S. pombe malic enzyme, which can use only NAD+ (27), malic enzyme from S. cerevisiae has been reported to use both NAD+ and NADP+ as electron acceptors (13). Furthermore, the Km value of the S. cerevisiae enzyme for malate was found to be over 15-fold higher than that of the S. pombe malic enzyme (13, 27). Despite these differences in cofactor specificity, substrate affinity, and, probably, localization, the amino acid sequences of the two yeast enzymes show significant similarities to each other (Fig. 2). Interestingly, the yeast enzymes are more closely related to a group of eubacterial malic enzymes (cluster II) than to those of higher eukaryotes, like plants (cluster III, including one fungal enzyme, and cluster V), animals and humans (cluster IV), and human parasites (cluster VI). Cluster VII comprises another group of eubacterial malic enzyme-related proteins which are, however, only very distantly related to the yeast enzymes. Intriguingly, within the individual clusters are found cytosolic as well as mitochondrial or chloroplast isoenzymes, which can be either NAD+ or NADP+ dependent. This finding indicates that sequence homologies among malic enzymes from various organisms are primarily determined by phylogenetic relationships rather than being the result of functional constraints related to catalytic properties and intracellular localization.
Regulation and physiological function of malic enzyme.
The MAE1 gene is expressed at low but constitutive levels during cultivation on the carbon sources glucose, ethanol, and acetate. Moreover, YKL029c/MAE1 was recently found to be constitutively transcribed during batch growth in a yeast extract-peptone medium from 2% glucose until glucose exhaustion (8). Although a three- to fourfold induction of MAE1 was observed under anaerobic conditions, a database search (36) with the promoter sequence of the MAE1 gene did not reveal any significant transcription factor binding sites.
The regulatory pattern of malic enzyme suggests a specific function in anaerobic metabolism, e.g., in the provision of intramitochondrial NADPH or pyruvate. However, the growth properties of an mae1 null mutant indicated that any such function was not essential under the experimental conditions tested. Growth of the mae1 mutants on glucose under aerobic conditions was not affected, and also in anaerobic, glucose-limited chemostat cultures of wild-type and mae1 null strains (D = 0.10 h−1), yields of biomass and ethanol were the same (data not shown).
A cyclic reaction involving pyruvate carboxylase, NAD+-dependent malate dehydrogenase, a mitochondrial dicarboxylate carrier, and (NADP+-linked) malic enzyme could act as a transhydrogenase system. Such a system could theoretically convert part of the NADH formed in biosynthesis (33) to NADPH, a cofactor for biosynthetic reactions. However, specific production rates of glycerol, which is the main redox sink for NADH under anaerobic conditions (33), were the same in anaerobic chemostat cultures of an mae1 null mutant and the isogenic wild type (data not shown).
Our results demonstrate that the mitochondrial malic enzyme can meet the biosynthetic requirement of S. cerevisiae for pyruvate during growth on ethanol or acetate. Alternatively, pyruvate can be provided by the Pyk1p isoenzyme of cytosolic pyruvate kinase. The recently discovered Pyk2p isoenzyme (3) alone could not sustain growth on ethanol. This is surprising, as its lack of activation by fructose-1,6-bisphosphate and relatively high transcript level during growth on ethanol (3) are both compatible with a role of Pyk2p during gluconeogenic growth, when intracellular fructose-1,6-bisphospate concentrations are low. Conversely, the kinetic properties of Pyk1p suggest a low in vivo activity during gluconeogenic growth, thus questioning its qualification in the provision of pyruvate (2). The higher growth rate on ethanol if not only Pyk1p but also Pyk2p is expressed (Table 2; compare EBY.D152 and EBY.D155) may reflect an in vivo interaction of the isoenzymes. Such an interaction (e.g., via formation of heteromers) might also explain the slightly different kinetics of pyruvate kinase activity in extracts of wild-type cells and pyk2 null mutants (3).
At least one additional S. cerevisiae enzyme activity described in the literature could also provide pyruvate for biosynthetic purposes. Serine dehydratase (encoded by the CHA1 gene) [4]) converts serine to pyruvate. The requirement for induction by serine (5) or other regulatory constraints may explain why this enzyme cannot meet the requirement for pyruvate of pyk1Δ pyk2Δ mae1Δ mutants during growth on ethanol media that contain ammonium salts as the nitrogen source. Suppressor mutants observed upon prolonged incubation of ethanol plates of pyk1Δ pyk2Δ mae1Δ mutants (2a) may represent regulatory mutants expressing serine dehydratase in media lacking serine.
Conclusions and outlook.
In this study, a clear phenotype of mae1 null mutants became apparent only when also PYK1 was inactivated. Thus, as in the other eukaryotic organisms investigated in the literature, the physiological function of malic enzyme in wild-type S. cerevisiae remains enigmatic. The identification of the MAE1 gene in S. cerevisiae and the accessibility of this yeast to molecular genetic and physiological investigations make this organism an interesting model system for further studies on this intriguing enzyme. Moreover, the availability of an Mae1p-overproducing strain will facilitate enzyme purification and thus a more detailed enzymological characterization of malic enzyme than was hitherto possible.
ACKNOWLEDGMENTS
We thank C. P. Hollenberg for his kind support and J. P. van Dijken for critical reading of the manuscript. We are grateful to K. Freidel for help in performing the Northern analysis and to M. A. H. Luttik for help with the subcellular fractionation experiments.
REFERENCES
- 1.Allison D S, Schatz G. Artificial mitochondrial presequences. Proc Natl Acad Sci USA. 1986;83:9011–9015. doi: 10.1073/pnas.83.23.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boles E. Pyruvate kinase. In: Entian K-D, Zimmermann F K, editors. Yeast sugar metabolism: biochemistry, genetics, biotechnology and applications. Lancaster, Pa: Technomic Publishing Co.; 1997. pp. 171–186. [Google Scholar]
- 2a.Boles, E. Unpublished data.
- 3.Boles E, Schulte F, Miosga T, Freidel K, Schlüter E, Zimmermann F K, Hollenberg C P, Heinisch J J. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J Bacteriol. 1997;179:2987–2993. doi: 10.1128/jb.179.9.2987-2993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornaes C, Petersen J G, Holmberg S. Serine and threonine catabolism in Saccharomyces cerevisiae: the CHA1 polypeptide is homologous with other serine and threonine dehydratases. Genetics. 1992;131:531–539. doi: 10.1093/genetics/131.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornaes C, Ignjatovic M W, Schjerling P, Kielland-Brandt M C, Holmberg S A. A regulatory element in the CHA1 promoter which confers inducibility by serine and threonine on Saccharomyces cerevisiae genes. Mol Cell Biol. 1993;13:7604–7611. doi: 10.1128/mcb.13.12.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruinenberg P M, van Dijken J P, Kuenen J G, Scheffers W A. Oxidation of NADH and NADPH by mitochondria from the yeast Candida utilis. J Gen Microbiol. 1985;131:1043–1051. [Google Scholar]
- 7.Burke R L, Tekamp-Olson P, Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983;258:2193–2201. [PubMed] [Google Scholar]
- 8.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 9.Distel B, van der Leij I, Kos W. Peroxisome isolation. In: Evans I, editor. Methods in molecular biology. Totowa, N.J: Humana Press; 1996. pp. 133–138. [DOI] [PubMed] [Google Scholar]
- 10.Dohmen R J, Strasser A W M, Höner C B, Hollenberg C P. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast. 1991;7:691–692. doi: 10.1002/yea.320070704. [DOI] [PubMed] [Google Scholar]
- 11.Douma A C, Veenhuis M, de Koning W, Evers M, Harder W. Dihydroxyacetone synthase is localized in the peroxisomal matrix of methanol-grown Hansenula polymorpha. Arch Microbiol. 1985;143:237–243. [Google Scholar]
- 12.Fernández M J, Medrano L, Ruiz-Amil M, Losada M. Regulation and function of pyruvate kinase and malate enzyme in yeast. Eur J Biochem. 1967;3:11–18. doi: 10.1111/j.1432-1033.1967.tb19493.x. [DOI] [PubMed] [Google Scholar]
- 13.Fuck E, Stärk G, Radler F. Äpfelsäurestoffwechsel bei Saccharomyces. II. Anreicherung und Eigenschaften eines Malatenzyms. Arch Mikrobiol. 1973;89:223–231. [PubMed] [Google Scholar]
- 14.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuczynski J T, Radler F. The anaerobic metabolism of malate of Saccharomyces bailii and the partial purification of malic enzyme. Arch Microbiol. 1982;131:266–270. doi: 10.1007/BF00405891. [DOI] [PubMed] [Google Scholar]
- 16.McNally T, Purvis I J, Fothergill-Gilmore L A, Brown A J P. The yeast pyruvate kinase gene does not contain a string of non-preferred codons: revised nucleotide sequence. FEBS Lett. 1989;247:312–316. doi: 10.1016/0014-5793(89)81359-6. [DOI] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18.Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5788. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polakis E S, Bartley W. Changes in the enzyme activities of Saccharomyces cerevisiae during aerobic growth on different carbon sources. Biochem J. 1965;97:284–297. doi: 10.1042/bj0970284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Postma E, Verduyn C, Scheffers W A, van Dijken J P. Enzymic analysis of the Crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol. 1989;53:468–477. doi: 10.1128/aem.55.2.468-477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pronk J T, Steensma H Y, van Dijken J P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schomburg D, Stephan D. Enzyme handbook. 9. Class 1.1 oxidoreductases EC 1.1.1.1-EC 1.1.1.149. Berlin, Germany: Springer Verlag; 1995. [Google Scholar]
- 25.Sherman F. Guide to yeast genetics and molecular biology. Getting started with yeast. Methods Enzymol. 1990;194:3–20. [Google Scholar]
- 26.Stucka R, Dequin S, Salmon J M, Gancedo C. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol Gen Genet. 1991;229:307–315. doi: 10.1007/BF00272171. [DOI] [PubMed] [Google Scholar]
- 27.Temperli A, Künsch U, Mayer K, Busch I. Reinigung und Eigenschaften der Malatdehydrogenase (decarboxylierend) aus Hefe. Biochim Biophys Acta. 1965;110:630–632. [PubMed] [Google Scholar]
- 28.Umbarger H E. Amino acid biosynthesis and its regulation. Annu Rev Biochem. 1978;47:533–606. doi: 10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]
- 29.Verduyn C, Postma E, Scheffers W A, van Dijken J P. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol. 1990;136:395–403. doi: 10.1099/00221287-136-3-395. [DOI] [PubMed] [Google Scholar]
- 30.Verduyn C, Postma E, Scheffers W A, van Dijken J P. Effect of benzoic acid on metabolic fluxes in yeast: a continuous-culture study on the regulation of respiration and fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 31.Viljoen M, Subden R E, Krizus A, van Vuuren H J J. Molecular analysis of the malic enzyme gene (mae2) of Schizosaccharomyces pombe. Yeast. 1994;10:613–624. doi: 10.1002/yea.320100506. [DOI] [PubMed] [Google Scholar]
- 32.Volschenk H, Viljoen M, Grobler J, Petzold B, Bauer F, Subden R E, Young R A, Lonvaud A, Denayrolles M, van Vuuren H J J. Engineering pathways for malate degradation in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:253–257. doi: 10.1038/nbt0397-253. [DOI] [PubMed] [Google Scholar]
- 33.van Dijken J P, Scheffers W A. Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol Rev. 1986;32:199–224. [Google Scholar]
- 34.von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 36.Wingender E, Kel A E, Kel O V, Karas H, Heinemeyer T, Dietze P, Knüppel R, Romaschenko A G, Kolchanov N A. TRANSFAC, TRRD and COMPEL: towards a federated database system on transcriptional regulation. Nucleic Acids Res. 1997;25:265–268. doi: 10.1093/nar/25.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]