Abstract
Posttranscriptional modification is common to many types of RNA, but the majority of information concerning structure and function of modification is derived principally from tRNA. By contrast, less is known about modification in rRNA in spite of accumulating evidence for its direct participation in translation. The structural identities and approximate molar levels of modifications have been established for 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfactaricus by using combined chromatography-mass spectrometry-based methods. Modification levels are exceptionally high for prokaryotic organisms, with approximately 38 modified sites in 16S rRNA and 50 in 23S rRNA for cells cultured at 75°C, compared with 11 and 23 sites, respectively, in Escherichia coli. We structurally characterized 10 different modified nucleosides in 16S rRNA, 64% (24 residues) of which are methylated at O-2′ of ribose, and 8 modified species in 23S rRNA, 86% (43 residues) of which are ribose methylated, a form of modification shown in earlier studies to enhance stability of the polynucleotide chain. From cultures grown at progressively higher temperatures, 60, 75, and 83°C, a slight trend toward increased ribose methylation levels was observed, with greatest net changes over the 23°C range shown for 2′-O-methyladenosine in 16S rRNA (21% increase) and for 2′-O-methylcytidine (24%) and 2′-O-methylguanosine (22%) in 23S rRNA. These findings are discussed in terms of the potential role of modification in stabilization of rRNA in the thermal environment.
Although most types of RNA are modified by enzymatic reactions following transcription, knowledge of the effects of posttranscriptional modifications on RNA structure, and thus ultimately on function, are drawn almost entirely from studies of tRNA (3, 4, 59) and of nucleoside or oligonucleotide models (15). By contrast, information on the structural identities (11) and roles of modifications in rRNA are much more limited, in spite of substantial evidence for the importance of both large and small ribosomal subunit RNAs in protein synthesis (23) and the observation that modified sites in E. coli are clustered at functional centers of the RNAs (5). Exceptions to these limitations have been the extensive work on mapping of modifications in some eukaryotic rRNAs, principally vertebrates and yeasts (34), and studies of the distribution and possible functions of the common modified nucleoside pseudouridine in rRNA from different phylogenetic sources (40, 41). Perhaps the best-documented conclusions regarding functional roles for modifications in rRNA are that they are required for subunit assembly and maturation (13, 14, 22, 44, 47, 53) and for fidelity of decoding (39). Otherwise, putative functions of rRNA modification, although interesting, remain in general speculative (31, 32, 40). Knowledge of posttranscriptional modifications in archaeal rRNA is particularly limited, although modification patterns in RNase T1 fragments in archaeal 16S rRNA in earlier work by Woese and colleagues (e.g., references 55 to 57) have served to indicate likely sites of conserved modifications and have shown the archaeal thermophiles to be more highly modified (56) than the eubacteria. 7-Methylguanosine (60) and N6,N6-dimethyladenosine (20) were identified in archaeal 16S rRNA. Otherwise, with the exception of 5S rRNAs from Pyrodictium occultum and Sulfolobus solfataricus (7) and pseudouridines in 23S rRNA from Halobacter halobium (41), there are no previous structure assignments for modified nucleosides in any archaeal rRNAs, nor is there information concerning the structural diversity of archaeal modifications in a phylogenetic sense.
This study was undertaken to establish the structural identities and approximate molar levels of posttranscriptional modifications in 16S and 23S rRNAs from the archaeal hyperthermophile S. solfataricus. Of potential relevance to rRNA, earlier studies of tRNA had revealed the possibility of changes in modification levels of specific modifications in response to cellular stress (9) and in particular temperature. An increase of methylation levels in tRNA was observed in response to culture temperature in the bacterial thermophile Bacillus stearothermophilus (57°C optimal for cell growth) (1), and a well-defined response in 5-methyl-2-thiouridine content and melting temperature (Tm) in tRNA from Thermus thermophilus (75°C optimum) was found to result from culture temperature changes (54). Experiments on unfractionated tRNA from the archaeal hyperthermophile Pyrococcus furiosus grown at 75, 85, and 100°C revealed progressively increasing amounts of three families of stabilizing tRNA nucleosides which were attributed to contribute to an exceptional Tm of 97°C (29). We therefore also examined the effects of cell culture temperature at 60, 75, and 83°C on modification levels of rRNA in S. solfataricus.
MATERIALS AND METHODS
Cells.
S. solfataricus P2 (ATCC 35092) was grown in Brock’s basal salts medium (6) adjusted to pH 3.4 with 9 N H2SO4. The medium was supplemented with yeast extract and sucrose to final concentrations of 0.1 and 0.2%, respectively. Cultures were grown aerobically at 60, 75, and 83°C in 500-ml batches, using an Innova 3000 shaker bath (New Brunswick Scientific, Edison, N.J.) rotating at 170 rpm filled with a mixture of ethylene glycol-H2O (1:1, vol/vol). Cell stocks for the 60 and 83°C cultures were produced from the 75°C optimum culture by changing the temperature of successive cultures in 3- to 5-degree increments. Growth phase was monitored by turbidity measurements at 600 nm. Cultures were harvested in the late exponential or early stationary phase by centrifugation at 7,000 rpm for 10 min. Multiple isolates from each of the three temperatures were combined, and cell pellets were stored at −20°C.
Isolation of rRNA.
Isolation procedures were adapted from standard protocols (38, 49). Frozen cells were ground with an equal amount (gram [wet weight]/gram) of alumina (Sigma type A-5) until a creamy paste was formed. The paste was diluted to approximately 0.3 g/ml with a buffer containing 20 mM Tris HCl (pH 7.5), 100 mM NH4Cl, 10 mM MgCl2, and 6 mM 2-mercaptoethanol followed by treatment with RNase-free DNase (5 μg/g of cell paste). After occasional grinding over 10 min, the alumina and cell debris were removed by centrifugation at 15,000 rpm for 50 min in a Beckman J2-21 centrifuge. Ribosomes were pelleted from the clear supernatant at 47,000 rpm for 3.5 h in a Beckman Ti 50 rotor. The supernatant was discarded; ribosome pellets were rinsed briefly with high-salt buffer (10 mM Tris HCl [pH 7.5], 500 mM NH4Cl, 10 mM MgCl2, 6 mM 2-mercaptoethanol) and resuspended in the same buffer. This suspension was layered over an equal volume of 20% sucrose and recentrifuged under the same conditions. The resulting 70S ribosome pellet was suspended in 10 mM Tris HCl (pH 7.5)–30 mM NH4Cl–10 mM MgCl2–6 mM 2-mercaptoethanol–1% NaCl–0.5% sodium dodecyl sulfate and then extracted with phenol. RNA was separated into 23S, 16S, and 5S RNAs by two successive 10 to 30% sucrose gradient centrifugations in 10 mM Tris HCl (pH 7.5)–50 mM KCl–1% methanol, using an SW28 rotor.
Enzymatic digestion of rRNA.
rRNA was hydrolyzed to nucleosides, using nuclease P1 (Gibco BRL, Gaithersburg, Md.), snake venom phosphodiesterase I (Sigma, St. Louis, Mo.), and bacterial alkaline phosphatase (Calbiochem, La Jolla, Calif.) as described by Crain (10). Digests were stored at −20°C until analysis by liquid chromatography-electrospray mass spectrometry (LC-MS).
LC-MS.
Analysis of nucleoside mixtures from rRNA digests was performed on a Hewlett-Packard 1090 series II liquid chromatograph (Hewlett-Packard Co., Palo Alto, Calif.) with a diode array UV detector (190 to 320 nm), directly interfaced to a Quattro II triple-quadrupole mass spectrometer (Micromass, Manchester, England) equipped with an electrospray ionization source. High-performance liquid chromatography (HPLC) separations were achieved by using a Supelcosil LC-18-S reversed-phase column (2.1 by 250 mm, 5-μm-diameter particles; Supelco, Bellefonte, Pa.) and a Supelguard LC-18-S guard column (2.1 by 20 mm; Supelco) thermostatted to 40°C, at a flow rate of 0.3 ml/min. Elution solvents consisted of 5 mM ammonium acetate (pH 6.0) (solvent A) and 40% (vol/vol) acetonitrile in water (solvent B). The gradient profile of Buck et al. (8) was altered to accommodate a lower buffer concentration (5 mM ammonium acetate [pH 6.0]) which is more compatible with electrospray ionization. UV data were acquired continuously, and mass spectra were recorded every 1.0 s during the 46-min chromatographic separation. The procedures and interpretation of data for qualitative LC-MS analysis of nucleosides in RNA hydrolysates have been previously described in detail (42).
LC-MS analysis with deuterium exchange.
The analysis of nucleoside mixtures was also carried out by LC-MS using deuterated solvents under conditions which would allow complete exchange of all labile hydrogen atoms with deuterium atoms (18). Procedures for the preparation of labeled HPLC solvents, including ND4COCH3, closely followed an earlier protocol (42). Complete equilibration of the column prior to sample analysis eliminated any variations in the degree of isotopic exchange.
Estimation of modified nucleoside molar ratios.
The number of modified residues present in 16S and 23S rRNAs was estimated on the basis of UV detection chromatograms generated from the LC-MS analysis of enzymatic hydrolyzates. The chromatographic peak areas were derived from a designated reference peak representing one of the major nucleosides (A, U, G, or C) or another modified nucleoside for which a standard molar response relationship had been previously established. Whenever possible, the measured numbers of modified residues were validated by an independent calculation using a second reference molar response. Experimental ratios were taken from HPLC peak areas from UV detection at 260 nm, while most of the standard molar ratios had been earlier measured at 254 nm. No significant error was introduced by using two different wavelengths except in the case of ac4C, which exhibits a substantial increase in molar absorptivity at 254 nm. For this nucleoside and for the partially resolved m2G peak (Fig. 1), all calculations were performed with 254-nm data. Standard molar response ratios of nucleosides were derived from the tabulated data of Gehrke and Kuo (21), or from LC-MS analysis of enzymatic digests of E. coli isoacceptor tRNAs, and of 16S and 23S rRNAs for which modification identities and levels have been well characterized (summarized in reference 11). Each value reported in Table 1 was established from the mean of three to five separate chromatographic analyses.
FIG. 1.
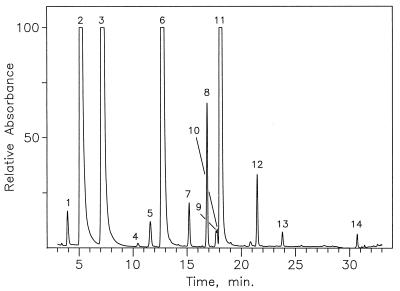
Determination of nucleosides from S. solfataricus 16S rRNA (75°C culture) by LC-MS analysis of an enzymatic digest, using UV detection at 260 nm. Nucleoside identities established from chromatographic retention times and mass spectra: 1, ψ; 2, C; 3, U; 4, m5C; 5, Cm; 6, G; 7, Um; 8, Gm; 9, m2G; 10, ac4C; 11, A; 12, Am; 13, m6A; 14, m26A. Unnumbered peaks were shown by corresponding mass spectra not to represent nucleosides.
TABLE 1.
Modified nucleosides from S. solfataricus 16S and 23S rRNAs
| rRNA | Nucleoside | Estimated no. of residuesa |
|---|---|---|
| 16S | Cm | 4.7 |
| Um | 4.6 | |
| Am | 4.4 | |
| Gm | 10.7 | |
| ψ | 4.8 | |
| m5C | ∼1 | |
| ac4C | ≥4b | |
| m2G | 1–2 | |
| m6A | 1–2 | |
| m26A | 1 | |
| 23S | Cm | 9.7 |
| Um | 9.5 | |
| Am | 11.1 | |
| Gm | 12.8 | |
| ψ | 3.7 | |
| m3U | ∼1 | |
| m5C | 1–2 | |
| ac4C | <1b | |
| m6A | —c | |
| m26A | — | |
| m2G | — |
75°C culture.
Level possibly influenced by chemical degradation. See text for discussion.
—, Measured as less than 0.2 residues each; judged to be probable contaminant. See text for discussion.
Nomenclature.
Nucleoside abbreviation symbols and names used are as listed in reference 33.
RESULTS
Determination of modified nucleosides in S. solfataricus 16S and 23S rRNAs.
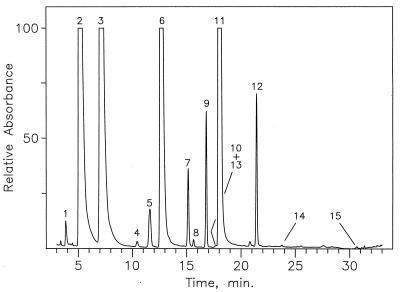
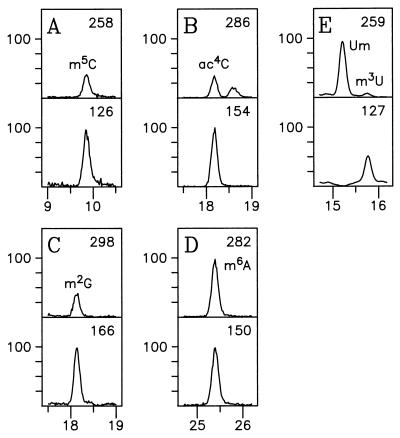
Chromatographic separation of nucleosides produced by complete enzymatic hydrolysis of 16S and 23S rRNAs are shown in Fig. 1 and 2, respectively. Nucleoside identities as indicated were established primarily from relative retention times and from electrospray ionization mass spectra recorded during passage of the HPLC effluent from the UV detector directly into the mass spectrometer (42). The retention times observed differed slightly from cataloged values, which were measured by using higher concentrations of ammonium acetate buffer (0.25 M [42] versus 5 mM in the present study). Evidence of trace cross-contamination between rRNAs was shown by the presence of substoichiometric impurities as indicated in Fig. 2. Of particular importance is the absence of tRNA in the rRNA isolates, which was established by the absence of characteristic tRNA nucleosides, in particular N6-threonylcarbamoyladenosine (t6A). Examples of the most difficult cases of identification of rRNA nucleosides are represented in Fig. 3, in which mass spectrometer ion signals were tracked chromatographically, with elution times measured within the approximate expected values (42). In both Fig. 1 and Fig. 2, two cases were encountered in which the assignments shown were required to be verified by the method of LC-MS analysis in deuterated mobile phase (18) in order to unambiguously distinguish base-methylated nucleoside isomers having similar chromatographic retention times. Those cases were N4-methyl versus 5-methylcytidine (four versus five deuterium atoms incorporated in the neutral molecule) and 2-methyl versus N6-methyladenosine (five versus four deuterium atoms incorporated), all of which were readily distinguished from their respective mass spectra, following deuterium exchange. In the case of m6A in 16S rRNA (Fig. 1), further confirmation of the site of adenosine methylation as N6 was gained by HPLC coinjection of the rRNA digest with authentic m6A (data not shown). The structures of modified nucleosides from both rRNAs characterized by LC-MS in this study are shown in reference 33 or may be viewed over the World Wide Web at http://www-medlib.med.utah.edu/RNAmods/RNAmods.html.
FIG. 2.
Chromatographic separation of nucleosides from S. solfataricus 23S rRNA (75°C culture) by LC-MS analysis of an enzymatic digest, using UV detection at 260 nm. Nucleoside identities established from chromatographic retention times and mass spectra: 1, ψ; 2, C; 3, U; 4, m5C; 5, Cm; 6, G; 7, Um; 8, m3U; 9, Gm; 10, ac4C; 12, Am. Substoichiometric impurities: 13(m2G), 14(m6A), and 15(m26A). Unnumbered peaks were shown by corresponding mass spectra not to represent nucleosides.
FIG. 3.
Identification of modified nucleosides in 16S rRNA (A to D) and 23S rRNA (E), using selected ion profiles for the protonated molecule (top sections) and protonated base (bottom sections) ions. (A) 5-Methylcytidine; (B) N4-acetylcytidine; (C) N2-methylguanosine; (D) N6-methyladenosine; (E) 2′-O-methyluridine (15.3 min) and 3-methyluridine (14.8 min). Relative ion abundance values (ordinates) have been normalized for each compound to 100% in panels A to D and to 100% for the most abundant m/z 259 signal in panel E.
Estimation of rRNA modification levels.
Approximate molar numbers of all posttranscriptionally modified nucleosides detected in digests of 16S and 23S rRNAs are given in Table 1. The abundances of ribose-methylated nucleosides (Cm, Um, Am, and Gm) were estimated from comparisons of HPLC UV detection peak areas compared with those of the parent unmodified nucleosides (C, U, A, and G) having the same chromophore, where numbers of residues are known from gene sequence data for 16S (17) and 23S (35) rRNAs. Through use of reference standard calibration curves of concentration versus HPLC peak area, care was taken to avoid nonlinear parts of the curve at higher concentrations to avoid deviations from Beer’s law relationships. Using the Fisons-Micromass data system, the useful region corresponded to responses for the major nucleosides in rRNA digests of fewer than 105 detector counts (arbitrary area units). Although the use of UV absorbance ratios is considered to be an accurate means of determining nucleoside molar ratios (21), our previous experience coupled with data from the present study suggests that values for the principal modified nucleosides in Table 1 (Cm, Um, Am, Gm, and ψ) are generally not accurate to more than ±10 to 15% of the values shown. For example, uncertainty in the estimated number of Cm residues, 4.7, would be approximately ±0.47 to 0.71 residues. The finding of a fractional number of residues represents a combination of experimental error and the possibility that some sites are substoichiometrically modified. Additional uncertainties prevail for nucleosides present at levels of only one or two residues per rRNA molecule, attributed principally to run-to-run variations and uncertainties in baseline positions used for peak area calculations. Hence, the values given in Table 1 for ac4C and the base-methylated nucleosides in general are much less reliable than for the principal modified nucleosides, even though their structural identities are certain. N4-Acetylcytidine levels have added uncertainty due to the possibility of slow chemical degradation (by deacylation) after rRNA isolation, so that an outside minimum of four residues was obtained for 16S rRNA, and its presence in 23S rRNA is considered tentative. In 23S rRNA, only trace amounts of m6A, m26A, and m2G were observed, and their detection is judged to likely result from contamination by other RNAs. In summary, from the 75°C culture isolates, approximately 38 posttranscriptionally modified residues were found in 16S rRNA, corresponding to 2.5% of total nucleosides, and 50 modified residues, or 1.7%, were found in 23S rRNA.
Variations in modified nucleoside content with cell culture temperature.
The optimal growth temperature for S. solfataricus P2 is approximately 75 to 80°C (46). The effects of cell culture temperature on each type of modified nucleoside were studied at culture temperatures 60, 75, and 83°C, which empirically included the lower and upper temperatures at which reasonable cell masses could be accumulated. Qualitatively, growth rate was slowest at 60°C and somewhat faster at 83°C, but with both rates slower than at 75°C. Relative molar values were obtained at each of the three culture temperatures for the five principal modified nucleosides for 16S and 23S rRNAs (Table 2). Measurements to establish trends for the less common modified nucleosides (Table 1) were considered insufficiently accurate to assess the effects of temperature. Further, a slight effect of unknown magnitude on modification levels may result from pooling of cells harvested at slightly different phases of their growth curves, although we are aware of no such previously reported effect. The fractional nature of molar values reported in Table 2 are a consequence of two factors: limitations in the accuracy and precision of the measurement and the fact that partial modification at a given site may occur in conjunction with variations in culture temperature (see below).
TABLE 2.
Variations in modified nucleoside content of S. solfataricus 16S and 23S rRNAs with culture temperature
| rRNA | Nucleoside | No. of residues
|
% Net changea | ||
|---|---|---|---|---|---|
| 60°C | 75°C | 83°C | |||
| 16S | Cm | 4.6 | 4.7 | 4.9 | +8.2 |
| Um | 4.1 | 4.6 | 4.6 | +13 | |
| Am | 4.0 | 4.4 | 4.8 | +21 | |
| Gm | 9.3 | 10.7 | 9.7 | +5.1 | |
| ψ | 3.8 | 4.8 | 3.9 | +1.9 | |
| 23S | Cm | 8.4 | 9.7 | 10.5 | +24 |
| Um | 9.7 | 9.5 | 9.5 | −1.0 | |
| Am | 10.1 | 11.1 | 10.9 | +7.9 | |
| Gm | 12.0 | 12.8 | 14.7 | +22 | |
| ψ | 3.3 | 3.7 | 3.8 | +14 | |
Net change from 60 to 83°C; values rounded from three significant figures.
The general relationship observed in both rRNAs was an increase in modification levels with culture temperature (with the exception of 2′-O-methyluridine in 23S rRNA). The clearest nucleoside composition changes within the accuracy and precision of the method were observed for Am in 16S rRNA (Table 2), with a smooth transition of +10% at each temperature increase (21% net increase), and for the ribose-methylated nucleosides Cm (24% net increase) and Gm (22% net increase) in 23S rRNA. In spite of the general upward trend in modification level with temperature, the remaining apparent increases, including 14% for ψ in 23S rRNA are, individually, judged to be of uncertain significance. In particular, net changes of several percent or less (ψ in 16S rRNA and Um in 23S rRNA) are interpreted as representing no change. The meaning of larger increases (>20%) in terms of individual sites of modification in rRNA is considered in Discussion.
DISCUSSION
The posttranscriptional modification levels in rRNA from S. solfataricus are the highest thus far reported for prokaryotic organisms and are probably also high for other members of the archaeal kingdom Crenarchaeota, most members of which are thermophiles (51). The principal modified residues in both 16S and 23S rRNA are ribose methylated at O-2′ (constituting 64 to 86% of all modifications) and may play a functional role in structural stabilization. The latter possibility is consistent with a discernible increase in ribose methylation levels with cell culture temperature, discussed below.
S. solfataricus 16S rRNA contains approximately 38 posttranscriptionally modified nucleosides when cultured at 75°C (Table 1), compared with 11 in E. coli (2, 28), which is the most extensively studied bacterium in this regard. The Sulfolobus 23S rRNA has a similarly high level, ∼50, compared with 23 in E. coli (30). It is notable that while 5S rRNA in mesophiles is rarely modified, a previous study revealed the 5S rRNA of S. solfataricus to contain one residue of 2′-O-methylcytidine (7), a modification which has been shown to confer conformational stability (26). The majority of modified sites in S. solfataricus 16S and 23S RNAs are methylated at O-2′ in ribose, in sharp distinction to E. coli, which contains only one such methyl group in 16S rRNA and three in 23S rRNA. The present data do not permit the conclusion that a direct role is played by these residues in secondary and tertiary stabilization of rRNA. However, three lines of evidence are clearly consistent with this possibility. First, extensive studies using nuclear magnetic resonance spectroscopy on nucleoside and oligonucleotide models have demonstrated that 2′-O-methylation results in thermodynamic stabilization of the C3′-endo sugar conformation in order to avoid base–O-2′ steric interactions in the alternate C2′-endo conformation (15) in both pyrimidines (27) and purines (25). This ribose modification stabilizes the A-type helical conformation found in RNA, resulting in enhanced regional rigidity (27) and higher Tm (12, 24), and thus is expected to mediate the effects of intrinsic dynamic motion in the thermophilic environment. The second line of evidence for consideration comes from earlier studies of unfractionated tRNA from the moderate bacterial thermophile B. stearothermophilus (1) and the archaeal hyperthermophile P. furiosus (29), in which increased ribose methylation levels resulted from elevations in culture temperature. It is recognized that alternate interpretations of the foregoing results are possible (and are not mutually exclusive): namely, ribose methylation to prevent phosphodiester bond hydrolysis (which requires a free 2′-hydroxyl) at high temperature or to prevent adventitious nuclease cleavages associated with an increased population of open tertiary structures at higher temperatures (1). However, in the case of P. furiosus (29) and other organisms (16, 45), overall tRNA modification was shown to result in higher Tm values, in addition to and independent of the G-C content of stem regions. Additionally, in the present study, higher culture temperature resulted in a discernible increase in ribose methylation levels in both rRNAs, a finding not previously reported for rRNA.
Although temperature-induced increases in ribose methylation of 20% or more (Table 2) are considered significant, the present study does not address the interesting question as to whether the increases are uniform throughout sites of modification in each molecule or are concentrated at functionally selective sites. This issue will ultimately require knowledge of specific sites of modification, which bears on the role of ribose methylation. Unfortunately, this issue was not addressed in previous work on temperature dependence of ribose methylation in thermophile tRNA, which was carried out with total digests of unfractionated tRNA (1, 29).
The modified nucleosides identified in 16S and 23S rRNAs (Table 1) constitute the first report of the major ribose-methylated nucleosides Cm, Um, and Gm in any prokaryotic small subunit rRNA and the first identification of Am in noneukaryotic rRNA (33). Similarly, the base-methylated nucleosides m5C, m6A, and m2G were not previously found in archaeal rRNA; however, in general we interpret these observations as reflecting the absence of previous structural studies on archaeal rRNA at the nucleoside level rather than attributing their presence necessarily to a unique property of Sulfolobus. The finding of pseudouridines in both rRNAs was anticipated, in view of its common occurrence in most types of RNA (33, 40). The occurrence of a single residue of N6,N6-dimethyladenosine in 16S rRNA is apparently not unique in the archaea (56), but the presence of two residues (at positions 1518 and 1519 [E. coli numbering system]) is more common, in all three phylogenetic domains (52).
As greater knowledge of the identities and distributions of posttranscriptional modifications in rRNA is gained, it will be interesting to consider the apparent differences in modification levels between tRNA and rRNA for a given organism. Results reported here show that levels of rRNA modification are about four- to fivefold lower than in tRNA (50), qualitatively similar to the difference observed for E. coli (21, 50). Particularly from the standpoint of thermal stabilization, one may speculate that one element of this apparently characteristic difference lies in the degree of structural support afforded by the ribosomal proteins, which interact extensively with rRNA (37). By contrast, the significantly smaller tRNA molecule is much more self-sufficient in terms of maintenance of the conserved L structure which is enforced in part by numerous modifications (3, 4, 43).
From the standpoint of nucleoside modifications in tRNA, the archaea are more similar to the eukaryotes than to bacteria (36), and it is therefore of note that even the thermophilic archaeon Sulfolobus has considerably lower levels of modification than eukaryotes (although our detailed knowledge base for eukaryotes is largely restricted to vertebrates [34, 40]). Comparison of modification patterns in rRNA from within the archaea are probably more relevant in a phylogenetic sense. It will therefore be of interest to examine mesophilic organisms from both major kingdoms of the domain Archaea (the Euryarchaeota and Crenarchaeota [58]) to further address the thesis that the nature and levels of rRNA modification found in S. solfataricus are to a significant extent devoted to RNA stabilization at high temperatures.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM29812 from the National Institute of General Medical Sciences.
We are grateful to W. F. Doolittle, Dalhousie University, for the 16S rRNA gene sequence for S. solfataricus P2 and to M. Schenk, Dalhousie University, for starting cell cultures and instructions regarding their growth. Figures were prepared by S. C. Pomerantz.
REFERENCES
- 1.Agris P F, Koh P, Söll D. The effect of growth temperature on the in vivo ribose methylation of Bacillus stearothermophilus. Arch Biochem Biophys. 1973;154:277–282. doi: 10.1016/0003-9861(73)90058-1. [DOI] [PubMed] [Google Scholar]
- 2.Bakin A, Kowalak J A, McCloskey J A, Ofengand J. The single pseudouridine residue in Escherichia coli 16S RNA is located at position 516. Nucleic Acids Res. 1994;18:3681–3684. doi: 10.1093/nar/22.18.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björk G, Ericson J U, Gustafsson C E D, Hagervall T G, Jönsson Y H, Wikström P M. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- 4.Björk G. The role of modified nucleosides in tRNA interactions. In: Hatfield D L, Lee B J, Pirtle R M, editors. Transfer RNA in protein synthesis. Boca Raton, Fla: CRC Press; 1992. pp. 23–85. [Google Scholar]
- 5.Brimacombe R, Mitchell P, Osswald M, Stade K, Bochkariov D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 1993;7:161–167. doi: 10.1096/fasebj.7.1.8422963. [DOI] [PubMed] [Google Scholar]
- 6.Brock T D, Brock K M, Belly R T, Weiss R L. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 7.Bruenger E, Kowalak J A, Kuchino Y, McCloskey J A, Mizushima H, Stetter K O, Crain P F. 5S rRNA modification in the hyperthermophilic archaea Sulfolobus solfataricus and Pyrodictium occultum. FASEB J. 1993;7:196–200. doi: 10.1096/fasebj.7.1.8422966. [DOI] [PubMed] [Google Scholar]
- 8.Buck M, Connick M, Ames B N. Complete analysis of tRNA-modified nucleosides by high performance liquid chromatography: the 29 modified nucleosides of Salmonella typhimurium and Escherichia coli tRNA. Anal Biochem. 1983;129:1–13. doi: 10.1016/0003-2697(83)90044-1. [DOI] [PubMed] [Google Scholar]
- 9.Buck M, Ames B N. A modified nucleotide in tRNA as a possible regulator of aerobiosis: synthesis of cis-2-methyl-thioribosylzeatin in the tRNA of Salmonella. Cell. 1984;36:523–531. doi: 10.1016/0092-8674(84)90245-9. [DOI] [PubMed] [Google Scholar]
- 10.Crain P F. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 11.Crain P F, McCloskey J A. The RNA modification database—1998. Nucleic Acids Res. 1998;26:196–197. doi: 10.1093/nar/26.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cummins L L, Owens S R, Risen L M, Lesnick E A, Freier S M, McGee D, Guinosso C J, Cook P D. Characterization of fully modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham P R, Weitzmann C J, Nègre D, Sinning J G, Frick V, Nurse K, Ofengand J. In vitro analysis of the role of rRNA in protein synthesis: site-specific mutation and methylation. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C: ASM Press; 1990. pp. 243–252. [Google Scholar]
- 14.Cunningham P R, Richard R B, Weitzmann C J, Nurse K, Ofengand J. The absence of modified nucleotides affects both in vitro assembly and in vitro function of the 30S-ribosomal subunit of Escherichia coli. Biochimie. 1991;73:789–796. doi: 10.1016/0300-9084(91)90058-9. [DOI] [PubMed] [Google Scholar]
- 15.Davis D R. Biophysical and conformational properties of modified nucleosides in RNA (nuclear magnetic resonance studies) In: Grosjean H, Benne R, editors. Modification and editing of RNA: the alteration of RNA structure and function. Washington, D.C: ASM Press; 1998. pp. 85–102. [Google Scholar]
- 16.Derrick W B, Horowitz J. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: evidence for stable base modification-dependent conformers. Nucleic Acids Res. 1993;21:4948–4953. doi: 10.1093/nar/21.21.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doolittle, W. F. (Dalhousie University). 1996. Personal communication.
- 18.Edmonds C G, Pomerantz S C, Hsu F F, McCloskey J A. Thermospray liquid chromatography/mass spectrometry in deuterium oxide. Anal Chem. 1988;60:2314–2317. doi: 10.1021/ac00171a034. [DOI] [PubMed] [Google Scholar]
- 19.Edmonds C G, Crain P F, Gupta R, Hashizume T, Hocart C H, Kowalak J A, Pomerantz S C, Stetter K O, McCloskey J A. Posttranscriptional modification of tRNA in thermophilic archaea (archaebacteria) J Bacteriol. 1991;173:3138–3148. doi: 10.1128/jb.173.10.3138-3148.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox G E, Magrum L J, Balch W E, Wolfe R S, Woese C R. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc Natl Acad Sci USA. 1977;74:4537–4541. doi: 10.1073/pnas.74.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gehrke C W, Kuo K C. Ribonucleoside analysis by reversed-phase high performance liquid chromatography. J Chromatogr A. 1990;45:A3–A64. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- 22.Green R, Noller H F. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA. 1996;2:1011–1021. [PMC free article] [PubMed] [Google Scholar]
- 23.Green R, Noller H F. Ribosomes and translation. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 24.Inoue H, Hayase Y, Imura A, Iwai S, Miura K, Ohtsuka E. Synthesis and hybridization studies on two complementary nona(2′-O-methyl) ribonucleotides. Nucleic Acids Res. 1987;15:6131–6148. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai G, Ue H, Yasuda M, Sakamoto K, Hashizume T, McCloskey J A, Miyazawa T, Yokoyama S. Relation between functions and conformational characteristics of modified nucleosides found in tRNAs. Nucleic Acids Symp Ser. 1991;25:49–50. [PubMed] [Google Scholar]
- 26.Kawai G, Hashizume T, Yasuda M, Miyazawa T, McCloskey J A. Conformational rigidity of N4-acetyl-2′-O-methylcytidine found in tRNA of extremely thermophilic archaebacteria (archaea) Nucleosides Nucleotides. 1992;11:759–771. [Google Scholar]
- 27.Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, Iimori T, Watanabe T, Miyazawa T, Yokoyama S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- 28.Kowalak J A, Pomerantz S C, Crain P F, McCloskey J A. A novel method for the determination of posttranscriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 1993;21:4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalak J A, Dalluge J J, McCloskey J A, Stetter K O. Role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry. 1994;33:7869–7876. doi: 10.1021/bi00191a014. [DOI] [PubMed] [Google Scholar]
- 30.Kowalak J A, Bruenger E, McCloskey J A. Posttranscriptional modification of the central loop of domain V in E. coli 23S ribosomal RNA. J Biol Chem. 1995;270:17758–17764. doi: 10.1074/jbc.270.30.17758. [DOI] [PubMed] [Google Scholar]
- 31.Lane B G, Ofengand J, Gray M W. Pseudouridine in the large subunit (23S-like) ribosomal RNA: the site of peptidyl transferase in the ribosome? FEBS Lett. 1992;302:1–4. doi: 10.1016/0014-5793(92)80269-m. [DOI] [PubMed] [Google Scholar]
- 32.Lane B G, Ofengand J, Gray M W. Pseudouridine and O2′-methylated nucleosides. Significance of their selective occurrence in rRNA domains that function in ribosome-catalyzed synthesis of the peptide bond in proteins. Biochimie. 1995;77:7–15. doi: 10.1016/0300-9084(96)88098-9. [DOI] [PubMed] [Google Scholar]
- 33.Limbach P A, Crain P F, McCloskey J A. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maden B E H. The numerous modified nucleosides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 35.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCloskey J A. Nucleoside modification in archaebacterial transfer RNA. Syst Appl Microbiol. 1986;7:246–252. [Google Scholar]
- 37.Mueller F, Brimacombe R. A new model for the three-dimensional folding of Escherichia coli 16S ribosomal RNA. II. The RNA-protein interaction data. J Mol Biol. 1997;271:545–565. doi: 10.1006/jmbi.1997.1211. [DOI] [PubMed] [Google Scholar]
- 38.Nierhaus K H. Reconstitution of ribosomes. In: Spedding G, editor. Ribosomes and protein synthesis. A practical approach. Oxford, England: Oxford University Press; 1990. pp. 161–189. [Google Scholar]
- 39.O’Connor M, Thomas C L, Zimmerman R A, Dahlberg A E. Decoding fidelity at the ribosomal A and P sites: influence of mutations in three different regions of the decoding domain in 16S RNA. Nucleic Acids Res. 1997;25:1185–1193. doi: 10.1093/nar/25.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ofengand J, Bakin A, Wrzesinski J, Nurse K, Lane B G. The pseudouridine residues of ribosomal RNA. Biochem Cell Biol. 1995;73:915–924. doi: 10.1139/o95-099. [DOI] [PubMed] [Google Scholar]
- 41.Ofengand J, Bakin A. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria, and chloroplasts. J Mol Biol. 1997;266:246–268. doi: 10.1006/jmbi.1996.0737. [DOI] [PubMed] [Google Scholar]
- 42.Pomerantz S C, McCloskey J A. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 1990;193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- 43.Quigley G J, Rich A. Structural domains of transfer RNA molecules. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- 44.Rydén-Aulin M, Shaoping Z, Kylsten P, Isaksson L A. Ribosome activity and modification of 16S RNA are influenced by deletion of ribosomal protein S20. Mol Microbiol. 1993;7:983–992. doi: 10.1111/j.1365-2958.1993.tb01190.x. [DOI] [PubMed] [Google Scholar]
- 45.Sampson J R, Uhlenbeck O C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schenk, M. (Dalhousie University). 1996. Personal communication.
- 47.Sirum-Connolly K, Mason T L. Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science. 1993;262:1886–1889. doi: 10.1126/science.8266080. [DOI] [PubMed] [Google Scholar]
- 48.Sirum-Connolly K, Peltier J M, Crain P F, McCloskey J A, Mason T L. Implications of a functional large ribosomal RNA with only three modified nucleotides. Biochimie. 1995;77:30–39. doi: 10.1016/0300-9084(96)88101-6. [DOI] [PubMed] [Google Scholar]
- 49.Spedding G. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles. In: Spedding G, editor. Ribosomes and protein synthesis. A practical approach. Oxford, England: Oxford University Press; 1990. pp. 1–29. [Google Scholar]
- 50.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stetter K O. Life at the upper temperature border. In: Trân Thanh Vân J K, Mounolou J C, Schneider J, McKay C, editors. Frontiers of life. Gif-sur-Yvette, France: Editions Frontières; 1992. pp. 195–219. [Google Scholar]
- 52.Van Kippenberg P H, Van Kimmenade J M A, Heus H A. Phylogeny of the conserved 3′ terminal structure of the RNA of small ribosomal subunits. Nucleic Acids Res. 1984;12:2595–2604. doi: 10.1093/nar/12.6.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaughan M H, Soeiro R, Warner J R, Darnell J E. Role of RNA methylation in rRNA maturation. Proc Natl Acad Sci USA. 1967;58:1527–1534. doi: 10.1073/pnas.58.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe K, Shinma M, Oshima T, Nishimura S. Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem Biophys Res Commun. 1976;72:1137–1144. doi: 10.1016/s0006-291x(76)80250-1. [DOI] [PubMed] [Google Scholar]
- 55.Woese C R, Gutell R, Gupta R, Noller H F. Detailed analysis of the higher order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983;47:621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woese C R, Gupta R, Hahn C M, Zillig W, Tu J. The phylogenetic relationships of three sulfur dependent archaebacteria. Syst Appl Microbiol. 1984;5:97–105. doi: 10.1016/s0723-2020(84)80054-5. [DOI] [PubMed] [Google Scholar]
- 57.Woese C R, Olsen G J. Archaebacterial phylogeny: perspectives on the urkingdoms. Syst Appl Microbiol. 1986;7:161–177. doi: 10.1016/s0723-2020(86)80001-7. [DOI] [PubMed] [Google Scholar]
- 58.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yokoyama S, Nishimura S. Modified nucleosides and codon recognition. In: Söll D, RajBhandary U L, editors. tRNA: structure, biosynthesis, and function. Washington, D.C: ASM Press; 1995. pp. 207–223. [Google Scholar]
- 60.Zueva V S, Mankin A S, Bogdanov A A, Thurlow D L, Zimmerman R A. Occurrence and location of 7-methylguanine residues in small-subunit ribosomal RNAs from eubacteria, archaebacteria and eukaryotes. FEBS Lett. 1985;188:233–238. [Google Scholar]