Abstract
The xylR and xylS genes are divergent and control transcription of the TOL plasmid catabolic pathways for toluene metabolism. Four promoters are found in the 300-bp intergenic region: Pr1 and Pr2 are constitutive ς70-dependent tandem promoters that drive expression of xylR, while expression of the xylS gene is driven from Ps2, a constitutive ς70-dependent promoter, and by the regulatable ς54 class Ps1 promoter. In Ps1 the XylR targets (upstream activator sequences [UASs]) overlap the Pr promoters, and two sites for integration host factor (IHF) binding are located at the region from positions −2 to −30 (−2/−30 region) and the −137/−156 region, the latter overlapping the Pr promoters. When the XylR protein binds to the UASs in the absence of effector, it represses expression from Pr promoters. In the XylR-plus background and in the absence of an effector, the level of expression from Ps1 is low, although detectable, whereas Ps2 is active. In this background and in the presence of an effector, XylR increases autorepression. In a ς54-deficient Pseudomonas putida background, no expression occurred from Ps1 regardless of the presence of an effector. However, in the presence of an effector, the amount of RNA produced from Pr promoters was almost undetectable. This finding suggests that when no transcription occurred at the Ps1 promoter, clearance of XylR from the UASs was almost negligible. In this background, expression from Ps2 was very high regardless of the presence of an effector; this finding suggests that RNA polymerase containing ς54 modulates expression from the downstream Ps2 ς70-dependent promoter. In a P. putida IHF-minus background and in the presence of effector, Ps1 expression was the highest found; in contrast, the basal levels of this promoter were the lowest observed. This finding suggests that IHF acts in vivo as a repressor of the ς54-dependent Ps1 promoter. In an IHF-deficient host background, expression from Ps2 in the presence of effector was negligible. Thus, binding of RNA polymerase containing ς54 at the upstream promoter may modulate expression from the Ps2 promoter.
The xyl genes of the Pseudomonas putida TOL plasmid encode the genetic information required for the degradation of toluene and related aromatic compounds. The upper and meta pathway operons, expressed from the Pu and Pm promoters, respectively, comprise the xyl structural genes, whereas the xylR and xylS genes, expressed from the Pr and Ps promoters, respectively, encode the regulatory proteins of the catabolic operons (see reference 29 for a review).
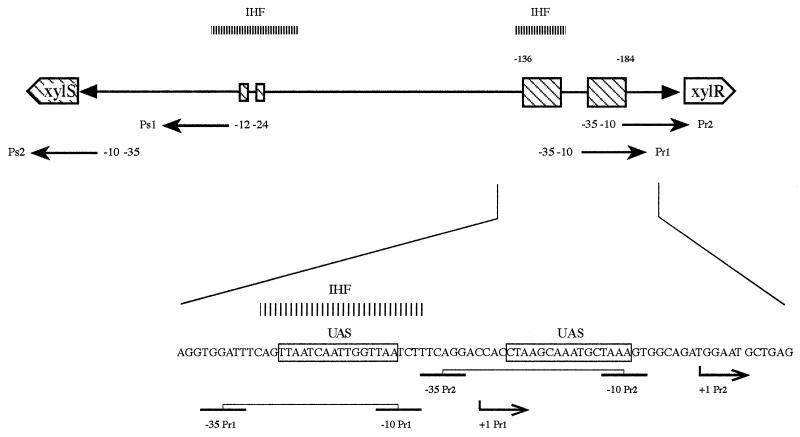
The XylR protein is the master regulator in the control of TOL plasmid catabolic operons for the metabolism of toluene (12, 16, 18, 27, 30). Expression of XylR occurs from two tandem promoters, one distal (Pr1) and one proximal (Pr2). Expression from these promoters is high regardless of the growth phase and growth conditions (17, 23). The xylS gene is expressed from a single promoter or from two tandem promoters, depending on the growth conditions (10). In the absence of aromatic hydrocarbons, the xylS gene is mainly expressed at low constitutive levels from a ς70-dependent promoter called Ps2 (10). In the presence of toluene, active XylR protein bound to target upstream activator sequences (UASs) stimulates transcription from the ς54-dependent Ps1 promoter from a distance, without varying the transcription level of Ps2 (10). Interactions between the activator bound to cognate UASs located at around positions −136 to −184 and the ς54-containing RNA polymerase, bound to the region from positions −12 to −24 (the −12/−24 region), require looping out of the intervening DNA (1, 5, 21, 22, 33).
The xylR and xylS genes are transcribed divergently (see reference 29 for a review) such that the target UASs for XylR protein in the xylS gene Ps1 promoter overlap the xylR promoters: the target inverted repeats of XylR in Ps1, located between −136 and −154 (UAS1), overlap the RNA polymerase recognition site of Pr1 between −11 and −26. The XylR target in UAS2, located between −169 and −184, overlaps Pr1 between +5 and +20 and overlaps Pr2 between −24 and −19 (Fig. 1) (11, 13, 14, 25). In assays with fusions of Pr to ′lacZ in Escherichia coli, expression from Pr promoters was about two- to threefold higher in the isogenic XylR-minus background than in the XylR-plus background, which suggested that XylR controls its own synthesis (14, 17, 19).
FIG. 1.
Organization of the xylR-xylS intergenic region. The Ps1 promoter includes UASs (large boxes) for the master activator of the system, XylR; the −12/−24 sequences recognized by ς54-containing RNA polymerase (small boxes); and two potential IHF binding sites (shaded bars). Also indicated are consensus ς70-containing RNA polymerase-binding sites (−10 to −35) and the transcription initiation point (+1) for the xylR gene Pr tandem promoters Pr1 and Pr2 and for the xylS gene Ps2 promoter. The nucleotide sequence of the Pr promoter region that overlaps the Ps1 UASs is shown in detail.
In the DNA region comprising the tandem divergent Pr and Ps promoters, Holtel et al. (13, 14) found two integration host factor (IHF) binding sites at −2 to −30 and at −137 to −156 with respect to the Ps1 transcription point (Fig. 1) in in vitro protection DNase I footprinting assays. The IHF site at −137 to −156 corresponds to −8 to −27 of Pr1, i.e., the zone showing no significant overlap with the Pr2 region (Fig. 1). In spite of this in vitro finding, the in vivo expression from the xylR and xylS promoters in IHF-minus and IHF-plus E. coli backgrounds, determined as β-galactosidase activity by using independent fusions of Ps and Pr to ′lacZ, was found not to be significantly affected. It was, therefore, suggested that these IHF binding sites were irrelevant for transcriptional control of Pr and Ps promoters (13, 14).
XylR autoregulation studies have been carried out in E. coli by using fusions of the Pr promoters to lacZ, an approach which does not make it possible to distinguish the repressive effect of XylR on each of the single Pr promoters. Moreover, in these studies expression from the tandem divergent Ps1 and Ps2 promoters was overlooked. We therefore investigated the autoregulation of Pr promoters and expression from Ps promoters in different P. putida genetic backgrounds, with the promoters in their natural location in the TOL plasmid.
MATERIALS AND METHODS
Strains and culture conditions.
The strains and plasmids used in this study are listed in Table 1. Bacteria were grown on M9 minimal medium with 10 mM succinate as a carbon source (20), supplemented with 1/20 LB to avoid differences in growth between strains with different mutations. Duplicate cultures (20 ml) were prepared by diluting overnight cultures to an initial turbidity at 660 nm of 0.4 and incubating them at 30°C for 2 h with continuous shaking. Then benzyl alcohol was added to one of the cultures to a final concentration of 5 mM. Cells were incubated for 2 h, and samples were taken for mRNA analyses (see below). When required, the following antibiotics were added to the concentrations (in micrograms per milliliter) indicated: ampicillin, 100; chloramphenicol, 30; kanamycin, 50; piperacillin, 90; and streptomycin, 50.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| P. putida | ||
| KT2440 | hsdMR Ben+ | 9 |
| KT2440-rpoN | KT2440, rpoN::Km | 20 |
| KT2440-IHF3 | KT2440, ihfA::Km | This study |
| E. coli | ||
| CC118 pir | recA pir lysogen | 25 |
| HB101 | recA Smr | 4 |
| Plasmids | ||
| pRJS1 | ihfA::Km at pEX100T | 6 |
| pRK600 | ColE1, RK2-Mob+, RK2-Tra+, Cmr | This laboratory |
| pUNφ19 | pUC19 derivative, pRP4 Mob+, Apr | This study |
| pUNxylR::Ω | pUNφ19 derivative bearing xyl1::Ω, Apr, Smr | This study |
| pWW0 | Tol+, xyl1+, xyl1+ | 35 |
| pWWΩXylR | pWW0::xylR::ΩSm | This study |
Abbreviations: Ben+ and Tol+, degradation of benzoate and toluene, respectively; Rifr, Smr, Apr, and Kmr, resistance to rifampin, streptomycin, ampicillin, and kanamicin, respectively.
DNA techniques.
DNA preparation, digestion with restriction enzymes, analysis by agarose gel electrophoresis, isolation of DNA fragments, ligations, labeling, and sequencing reactions were done according to standard procedures (3, 32).
Transfer of the TOL plasmid between different P. putida strains.
Rifampin-resistant P. putida KT2440 mutant strains carrying the wild-type or mutant TOL plasmids were obtained by direct transfer mobilization of the TOL plasmid from the rifampin-sensitive P. putida KT2440 to the recipient strain and counterselection of transconjugants in selective medium.
Reverse genetic construction of TOL plasmid derivatives.
Plasmid pWWΩXylR was constructed as follows. The 2-kb HpaI fragment from TOL plasmid pWW0 bearing the whole xylR gene was cloned at the SmaI site of pUNφ19. The xylR gene was knocked out by inserting the omega interposon, which carried the streptomycin-spectinomycin resistance gene (8) at the single PstI site within the xylR coding region. This plasmid, called pUNxylR::Ω, was transformed in E. coli HB101. A triparental mating involving P. putida KT2440(pWW0) as the recipient strain, HB101(pUNxylR::Ω) as the donor strain, and HB101(pRK600) as a helper strain was done. P. putida transconjugants in which the xylR::Ω mutant had recombined with the wild-type gene were selected as streptomycin resistant in minimal medium with benzoic acid as the sole C source. The candidate mutant TOL plasmids were transferred through direct mating to wild-type P. putida KT2442 and checked by Southern hybridization for the absence of wild-type copies of the gene. One of the mutant plasmids, called pWWΩXylR, was kept for further studies.
Reverse genetic construction of an IHF-deficient P. putida strain.
Strain P. putida KT2440::IHF3 was obtained through recombination of wild-type P. putida KT2440 with plasmid pJRS1 (6), which carries the ihfA gene of P. putida with an internal sequence replaced by a kanamycin resistance cassette. Correct recombination of the IHF::Km DNA in the chromosome of the host strain was checked by PCR and Southern hybridization (not shown).
RNA preparation, analysis, and primer extension.
RNA was extracted with a modification of the guanidinium isothiocyanate-phenol method (24). The RNA concentration was determined by measuring A260. Hybridization of the 35P-5′-end-labeled single-stranded DNA primer (105 cpm) complementary to the mRNA transcript produced from Pr and Ps promoters and primer extension with avian myeloblastosis virus reverse transcriptase were done as described previously (24). In all assays 20 μg of total RNA was used as the template. The oligonucleotides used in the reverse primer extension reactions were 5′-ACGGATCTGGCTGCTAAGGTCTTGC-3′ for the Pr promoters and 5′-GAGACTGCATAGGGCTCGGCGTGG-3′ for the Ps promoters. cDNA products were analyzed in a urea-polyacrylamide sequencing gel, and the intensities of the bands in the autoradiography were densitometrically determined. All assays were done at least three times, without significant differences in the detected levels of the different extended products. Characteristic assays are described in Results.
RESULTS
Autoregulation of XylR increases in the presence of pathway substrates.
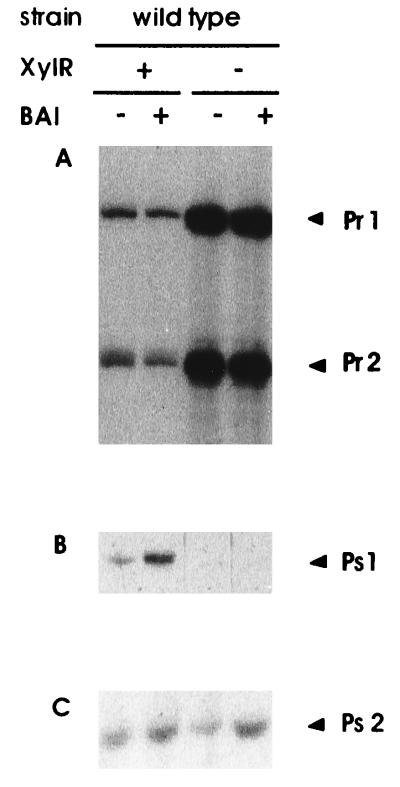
To evaluate expression from Pr1, Pr2, Ps1, and Ps2 in P. putida, we determined transcription from these promoters in the wild-type P. putida strain KT2440 bearing the archetypal TOL plasmid pWW0 and its xylR mutant pWWΩXylR. Cells were grown on succinate minimal medium with and without benzyl alcohol. In the absence of the aromatic hydrocarbon in P. putida(pWW0), we observed strong expression from Pr1 and Pr2 (Fig. 2A), measurable levels of expression from Ps2 (Fig. 2C), and low but detectable levels from Ps1 (Fig. 2B), in accordance with previous findings (10, 17, 18, 23). In the presence of benzyl alcohol transcription from Pr promoters decreased to one-half to one-fourth the levels found in the absence of the aromatic hydrocarbon (Fig. 2A). In contrast, high expression from Ps1 was found and expression from Ps2 was unaltered (Fig. 2B and C). These results suggest that autoregulation of Pr promoters is sensitive to the addition of a pathway substrate.
FIG. 2.
Autoregulation of xylR gene expression and xylS gene expression in wild-type P. putida bearing wild-type or xylR-knocked out TOL plasmid. P. putida KT2440(pWW0) and P. putida KT2440(pWWΩXylR) were grown on minimal medium as described in Materials and Methods. Overnight cultures were diluted in duplicate to reach a turbidity of 0.4 at 660 nm and were incubated at 30°C with shaking. Two hours later 5 mM benzyl alcohol was added to one of the cultures, and growth was continued for 2 h at 30°C. After this time samples were withdrawn from both cultures for mRNA analysis as described in Materials and Methods. Two sets of reactions were carried out in parallel with primers to detect mRNAs from Pr promoters and Ps promoters. The resulting cDNAs were run in independent sequencing gels. (A) Pr1 and Pr2 indicate 208- and 180-bp cDNAs, respectively; (B) Ps1 indicates a 208-bp cDNA; (C) Ps2 indicates a 77-bp cDNA.
When P. putida bore the truncated xylR gene in pWWΩXylR, the level of expression from Pr1 and Pr2 was about 10-fold higher than in the isogenic xylR-plus background. The presence of the aromatic alcohol did not influence these high levels of expression (Fig. 2A). In the xylR-minus background, no expression from Ps1 was found (Fig. 2B). The level of expression from Ps2 was similar to that found in the isogenic xylR-plus background (Fig. 2C).
Autoregulation of XylR and expression from Ps promoters in a P. putida ς54-deficient background.
In ς54-dependent promoters, transcription stimulation requires a loop to allow the regulator bound at a distance to its UAS targets to interact with RNA polymerase containing ς54 bound at −12 to −24 (5, 21, 33). Transcription from the Ps1 promoter is ς54 and XylR dependent (12). In a heterologous E. coli background, it was suggested that autoregulation of XylR might be mediated by ς54 (11); however, no conclusive evidence was provided for this possibility. This prompted us to analyze the expression from Ps1, Ps2, Pr1, and Pr2 in a ς54-deficient P. putida background both in the absence and in the presence of benzyl alcohol and with and without XylR. The results were compared with those obtained in the isogenic ς54-proficient P. putida strain described above.
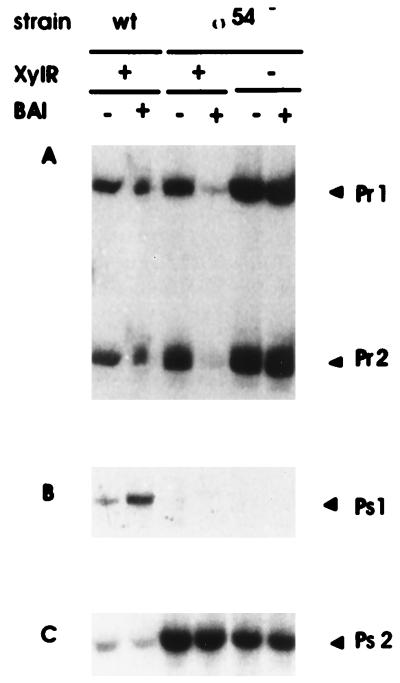
In the ς54-deficient P. putida strain bearing the wild-type pWW0 plasmid, transcription from Pr1 and Pr2 was slightly (twofold) higher than in the ς54-proficient wild-type host background (Fig. 3A). Autoregulation in response to XylR effectors was strongly increased (Fig. 3A). In the ς54-deficient background, there was no transcription from Ps1 (Fig. 3B); however, expression from the Ps2 promoter was 10-fold higher than in the isogenic ς54-proficient background (Fig. 3C).
FIG. 3.
Autoregulation of xylR gene expression and expression from the Ps promoters in ς54-deficient and -proficient P. putida hosts. Overnight cultures of strains P. putida KT2440(pWW0), P. putida KT2440::rpoN(pWW0), and P. putida KT2440::rpoN(pWWΩXylR) were cultured and treated as described in the legend to Fig. 2. Two sets of reactions were carried out in parallel with primers to detect mRNAs from Pr promoters and Ps promoters. The resulting cDNAs were run in independent sequencing gels. (A) Pr1 and Pr2 indicate 208- and 180-bp cDNAs, respectively; (B) Ps1 indicates a cDNA of 208 bp; (C) Ps2 indicates a cDNA of 77 bp.
In the P. putida ς54-minus background, expression from Pr1, Pr2, and Ps1 in pWWΩXylR followed the pattern described above for the ς54-proficient background (Fig. 2A): expression from Pr1 and Pr2 was much higher than in the XylR-plus background, and there was no transcription from Ps1. In a P. putida ς54-minus background with pWWΩXylR, the pattern of expression was similar to that of pWW0 in this host background: extremely high levels of expression from Ps2 (Fig. 3C), in contrast with the results in the P. putida ς54-plus background. Therefore, XylR is clearly the main control element governing expression from Ps1, Pr1, and Pr2, while expression from the Ps2 promoter may be influenced by occupancy and expression of the upstream Ps1.
Autoregulation of xylR promoters and activation of transcription from the Ps promoters in a P. putida IHF-minus background.
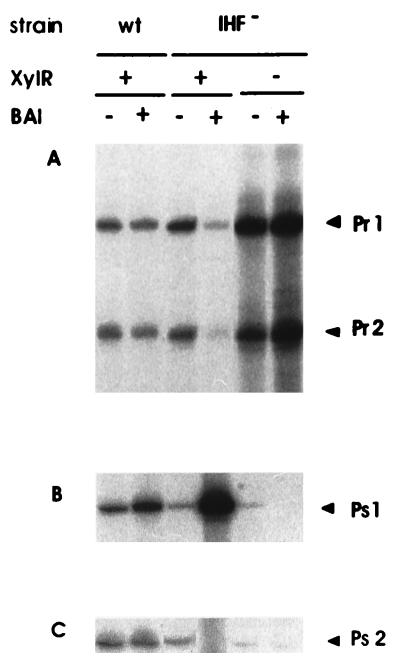
The fact that one of the two IHF sites in the Pr-Ps intergenic region overlaps the XylR targets and that the other IHF site overlaps the −12/−24 region of the Ps1 promoter prompted us to generate an IHF mutant of P. putida as described in Materials and Methods. We then studied autoregulation of xylR and expression from the xylS promoters in the XylR plus and minus backgrounds in the presence and in the absence of benzyl alcohol. The results were compared with those reported above for the wild-type P. putida host strain. In the IHF-minus XylR-plus background in the absence of effector, the level of expression from Pr1 and Pr2 was slightly higher than that in the IHF-plus background, while in the presence of an effector, expression from both promoters decreased strongly (Fig. 4A). In an IHF-minus XylR-minus background, the expression from Pr1 and Pr2 was again extremely high (Fig. 4A), as in the isogenic IHF-plus background (Fig. 4A). This finding suggests that IHF does not alter the pattern of derepresion of Pr1 and Pr2 in the absence of XylR.
FIG. 4.
Autoregulation of xylR gene expression and expression from the Ps promoters in IHF-deficient and -proficient P. putida strains. Overnight cultures of strains P. putida KT2440(pWW0), P. putida KT2440::IHF3(pWW0), and P. putida KT2440::IHF3 (pWWΩXylR) were cultured and treated as described in the legend to Fig. 2. Two sets of reactions were carried out in parallel with primers to detect mRNAs from Pr promoters and Ps promoters. The resulting cDNAs were run in independent sequencing gels. (A) Pr1 and Pr2 indicate 208- and 180-bp cDNAs, respectively; (B) Ps1 indicates a cDNA of 208 bp; (C) Ps2 indicates a cDNA of 77 bp.
We also examined expression from Ps1 and Ps2 in the IHF-minus xylR-plus background. The basal level of Ps1 decreased with respect to that in the wild-type strain, whereas in the presence of an effector, expression from the Ps1 promoter was up to 20-fold greater (Fig. 4B). No expression from Ps1 was observed in the IHF-minus background in the absence of XylR. These results suggest that IHF is involved in the modulation of Ps1 expression.
The level of expression of Ps2 in the IHF-minus xylR-plus background in the absence of an effector was low, as in the wild-type P. putida host background (Fig. 4C). In this host background in the presence of an effector, expression from Ps2 was almost negligible (Fig. 4C). These results indicate that the host IHF background influences expression from Ps2.
DISCUSSION
The XylR and XylS proteins are involved in transcriptional control of the P. putida TOL plasmid catabolic pathways for toluene and xylene metabolism (10, 18, 30). The level of these proteins is finely modulated in vivo to achieve a critical feature: silencing of catabolic operons in the absence of effectors, and a rapid and coordinated response of the catabolic operons to the presence of aromatic hydrocarbons.
This fine regulation occurs in the 300-bp intergenic region between the xylR and xylS genes, through the interplay of RNA polymerase with either ς70 or ς54, and with XylR and IHF, which govern the expression of Pr1/Pr2 and Ps1/Ps2 promoters.
All four promoters have good consensus sequences for binding of the RNA polymerase with ς70 (Pr1, Pr2, and Ps2) or with ς54 (Ps1), and therefore high levels of expression from all of them should be expected. However, we have shown in this study that the levels of expression normally observed in the wild-type strain for each promoter under the most favorable conditions in the laboratory are clearly below maximum levels. This finding indicates that expression from these promoters in vivo is optimized rather than maximized and that a repressive element (or elements) is involved in the maintenance of appropriate levels of expression. The levels of Pr1 and Pr2 obtained in the presence of XylR (wild-type condition) are 1/10 of the levels in the isogenic XylR-deficient background (Fig. 2). Induced levels of Ps1 promoter in the IHF-minus strain are about 10 times higher than in the wild-type strain (Fig. 4). Transcriptional activity from Ps2 is low in the wild-type strain, whereas it is strongly increased by the absence of ς54, both in the presence and in the absence of benzyl alcohol (Fig. 3).
Activation of the Ps1 promoter and autoregulation of XylR seem to be the two consequences of a single event, i.e., the binding of XylR to the UASs for Ps1 promoter that overlap the Pr1 and Pr2 promoters (Fig. 1), in agreement with the finding that XylR is consistently bound to target sequences (1, 22). Our results show that autoregulation is stronger in the presence of effector (Fig. 2). We propose two explanations for the finding: (i) binding of an effector to XylR may increase the affinity of the regulator for its binding sequence, thus reducing access of RNA polymerase containing ς70 to the Pr promoters; and (ii) effector binding to XylR may favor ATP binding and XylR oligomerization (27) such that the formation of a supramolecular complex limits the access of RNA polymerase containing ς70 to Pr promoters. To explain how Pr1, Pr2, and Ps1 all become active in the presence of an XylR effector, we suggest that in each round of transcription from the Ps1 promoter, the XylR regulator is transiently released from the UASs such that a certain equilibrium is established between occupied and nonoccupied UASs. In this situation, some transcription from both Pr1 and Pr2 promoters is allowed. Further support for this model is the finding that when transcription from Ps1 cannot occur because of the absence of ς54, in the presence of an effector the XylR protein bound to the target UASs is probably not released (or is released at a lower rate) so that access of RNA polymerase containing ς70 to either Pr1 or Pr2 is impeded. Pérez-Martín and de Lorenzo (27) suggested a cycle for XylR-dependent transcription stimulation from XylR-regulated promoters; in this cycle ATP binding was the driving force for XylR multimerization at the UASs, and ATP hydrolysis was sine qua non for initiation of transcription by RNA polymerase containing ς54, as is also the case with other regulators (34). Upon ATP hydrolysis, ADP is released and the multimer is dismantled, so that after regeneration of ATP a new cycle can start. The strong repression at Pr promoters in the ς54-deficient background could reflect blockage of the cycle because the XylR multimer is maintained at the UAS in the absence of appropriate transcriptional machinery. Consistent with this possibility is that in the presence of effector, very low levels of transcription from both Pr1 and Pr2 promoters were found in the ς54-deficient background (Fig. 3).
The expression from Ps1 in pWW0 in the isogenic IHF-plus and IHF-minus strains deserves attention. Figure 4 shows that the basal levels of Ps1 were higher in the IHF-plus background than in the IHF-minus background. Surprisingly, the highest levels of expression from Ps1 were found in the IHF-deficient mutant in the presence of effector-activated XylR protein. In the absence of IHF, these findings could reflect either or both of the following possibilities: better access of XylR to its binding site or facilitated access of RNA polymerase containing ς54 to the −12/−24 region of Ps1. In either case, transcription from Ps1 could occur (and in fact occurs) at a higher level. Regardless of the molecular mechanism behind this phenomenon, which requires further in vitro study, it is clear that IHF functions as a negative regulator of the expression of Ps1. This may be a consequence of both physical and structural hindrance, as the DNA bends induced by the IHF protein bound to two proximal sites can give rise to a highly ordered structure that may be involved in restriction of access to the corresponding promoters. The high level of expression from Ps1 in the IHF-minus background in the presence of effectors contrasts with the diminished expression from the TOL plasmid Pu promoter for the upper pathway in an IHF-deficient background (1, 6, 28). We should consider the homologies and differences between the two promoters: both Pu and Ps1 exhibit −12/−24 sequences for the binding of RNA polymerase containing ς54 and UASs for the binding of XylR. In contrast, the IHF binding site in Pu lies between the UASs and the −12/−24 box, whereas in the Ps1 promoter, two IHF binding sites are found, one located in the region that overlaps the binding sites for RNA polymerase containing ς54 (−2 to −30 in Ps1) and the other one overlapping the XylR UASs at −137 to −156 in Ps1. This second binding site also overlaps the Pr promoters (Fig. 1) (13, 14). The differences in organization between Pu and Ps1 may also be responsible for the finding that the Pu promoter is about fourfold stronger than Ps1 (23).
Another unexpected finding is the high level of expression from Ps2 in the absence of ς54, regardless of the presence of XylR and/or effector. These high levels of expression are found only in those situations where RNA polymerase is unable to bind to the Ps1 promoter. In agreement with this hypothesis is our finding that in an IHF-minus mutant, in which access of RNA polymerase containing ς54 to its binding site at Ps1 is facilitated, expression of Ps2 promoter was always greatly decreased. Further in vitro assays are needed to determine the specific binding constant of each of these proteins for the corresponding targets to establish whether a hierarchy of binding exists in the intergenic Pr and Ps promoters.
ACKNOWLEDGMENTS
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (AMB1038-CO2-01) and the Biotechnology Programme of the Commission of the European Communities (BIO4-CT97-2040). Exchange visits between GBF and CSIC were supported by the Acciones Integradas program between the two organizations.
We thank E. Santero for comments on the manuscript.
REFERENCES
- 1.Abril M A, Buck M, Ramos J L. Activation of the Pseudomonas TOL plasmid upper pathway operon. J Biol Chem. 1991;266:15832–15838. [PubMed] [Google Scholar]
- 2.Abril M A, Michán C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991. [Google Scholar]
- 4.Boyer H B, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;4:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 5.Buck M, Miller S, Drummond M, Dixon R. Upstream activators are present in the promoters of nitrogen fixation genes. Nature. 1986;320:374–378. [Google Scholar]
- 6.Calb R, Davidovitch A, Koby S, Giladi H, Goldenberg D, Margalit H, Holtel A, Timmis K N, Sánchez-Romero J M, de Lorenzo V, Oppenheim A B. Structure and function of the Pseudomonas putida integration host factor. J Bacteriol. 1996;178:6319–6326. doi: 10.1128/jb.178.21.6319-6326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgado A, Ramos J L. Genetic evidence for activation of the positive transcriptional regulator XylR, a member of the NtrC family of regulators, by effector binding. J Biol Chem. 1994;269:8059–8062. [PubMed] [Google Scholar]
- 8.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 9.Franklin F C H, Bagdasarian M, Bagdasarian M M, Timmis K N. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta-cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallegos M T, Marqués S, Ramos J L. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a ς70-dependent promoter or from ς70- and ς54-dependent tandem promoters according to the compound used for growth. J Bacteriol. 1996;178:2356–2361. doi: 10.1128/jb.178.8.2356-2361.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomada M, Inouye S, Imaishi H, Nakazawa A, Nakazawa T. Analysis of an upstream regulatory sequence required for activation of the regulatory gene xylS in xylene metabolism directed by the TOL plasmid of Pseudomonas putida. Mol Gen Genet. 1992;233:419–426. doi: 10.1007/BF00265439. [DOI] [PubMed] [Google Scholar]
- 12.Holtel A, Abril M A, Marqués S, Timmis K N, Ramos J L. Promoter-upstream activator sequences are required for expression of the xylS gene and upper-pathway operon on the Pseudomonas TOL plasmid. Mol Microbiol. 1990;4:1551–1556. doi: 10.1111/j.1365-2958.1990.tb02066.x. [DOI] [PubMed] [Google Scholar]
- 13.Holtel A, Goldenberg D, Giladi H, Oppenheim A B, Timmis K N. Involvement of IHF protein in expression of the Ps promoter of the Pseudomonas putida TOL plasmid. J Bacteriol. 1995;177:3312–3315. doi: 10.1128/jb.177.11.3312-3315.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtel A, Timmis K N, Ramos J L. Upstream binding sequences of the XylR activator protein and integration host factor in the xylS gene promoter region of the Pseudomonas TOL plasmid. Nucleic Acids Res. 1992;20:1755–1762. doi: 10.1093/nar/20.7.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inouye S, Nakazawa A, Nakazawa T. Molecular cloning of gene xylS of the TOL plasmid: evidence for positive regulation of the xylDEGF operon by xylS. J Bacteriol. 1981;148:413–418. doi: 10.1128/jb.148.2.413-418.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inouye S, Nakazawa A, Nakazawa T. Molecular cloning of regulatory gene xylR and operator-promoter regions of the xylABC and xylDEGF operon of the TOL plasmid. J Bacteriol. 1983;155:1192–1199. doi: 10.1128/jb.155.3.1192-1199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inouye S, Nakazawa A, Nakazawa T. Determination of the transcription initiation site and identification of the protein product of the regulatory gene XylR for xyl operons on the TOL plasmid. J Bacteriol. 1985;163:863–869. doi: 10.1128/jb.163.3.863-869.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inouye S, Nakazawa A, Nakazawa T. Expression of the regulatory gene xylS on the TOL plasmid is positively controlled by the xylR gene product. Proc Natl Acad Sci USA. 1987;84:5182–5186. doi: 10.1073/pnas.84.15.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inouye S, Nakazawa A, Nakazawa T. Nucleotide sequence of the regulatory gene xylR of the TOL plasmid from Pseudomonas putida. Gene. 1988;66:301–306. doi: 10.1016/0378-1119(88)90366-6. [DOI] [PubMed] [Google Scholar]
- 20.Köhler T, Harayama A, Ramos J L, Timmis K N. Involvement of Pseudomonas putida RpoN ς factor in regulation of various metabolic functions. J Bacteriol. 1989;171:4326–4333. doi: 10.1128/jb.171.8.4326-4333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of sigma-54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenzo V de, Herrero M, Metzke M, Timmis K N. An upstream xylR and IHF induced nucleoprotein complex regulates the sigma-54 dependent Pu promoter of TOL plasmid. EMBO J. 1991;10:1159–1167. doi: 10.1002/j.1460-2075.1991.tb08056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marqués S, Holtel A, Timmis K N, Ramos J L. Transcriptional induction kinetics from the promoters of the catabolic pathways of TOL plasmid pWW0 of Pseudomonas putida for metabolism of aromatics. J Bacteriol. 1994;176:2517–2524. doi: 10.1128/jb.176.9.2517-2524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marqués S, Ramos J L, Timmis K N. Analysis of the mRNA structure of the Pseudomonas putida meta-fission pathway operon around the transcription initiation point, the xylTE and the xylFJ region. Biochim Biophys Acta. 1993;1216:227–237. doi: 10.1016/0167-4781(93)90149-8. [DOI] [PubMed] [Google Scholar]
- 25.Pérez-Martín J, de Lorenzo V. The ς54-dependent promoter Ps of the plasmid of Pseudomonas putida requires HU for transcriptional activation in vivo by XylR. J Bacteriol. 1995;177:3758–3763. doi: 10.1128/jb.177.13.3758-3763.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Martín J, de Lorenzo V. The amino-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc Natl Acad Sci USA. 1995;92:9392–9396. doi: 10.1073/pnas.92.20.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez-Martín J, de Lorenzo V. ATP binding to the ς54-dependent activator XylR triggers a protein multimerization cycle catalyzed by UAS DNA. Cell. 1996;86:331–339. doi: 10.1016/s0092-8674(00)80104-x. [DOI] [PubMed] [Google Scholar]
- 28.Pérez-Martín J, Timmis K N, de Lorenzo V. Co-regulation by bent DNA: functional substitutions of the IHF site at the ς54-dependent promoter Pu of the upper-TOL operon by intrinsically curved sequences. J Biol Chem. 1994;269:22657–22662. [PubMed] [Google Scholar]
- 29.Ramos J L, Marqués S, Timmis K N. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid encoded regulators. Annu Rev Microbiol. 1997;51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 30.Ramos J L, Mermod N, Timmis K N. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage. Mol Microbiol. 1987;1:297–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- 31.Ramos J L, Stolz A, Reineke W, Timmis K N. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc Natl Acad Sci USA. 1986;83:8467–8471. doi: 10.1073/pnas.83.22.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Su W, Porter S, Kustu S, Echols H. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wedel A, Kustu S. The bacterial enhancer-binding protein NtrC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev. 1995;9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 35.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]