Abstract
Cell therapies are expected to increase over the next decade owing to increasing demand for clinical applications. Mesenchymal stromal cells (MSCs) have been explored to treat a number of diseases, with some successes in early clinical trials. Despite early successes, poor MSC characterization results in lessened therapeutic capacity once in vivo. Here, we characterized MSCs derived from bone marrow (BM), adipose tissue and umbilical cord tissue for sphingolipids (SLs), a class of bioactive lipids, using liquid chromatography/tandem mass spectrometry. We found that ceramide levels differed based on the donor’s sex in BM-MSCs. We detected fatty acyl chain variants in MSCs from all three sources. Linear discriminant analysis revealed that MSCs separated based on tissue source. Principal component analysis showed that interferon-γ–primed and unstimulated MSCs separated according to their SL signature. Lastly, we detected higher ceramide levels in low indoleamine 2,3-dioxygenase MSCs, indicating that sphingomyelinase or ceramidase enzymatic activity may be involved in their immune potency.
Keywords: adipose-derived mesenchymal stromal cells, bone marrow–derived mesenchymal stromal cells, cell manufacturing, immune potency, lipidomics, umbilical cord tissue–derived mesenchymal, stromal cells
Introduction
Cellular therapies have begun to revolutionize the medical field by leading to the development of cutting-edge therapeutics to treat various diseases such as cancer, while offering promising treatments to repair damaged or destroyed tissue. As a result, the cell therapeutic industry is expected to expand over the next few decades. In 2019, the global cell therapy market was valued at $755.4 million USD and is projected to be worth $11 billion USD by 2029 [1–4]. With the expected increase of cell therapies, scaling-up processes and manufacturing strategies have been expanded to meet clinical demand.
In particular, the manufacturing of MSCs has gained much interest owing to MSCs’ multipotent differentiation, immunomodulation, immune-suppression and pro-regenerative properties via secreted cytokines, extracellular vesicles (EVs) and trophic factors [5 8]. In recent years, preclinical and clinical trials have begun to investigate potential therapeutic effects of MSCs in vivo. As of 2021, more than 1,000 clinical trials have been conducted to investigate MSCs’ therapeutic potency. Studies have assessed MSC-based immunotherapies’ efficacy in treating diabetes mellitus, cardiovascular diseases, graft-versus-host disease, Crohn’s disease and various inflammatory diseases [9–11]. Despite promising preclinical results, factors such as culture conditions, donor history, donor variability, tissue source, manufacturing processes and cell isolation techniques affect MSC therapeutic efficacy in vivo, resulting in failed primary endpoints in clinical trials [12–14]. Minimal criteria for characterization of MSCs include adherence to plastic, expression of specific surface antigens, and multipotent differentiation potential in vitro [15]; however, the International Society for Cell & Gene Therapy (ISCT) has more recently called for improvements to MSC characterization by recommending the inclusion of functional assays and multi-omic profiling for additional means of characterization [16].
Metabolomics is an emerging field that assesses multiparametric metabolic responses of cells and tissue to external stimuli [17]. Moreover, high-throughput analytical techniques have greatly accelerated the field, allowing the collection of large and robust metabolomic datasets [18–20]. In particular, metabolomics has been used to explore manufacturing process parameters for culture condition optimization, while offering potential advantages such as culture standardization and cost-effectiveness [21–23]. Lipidomics, a sub-branch of metabolomics, is a powerful technique that combines high-throughput analytical methods, such as mass spectrometry (MS), and informatics to quantify and characterize cells’ lipid profile (or lipidome). Lipid components make up approximately one-third of all metabolites; thus, understanding their dynamic roles in biochemical signaling will allow researchers to prevent, diagnose and treat a wide range of human diseases [24]. With advances of MS technologies, researchers are able to investigate thousands of lipid species that may be involved in normal cell behavior and disease pathologies. This highlights the need for investigating lipid metabolism to help develop a clear understanding of their complex functions, not only in clinical settings but also in cell manufacturing.
Bioactive lipids are a class of lipids that participate in complex cellular and molecular processes that regulate cell homeostasis, cell fate, inflammation and many other biological processes [25]. Functionally, bioactive lipids respond to stimuli, allowing them to regulate downstream targets of interest. As a result, bioactive lipids are involved in regulatory cell circuits, which sets them apart from other lipid classes [26]. Sphingolipids (SLs) are a class of bioactive lipids that comprise a 18- to 20-carbon sphingosine backbone, a polar headgroup and a fatty acyl chain, offering unique functional roles in cell biology. SLs participate in various signaling pathways related to cell fate, proliferation, extracellular vesicle biogenesis, migration, differentiation, inflammation and other important processes [27,28]. Limited studies suggest that SLs sphingosine-1-phosphate (S1P) and ceramide-1-phosphate influence MSCs’ osteogenic differentiation, morphology and proliferation [29–31], but no studies have measured the broader network of bioactive SL metabolites in MSCs. In this study, we characterized MSCs’ sphingolipidome using liquid chromatography/tandem MS (LC-MS/MS). We obtained MSCs derived from bone marrow (BM), adipose tissue (AD) and umbilical cord tissue (UCT) from male and female donors and extracted their sphingolipids, as described in our previous work [32]. This study investigated how factors such as donor’s sex, culture conditions, cytokine priming and MSC tissue origin affect the MSC SL profile. SL characterization of sex-specific differences and MSC tissue origin have not been reported. These factors may play a role in MSC in vivo therapeutic efficacy and should be explored for critical quality attribute optimization in the cell manufacturing sector.
Results
LC-MS/MS detection of long chain bases and complex SLs in MSCs
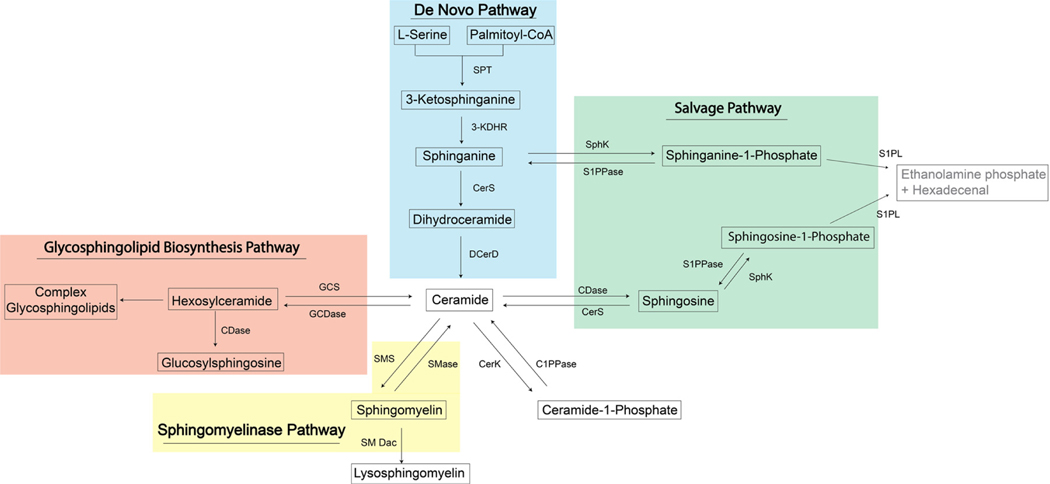
Sphingolipids are a structurally diverse class of membrane lipids that are composed of an 18- to 20-carbon amino-alcohol backbone, sphingosine (Sph), and are synthesized in the endoplasmic reticulum (ER) [33]. Sph and dihydrosphingosine, also referred to as sphinganine (Sa), are the basic building blocks of SLs in mammalian cells. This “sphingoid” backbone can be modified to produce a wide variety of SLs with distinct functions in cell signaling and structural adaptations of biological membranes (Figure 1, Table 1) [34]. N-acylation of sphingosine by ceramide synthases (CerS) generates ceramide (Cer), whereas phosphorylation of the C1-hydroxyl group of sphingosine by sphingosine kinase (SphK) produces S1P [35]. Cer can in turn serve as a precursor of more complex sphingolipids (CSLs), such sphingomyelins (SMs) and ceramide-1-phosphate.
Figure 1.
Sphingolipid metabolic pathway. 3-KDHR, 3-ketosphinganine reductase; C1PPase, ceramide-1-phosphate phosphatase; CDase, ceramidase; CerK, ceramide kinase; CerS, ceramide synthase; DES, dihydroceramide desaturase; GCS, glucosyl/galactosyl-ceramidase; S1PPase, sphingosine-1-phosphate phosphatase; SMase, sphingomyelinase; SMDac, sphingomyelin deacylase; SMS, sphingomyelin synthase; SphK, sphingosine kinase; SPT, serine palmitoyltransferase.
Table 1.
Sphingolipid functions.
| Lipid | Functions |
|---|---|
| Ceramide | Inflammation and cell cycle Pro-apoptotic, though apoptotic properties may depend on chain length; in the skin, only ceramides with fatty acid chains shorter than 20 carbons are apoptotic [7,80] Pro-inflammatory: activates NF-κB and group IV cPLA2 (enzyme that cleaves arachidonic from phospholipids for prostaglandin synthesis) [81] In MSCs Stimulates release of extracellular vesicles [82] |
| Sphingosine | Inflammation and cell cycle Inhibits cell proliferation and promotes apoptosis [8,83] May stimulate proinflammatory cytokine production by macrophages and monocytes [84,85] |
| Sphingosine-1-phosphate | Inflammation Exhibits both pro-inflammatory and anti-inflammatory properties; effects may vary by cell type or disease pathology [86] Promotes lymphocyte egress from secondary lymphoid organs [86,87] Activates NF-κB and promotes the release of inflammatory cytokines and chemokines, such as TNFα, interleukin-6, CCL5, and CXCL10 [80,81,86] Exerts anti-inflammatory effects in murine models of atopic dermatitis [81] Protects cardiomyocytes from apoptosis after ischemia [80] In MSCs Stimulates ASC and BM-MSC migration [88] Enhances ASC engraftment and cardioprotective properties after myocardial infarction [89] Causes UC-MSCs to upregulate expression of pro-angiogenic genes and more effectively suppress TNFα production by macrophages [88] MSC EVs dose-dependently increase S1Pin HK-2 cells by activating sphingosine kinase 1 [90] |
| Sphingomyelin | Inflammation Promotes NF-κB activation, CCL5 expression, and recruitment of CD14, Toll-like receptor4, and TNF receptors to the cell membrane in macrophages [91,92] Long-chain and very-long-chain SM levels correlate to increased levels of inflammatory cytokines in atherosclerotic plaques [93,94] Protects against cigarette smoke–induced airway inflammation and is associated with reduced risk for emphysema [95,96] In MSCs In BM-MSCs, SM levels increase with aging and promote senescence [97] |
| Sphinganine-1-phosphate | Inflammation Attenuates apoptosis and/or necroptosis during ischemia-reperfusion injury in the liver and kidneys [98] |
| Hexosylceramide | Inflammation Glucosylceramide (GlcCer) acts as a ligand on the surface of pathogens or damaged cells to stimulate macrophages. 24:1 GlcCer is more effective at activating macrophages than 16:0 and 18:0 GlcCer [99]. GlcCer causes an increase in major histocompatibility complex class II expression by dendritic cells and helps them activate T cells [99]. Galactosylceramide binds to CD1d on antigen-presenting cells and stimulates semi-invariant T cells and invariant NK cells. This leads to a production of signaling molecules that recruit other immune cells [100,101]. Inflammatory properties may be restricted to HexCer with fatty acid chains containing ≥16 carbons [100,102,103] In MSCs Promotes adipogenic differentiation and inhibits osteogenic differentiation[104] |
| Glucosylsphingosine | Inflammation Pro-inflammatory: binds to CD1dand activates NK cells [105] |
TNFα, tumor necrosis factor α.
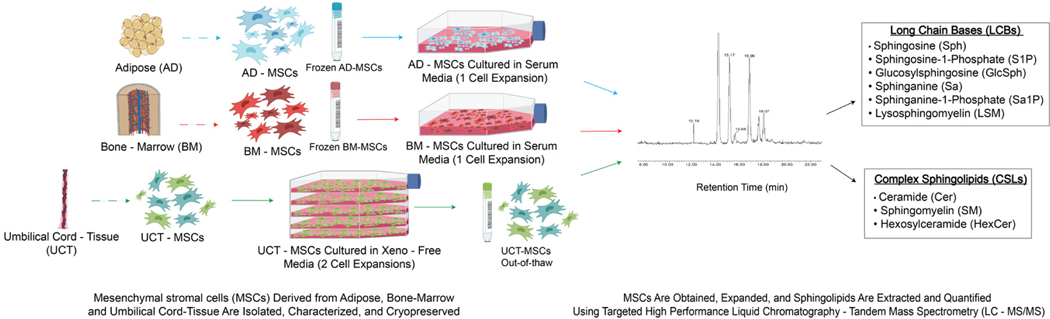
Precursor and product ions were input before MS analysis for targeted SL analysis. Long-chain bases (LCBs) and CSL amounts were normalized by total protein. We detected and measured the following LCBs: Sa, sphinganine-1-phosphate (Sa1P), Sph, glucosylsphingosine (GlcSph), S1P and lysosphingomyelin (LSM). We detected and measured the following CSLs: SM, Cer and hexosylceramide (HexCer) (Figure 2). The Benjamini–Hochberg (BH) method was used to reduce the false discovery rate in the lipidomic data. In each statistical comparison, the P values were adjusted using the BH method to reduce the number of false positives.
Figure 2.
Experimental overview.
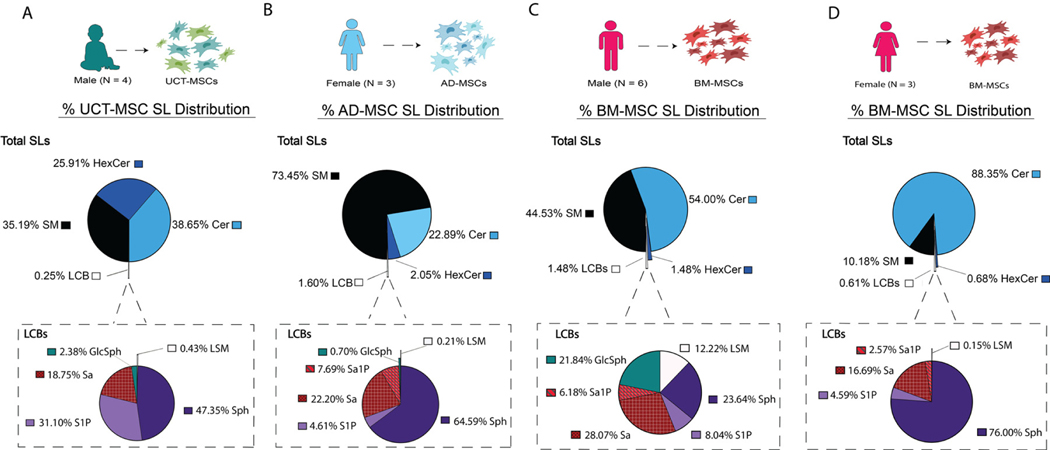
UCT-MSCs’ SL profile consisted of 99.75% CSLs and 0.25% LCBs. The CSL pool was comprised of Cer (38.65%), SM (35.19%) and HexCer (25.91%). When measuring the LCBs, Sa1P was not detected in UCT-MSCs. Sph and S1P made up majority of the LCB pool with 47.35% and 31.10%, respectively. Sa (18.75%), GlcSph (2.38%) and LSM (0.43%) made up the remaining detected LCBs (Figure 3A). AD-MSCs’ total sphingolipidome consisted of 98.04% CSLs and 1.60% LCBs. SM was the predominant CSL detected (73.45%), followed by Cer (22.89%) and HexCer (2.05%). Six LCB species were detected in AD-MSCs. Sph (64.59%) was the predominant LCB detected. The least detected LCB, LSM, made up only 0.21% of the LCB lipid pool. The remaining LCB pool was made up of Sa (22.20%), Sa1P (7.69%), GlcSph (0.70%) and S1P (4.61%) (Figure 3B). Regarding the SL distribution between sexes, male and female BM-MSCs had observable differences. Male BM-MSCs’ sphingolipidome consisted of 54% Cer, 44.53% SM, 1.48% HexCer and 1.48% LCBs. The LCBs consisted of 23.64% Sph, 8.04% S1P, 28.07% Sa, 6.18% Sa1P, 21.84% GlcSph and 12.22% LSM (Figure 3C). In contrast, the female BM-MSC sphingolipidome comprised 88.35% Cer, 10.18% SM, 0.68% HexCer and 0.61% LCBs. The LCB pool was made up of 16.69% Sa, 2.57% Sa1P, 76.00% Sph, and 0.15% LSM (Figure 3D).
Figure 3.
LC-MS/MS detects CSLs and LCBs in MSCs from all tissue origins. Pie chart illustrates the total SL distribution and the call-out pie charts illustrate the LCBs. (A) Percentage SL species distribution in UCT-MSC (n = 4). (B) Percentage SL species distribution of AD-MSCs (n = 3). (C) Percentage SL species distribution of male BM-MSC donors (n = 6). (D) Percentage SL species distribution of female BM-MSC donors (n = 3).
Sphingolipid Acyl Chain Variants
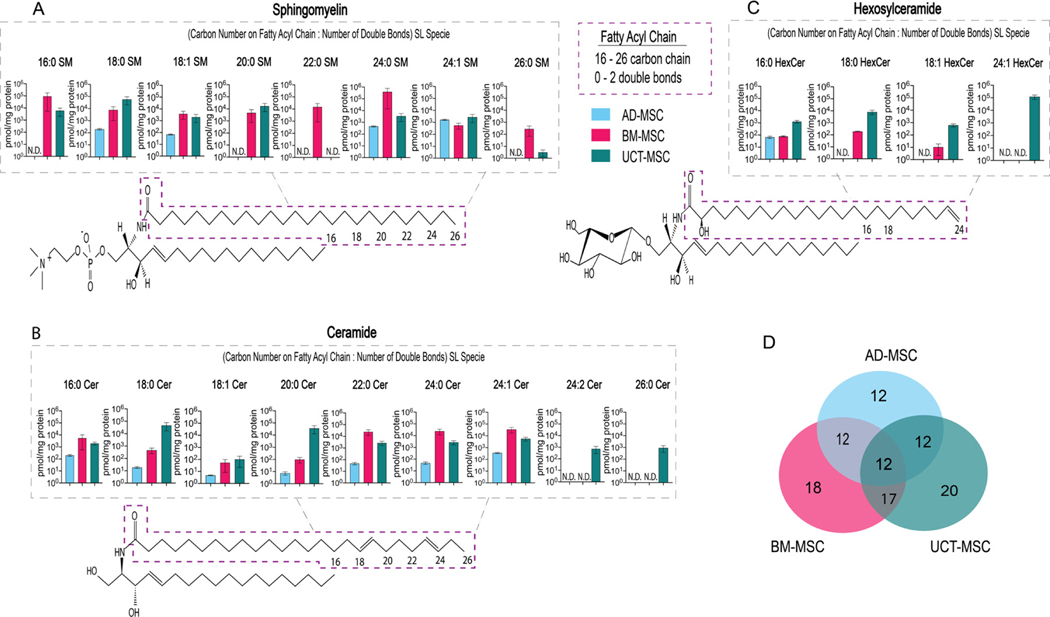
Recently, it has been shown that the functions of SLs differ owing to the length of their fatty acyl chains [36,37]. Fatty acyl chain variants have not been well characterized in MSCs derived from different tissues. To assess different SL fatty acyl chain variants in MSCs, SLs were extracted and quantified with LC-MS/MS. Fatty acyl chains ranging from 16 to 26 carbons in length and containing 0 to 2 double bonds were detected in MSCs from all three tissue sources.
Figure 4A illustrates the SM acyl chain variants detected in AD-, BM- and UCT-MSCs. In AD-MSCs, the only species detected were 18:0, 18:1, 24:0 and 24:1 SM. These species were present in roughly similar concentrations. On the other hand, 16:0, 20:0, 22:0, 24:0 and 26:0 SM were not detected. In BM-MSCs, all the SM species targeted by our LC-MS/MS method, except 24:2 SM, were present. Of those, 16:0, 22:0 and 24:0 SM were the most abundant. All species except 24:2 SM were also detected in UCT-MSCs, with 18:0 SM having the highest concentration. The other species were present in lower concentrations in UCT-MSCs.
Figure 4.
Fatty acyl chain length variants detected by LC-MS/MS in AD-, BM- and UCT-MSCs. Every sphingolipid consists of an 18- to 20-carbon sphingosine backbone, a polar head-group and a fatty acyl chain. The purple box highlights the fatty acyl chain location on SM, Cer and HexCer. (A) SM fatty acyl chain variants. (B) Cer fatty acyl chain variants. (C) HexCer fatty acyl chain variants. (D) Venn diagram of the number of fatty acyl chain lengths variants detected in the three tissue origins; 21 fatty acyl chain variants were measured. The overlap between tissue origins is the shared fatty acyl variants; N.D., not detected; AD-MSC (n = 3), BM-MSC (n = 9) and UCT-MSC (n = 4).
Figure 4B depicts the Cer acyl chain variants detected in MSCs from the three tissue sources. In AD-MSCs, low concentrations of most Cer species were detected, except for 24:2 and 26:0 Cer, which were not detected at all. In BM-MSCs, 22:0, 24:0 and 24:1 Cer were the most abundant, 16:0 and 18:0 Cer were present in moderate concentrations and 18:1 and 20:0 Cer were present in low concentrations compared with the most abundant species. 24:2 and 26:0 Cer were not detected. All the Cer species were detected in UCT-MSCs, with 18:0, 20:0 and 24:1 being the most abundant.
Figure 4C shows the HexCer species detected in MSCs. HexCers comprise a small fraction of the AD-MSCs’ lipidome. The only species detected in AD-MSCs was 16:0 HexCer. In BM-MSCs, three HexCer acyl chain variants, 16:0, 18:0 and 18:1, were detected. In UCT-MSCs, HexCers were abundant, and all the HexCer species targeted by the MS/MS method were detected.
Figure 4D shows the number of SL acyl chain length variants detected in AD-, BM- and UCT-MSCs. A total of 21 acyl chain metabolites were detected and measured by LC-MS/MS, illustrated in Figure 4A C. 12 of 21 metabolites were detected and measured in AD-MSCs. These 12 metabolites were also detected in both BM-MSCs and UCT-MSCs. 18 of 21 fatty acyl chain variants were measured in BM-MSCs. 20 of 21 fatty acyl chain variants were measured in UCT-MSCs. 17 common fatty acyl chain variants were measured in both BM- and UCT-MSCs, and 12 common fatty acyl chain variants were measured in MSCs from all three tissue sources.
Robustness of SL Concentrations Across Tissue Sources
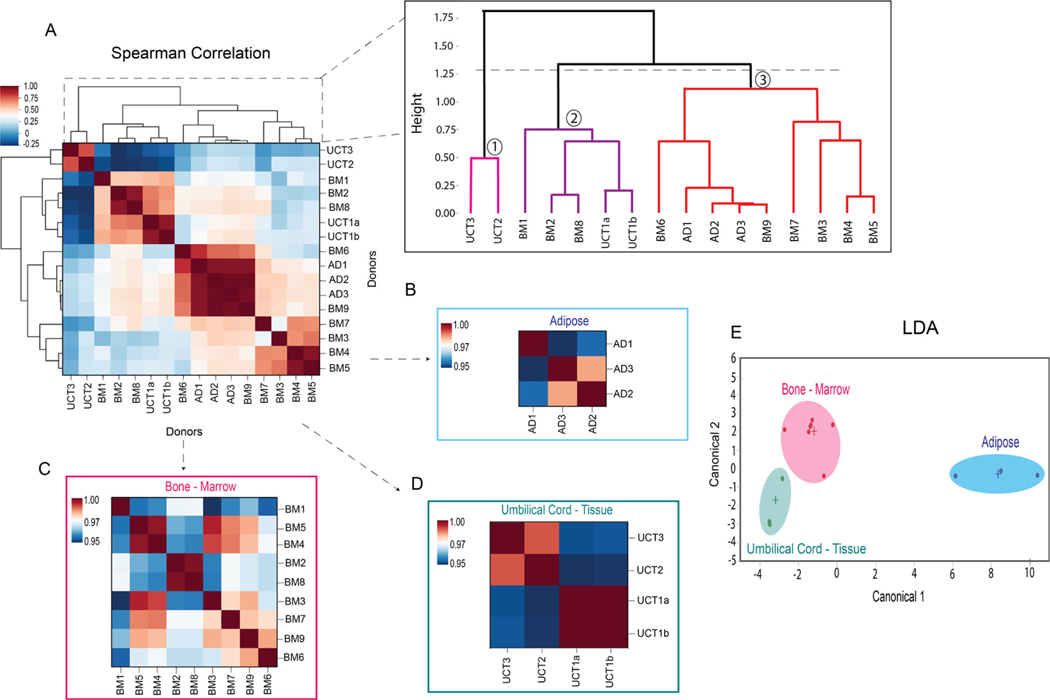
To investigate the relationship between SL concentration and MSC tissue of origin, a Spearman correlation heatmap with hierarchical clustering was generated. The heatmap illustrates correlations among donors based on each donor’s SL profile. Hierarchical clustering was performed on the correlation matrix to partition donors into clusters based on their correlations with other donors. Clusters were generated by “cutting” the dendrogram at the dotted line shown in Figure 5A, according to Python’s default settings.
Figure 5.
Heatmap Spearman correlations with hierarchical clustering and LDA of MSC tissue sources. (A) Spearman’s correlation heatmap of MSC donors from the three tissue origins. (B) Spearman’s correlation heatmap of AD-MSC donors. (C) Spearman’s correlation heatmap of BM-MSC donors. (D) Spearman’s correlation heatmap of UCT-MSC donors. (E) LDA of MSCs donors from the three tissue origins. AD-MSC (n = 3), BM-MSC (n = 9) and UCT-MSC (n = 4).
Hierarchical clustering grouped the MSC donors into three clusters (Figure 5A). Cluster 1 contained two UCT-MSC donors: UCT2 and UCT3. These donors had a similar lipidomic profile, as indicated by the high correlation between them (r > 0.7) in the heatmap. Moreover, they were very dissimilar from the other donors, as their clade was the first to split off from the rest of the dendrogram. Cluster 2 contained three BM-MSC donors, BM1, BM2 and BM8, and one UCT-MSC donor, UCT1a and UCT1b, which came from the same donor but were processed at different times. Donors BM2 and BM8 were very similar to each other (r = 0.94) and moderately similar to UCT1 (r = 0.64 to. 0.68). In contrast, BM1 was more different from the rest of the donors in the cluster. Finally, cluster 3 was quite heterogeneous. It contained all the AD-MSC donors as well as the remaining BM-MSC donors. This cluster could be further subdivided into two groups. The first contained all the AD-MSCs (AD1, AD2 and AD3) and BM3, all of which (except for AD1) were closely correlated. The second contained BM5, BM4, BM7, BM9 and BM6, which were also closely correlated to each other.
The clustering patterns reveal considerable heterogeneity within MSC donors derived from the three tissue sources. Within each cluster, donors from the same tissue of origin were usually grouped more closely to one another than to donors from other tissues of origin, which supports the differences in sphingolipid concentrations observed between tissue sources in Figure 3. Nonetheless, the BM and UCT donors were split among several clusters, which reveals considerable heterogeneity within these tissues of origin. The BM donors were divided into two distinct clusters; one of them (cluster 2) was more similar to UCT donors, and the other (cluster 3) was more similar to AD donors. In addition, the UCT donors were also partitioned into two clusters. The first cluster (cluster 1) was dissimilar from all other donors, and the second cluster (cluster 2) shared some characteristics with BM-MSCs. This suggests that underlying factors could influence the lipidome of MSCs and therefore drive the correlations and clustering between MSC donors. For instance, sex differences could contribute to the clustering, since all the BM and UCT donors in cluster 2 were male and all the BM and AD donors in cluster 3 were female. Additionally, differences in indoleamine 2,3-dioxygenase (IDO) activity could contribute to the heterogeneity among BM donors. MSCs from BM donors in cluster 2 had lower IDO activity (<20 pg kynurenine [kyn]/cell/day), whereas MSCs from the majority of BM donors in cluster 3 had high IDO activity (≈30 pg kyn/cell/day or higher).
When comparing MSCs from the same tissue source, the MSCs were all strongly correlated with one another, illustrated by warmer colors and high correlation coefficient values (≥0.95) (Figure 5B–D). To gain insight into whether the MSCs can be classified into their tissue origin based on their SL profiles, linear discriminant analysis (LDA) was performed. Clear separation occurred between the donors’ three tissue sources. It can be inferred that donor similarities in the SL compositions led to class separation based on tissue source (Figure 5E).
Differences in High- and Low-IDO BM-MSCs
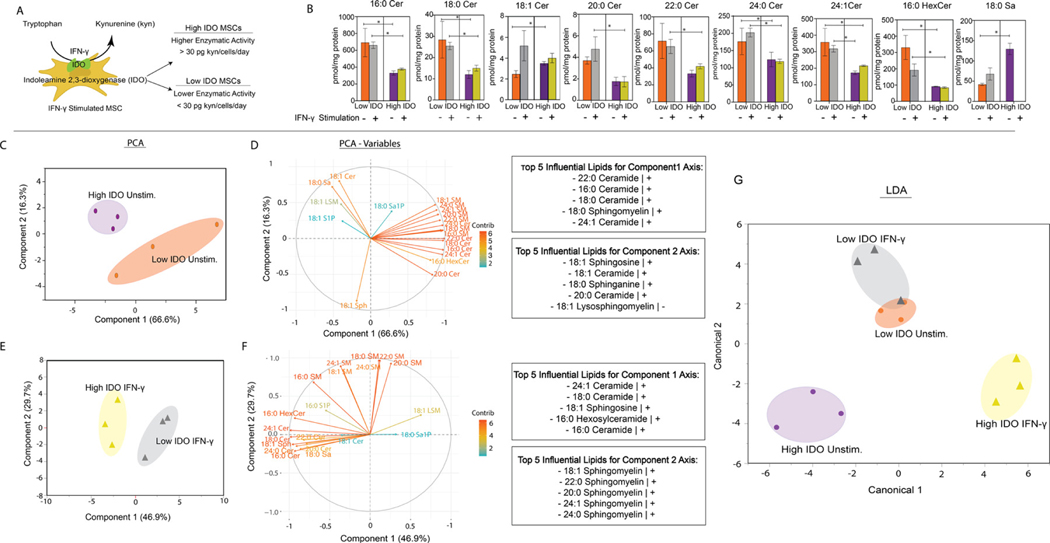
IDO activity is a common benchmark of immunomodulatory potency. This enzyme catalyzes the conversion of tryptophan to kynurenine, which is an immunosuppressive metabolite, inhibiting pro-inflammatory immune cell activity [38]. We stimulated MSCs from two BM donors (BM1 and BM6) with interferon-γ (IFN-γ), which resulted in an increase of IDO enzymatic activity [39]. Based on the IDO activity values provided by the manufacturer, principal component analysis (PCA) was performed on the nine reported IDO values. The donors were grouped based on IDO value. The average IDO value was used as a threshold to distinguish between high- and low-IDO MSCs. Donor BM1 was classified as low IDO, as its IDO activity was below the mean (30 pg kyn/cell/day), and donor BM6 was classified as high IDO, as its IDO activity was above the mean (Figure S1, Figure 6A).
Figure 6.
Unstimulated and IFN-γ–stimulated BM-MSC SL profile. (A) Schematic illustrating IDO converting tryptophan to kynurenine (kyn), an immunosuppressive metabolite. (B) SL concentrations of unstimulated and IFN-γ–primed MSCs. (C) PCA of high and low IDO unstimulated (Unstim) MSC groups. (D) PCA variable plot of MSC groups. (E) PCA of high and low IDO IFN-γ–primed groups. (F) PCA variable plot of IFN-γ groups. (G) LDA of high and low MSCs from Unstim and IFN-γ groups. *Statistical significance (P < .05). Mann–Whitney U test was used to determine significance. All groups were measured in triplicates (n = 3).
We investigated the relationship between IDO activity levels and the MSC sphingolipidome by comparing LC-MS/MS analysis profiles of unstimulated and IFN-γ–primed high- and low-IDO BM-MSCs. PCA was conducted to determine whether BM-MSCs can separate into high- and low-IDO groups based on their SL profiles. When measuring SL species, greater concentrations of acyl chain Cer species (16:0, 18:0, 20:0, 22:0, 24:0 and 24:1) and 16:0 HexCer were detected in both low-IDO MSC treatment groups compared with both high-IDO MSC treatment groups. Unstimulated high IDO had the greatest concentration of 18:0 Sa; however, Sa was not detected in high-IDO IFN-γ–treated MSCs. When looking at the effect of priming MSCs with IFN-γ on the SL profile, low-IDO MSCs had greater concentrations of 18:1, 20:0 and 24:0 Cer and 18:0 Sa than the low-IDO unstimulated group. The remaining SL species had comparable levels, with the exception of 16:0 HexCer, for which greater amounts of SLs were detected in the unstimulated group. In high-IDO MSCs, unstimulated and IFN-γ–primed MSCs had comparable concentrations of SLs, with the exception of Sa (Figure 6B). IFN-γ priming did not significantly alter SM levels in high- or low-IDO groups.
To understand the relationship between SL species detected in high- and low-IDO BM-MSCs, PCA was conducted. The PCA score plot of unstimulated MSCs shows a clear separation between high- and low-IDO groups. The separation was primarily due to the first principal component (PC1), which explains 66% of the variance in the data, and the second principal component (PC2), which explains 16% of the variance (Figure 6C). The lipids that most strongly contributed to the separation along PC1 consist predominantly of ceramide species (22:0, 16:0, 18:0 and 24:1 Cer), as well as one sphingomyelin species (18:0 SM) (Figure 6D). The lipids with the greatest influence on separation along PC2 include 18:1 Cer, 18:0 Sa, 18:1 Sph, 20:0 Cer and 18:1 LSM. All the lipids except for 18:1 LSM correlate positively along the PC2 axis. These results suggest possible differences in ceramide and LCB metabolism between high- and low-IDO MSCs in the unstimulated state. When conducting PCA on IFN-γ–primed high- and low-IDO groups, separation was primarily due to PC1, with 46.9% of the variance in the data, and PC2, with 29.7% (Figure 6E). The lipids that most strongly contributed to the separation along PC1 consisted of 24:1 Cer, 18:0 Cer, 18:1 Sph, 16:0 HexCer and 16:0 Cer. The lipids with the greatest influence on separation along PC2 were predominantly SM species (18:1, 22:0, 20:0, 24:1 and 24:0 SM) (Figure 6F). This suggests differences in ceramide and sphingomyelin metabolism between low- and high-IDO MSCs after IFN-γ priming.
Furthermore, we wanted to assess whether SLs can be used as predictor features to classify MSCs into groups based on their IDO activity. We used LDA, which is a supervised machine learning algorithm that can perform dimensionality reduction and classification. When used for dimensionality reduction, LDA relies on the class labels of each observation and projects the data onto an axis that maximizes the separation between the classes [40]. LDA is increasingly being used in biomedical applications to differentiate between healthy and diseased states or to classify various cell states within the same cell line [41–43]. We used SL concentrations as the input features and the IDO activity + IFN-γ treatment (low-IDO unstimulated, low-IDO IFN-γ primed, high-IDO unstimulated and high-IDO IFN-γ primed) as the class labels in LDA. The LDA plot showed separation between high-IDO unstimulated and IFN-γ groups. Interestingly, the low-IDO unstimulated and low-IDO IFN-γ did not have much separation, possibly because of similar SL profiles (Figure 6G).
Discussion
MSCs have been shown to be therapeutic in preclinical and early clinical trials in treating acute and chronic inflammatory diseases. Despite their therapeutic potential, discrepancies in MSC characterization have limited further clinical application. Our research is motivated by a growing appreciation of how local accumulation of specific sphingolipids (SLs) either directly or indirectly regulates multiple cellular properties that may predict stem cell potency, including mechanics of plasma and nuclear membrane, mitochondrial activity and cell death mechanisms [35,44,45]. SLs are a highly diverse family of molecules that are important building blocks of eukaryotic membranes [33]. Before this work, we demonstrated that modulating novel S1P and S1P receptor 3 (S1PR3) signaling mobilizes both hematopoietic stem cells and MSCs to enhance ectopic bone growth, further emphasizing the importance of understanding SL functions [46,47]. In this study, we performed LC-MS/MS to characterize AD-, BM- and UCT-MSC sphingolipid metabolism profiles.
Demographic features have not been fully explored as it pertains to MSC lipidomic characterization. It is not well understood how factors such as donor sex affect MSC SL metabolism. When comparing male and female MSC Cer levels, females possess greater levels of Cer. Generally, Cer, a central intermediate, regulates cell fate by activating the apoptosis regulator proteins, Bcl-2-associated X and Bcl-2 homologous antagonist, leading mitochondrial outer membrane permeabilization and apoptosis [48]. This internal feedback system illustrates SL complex interconnectivity and signaling. It is also interesting to speculate that differences in sphingolipid profiles are a result of differences in body composition of male and female donors and/or differing hormonal influences on cell properties [49–51]. Li et al. [52] recently reported a relationship between Cer and estrogen, showing decreased CerS and sphingomyelinase activity in ovariectomized rats. This resulted in perimenopausal hypertension, suggesting possible changes in Cer homeostasis among female donor cells [52]. As we detected SL differences in BM-MSCs from male and female donors, additional studies and analysis are needed to investigate the effect of sex on the MSC sphingolipidome.
Treating MSCs with IFN-γ activates their anti-inflammatory functions, thus priming them for immunomodulation [53]. IFN-γ increases MSCs’ IDO enzymatic activity, which catalyzes the rate-limiting step in the conversion of the amino acid tryptophan into kynurenine. The depletion of tryptophan (or the accumulation of kynurenine) inhibits T cell effector function, resulting in T cell death [54]. MSCs with higher IDO activity (≥30 pg kyn/cell/day) are more effective in regulating immune responses than MSCs with lower IDO activity (<30 pg kyn/cell/day) [55]. However, variability in immune modulatory potency among donors based on IDO assays remains a clinical hurdle [56]. When investigating the effects of IFN-γ on MSCs, Campos et al. [31] uncovered increases in very long chain (34- to 42-carbon chain) SM species after priming. However, those studies did not solely focus on MSC sphingolipidome characterization. Assessing the sphingolipid profile of unstimulated and IFN-γ–primed BM-MSCs uncovered differences in Cer species and Sa between low- and high-IDO MSCs. Cer acts as a secondary messenger in signaling pathways relating to immune responses such as cytokine production and cell activation [57]. Jrad-Lamine et al. [58] showed that mice lacking the expression of Ido1 gene possessed higher levels of Cer than wild-type mice. Our findings show a similar trend, where low-IDO MSCs have higher Cer levels. It remains unclear whether and how changes in Cer signaling may influence immunomodulatory properties or paracrine activity of MSCs directly, but these results do raise the exciting possibility that pharmacologic or genetic inhibition of enzymes resulting in Cer accumulation, such as sphingomyelinase, may be effective in culture to preserve or enhance the therapeutic potency of MSCs. Further studies will need to be conducted to understand the role of Cer in low-IDO MSCs and associated implications on their in vivo immune modulatory properties.
Circulating lipids, such as cholesterols and triglycerides, have been the gold standard for understanding the progression of diseases such as hypertension, diabetes and cardiovascular diseases [59,60]. However, SLs in particular have a distinct role in which they form cellular membranes together with glycerolipids and sterols. This allows regulation of sorting and trafficking of membrane content and regulation of cellular morphology [61,62]. Sphingomyelins (SMs) are the most abundant SL in the membrane of mammalian cells, where they facilitate integral protein recruitment via SM cholesterol interactions, lipid raft formation, regulation of endocytosis, ion channel flux regulation and modulation of the biophysical properties of the cell membrane [63,64]. We detected SMs primarily in AD-MSCs. In vivo, adipocytes rely on lipid metabolism to regulate their unique functions. Structural membrane SLs such as SM, Cer and S1P regulate intracellular mechanistic roles influencing adipocyte morphology and lipid droplet biogenesis. Alexaki et al. [65] also demonstrated that introducing perturbations to the SL de novo pathway resulted in lower SM levels and a hypertrophic phenotype common to lipodystrophies and apoptotic adipocytes. It can be inferred from our analysis that MSCs derived from mature tissue sources such as AD-MSCs may retain characteristic SL profiles and cellular properties after isolation from adipose tissue. Previous lipidomic analyses of white adipose tissue wherein differing types of fat were derived from either the visceral or subcutaneous fat sources have shown that the tissues themselves retain a distinct “fingerprint” of lipid concentration [66]; thus, our findings imply that characterization methods targeting lipid metabolism in general and bioactive lipids in particular are a promising approach to assess therapeutic quality and potency of cell therapies.
SLs play an essential role in multiple stages during prenatal development and tissue maturation. Glycosphingolipids along with S1P participate in endocytosis of nutrients in the intestines, skin barrier permeability and homeostasis, and neural development [67–70]. In our study, we found HexCer and S1P had higher concentrations detected in UCT-MSCs. These distributions may be attributable to their role in developmental maturity of umbilical cord tissue, where cells derived from more primitive tissue sources retain characteristic SL metabolic profiles. Biosynthesis of individual fatty acyl chain variants of SL species may also be predictive of tissue source and cell potency. Six (dihydro) CerS isoforms have shown fatty acyl-CoA substrate specificity, leading to variations of the ceramide molecule based on saturation (saturated or unsaturated) or length (C14 to C26) of the N-acyl chain. For example, CerS1 produces C18 Cer species with stearoyl CoA as a substrate, whereas CerS2 is responsible for producing very long chain fatty acyl Cer species (C20 to C26) [71]. CerS3 prefers middle and long chain fatty acyl CoAs to generate large structural SLs [72], whereas CerS5 and CerS6 prefer palmitoyl-CoA substrate to generate C16 Cer species [73,74]. Therefore, MS-based determination of Cer acyl chain variations in cultured MSCs may elucidate which isoforms of CerS are most active. Interestingly, CerS2 and CerS3 expression is elevated in more primitive cells, including the placenta (CerS2) and testis (CerS3), and more weakly expressed in more mature tissues [74,75]. These findings suggest that generation of SL profiles may reveal novel enzymatic targets associated with cell maturity changes in culture. As specific acyl chain variants of Cer affect diverse biological processes including embryo development, macrophage differentiation and tumor progression [76], it is possible that by elucidating the role of CerS isoforms on cultured MSCs, we may determine whether generation of acyl chain variants of SLs only correlate with immunomodulatory properties or influence MSC properties directly.
Culture parameters such as media formulations may influence cell properties and the lipid profile. In this study, BM- and AD-MSCs were cultured using standard process medium used in industrial cell manufacturing settings, whereas UCT-MSCs were cultured in serum-free conditions for clinical applications. Despite culture differences, observable differences were noted between AD- and BM-MSCs cultured in the same medium. Overall, this warrants further investigation of media formulation’s influence on MSC properties and the sphingolipid profile.
In conclusion, we used state-of-the-art LC-MS/MS sphingolipidomic approaches established at Georgia Tech/Emory [77–79] to study how variations in the concentrations of regulatory SL metabolites may be used to characterize the quality and potency of cultured MSCs. Multivariate analysis of lipidomic network profiles yielded valuable insight into tissue origin and immune modulation of the cells. This analysis also elucidates particular enzymatic targets in the sphingolipid metabolic network that may be targeted to minimize undesired changes in membrane composition during culture to preserve or enhance cell quality.
Methods
UCT-MSC isolation and expansion
Detailed UCT-MSC isolation was described elsewhere [106]. We used UCT-MSC donors UCT1a, UCT1b, UCT2 and UCT3 for lipidomic analysis. UCT1a and UCT1b were obtained from the same donor. All the UCT-MSCs were collected from the umbilical cords of male babies and expanded up to passage 2 (P2) by the Department of Pediatrics at Duke University. For a typical expansion of UCT-MSCs, cryopreserved P0 vials were thawed at 37°C in a water bath for ≤2 min and quickly transferred to a 15-mL sterile tube containing xeno-free serum-free media (XS-FM, Prime-XV, Irvine Scientific) and gently mixed with the media by pipetting up and down. Cell viabilities and counts were measured using the acridine orange-propidium iodide method with a NucleoCounter (Chemometec). Then, the cells were suspended in XS-FM media and plated in a HYPER flask (1720-cm2 surface area; Corning). The UCT-MSCs were expanded for 5 to 7 days, harvested and cryopreserved as P1. Using a similar protocol, P1 to P2 expansion was performed, and the cryopreserved P2 (PDL: 13.0) UCT-MSC vials were shipped to Georgia Institute of Technology for characterization.
BM- and AD-MSC culture
Nine human BM-MSCs, six male donors (BM1, BM2, BM3, BM6, BM7, BM8) and three female donors (BM4, BM5, BM9) and three AD-MSCs from female donors were purchased from RoosterBio (Frederick, MD). Upon purchase, the BM-MSC population-double level (PDL) ranged from 6.3 to 9.4. AD-MSCs’ PDL ranged from 7.81 to 8.91. These cells were considered P0. The MSCs were thawed and expanded by protocols provided by RoosterBio. Cells were cultured in Rooster-Nourish medium (High Performance Media Kit, KT-001) containing serum-derived RoosterBooster in T-225 flasks at an approximate density of 3,333 cells/cm2. Cells were grown to 80% confluence (3 to 4 days) and then harvested the next day per manufacturer protocol. Cells were lifted from the flasks by TrypLE Express (Invitrogen), and the enzyme was neutralized with spent media before collecting the cells by centrifugation. Cells were frozen in Cryostor CS5 cryopreservation media for future experiments and stored in the vapor phase of liquid nitrogen. Cells were frozen as P1.
IFN-γ treatment
P1 BM-MSC donor samples, BM1 and BM6, were thawed and seeded in T-225 flasks at an approximate density of 3,333 cells/cm2. Cells were expanded until they reached ~90% confluence in Rooster-Nourish media. Cells were then passaged following manufacturing protocols and plated in 6-well plates (9 cm2) for IFN-γ treatment (? 500,000 cells/well) and allowed to attach overnight. The medium was aspirated, and Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, MA) with 2% fetal bovine serum was added. The cells were incubated with 50 ng/mL of IFN-γ for 24 hours. After 24-hour IFN-γ incubation, the MSCs were detached with a cell scraper and pelleted for sphingolipid extraction.
Sphingolipid extraction
SLs were extracted from MSCs using a modified procedure described elsewhere [107]. LCBs and CSLs were extracted separately owing to their different physical properties. Two aliquots from each MSC sample were taken for LCB analysis and CSL analysis. An aliquot was also kept for total protein quantification using a BCA protein assay. 1.5 mL of a 2:1 mixture of methanol:methylene chloride was added to each sphingoid base sample and 1.5 mL of a 2:1 mixture of methanol:chloroform was added to each complex sphingolipid sample. Next, 50 pmol of internal standard mixture (Avanti Polar Lipids) was added to each sample. Samples were incubated overnight at 48° C to extract lipids. Next, 150 μL of 1 M KOH in methanol was added to each sample and the samples were incubated at 37°C for 2 h. This is to cleave the ester bonds of contaminating glycerophospholipids. After incubating, 5 μL of glacial acetic acid was added to all samples to neutralize the KOH. pH was checked using pH strips. 1 mL of chloroform and 2 mL of deionized water were added to all complex sphingolipid samples to induce phase separation. Sphingoid base and complex sphingolipid samples were centrifuged at 1400g for 8 min to pellet cell debris. For sphingoid base samples, the supernatant was transferred to a new glass tube. For complex sphingolipid samples, the bottom chloroform phase was transferred to a new glass tube. For sphingoid base samples, 0.5 mL of the 2:1 methanol:methylene chloride mixture was added to the cell debris in the original glass tubes. For complex sphingolipid samples, 1 mL of chloroform was added to the cell debris in the original glass tubes. The original tubes were all centrifuged at 1400 xg for 8 minutes. For sphingoid base samples the second supernatant was added to the first supernatant. For complex sphingolipid samples, the second bottom phase was added to the first bottom phase. Remaining cell debris was discarded. Organic solvents were removed by vacuum drying overnight in a Savant SpeedVac (Thermo Fisher). The dried SLs were stored in a −20°C freezer until analysis.
Preparation of samples for sphingolipid analysis
Dried LCBs samples were resuspended in 300 μL of a 3:2 mixture of mobile phase A1:mobile phase B1 solvent. Mobile phase A1 consisted of 58:41:1 methanol:water:formic acid and 5 mM ammonium formate. Mobile phase B1 consisted of 99:1 methanol:formic acid and 5 mM ammonium formate. Dried complex sphingolipid samples were resuspended in 300 μL of mobile phase A2. Mobile phase A2 consisted of 97:21:1 acetonitrile:methanol:formic acid and 5 mM ammonium formate. Mobile phase B2 consisted of 89:6:5 methanol: water:formic acid and 50 mM triethylammonium acetate. Resuspended samples were centrifuged at 18,000g for 10 min to remove any remaining cell debris. Total protein amount in each sample was determined for normalization using a Pierce BCA Protein Assay (Thermo Fisher Scientific) following the recommended procedure from the manufacturer. The top 200 μL was transferred to a MS autosampler tube.
Sphingolipid LC-MS/MS analysis
Sphingolipids were analyzed using Micromass Quattro LC in multiple reaction mode. Fragmented ions were detected with product ion scan. The LCB samples were separated using a 2.1 (internal diameter) × 150 mm Phenomenex C18 column and a binary solvent system at a flow rate of 300 μL/min. Before injection, the column was equilibrated with 100% mobile phase A1. After injection, the solvent composition was held at 100% A1 for 5 min, followed by a linear gradient to 100% B1 over 15 min. The solvent composition was held at 100% B1 for 5 min, dropped back to 100% A over 1 min and held at 100% A for 4 min. The CSL samples were separated using a 2.1 (internal diameter) × 150 mm Supelcosil NH2 column and a binary solvent system at a flow rate of 300 μL/min. Before injection, the column was equilibrated with 100% mobile phase A2. After injection, the solvent composition was held at 100% A for 5 min, followed by a linear gradient to 100% B2 for 1 min. The solvent composition was held at 100% B for 14 min, dropped back to 100% A over 1 min and held at 100% A2 for 9 min. Because glucosylceramide and galactosylceramide are not separated by this chromatography method, their concentrations are reported together as hexosylceramide. Internal standards were run to assess retention time drift during MS analysis.
Quantification and Statistical Analysis
Statistical analysis was conducted in Prism 8.0 (GraphPad Software, La Jolla, CA), JMP Pro 15, RStudio (PBC, Boston, MA) and Python 3.7. Comparisons between experimental groups were carried out in Python using Mann–Whitney U tests because of the non-Gaussian nature of the data. The results are presented as mean ± SEM on bar plots generated in Prism. Wide LDA and PCA were conducted in JMP Pro 15. Graphical illustrations of PCA variable loading plots were generated in RStudio. Hierarchical clustering of MSC donors was conducted using Python’s Seaborn and SciPy packages. First, a Spearman correlation matrix was generated to show correlations among all the MSC donors based on their sphingolipid profiles. Next, hierarchical clustering using the single linkage method (default) was performed on the correlation matrix to group donors into clusters based on their correlations to other donors. Correlations between donors and clustering results were visualized on both a stand-alone dendrogram and a clustered Spearman correlation heatmap. Heatmaps were generated using Python’s Matplotlib and Seaborn visualization packages. Finally, false discovery rate testing was performed using the original Benjamini–Hochberg method in Prism.
Supplementary Material
Acknowledgments
This work was funded by The Marcus Foundation, the Marcus Center for Therapeutic Cell Characterization and Manufacturing, The Georgia Tech Foundation, and the Georgia Research Alliance. Krishnendu Roy was supported by the National Science Foundation Engineering Research Center for Cell Manufacturing Technologies (NSF EEC 1648035). S’Dravious DeVeaux was supported by the NGMS-sponsored Cell and Tissue Engineering NIH Biotechnology Training Grant (T32 GM-008433). We thank Research Coordinator, David Bostwick, in Parker H. Petit Institute for Bioengineering and Biosciences mass spectrometry core at Georgia Tech for his assistance with mass spectrometry method development. We thank Kuang-Drew Li and Sami Belhareth for preliminary lipidomic analysis. Graphics used for figure illustrations were created using Biorender.com. The graphics were exported with a paid Biorender.com subscription.
Footnotes
Declaration of Competing Interest
The authors declare no competing interest.
Supplementary materials
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.jcyt.2021.12.009.
References
- [1].Pereira Chilima TD, Moncaubeig F, Farid SS. Impact of allogeneic stem cell manufacturing decisions on cost of goods, process robustness and reimbursement. Biochemical Engineering Journal 2018;137:132–51. [Google Scholar]
- [2].Mason C, Brindley DA, Culme-Seymour EJ, Davie NL. Cell therapy industry: billion dollar global business with unlimited potential. Regen Med 2011;6(3): 265–72. [DOI] [PubMed] [Google Scholar]
- [3].Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Sci Transl Med 2013;5(179):179ps7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rao M, Mason C, Solomon S. Cell therapy worldwide: an incipient revolution. Regen Med 2015;10(2):181–91. [DOI] [PubMed] [Google Scholar]
- [5].Wang M, Yuan Q, Xie L. Mesenchymal Stem Cell-Based Immunomodulation: Properties and Clinical Application. Stem Cells International 2018;2018:3057624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cosenza S, Toupet K, Maumus M, Luz-Crawford P, Blanc-Brude O, Jorgensen C, Noël D. Mesenchymal stem cells-derived exosomes are more immunosuppressive than microparticles in inflammatory arthritis. Theranostics 2018;8 (5):1399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Blaess M, Deigner HP. Derailed Ceramide Metabolism in Atopic Dermatitis (AD): A Causal Starting Point for a Personalized (Basic) Therapy. Int J Mol Sci 2019;16 (20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ahn EH, Lee MB, Seo DJ, Lee J, Kim Y, Gupta K. Sphingosine Induces Apoptosis and Down-regulation of MYCN in PAX3-FOXO1-positive Alveolar Rhabdomyo-sarcoma Cells Irrespective of TP53 Mutation. Anticancer Res 2018;38(1):71–6. [DOI] [PubMed] [Google Scholar]
- [9].Galipeau J. Mesenchymal Stromal Cells for Graft-versus-Host Disease: A Trilogy. Biology of Blood and Marrow Transplantation 2020;26(5):e89–91. [DOI] [PubMed] [Google Scholar]
- [10].Sanjucta A, Sayani M, Dwaipayan S. Mesenchymal Stem Cell as a Potential Therapeutic for Inflammatory Bowel Disease-Myth or Reality? Current Stem Cell Research & Therapy 2017;12(8):644–57. [DOI] [PubMed] [Google Scholar]
- [11].Bagno L, Hatzistergos KE, Balkan W, Hare JM. Mesenchymal Stem Cell-Based Therapy for Cardiovascular Disease: Progress and Challenges. Mol Ther 2018;26 (7):1610–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sisakhtnezhad S, Alimoradi E, Akrami H. External factors influencing mesenchymal stem cell fate in vitro. Eur J Cell Biol 2017;96(1):13–33. [DOI] [PubMed] [Google Scholar]
- [13].Phinney DG, Galipeau J. Manufacturing mesenchymal stromal cells for clinical applications: A survey of Good Manufacturing Practices at US academic centers. Cytotherapy 2019;21(7):782–92. [DOI] [PubMed] [Google Scholar]
- [14].Mendicino M, Bailey Alexander M, Wonnacott K, Puri Raj K, Bauer Steven R. MSC-Based Product Characterization for Clinical Trials: An FDA Perspective. Cell Stem Cell 2014;14(2):141–5. [DOI] [PubMed] [Google Scholar]
- [15].Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006;8(4):315–7. [DOI] [PubMed] [Google Scholar]
- [16].Viswanathan S, Shi Y, Galipeau J, Krampera M, Leblanc K, Martin I, Nolta J, Phinney DG, Sensebe L. Mesenchymal stem versus stromal cells: International Society for Cell & Gene Therapy (ISCT®) Mesenchymal Stromal Cell committee position statement on nomenclature. Cytotherapy 2019;21(10):1019–24. [DOI] [PubMed] [Google Scholar]
- [17].McCartney A, Vignoli A, Biganzoli L, Love R, Tenori L, Luchinat C, Di Leo A. Metabolomics in breast cancer: A decade in review. Cancer Treatment Reviews 2018;67:88–96. [DOI] [PubMed] [Google Scholar]
- [18].Gowda GAN, Djukovic D. Overview of mass spectrometry-based metabolomics: opportunities and challenges. Methods Mol Biol 2014;1198:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ren J-L, Zhang A-H, Kong L, Wang X-J. Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC Advances 2018;8 (40):22335–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Emwas A-H, Roy R, McKay RT, Tenori L, Saccenti E, Gowda GAN, Raftery D, Alahmari F, Jaremko L, Jaremko M, Wishart DS. NMR Spectroscopy for Metabolomics Research. Metabolites 2019;9(7):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].León Z, García-Cañaveras JC, Donato MT, Lahoz A. Mammalian cell metabolomics: experimental design and sample preparation. Electrophoresis 2013;34 (19):2762–75. [DOI] [PubMed] [Google Scholar]
- [22].Kalluri U, Naiker M, Myers MA. Cell culture metabolomics in the diagnosis of lung cancer-the influence of cell culture conditions. J Breath Res 2014;8 (2):027109. [DOI] [PubMed] [Google Scholar]
- [23].Muschet C, Möller G, Prehn C, de Angelis MH, Adamski J, Tokarz J. Removing the bottlenecks of cell culture metabolomics: fast normalization procedure, correlation of metabolites to cell number, and impact of the cell harvesting method. Metabolomics 2016;12(10). 151–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].O’Donnell VB, Ekroos K, Liebisch G, Wakelam M. Lipidomics: Current state of the art in a fast moving field. Wiley Interdiscip Rev Syst Biol Med 2020;12(1):e1466. [DOI] [PubMed] [Google Scholar]
- [25].Leuti A, Fazio D, Fava M, Piccoli A, Oddi S, Maccarrone M. Bioactive lipids, inflammation and chronic diseases. Advanced Drug Delivery Reviews 2020. [DOI] [PubMed] [Google Scholar]
- [26].Musso G, Cassader M, Paschetta E, Gambino R. Bioactive lipid species and metabolic pathways in progression and resolution of nonalcoholic steatohepatitis. Gastroenterology 2018;155(2):282–302. e8. [DOI] [PubMed] [Google Scholar]
- [27].Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nature reviews Molecular cell biology 2018;19(3):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Verderio C, Gabrielli M, Giussani P. Role of sphingolipids in the biogenesis and biological activity of extracellular vesicles. J Lipid Res 2018;59(8):1325–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Marycz K, Śmieszek A, Jeleń M, Chrząstek K, Grzesiak J, Meissner J. The effect of the bioactive sphingolipids S1P and C1P on multipotent stromal cells–new opportunities in regenerative medicine. Cellular and Molecular Biology Letters 2015;20(3):510–33. [DOI] [PubMed] [Google Scholar]
- [30].Marycz K, Krzak J, Maredziak M, Tomaszewski KA, Szczurek A, Moszak K. The influence of metal-based biomaterials functionalized with sphingosine-1-phosphate on the cellular response and osteogenic differentaion potenial of human adipose derived mesenchymal stem cells in vitro. J Biomater Appl 2016;30 (10):1517–33. [DOI] [PubMed] [Google Scholar]
- [31].Campos AM, Maciel E, Moreira ASP, Sousa B, Melo T, Domingues P, Curado L, Antunes B, Domingues MRM, Santos F. Lipidomics of Mesenchymal Stromal Cells: Understanding the Adaptation of Phospholipid Profile in Response to Pro-Inflammatory Cytokines. Journal of Cellular Physiology 2016;231(5):1024–32. [DOI] [PubMed] [Google Scholar]
- [32].Ogle ME, Sefcik LS, Awojoodu AO, Chiappa NF, Lynch K, Peirce-Cottler S, Botchwey EA. Engineering in vivo gradients of sphingosine-1-phosphate receptor ligands for localized microvascular remodeling and inflammatory cell positioning. Acta Biomaterialia 2014;10(11):4704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol 2008;9(2):139–50. [DOI] [PubMed] [Google Scholar]
- [34].Gupta S, Maurya MR, Merrill AH Jr., Glass CK, Subramaniam S. Integration of lipidomics and transcriptomics data towards a systems biology model of sphingolipid metabolism. BMC Syst Biol 2011;5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hernandez-Corbacho MJ, Salama MF, Canals D, Senkal CE, Obeid LM. Sphingolipids in mitochondria. Biochim Biophys Acta 2017;1862(1):56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Montgomery MK, Brown SH, Lim XY, Fiveash CE, Osborne B, Bentley NL, Braude JP, Mitchell TW, Coster AC, Don AS. Regulation of glucose homeostasis and insulin action by ceramide acyl-chain length: A beneficial role for very long-chain sphingolipid species. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids 2016;1861(11):1828–39. [DOI] [PubMed] [Google Scholar]
- [37].Wattenberg BW. The long and the short of ceramides. Journal of Biological Chemistry 2018;293(25):9922–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mándi Y, Vécsei L. The kynurenine system and immunoregulation. Journal of neural transmission 2012;119(2):197–209. [DOI] [PubMed] [Google Scholar]
- [39].Jürgens B, Hainz U, Fuchs D, Felzmann T, Heitger A. Interferon-gamma-triggered indoleamine 2,3-dioxygenase competence in human monocyte-derived dendritic cells induces regulatory activity in allogeneic T cells. Blood 2009;114 (15):3235–43. [DOI] [PubMed] [Google Scholar]
- [40].Kim S-J, Magnani A, Boyd S. Robust fisher discriminant analysis. Advances in neural information processing systems 2006: 659–66. [Google Scholar]
- [41].Marchevsky AM, Tsou JA, Laird-Offringa IA. Classification of individual lung cancer cell lines based on DNA methylation markers: use of linear discriminant analysis and artificial neural networks. The Journal of Molecular Diagnostics 2004;6(1):28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Al-Dulaimi K, Chandran V, Nguyen K, Banks J, Tomeo-Reyes I. Benchmarking HEp-2 specimen cells classification using linear discriminant analysis on higher order spectra features of cell shape. Pattern Recognition Letters 2019;125:534–41. [Google Scholar]
- [43].Tang M, Xia L, Wei D, Yan S, Du C, Cui H-L. Distinguishing different cancerous human cells by raman spectroscopy based on discriminant analysis methods. Applied Sciences 2017;7(9):900. [Google Scholar]
- [44].Spincemaille P, Cammue BP, Thevissen K. Sphingolipids and mitochondrial function, lessons learned from yeast. Microbial Cell 2014;1(7):210–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Patwardhan GA, Beverly LJ, Siskind LJ. Sphingolipids and mitochondrial apoptosis. J Bioenerg Biomembr 2016;48(2):153–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Selma JM, Das A, Awojoodu AO, Wang T, Kaushik AP, Cui Q, Song H, Ogle ME, Olingy CE, Pendleton EG, Tehrani KF, Mortensen LJ, Botchwey EA. Novel Lipid Signaling Mediators for Mesenchymal Stem Cell Mobilization during Bone Repair. Cell Mol Bioeng 2018;11(4):241–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ogle ME, Olingy CE, Awojoodu AO, Das A, Ortiz RA, Cheung HY, Botchwey EA. Sphingosine-1-Phosphate Receptor-3 Supports Hematopoietic Stem and Progenitor Cell Residence Within the Bone Marrow Niche. Stem Cells 2017;35 (4):1040–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Dadsena S, Bockelmann S, Mina JGM, Hassan DG, Korneev S, Razzera G, Jahn H, Niekamp P, Müller D, Schneider M, Tafesse FG, Marrink SJ, Melo MN, Holthuis JCM. Ceramides bind VDAC2 to trigger mitochondrial apoptosis. Nature Communications 2019;10(1):1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mielke MM, Bandaru VVR, Han D, An Y, Resnick SM, Ferrucci L, Haughey NJ. Demographic and clinical variables affecting mid-to late-life trajectories of plasma ceramide and dihydroceramide species. Aging cell 2015;14(6):1014–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Norheim F, Bjellaas T, Hui ST, Krishnan KC, Lee J, Gupta S, Pan C, Hasin-Brumshtein Y, Parks BW, Li DY. Genetic, dietary, and sex-specific regulation of hepatic ceramides and the relationship between hepatic ceramides and IR. J Lipid Res 2018;59(7):1164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Krishnan KC, Mehrabian M, Lusis AJ. Sex differences in metabolism and cardio-metabolic disorders. Current opinion in lipidology 2018;29(5):404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li Y, Zhang W, Li J, Sun Y, Yang Q, Wang S, Luo X, Wang W, Wang K, Bai W, Zhang H, Qin L. The imbalance in the aortic ceramide/sphingosine-1-phosphate rheostat in ovariectomized rats and the preventive effect of estrogen. Lipids in Health and Disease 2020;19(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Polchert D, Sobinsky J, Douglas G, Kidd M, Moadsiri A, Reina E, Genrich K, Mehrotra S, Setty S, Smith B, Bartholomew A. IFN-gamma activation of mesenchymal stem cells for treatment and prevention of graft versus host disease. Eur J Immunol 2008;38(6):1745–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhai L, Bell A, Ladomersky E, Lauing KL, Bollu L, Sosman JA, Zhang B, Wu JD, Miller SD, Meeks JJ, Lukas RV, Wyatt E, Doglio L, Schiltz GE, McCusker RH, Wainwright DA. Immunosuppressive IDO in Cancer: Mechanisms of Action, Animal Models, and Targeting Strategies. Frontiers in Immunology 2020;11:1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mbongue JC, Nicholas DA, Torrez TW, Kim N-S, Firek AF, Langridge WHR. The Role of Indoleamine 2, 3-Dioxygenase in Immune Suppression and Autoimmunity. Vaccines (Basel) 2015;3(3):703–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gray A, Schloss RS, Yarmush M. Donor variability among anti-inflammatory preactivated mesenchymal stromal cells. Technology (Singap World Sci) 2016;4 (3):201–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ballou LR, Laulederkind SJ, Rosloniec EF, Raghow R. Ceramide signalling and the immune response. Biochimica et Biophysica Acta (BBA)-Lipids and Lipid Metabolism 1996;1301(3):273–87. [DOI] [PubMed] [Google Scholar]
- [58].Jrad-Lamine A, Henry-Berger J, Damon-Soubeyrand C, Saez F, Kocer A, Janny L, Pons-Rejraji H, Munn DH, Mellor AL, Gharbi N, Cadet R, Guiton R, Aitken RJ, Drevet JR. Indoleamine 2,3-dioxygenase 1 (ido1) is involved in the control of mouse caput epididymis immune environment. PLoS One 2013;8(6). e66494-e66494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zanoni P, Khetarpal SA, Larach DB, Hancock-Cerutti WF, Millar JS, Cuchel M, Der-Ohannessian S, Kontush A, Surendran P, Saleheen D, Trompet S, Jukema JW, De Craen A, Deloukas P, Sattar N, Ford I, Packard C, Majumder AaS, Alam DS, Di Angelantonio E, Abecasis G, Chowdhury R, Erdmann J, Nordestgaard BG, Nielsen SF, Tybjærg-Hansen A, Schmidt RF, Kuulasmaa K, Liu DJ, Perola M, Blankenberg S, Salomaa V, Männistö S, Amouyel P, Arveiler D, Ferrieres J, Müller-Nurasyid M, Ferrario M, Kee F, Willer CJ, Samani N, Schunkert H, Butterworth AS, Howson JMM, Peloso GM, Stitziel NO, Danesh J, Kathiresan S, Rader DJ. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science 2016;351(6278):1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Toth PP, Granowitz C, Hull M, Liassou D, Anderson A, Philip S. High triglycerides are associated with increased cardiovascular events, medical costs, and resource use: a real-world administrative claims analysis of statin-treated patients with high residual cardiovascular risk. Journal of the American Heart Association 2018;7(15):e008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kraft ML. Sphingolipid Organization in the Plasma Membrane and the Mechanisms That Influence It. Front Cell Dev Biol 2016;4:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zeidan YH, Jenkins RW, Hannun YA. Remodeling of cellular cytoskeleton by the acid sphingomyelinase/ceramide pathway. J Cell Biol 2008;181(2):335–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Slotte J. Molecular properties of various structurally defined sphingomyelins – Correlation of structure with function. Progress in Lipid Research 2013;52 (2):206–19. [DOI] [PubMed] [Google Scholar]
- [64].Slotte JP. Biological functions of sphingomyelins. Progress in Lipid Research 2013;52(4):424–37. [DOI] [PubMed] [Google Scholar]
- [65].Alexaki A, Clarke BA, Gavrilova O, Ma Y, Zhu H, Ma X, Xu L, Tuymetova G, Larman BC, Allende ML. De novo sphingolipid biosynthesis is required for adipocyte survival and metabolic homeostasis. Journal of Biological Chemistry 2017;292 (9):3929–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Pradas I, Huynh K, Cabre R, Ayala V, Meikle PJ, Jove M, Pamplona R. Lipidomics Reveals a Tissue-Specific Fingerprint. Front Physiol 2018;9:1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jennemann R, Kaden S, Sandhoff R, Nordström V, Wang S, Volz M, Robine S, Amen N, Rothermel U, Wiegandt H. Glycosphingolipids are essential for intestinal endocytic function. Journal of Biological Chemistry 2012;287(39): 32598–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Popovic ZV, Rabionet M, Jennemann R, Krunic D, Sandhoff R, Gröne H-J, Porubsky S. Glucosylceramide synthase is involved in development of invariant natural killer T cells. Frontiers in immunology 2017;8:848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Jennemann R, Sandhoff R, Wang S, Kiss E, Gretz N, Zuliani C, Martin-Villalba A, Jäger R, Schorle H, Kenzelmann M. Cell-specific deletion of glucosylceramide synthase in brain leads to severe neural defects after birth. Proceedings of the National Academy of Sciences 2005;102(35):12459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhao Z, Chen Z, Zhao X, Pan F, Cai M, Wang T, Zhang H, Lu JR, Lei M. Sphingosine-1-phosphate promotes the differentiation of human umbilical cord mesenchymal stem cells into cardiomyocytes under the designated culturing conditions. Journal of Biomedical Science 2011;18(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH Jr., Futerman AH. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J Biol Chem 2008;283(9):5677–84. [DOI] [PubMed] [Google Scholar]
- [72].Gault CR, Obeid LM, Hannun YA. An overview of sphingolipid metabolism: from synthesis to breakdown. Adv Exp Med Biol 2010;688:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Riebeling C, Allegood JC, Wang E, Merrill AH Jr., Futerman AH. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J Biol Chem 2003;278(44):43452–9. [DOI] [PubMed] [Google Scholar]
- [74].Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J 2005;390(Pt 1):263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Soupene E, Serikov V, Kuypers FA. Characterization of an acyl-coenzyme A binding protein predominantly expressed in human primitive progenitor cells. J Lipid Res 2008;49(5):1103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Cartier A, Hla T. Sphingosine 1-phosphate: Lipid signaling in pathology and therapy. Science 2019;366:6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Merrill AH Jr., Wang E, Mullins RE, Jamison WC, Nimkar S, Liotta DC. Quantitation of free sphingosine in liver by high-performance liquid chromatography. Anal Biochem 1988;171(2):373–81. [DOI] [PubMed] [Google Scholar]
- [78].Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, Kelly S, Allegood JC, Liu Y, Peng Q, Ramaraju H, Sullards MC, Cabot M, Merrill AH Jr.. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta 2006;1758(12):1864–84. [DOI] [PubMed] [Google Scholar]
- [79].Merrill AH Jr., Sullards MC. Opinion article on lipidomics: Inherent challenges of lipidomic analysis of sphingolipids. Biochim Biophys Acta 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hait NC, Maiti A. The role of sphingosine-1-phosphate and ceramide-1-phosphate in inflammation and cancer. Mediators of inflammation 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Gomez-Munoz A, Presa N, Gomez-Larrauri A, Rivera I-G, Trueba M, Ordonez M. Control of inflammatory responses by ceramide, sphingosine 1-phosphate and ceramide 1-phosphate. Progress in lipid research 2016;61:51–62. [DOI] [PubMed] [Google Scholar]
- [82].Yuan X, Logan TM, Ma T. Metabolism in Human Mesenchymal Stromal Cells: A Missing Link Between hMSC Biomanufacturing and Therapy? Frontiers in immunology 2019: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Pandey S, Banks KM, Kumar R, Kuo A, Wen D, Hla T, Evans T. Sphingosine Kinases Protect Murine ESCs from Sphingosine-induced Cell Cycle Arrest. STEM CELLS 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Puig N, Estruch M, Jin L, Sanchez-Quesada JL, Benitez S. The Role of Distinctive Sphingolipids in the Inflammatory and Apoptotic Effects of Electronegative LDL on Monocytes. Biomolecules 2019;9(8):300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Luheshi NM, Giles JA, Lopez-Castejon G, Brough D. Sphingosine regulates the NLRP3-inflammasome and IL-1β release from macrophages. Eur J Immunol 2012;42(3):716–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Aoki M, Aoki H, Ramanathan R, Hait NC, Takabe K. Sphingosine-1-phosphate signaling in immune cells and inflammation: roles and therapeutic potential. Mediators of inflammation 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Obinata H, Hla T. Sphingosine 1-phosphate and inflammation. International immunology 2019;31(9):617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Lidgerwood GE, Pitson SM, Bonder C, Pebay A. Roles of lysophosphatidic acid and sphingosine-1-phosphate in stem cell biology. Progress in lipid research 2018;72:42–54. [DOI] [PubMed] [Google Scholar]
- [89].Chen R, Cai X, Liu J, Bai B, Li X. Sphingosine 1-phosphate promotes mesenchymal stem cell-mediated cardioprotection against myocardial infarction via ERK1/2-MMP-9 and Akt signaling axis. Life sciences 2018;215:31–42. [DOI] [PubMed] [Google Scholar]
- [90].Yuan X, Li D, Chen X, Han C, Xu L, Huang T, Dong Z, Zhang M. Extracellular vesicles from human-induced pluripotent stem cell-derived mesenchymal stromal cells (hiPSC-MSCs) protect against renal ischemia/reperfusion injury via delivering specificity protein (SP1) and transcriptional activating of sphingosine kinase 1 and inhibiting necroptosis. Cell Death & Disease 2017;8(12):3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Prymas K, Świątkowska A, Traczyk G, Ziemlińska E, Dziewulska A, Ciesielska A, Kwiatkowska K. Sphingomyelin synthase activity affects TRIF-dependent signaling of Toll-like receptor 4 in cells stimulated with lipopolysaccharide. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids 2020;1865 (2):158549. [DOI] [PubMed] [Google Scholar]
- [92].Sakamoto H, Yoshida T, Sanaki T, Shigaki S, Morita H, Oyama M, Mitsui M, Tanaka Y, Nakano T, Mitsutake S. Possible roles of long-chain sphingomyelines and sphingomyelin synthase 2 in mouse macrophage inflammatory response. Biochemical and biophysical research communications 2017;482(2):202–7. [DOI] [PubMed] [Google Scholar]
- [93].Norris GH, Blesso CN. Dietary and endogenous sphingolipid metabolism in chronic inflammation. Nutrients 2017;9(11):1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Edsfeldt A, Dunér P, Ståhlman M, Mollet IG, Asciutto G, Grufman H, Nitulescu M, Persson AF, Fisher RM, Melander O. Sphingolipids contribute to human atherosclerotic plaque inflammation. Arteriosclerosis, thrombosis, and vascular biology 2016;36(6):1132–40. [DOI] [PubMed] [Google Scholar]
- [95].Gupta G, Baumlin N, Poon J, Ahmed B, Chiang Y-P, Railwah C, Kim MD, Rivas M, Goldenberg H, Elgamal Z. Airway resistance caused by sphingomyelin synthase 2 insufficiency in response to cigarette smoke. American journal of respiratory cell and molecular biology 2020;62(3):342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bowler RP, Jacobson S, Cruickshank C, Hughes GJ, Siska C, Ory DS, Petrache I, Schaffer JE, Reisdorph N, Kechris K. Plasma sphingolipids associated with chronic obstructive pulmonary disease phenotypes. American journal of respiratory and critical care medicine 2015;191(3):275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Khayrullin A, Krishnan P, Martinez-Nater L, Mendhe B, Fulzele S, Liu Y, Mattison JA, Hamrick MW. Very long-chain C24: 1 ceramide is increased in serum extra-cellular vesicles with aging and can induce senescence in bone-derived mesenchymal stem cells. Cells 2019;8(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Park SW, Kim M, Chen SW, Brown KM, D D’Agati V, Lee HT. Sphinganine-1-phosphate protects kidney and liver after hepatic ischemia and reperfusion in mice through S1P 1 receptor activation. Laboratory investigation 2010;90(8): 1209–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Nagata M, Izumi Y, Ishikawa E, Kiyotake R, Doi R, Iwai S, Omahdi Z, Yamaji T, Miyamoto T, Bamba T. Intracellular metabolite β-glucosylceramide is an endogenous Mincle ligand possessing immunostimulatory activity. Proceedings of the National Academy of Sciences 2017;114(16):E3285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Jeong Y-H, Kim Y, Song H, Chung YS, Park SB, Kim H-S. Anti-inflammatory effects of α-galactosylceramide analogs in activated microglia: involvement of the p38 MAPK signaling pathway. PLoS One 2014;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].von Gerichten J, Lamprecht D, Opálka L, Soulard D, Marsching C, Pilz R, Sencio V, Herzer S, Galy B, Nordström V. Bacterial immunogenic α-galactosylceramide identified in the murine large intestine: dependency on diet and inflammation. J Lipid Res 2019;60(11):1892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Apostolopoulou M, Gordillo R, Koliaki C, Gancheva S, Jelenik T, De Filippo E, Herder C, Markgraf D, Jankowiak F, Esposito I. Specific hepatic sphingolipids relate to insulin resistance, oxidative stress, and inflammation in nonalcoholic steatohepatitis. Diabetes Care 2018;41(6):1235–43. [DOI] [PubMed] [Google Scholar]
- [103].Zhang J-Y, Qu F, Li J-F, Liu M, Ren F, Zhang J-Y, Bian D-D, Chen Y, Duan Z-P, Zhang J-L. Up-regulation of plasma hexosylceramide (d18: 1/18: 1) contributes to genotype 2 virus replication in chronic hepatitis C: A 20-year cohort study. Medicine 2016;23(95). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Jang HJ, Lim S, Kim JM, Yoon S, Lee CY, Hwang HJ, Shin JW, Shin KJ, Kim HY, Park KI. Glucosylceramide synthase regulates adipo-osteogenic differentiation through synergistic activation of PPARγ with GlcCer. The FASEB Journal 2020;34 (1):1270–87. [DOI] [PubMed] [Google Scholar]
- [105].Murugesan V, Chuang WL, Liu J, Lischuk A, Kacena K, Lin H, Pastores GM, Yang R, Keutzer J, Zhang K. Glucosylsphingosine is a key biomarker of Gaucher disease. American journal of hematology 2016;91(11):1082–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Sun JM, Dawson G, Franz L, Howard J, McLaughlin C, Kistler B, Waters-Pick B, Meadows N, Troy J, Kurtzberg J. Infusion of human umbilical cord tissue mesenchymal stromal cells in children with autism spectrum disorder. Stem Cells Transl Med 2020;9(10):1137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Shaner RL, Allegood JC, Park H, Wang E, Kelly S, Haynes CA, Sullards MC, Merrill AH Jr.. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J Lipid Res 2009;50 (8):1692–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.