Abstract
Chitinase production in Streptomyces lividans is induced by chitin and repressed in the presence of glucose. A mutant of S. lividans TK24, strain G015, which was defective in glucose repression of chitinase production, was obtained by screening colonies for zones of clearing on colloidal chitin agar plates containing 1.0% (wt/vol) glucose. The transcriptional analysis of chiA in G015 with xylE, which encodes catechol 2,3-dioxygenase, as a reporter gene showed that the transcription from the chiA promoter of S. lividans TK24 occurred regardless of the presence of glucose. G015 was resistant to 2-deoxyglucose (2-DOG) and did not utilize glucose as a sole carbon source. When a DNA fragment containing glkA, a gene for glucose kinase, of Streptomyces coelicolor A3(2) was introduced into strain G015 on a low-copy-number plasmid, the sensitivity to 2-DOG, the ability to utilize glucose, and the glucose repression of chitinase production were restored. These results indicate that glkA is involved in glucose repression of chitinase production in S. lividans TK24.
Streptomyces species are saprophytic soil bacteria and are well known as decomposers of chitin, which is hydrolyzed by chitinase (EC 3.2.1.14). Many chitinase genes of Streptomyces have been cloned and sequenced (3, 7, 18–20, 23–25, 29). Their expression is induced by chitin and repressed by glucose (7, 19, 25). A pair of 12-bp direct repeats present in the promoter regions of all the known chitinase genes from Streptomyces (20) are implicated in the regulation of chitinase gene expression (5, 22). This suggests that there exists a common regulatory mechanism for induction and repression of chitinase genes in Streptomyces. Recently, reg1, a regulatory gene for amylase production in Streptomyces lividans, has been reported to be involved in the regulation of chitinase production as well (21). However, the mechanism of glucose repression of chitinase gene expression in Streptomyces is still not understood.
Mutants of Streptomyces coelicolor A3(2) that are resistant to 2-deoxyglucose (2-DOG) (8) are defective in glucose kinase activity (27) and relieved of glucose repression of several catabolite pathways, including sugar utilization (8), glycerol utilization (27), agarase production (10), and amylase production (31). These data indicate that glucose kinase plays a central role in glucose repression in S. coelicolor A3(2). The glkA gene of S. coelicolor A3(2), encoding glucose kinase (2), restores 2-DOG sensitivity and glucose repression of dagAp4 transcription (2) in 2-DOG-resistant mutants. However, Ingram and Westpheling (13) reported that glkA is not required for glucose repression of the chi63 promoter of Streptomyces plicatus (25) in a ccrA1-glkA double mutant of S. coelicolor A3(2).
We isolated mutants of S. lividans TK24 defective in glucose repression of chitinase production by using colloidal chitin agar plates containing glucose. The mutants made clear zones around their colonies, while their parental strain, TK24, did not. G015, one of the mutants defective in glucose repression of chitinase production, was resistant to 2-DOG and did not utilize glucose as a sole carbon source, suggesting that it contained the mutation in glkA. After studying this mutant, we report here that glkA is involved in glucose repression of chitinase production in S. lividans.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are shown in Table 1. Plasmid pEMX151, a derivative of the low-copy-number plasmid pXE4 (11), was used for transcriptional analysis of the chiA promoter from S. lividans. The BamHI-HindIII fragment of pXE4 located upstream of the xylE reporter gene was replaced with a fragment of about 1 kb containing the DNA sequence extending from ca. 500 bp upstream to ca. 500 bp downstream of the transcription initiation site of chiA (17). pXE4 was used to construct pGA01 and pGA02. A 1.2-kb fragment including the coding region and one of the two promoters of glkA of S. coelicolor A3(2) was generated by digesting pIJ2423 (2) with BglII, ligated to the BamHI site of pXE4, and introduced into Escherichia coli XL1-Blue by transformation. The orientations of glkA were determined from the digestion patterns with HindIII. The glkA gene was inserted in the same orientation as the xylE reporter gene in pGA01 and in the opposite orientation in pGA02. pGAH01 was constructed by ligating the 1.2-kb BglII fragment containing glkA into the BamHI site of the high-copy-number vector pIJ486 and introduced into S. lividans TK24 by transformation.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains or plasmid | Genotype and characteristics | Reference |
|---|---|---|
| S. lividans 66 | ||
| TK24 | SCP1− SCP2−a | 9 |
| G015 | SCP1− SCP2−glkA; a derivative of TK24 mutagenized by UV irradiation | This study |
| S. coelicolor A3(2) J1668 | hisA1 uraA1 strA1 pgl NF SCP2−glkΔ119; a spontaneous mutant of J1508 (10) | 6 |
| E. coli XL1-Blue | supE44 hsdR17 recA1 endA1 gyrA4 thi relA1 lac F′[proAB+ lacIqlacZ ΔM15::Tn10 (Tetr)] | 26 |
| Plasmids | ||
| pXE4 | Low-copy-number plasmid with xylE as a reporter gene | 11 |
| pEMX151 | pXE4 containing a 1.0-kb insert with the chiA promoter | 17 |
| pGA01 | pXE4 containing a 1.2-kb insert with glkA of S. coelicolor A3(2); the orientation of glkA is the same as that of xylE | This study |
| pGA02 | pXE4 containing a 1.2-kb insert with glkA of S. coelicolor A3(2); the orientation of glkA is opposite to that of xylE | This study |
| pIJ486 | High-copy-number plasmid | 9 |
| pGAH01 | pIJ486 containing a 1.2-kb insert with glkA of S. coelicolor A3(2) | This study |
| pIJ2423 | pIJ2925 (13a) containing a 1.2-kb insert with glkA of S. coelicolor A3(2) | 2 |
SCP2−, the strain can be introduced with pXE4.
Manipulation of DNA.
Protoplast preparation and transformation of S. lividans was performed according to the method of Hopwood et al. (9). Plasmids were prepared by the alkaline lysis method (9, 26). All procedures involving recombinant DNA were carried out as described by Sambrook et al. (26).
Isolation of S. lividans mutants.
Spores of S. lividans TK24 were irradiated with UV light (wavelength, 302 nm) to 0.4% survival and spread in the dark (9) on ISCG agar medium [0.15% (wt/vol) colloidal chitin, 5 mM MgSO4, 15 mM (NH4)2SO4, 25 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES; pH 7.2), 0.5 mM NaH2PO4, 0.5 mM K2HPO4, 0.1% (vol/vol) trace element solution (0.1 g each of ZnSO4-7H2O, FeSO4-7H2O, MnCl2-4H2O, and NaCl per liter), 1.0% (wt/vol) glucose, and 1.5% (wt/vol) agar]. Colloidal chitin was prepared by the method of Lingappa and Lockwood (16). After 4 weeks of incubation at 30°C in the dark, colonies with clear zones around them were picked up and streaked on SFM agar medium (30).
2-DOG sensitivity and glucose utilization.
To test resistance to 2-DOG, spores formed on SFM agar medium were spotted on NMMB agar medium (8) containing 10 mM arabinose with or without 100 mM 2-DOG. For G015 containing derivatives of pXE4, the agar media were supplemented with 50 μg of thiostrepton/ml. After incubation for 5 days at 30°C, resistance to 2-DOG was judged from the sizes of the colonies. The ability to utilize glucose was determined from growth on NMMB agar medium containing 1.0% (wt/vol) glucose or in SMM liquid medium without polyethylene glycol 1000 and casamino acids (28) but containing 1.0% (wt/vol) glucose as a sole carbon source. On the agar plates, the sizes of colonies of G015 and its derivatives were compared with those of TK24 and its derivatives, respectively. Agar medium supplemented with 50 μg of thiostrepton/ml was used for G015 containing derivatives of pXE4. Germinated spores (9) were inoculated and grown at 30°C with shaking at 200 rpm. One-milliliter portions of the culture fluid were sampled periodically, and the mycelia were harvested by centrifugation. Growth was monitored by the protein content of the mycelia, measured as follows. The pellet was washed with distilled water (MiliQ), resuspended in MiliQ, and sonicated on ice. One-half volume of 3 N NaOH was added to the sonicated lysate, and after incubation for 30 min at 100°C, the lysate was centrifuged at 10,000 rpm (M150; SAKUMA) (ca. 7,000 × g) for 10 min at room temperature. The protein concentration of the supernatant was measured by the method of Bradford (4) with bovine plasma gamma globulin as the protein standard, and the amount of protein per milliliter of culture was calculated.
Chitinase assay.
Twenty milliliters of Luria-Bertani (LB) liquid medium (26) was inoculated with germinated spores and incubated at 30°C for 48 h with shaking at 150 rpm. For strains containing pXE4 derivatives or pIJ486 derivatives, the medium was supplemented with 2 or 5 μg of thiostrepton/ml, respectively. The culture was divided into three aliquots. Mycelia in each aliquot were harvested by centrifugation and washed with 10 ml of YE medium (0.7 g of K2HSO4, 0.3 g of KH2SO4, 0.5 g of MgSO4, 0.01 g of FeSO4, 0.3 g of NH4NO3, and 1.0 g of yeast extract per liter). The mycelia were resuspended in an equal volume of YE medium, YE medium with 0.05% (wt/vol) colloidal chitin, or YE medium with 0.05% (wt/vol) colloidal chitin and 1.0% (wt/vol) glucose. The protein content of 1 ml of culture was measured as described above, and the inocula were adjusted to about 100 μg of protein per ml. The cultures were grown at 30°C with shaking. A 0.5-ml portion of the culture fluid was sampled periodically and stored at −80°C. The samples were thawed and centrifuged, and chitinase activity was measured in the culture supernatant as described previously (19), using the fluorogenic substrate 4-methylumbelliferyl-N,N′,-diacetyl chitobioside [4MU-(GlcNAc)2] or 4-methylumbelliferyl-N,N′,N"-triacetyl chitotrioside [4MU-(GlcNAc)3] (Sigma). One unit of chitinase activity was defined as described by Miyashita et al. (19). Chitinase activity was expressed in units per microgram of mycelial protein.
Catechol dioxygenase assay.
The transformants TK24(pEMX151) and G015(pEMX151) were cultured as described in “Chitinase assay” above with the exception that G015(pEMX151) was precultured in LB medium containing 2 μg of thiostrepton/ml for 72 h. The longer preculture period of G015(pEMX151) was due to the slower growth rate of G015(pEMX151) compared to that of TK24(pEMX151). The cell extracts were prepared, and catechol 2,3-dioxygenase activity was measured spectrophotometrically as described previously (12, 32). Catechol 2,3-dioxygenase activity was expressed as the rate of increase in optical density at 375 nm per minute per milligram of protein.
Glucose kinase assay.
Glucose kinase activity measurements were done as described by Angell et al. (2).
Southern hybridization.
Total DNA was prepared as described by Hopwood et al. (9). Two micrograms of the DNA was digested with BclI, electrophoresed in 0.8% (wt/vol) agarose gel, and transferred to a Hybond N+ membrane (Amersham). The 1.2-kb BglII fragment containing glkA from S. coelicolor A3(2), which was prepared from pIJ2423, was used as a probe. Hybridization was performed with digoxigenin (Boehringer Mannheim) by following the manufacturer’s instructions.
RESULTS
G015 is defective in glucose repression of chitinase production.
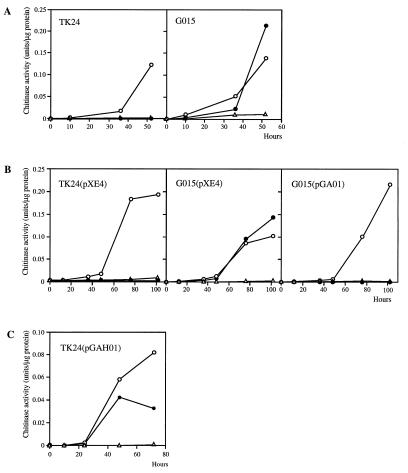
To isolate mutants of S. lividans TK24 defective in glucose repression of chitinase production, UV-irradiated spores were spread on ISCG agar medium. After 3 weeks of incubation at 30°C, several colonies among approximately 5,000 had clear zones around them and one, G015, was resistant to 2-DOG (see below). The phenotype of G015 on ISCG medium was confirmed after single-colony purification. Chitinase production by G015 was then compared with that of TK24 in liquid medium. Chitinase production by G015 was induced by the addition of 0.05% (wt/vol) colloidal chitin regardless of the presence of glucose, whereas TK24 did not produce chitinase in the presence of glucose (Fig. 1A). These results indicated that G015 was defective in glucose repression of chitinase production.
FIG. 1.
Chitinase activity induced by colloidal chitin in the presence or absence of glucose in S. lividans TK24 and G015 (A); in TK24(pXE4), G015(pXE4), and G015(pGA01) (B); and in TK24(pGAH01) (C). Chitinase activity was measured with 4MU-(GlcNAc)2 for panel A and 4MU-(GlcNAc)3 for panels B and C. At time zero, cultures grown in LB medium at 30°C for 48 h were divided into three aliquots, and the mycelia in each aliquot were suspended in YE medium (open triangles), YE medium plus 0.05% (wt/vol) colloidal chitin (open circles), or YE medium plus 0.05% (wt/vol) colloidal chitin plus 1.0% (wt/vol) glucose (solid circles).
Glucose repression of chiA transcription is not observed in G015.
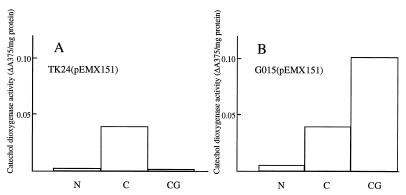
To investigate the regulation of chitinase gene expression in G015, the levels of transcription from the chiA promoter in TK24 and G015 were compared by using xylE, which encodes catechol 2,3-dioxygenase, as a reporter gene. As shown in Fig. 2, in G015(pEMX151) transcription from the chiA promoter occurred in the presence of colloidal chitin regardless of the presence of glucose, whereas glucose strongly repressed the transcription of chiA in TK24(pEMX151). The level of expression of the chiA promoter in G015(pEMX151) in the presence of colloidal chitin and glucose was 2 to 3 times greater than that observed in the presence of colloidal chitin alone.
FIG. 2.
Catechol dioxygenase activity of S. lividans TK24(pEMX151) (A) and G015(pEMX151) (B). Catechol dioxygenase activity was measured 45 h after subculturing (as described in the legend to Fig. 1) from LB medium supplemented with 2 μg of thiostrepton/ml. ΔA375 indicates the increase in optical density per minute at 375 nm. N, YE medium; C, YE medium plus 0.05% (wt/vol) colloidal chitin; CG, YE medium plus 0.05% (wt/vol) colloidal chitin plus 1.0% (wt/vol) glucose.
G015 was resistant to 2-DOG and did not utilize glucose.
G015 grew on NMMB agar medium containing 10 mM arabinose regardless of the presence of 100 mM 2-DOG, whereas TK24 did not grow in the presence of 2-DOG. On NMMB agar medium containing 1.0% (wt/vol) glucose as a sole carbon source, G015 grew poorly compared to TK24. In SMM liquid medium containing 1.0% (wt/vol) glucose as a sole carbon source, G015 did not grow at all. The resistance to 2-DOG and the defect in glucose utilization suggested that the glkA gene was defective in G015 as shown in S. coelicolor A3(2) (2).
glkA restored 2-DOG sensitivity, glucose utilization, and glucose repression in G015.
Angell et al. (2) showed that integration of a wild-type glkA gene into the chromosome of a 2-DOG-resistant mutant of S. coelicolor A3(2) restored 2-DOG sensitivity, glucose utilization, and glucose repression of dagAp4. To examine whether the glkA gene restored these three phenotypes in G015, the glkA gene of S. coelicolor A3(2) was introduced into G015. A low-copy-number plasmid, pGA01 (a derivative of pXE4), that contained glkA was constructed. G015 and TK24 were transformed with pGA01 or pXE4, and G015(pXE4), G015(pGA01), and TK24(pXE4) were obtained. The glucose kinase activity of G015(pXE4) was 1/10 that of TK24(pXE4), while G015(pGA01) had approximately the same level as TK24(pXE4) (Table 2). In contrast to G015(pXE4), G015(pGA01) grew well on NMMB agar medium containing 1.0% (wt/vol) glucose and showed no growth in the presence of 100 mM 2-DOG, like TK24(pXE4) (Table 2). These data indicate that G015 is a glucose kinase mutant of S. lividans. Moreover, G015(pGA01) did not produce chitinase when glucose was supplied together with colloidal chitin (Fig. 1B). When pGA02, with glkA in the opposite orientation, was introduced into G015, the same results were obtained (data not shown). Thus, glkA restores glucose repression of chitinase production in G015, and it appears that glkA is involved in glucose repression of chitinase production in S. lividans TK24.
TABLE 2.
Glucose kinase activity, glucose utilization, and resistance to 2-DOG of transformants of S. lividans TK24 and G015
| Strain | Glucose kinase activity (U) | Glucose utilizationa | Resistance to 2-DOGb |
|---|---|---|---|
| TK24(pXE4) | 23 | + | S |
| G015(pXE4) | 2 | − | R |
| G015(pGA01) | 19 | + | S |
Glucose utilization was estimated by growth on NMMB agar medium containing 1.0% (wt/vol) glucose and 50 μg of thiostrepton/ml. +, good growth; −, significantly poorer growth compared to that of TK24(pXE4).
Resistance to 2-DOG was judged from growth on NMMB agar medium containing 10 mM arabinose, 100 mM 2-DOG, and 50 μg of thiostrepton/ml. S, sensitive to 2-DOG; R, resistant to 2-DOG.
Introduction of glkA on a high-copy-number plasmid relieves glucose repression of chitinase production in S. lividans.
Introduction of glkA on a high-copy-number plasmid relieves glucose repression in S. coelicolor A3(2) (1, 14). The same phenomenon might be expected in S. lividans if glkA is involved in glucose repression of chitinase production in that species. To assess this, the high-copy-number plasmid pGAH01, a derivative of pIJ486 with a 1.2-kb BglII fragment containing glkA of S. coelicolor A3(2), was constructed and introduced into S. lividans TK24 by protoplast transformation. Chitinase production in the presence of glucose was measured in liquid culture. Chitinase production in TK24(pGAH01) was relieved of glucose repression (Fig. 1C). This result strongly supports the involvement of glkA in glucose repression of chitinase production.
DISCUSSION
G015, a mutant of S. lividans TK24 that was defective in glucose repression of chitinase production was isolated. G015 showed 2-DOG resistance, no growth in medium that contained glucose as a sole carbon source, and low glucose kinase activity. The glkA gene of S. coelicolor A3(2) restored 2-DOG sensitivity, glucose utilization, and glucose kinase activity to approximately wild-type levels in G015 (Table 2). These data indicate that G015 possesses a mutated glucose kinase gene. The glk locus of S. coelicolor A3(2) is relatively unstable, with a high spontaneous mutation frequency. Spontaneous 2-DOG-resistant mutants show deletion in the glk locus (6). However, Southern analysis revealed that glkA was not deleted in G015, and the signal obtained with G015 was the same as those in S. lividans TK24 and S. coelicolor A3(2) (data not shown). Thus, the mutation of glkA in G015 is likely to be a UV-induced point mutation.
The fact that a glk mutant of S. lividans is relieved of glucose repression of chitinase production suggests the involvement of glkA in glucose repression in S. lividans. Consistent with this, the glkA gene of S. coelicolor A3(2) restored glucose repression of chitinase production in G015 when cloned on a low-copy-number plasmid (Fig. 1B). Moreover, introduction of glkA on a high-copy-number plasmid resulted in the relief of glucose repression of chitinase production in S. lividans TK24 (Fig. 1C). This is also consistent with the relief of glucose repression of dagAp4 transcription when glkA is overexpressed in S. coelicolor A3(2) M145 (1).
The expression level of the chiA promoter in G015 (pEMX151) in the presence of colloidal chitin and glucose was about twice that observed on colloidal chitin alone (Fig. 2B). A similar stimulatory effect from loss of glkA was observed for dagAp4 transcription in S. coelicolor (1). Hodgson (8) showed that most of the glk mutants of S. coelicolor A3(2) which do not utilize glucose as a sole carbon source can transport glucose. Considering that G015 is a glk mutant which cannot utilize glucose, although it is a derivative of S. lividans TK24, it is conceivable that the process of glucose transport or glucose itself stimulates the expression of chiA in G015.
Ingram and Westpheling (13) reported that glkA of S. coelicolor A3(2) is not required for glucose repression of the chi63 promoter of S. plicatus (25), based on a ccrA1-glkA double mutant of S. coelicolor A3(2). This is in contrast to the conclusion obtained in this study of S. lividans. To confirm the involvement of the glkA gene in glucose repression of chitinase production in S. coelicolor, we examined the native chitinase production of a glk mutant of S. coelicolor A3(2) in liquid medium that contained colloidal chitin with or without glucose. Chitinase production in J1668, a glkA deletion mutant of J1508 (10), was repressed in the presence of glucose in liquid media (data not shown), consistent with the finding of Ingram and Westpheling (13). Thus, it seems that the mechanism of glucose repression of chitinase production in S. coelicolor A3(2) is different from that in S. lividans, which is surprising given that they are very closely related species (15) and possess nearly identical chitinase genes (unpublished data).
ACKNOWLEDGMENTS
We are grateful to M. J. Bibb (John Innes Centre, Norwich, United Kingdom) for valuable discussions and for providing J1668 and pIJ2423.
This work was supported in part by a Grant-in-Aid (Bio Design Program 6201) from the Ministry of Agriculture, Forestry, and Fisheries of Japan.
REFERENCES
- 1.Angell S, Lewis C G, Buttner M J, Bibb M J. Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol Gen Genet. 1994;244:135–143. doi: 10.1007/BF00283514. [DOI] [PubMed] [Google Scholar]
- 2.Angell S, Schwarz E, Bibb M J. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol Microbiol. 1992;6:2833–2844. doi: 10.1111/j.1365-2958.1992.tb01463.x. [DOI] [PubMed] [Google Scholar]
- 3.Blaak H, Schnellmann J, Walter S, Henrissat B, Schrempf H. Characteristics of an exochitinase from Streptomyces olivaceoviridis, its corresponding gene, putative protein domains and relationship to other chitinases. Eur J Biochem. 1993;214:659–669. doi: 10.1111/j.1432-1033.1993.tb17966.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Delic I, Robbins P, Westpheling J. Direct repeat sequences are implicated in the regulation of two Streptomyces chitinase promoters that are subject to carbon catabolite control. Proc Natl Acad Sci USA. 1992;89:1885–1889. doi: 10.1073/pnas.89.5.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher S H, Bruton C J, Chater K F. The glucose kinase gene of Streptomyces coelicolor and its use in selecting spontaneous deletions for desired regions of the genome. Mol Gen Genet. 1987;206:35–44. doi: 10.1007/BF00326533. [DOI] [PubMed] [Google Scholar]
- 7.Fujii T, Miyashita K. Multiple domain structure in a chitinase gene (chiC) of Streptomyces lividans. J Gen Microbiol. 1993;139:677–686. doi: 10.1099/00221287-139-4-677. [DOI] [PubMed] [Google Scholar]
- 8.Hodgson D A. Glucose repression of carbon source uptake and metabolism in Streptomyces coelicolor A3(2) and its perturbation in mutants resistant to 2-deoxyglucose. J Gen Microbiol. 1982;128:2417–2430. [Google Scholar]
- 9.Hopwood D A, Bibb M J, Charter K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic Manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 10.Ikeda H, Seno E T, Bruton C J, Chater K F. Genetic mapping, cloning, and physiological aspects of the glucose kinase gene of Streptomyces coelicolor. Mol Gen Genet. 1984;196:501–507. doi: 10.1007/BF00436199. [DOI] [PubMed] [Google Scholar]
- 11.Ingram C, Brawner M, Youngman P, Westpheling J. xylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. J Bacteriol. 1989;171:6617–6624. doi: 10.1128/jb.171.12.6617-6624.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingram C, Delic I, Westpheling J. ccrA1: a mutation in Streptomyces coelicolor that affects the control of catabolite repression. J Bacteriol. 1995;177:3579–3586. doi: 10.1128/jb.177.12.3579-3586.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingram C, Westpheling J. The glucose kinase gene of Streptomyces coelicolor is not required for glucose repression of the chi63 promoter. J Bacteriol. 1995;177:3587–3588. doi: 10.1128/jb.177.12.3587-3588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Janssen G R, Bibb M J. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli clones. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 14.Kwakman J H J M, Postma P W. Glucose kinase has a regulatory role in carbon catabolite repression in Streptomyces coelicolor. J Bacteriol. 1994;176:2694–2698. doi: 10.1128/jb.176.9.2694-2698.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leblond P, Redenbach M, Cullum J. Physical map of the Streptomyces lividans 66 genome and comparison with that of the related strain Streptomyces coelicolor A3(2) J Bacteriol. 1993;175:3422–3429. doi: 10.1128/jb.175.11.3422-3429.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lingappa Y, Lockwood J L. Chitin media for selective isolation and culture of Actinomycetes. Phytopathology. 1962;52:317–323. [Google Scholar]
- 17.Miyashita, K., T. Fujii, and T. Kajiwara. Unpublished data.
- 18.Miyashita K, Fujii T. Nucleotide sequence and analysis of a gene (chiA) for chitinase from Streptomyces lividans 66. Biosci Biotechnol Biochem. 1993;57:1691–1698. doi: 10.1271/bbb.57.1691. [DOI] [PubMed] [Google Scholar]
- 19.Miyashita K, Fujii T, Sawada Y. Molecular cloning and characterization of chitinase genes from Streptomyces lividans 66. J Gen Microbiol. 1991;137:2065–2072. [Google Scholar]
- 20.Miyashita K, Fujii T, Watanabe W, Ueno H. Nucleotide sequence and expression of a gene (chiB) for a chitinase from Streptomyces lividans. J Ferment Bioeng. 1997;83:26–31. [Google Scholar]
- 21.Nguyen J, Francou F, Virolle M-J, Guérineau M. Amylase and chitinase genes in Streptomyces lividans are regulated by reg1, a pleiotropic regulatory gene. J Bacteriol. 1997;179:6383–6390. doi: 10.1128/jb.179.20.6383-6390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni X, Westpheling J. Direct repeat sequences in the Streptomyces chitinase-63 promoter direct both glucose repression and chitin induction. Proc Natl Acad Sci USA. 1997;94:13116–13121. doi: 10.1073/pnas.94.24.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohno T, Armand S, Hata T, Nikaidou N, Henrissat B, Mitsutomi M, Watanabe T. A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J Bacteriol. 1996;178:5065–5070. doi: 10.1128/jb.178.17.5065-5070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robbins P W, Albright C, Benfield B. Cloning and expression of a Streptomyces plicatus chitinase (chitinase-63) in Escherichia coli. J Biol Chem. 1988;263:443–447. [PubMed] [Google Scholar]
- 25.Robbins P W, Oberbye K, Albright C, Benfield B, Pero J. Cloning and high-level expression of chitinase-encoding gene of Streptomyces plicatus. Gene. 1992;111:69–76. doi: 10.1016/0378-1119(92)90604-n. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Seno E T, Chater K F. Glycerol catabolic enzymes and their regulation in wild-type and mutant strains of Streptomyces coelicolor A3(2) J Gen Microbiol. 1982;129:1403–1413. doi: 10.1099/00221287-129-5-1403. [DOI] [PubMed] [Google Scholar]
- 28.Strauch E, Takano E, Baylis H A, Bibb M J. The stringent response in Streptomyces coelicolor A3(2) Mol Microbiol. 1991;5:289–298. doi: 10.1111/j.1365-2958.1991.tb02109.x. [DOI] [PubMed] [Google Scholar]
- 29.Tsujibo H, Endo H, Minoura K, Miyamoto K, Inamori Y. Cloning and sequence analysis of the gene encoding a thermostable chitinase from Streptomyces thermoviolaceus OPC-520. Gene. 1993;134:113–117. doi: 10.1016/0378-1119(93)90183-4. [DOI] [PubMed] [Google Scholar]
- 30.van Wezel G P, White J, Young R, Postma P W, Bibb M J. Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor A3(2) is controlled by malR, a member of the lacI-galR family of regulatory genes. Mol Microbiol. 1997;23:537–549. doi: 10.1046/j.1365-2958.1997.d01-1878.x. [DOI] [PubMed] [Google Scholar]
- 31.Virolle M-J, Bibb M J. Cloning, characterization and regulation of an α-amylase gene from Streptomyces limosus. Mol Microbiol. 1988;2:197–208. doi: 10.1111/j.1365-2958.1988.tb00021.x. [DOI] [PubMed] [Google Scholar]
- 32.Zukowski M M, Gaffney D F, Speck D, Kauffmann M, Findeli A, Wisecup A, Lecocq J P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci USA. 1983;80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]