Abstract
Metastatic dissemination of solid tumors, a leading cause of cancer-related mortality, underscores the urgent need for enhanced insights into the molecular and cellular mechanisms underlying metastasis, chemoresistance, and the mechanistic backgrounds of individuals whose cancers are prone to migration. The most prevalent signaling cascade governed by multi-kinase inhibitors is the mitogen-activated protein kinase (MAPK) pathway, encompassing the RAS–RAF–MAPK kinase (MEK)–extracellular signal-related kinase (ERK) pathway. RAF kinase is a primary mediator of the MAPK pathway, responsible for the sequential activation of downstream targets, such as MEK and the transcription factor ERK, which control numerous cellular and physiological processes, including organism development, cell cycle control, cell proliferation and differentiation, cell survival, and death. Defects in this signaling cascade are associated with diseases such as cancer. RAF inhibitors (RAFi) combined with MEK blockers represent an FDA-approved therapeutic strategy for numerous RAF-mutant cancers, including melanoma, non-small cell lung carcinoma, and thyroid cancer. However, the development of therapy resistance by cancer cells remains an important barrier. Autophagy, an intracellular lysosome-dependent catabolic recycling process, plays a critical role in the development of RAFi resistance in cancer. Thus, targeting RAF and autophagy could be novel treatment strategies for RAF-mutant cancers. In this review, we delve deeper into the mechanistic insights surrounding RAF kinase signaling in tumorigenesis and RAFi-resistance. Furthermore, we explore and discuss the ongoing development of next-generation RAF inhibitors with enhanced therapeutic profiles. Additionally, this review sheds light on the functional interplay between RAF-targeted therapies and autophagy in cancer.
Subject terms: Drug development, Metastasis
Introduction
The mitogen-activated protein kinase (MAPK) pathway transmits extracellular signals from the membrane to intracellular destinations and is involved in various biological functions.1 The MAPK pathway is dysregulated in many RAS-associated cancers. RAS mutations result in the constitutive activation of the MAPK pathway, leading to uncontrolled cell proliferation and resistance to apoptosis-inducing drugs.2,3 Although many RAS inhibitors have been isolated and studied, the development of drugs targeting RAS is limited by a lack of well-defined druggable nooks and cavities on the RAS surface.4 However, interrupting signals between RAS and downstream effectors, such as the RAF–MAPK kinase (MEK)–extracellular signal-related kinase (ERK) pathway, could represent a new therapeutic strategy for RAS-driven cancers.5–7
The RAF protein family consists of three serine (Ser)/threonine (Thr) kinases (ARAF, BRAF, and CRAF) that act as mediators between membrane-bound RAS-GTPases and downstream kinases, such as MEK and ERK, in the MAPK signaling pathway.8 RAF proteins coordinate various cellular responses by regulating cytoplasmic and nuclear activities, such as cell cycle progression, proliferation, metabolism, migration, differentiation, and apoptosis.9,10 RAF is highly conserved in mammals, and RAF mutations are associated with many human cancers, including melanoma, breast cancer, ovarian cancer, colon cancer, thyroid cancer, and prostate cancer.11 Mutations in BRAF and RAS that dysregulate MAPK signaling are strongly associated with human malignancies.12 All members of the RAF family interact with RAS; however, this contact alone is insufficient to activate RAF. For example, several RAS mutants, such as RASV12Y32F and RASV12T35S are insufficient to activate RAF in vitro, suggesting that RAF kinase activation requires other factors.13 A recent study indicates that the activation of RAF necessitates dimerization, and exploring RAF activation is currently being viewed as a potential target for therapeutic intervention in several clinical contexts, including diverse cancer types.10
Numerous RAF inhibitors are considered potential therapeutic agents, eliciting high levels of responses in various RAF-mutant carcinomas.14,15 However, single-agent therapies targeting RAF have not resulted in significant long-term survival benefits due to the frequent development of drug resistance, often associated with mutational changes in MAPK components that result in the reactivation of the MAPK pathway.16,17 Combination therapeutic strategies using both RAF and MEK inhibitors may represent a more effective treatment strategy in patients with advanced or metastatic RAF-mutant carcinomas.18–21 Although this approach has demonstrated potential efficacy in preclinical studies, clinical testing has not demonstrated durable responses, and a single-arm study demonstrated that this strategy is associated with a predictable pattern of adverse effects due to the substantial inhibition of multiple paralogs.22,23 Identifying key downstream signals in the MAPK pathway is essential for minimizing paralog redundancy and cascade interactions, which may limit both the cancerous activity of RAF and drug toxicity in normal cells.
Autophagy, an intracellular catabolic process, may assist cancer cells in evading from RAFi, as many RAFi-resistant cells exhibit enhanced autophagic activity.24–26 Both preclinical and clinical data suggest that inhibiting both autophagy and MAPK pathway activity may serve as a novel and effective treatment strategy for BRAF and KRAS-mutant cancers.24,25,27 In particular, Chih-Shia Lee’s group showed that targeting both RAF and autophagy genes results in the best therapeutic outcomes. The inhibition of BRAF or CRAF, together with ATG7 inhibition, was found to be a viable treatment strategy for RAS-driven tumors.27 Understanding the mechanisms underlying RAFi-induced autophagy in the setting of recurrent somatic genetic alterations and RAF mutations could offer a “precision medicine” paradigm for diagnosing and treating tumors, including RAF-mutant tumors. This review focuses on potentially unique therapeutic approaches that target the basic components of RAF signaling and autophagy in RAS-dependent and RAS-independent cancers.
The discovery and major developments of RAS/RAF/MAPK in health and disease
The RAF/RAF/MEK/ERK signaling cascade is a well-established MAPK pathway in cell biology that governs several crucial cellular processes such as development, differentiation, proliferation, and death.28 With this cascade, various isoforms of RAS, RAF, MEK, and ERK exhibit differences in efficacy, function, and, notably, carcinogenic potential.
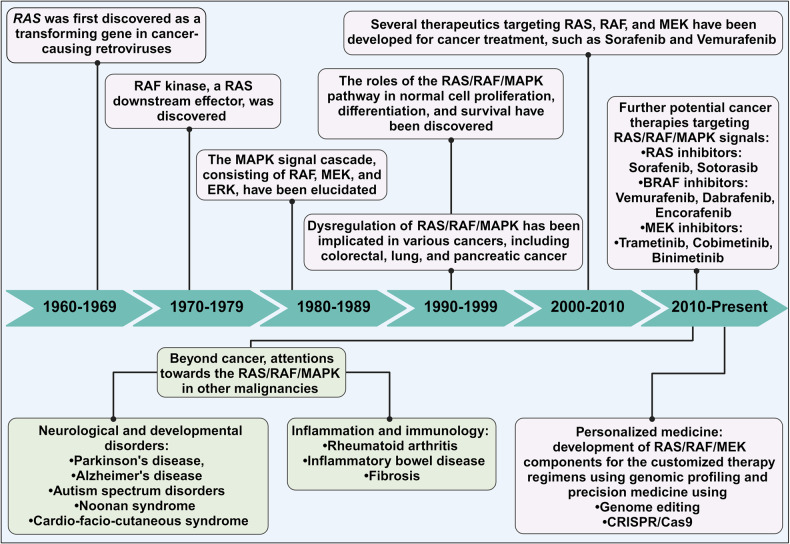
Our understanding of oncogenic potential began with the discovery of the highly carcinogenic Harvey murine sarcoma virus29 in 1964 and the Kirsten murine sarcoma virus30 in 1967. In the late 1970s and early 1980s, groundbreaking studies by Scolnick and colleagues identified the cellular origins of viral H-RAS and K-RAS genes31, and an avian homolog MH2 retrovirus32 in 1984, and an avian homolog MH2 retrovirus32 in 1984, respectively. Later, these two oncogenes are known as the first rapidly accelerated fibrosarcoma (RAF) gene with serine/threonine kinase activity.33 Subsequently, Raf-1 gene product (named as CRAF), a cellular counterpart of v-Raf, and other cellular counterparts such as C-Raf-1 and C-Raf-2 genes were cloned and sequenced in 1985.34 ARAF and BRAF, two additional members of the RAF family, were reported in 1986 and 1988, respectively.35 In 1988, MAPK, initially named microtubule-associated protein-2 protein kinase (MAP-2 kinase), was identified in mammalian cells36, and subsequently in yeast cells.37 MEK (mitogen-activated protein kinase kinase) and ERK (mitogen-activated protein kinase), both cytoplasmic protein kinases activated by mitogens, were discovered in the 1990s38. Further, RAF protein was functionally identified as a direct MEK activator39 in 1992 and a RAS effector in 1993.40 These findings marked the beginning of the MAPK cascade era, where it become evident that MAPK kinase signaling cascades play a pivotal role in initiating proliferative and oncogenic activities.41 In 2005, the U.S. Food and Drug Administration (FDA) approved Nexavar (Sorafenib), an oral multi-kinase inhibitor targeting the MAPK pathway, for the treatment of hepatocellular carcinoma (HCC), renal cell carcinoma (RCC), and thyroid carcinoma (TC).42 Subsequently, Sorafenib was proposed as a MAPK pathway inhibitor for malignant peripheral nerve sheath tumors (MPNSTs).43 Vemurafenib (Zelboraf), a potent BRAFV600E mutant inhibitor, was synthesized in early 2005 and received FDA approval for the treatment of metastatic and late-stage melanoma in 2011.44 Following this, FDA approval were granted for two BRAF inhibitors, Dabrafenib (Tafinlar) in 2013 and Encorafenib (Braftovi) in 2018. Trametinib (Mekinist) was approved in 2013 as a single-agent oral treatment for unresectable or metastatic melanoma in adult patients with BRAFV600E or BRAFV600K mutations.45 From 2014 to 2023, Trametinib, in combination with Dabrafenib, received FDA approval for the treatment of various solid tumors, including melanoma, non-small cell lung cancer (NSCLC), anaplastic thyroid cancer (ATC), and, low-grade glioma (LGG).46 In addition, two MEK inhibitors, Cotellic (Cobimetinib) and Mektovi (Binimetinib) were approved in 2015 and 2018, respectively, for the treatment of melanoma, either as a single-agent or in combination with other MAPK inhibitors.47 Ongoing preclinical and clinical investigations underscore the potential of the RAS/RAF/MAPK pathway as a significant therapeutic target, particularly in the era of precision medicine, with a focus on combination treatments.48 For example, a CRISPR/cas9 gene deletion study in lung cancer cells revealed that the deletion of KEAP1, in the presence of specific RAS/RAF/MAPK pathway inhibitors, alters cell metabolism and enables cells to proliferate without MAPK signaling.49 These major milestones are depicted in Fig. 1.
Fig. 1.
Historical events of the discovery and development of the RAS/RAF/MAPK pathway in health and diseases. The journey of the MAPK signal cascade commenced in 1960s with the groundbreaking discovery of the viral RAS gene. Subsequently, in 1992, the identification of RAF as both an upstream kinase activator of MEK and a RAS effector marked significant milestones. These pivotal findings culminated in the comprehensive definition of the entire MAPK signaling pathway. Over time, the MAPK signal emerged as a critical components in the development of therapeutic strategies for combating cancer. Each of these major milestones in the RAS/RAF/MAPK discovery is represented within its respective box. This figure was created with BioRender.com
Beyond cancer, ongoing studies aim to develop new treatments targeting RAS/RAF/MAPK for various disorders, including neurological, developmental, and metabolic diseases. Neurological disorders such as autism spectrum disorder (ASD), Parkinson’s disease (PD), Alzheimer’s disease (AD), Cardio-Facio-cutaneous (CFC), and Noonan syndromes (NS) have been linked to abnormalities in the regulation of the MAPK signaling pathway.50 Moreover, the RAS/RAF/MAPK pathway is gaining attention as a potential target for the development of novel anti-inflammatory drugs, with implications for conditions like rheumatic arthritis (RA)51, inflammatory bowel disease (IBD)52 and pulmonary fibrosis (PF).53
MAPK signaling
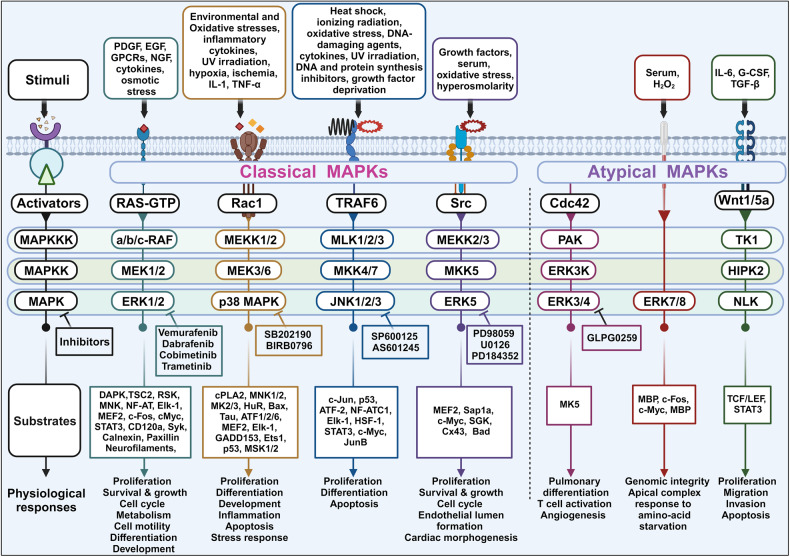
MAPKs are Ser/Thr kinases that play various roles in cellular responses to stimuli, including mitogens, osmotic stress, heat shock, and proinflammatory cytokines. MAPKs are involved in many cellular processes, such as proliferation, gene expression, differentiation, mitosis, cell survival, and apoptosis.54 Mammals possess four primary MAPKs: (1) ERK1/2, (2) c-Jun N-terminal kinase (JNK)1–3, (3) p38, and (4) ERK5.55–57 In addition to these four primary MAPKs, numerous atypical MAPKs (e.g., ERK 3/4, ERK 7/8, and Nemo-like kinase [NLK]) have been identified with less well-defined roles and unique mechanisms of activation.58–60 MAPK cascades consist of a signaling relay that is partially regulated by phosphorylation and typically involves three consecutive protein kinases: MEK kinase (MAPKKK), MEK, and MAPK (Fig. 2). MAPK cascades are activated by cell-surface receptors via cytoplasmic signaling proteins, and these signaling pathways are often dysregulated in human cancers. ERK1 and ERK2 are frequently investigated by researchers worldwide due to their critical roles in cell proliferation and survival. The JNK and p38 MAPK pathways primarily play roles in responding to cellular stress and regulating apoptosis. In contrast, the most extensively studied RAS/RAF/MAPK pathway holds a central position in governing cell proliferation and differentiation, serving as a vital component of the cellular signal transduction network. Consequently, proteins involved in the RAS/RAF/MAPK cascade have frequently been targeted in cancer drug discovery, leading to the clinical development of protein kinase inhibitors.61
Fig. 2.
Mitogen-activated protein kinase (MAPK) cascades and their physiological functions. All cascades consist of three-layered core-signaling pathways in which each kinase is consecutively activated, and MAPK components are highly conserved. The first layer consists of MAPK kinase kinases (MAPKKKs or MEKKs), which are activated by stimuli and phosphorylate and activate MAPK kinases (MAPKKs or MEKs). MAPKKs are dual-specificity kinases that can phosphorylate threonine or tyrosine residues to activate the terminal serine/threonine MAPK, leading to the activation of multiple cytoplasmic and nuclear proteins involved in various biological functions. This figure was created with BioRender.com
RAS/RAF/MAPK signaling: structure, upstream activator and downstream effectors
The ERK (MAPK) kinase plays a pivotal role within the RAS/RAF/MAPK signal transduction pathway, exerting control over various facets of cellular metabolism in cancer cells.62,63 Consequently, when it comes to development of anticancer drugs, the focus largely centers on three key upstream regulators and ERK protein in the ERK pathway: RAS (upstream activator of RAF), RAF (direct effector of RAS and activator of MEK), MEK (functioning as MAPKK), and ERK (as MAPK).
RAS/RAF/MAPK can be activate via two pathways: (1) a ligand-dependent pathway, in which ligands, such as growth factors, hormones, or cytokines, physically engage with receptors; and (2) a ligand-independent pathway, in which signaling is induced by a physical stressor, such as radiation, injury, or osmotic pressure.59 Aberrant RAF activation or mutations in upstream activators such as RAS or receptor tyrosine kinases (RTKs) can contribute to the development of malignancies in humans.64 Beyond RAS mutations, disruptions of RAS upstream components can also impact RAF activation. Receptors engaged by various growth factors, including TGF-α, EGF, VEGF, and platelet-derived growth factor-beta (PDGF-β), can instigate the canonical RAS–RAF–MEK–ERK pathway during various biological processes (Fig. 3a).65–67 Further investigations into RAF activation shed light on critical molecular mechanisms underlying cell proliferation, survival, and metastasis in cancer, particularly through the influence of the EGF receptor (EGFR) and small RAS-GTPases.68 Consequently, extensive research is currently underway to target RAF kinase as a promising avenue for the development of anticancer drugs.68,69
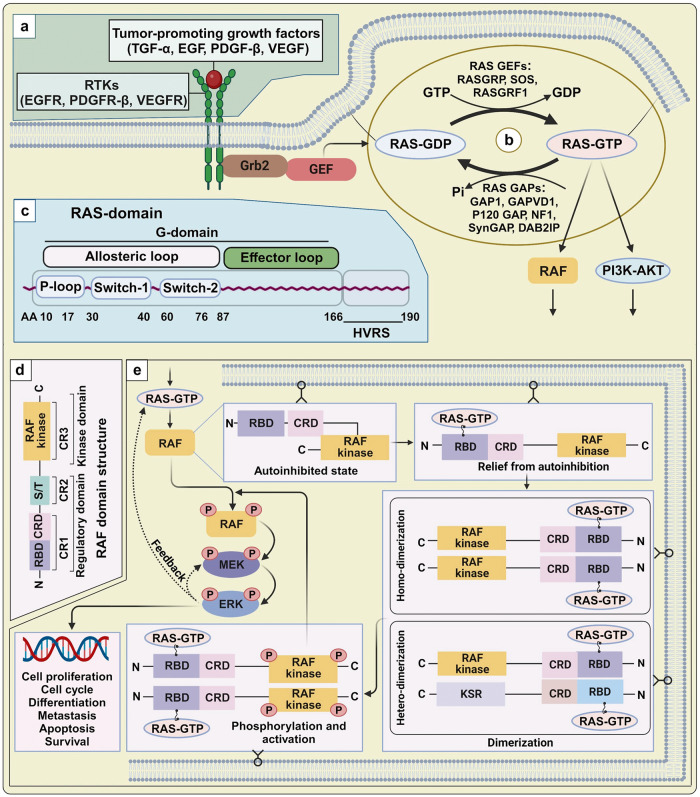
Fig. 3.
Structure and activation mechanism of RAS and RAF kinase in the RAS/RAF/MAPK signal cascade. a RAS upstream components. Various mitogens including TGF-α, EGF, VEGF, and PDGF-β bind to their own receptors and lead to RAS activation and subsequent stimulation of the MAPK pathway. b GTPase cycle. GEFs stimulates the transition of inactive RAS-GDP to active RAS-GTP, enabling to transmit the proliferation and differentiation signals through its downstream effectors. Subsequently, the active RAS can be quickly deactivated by the action of GAPs. c RAS domain. The effector lobe (1–86 a.a.), allosteric lobe (87–165 a.a.), and HVR (167–188/189 a.a.) are all parts of the structure of RAS proteins. The effector lobe contains switches I (30–40 a.a.) and II (60–76 a.a.) are involved in effector binding and GEF or GAP binding, respectively. d RAF domain structure. RAF proteins consist of three conserved regions (CR1, CR2, and CR3) or two functional domains: an N-terminal regulatory domain and a C-terminal catalytic domain. e RAF dimerization. In the absence of cellular stimulation, RAF tends to exist in the monomeric, autoinhibited state. Upon stimulation by RAS-GTP, the autoinhibitory domain is released, freeing the inactive kinase domain to form homo- or heterodimers (with kinase suppressor of RAS [KSR]). Dimerization triggers mutual phosphorylation of the dimer components, fully activating the kinase. Phosphorylation and activation of target proteins, such as MEK1 and MEK2, propagates the MAPK cascade, leading to ERK1/ERK2 activation. This figure was created with BioRender.com
RAS
RAS, a pivotal upstream protein in the RAF/MAPK pathway, holds the distinction of being the founding member of the extensive RAS superfamily of small GTPases.70 RAS is extremely prevalent, as RAS mutations are detected in approximately 30% of all tumors. RAS activity varies across different cancer types. For example, NRAS is activated in lymphoid and myeloid malignancies, whereas KRAS is highly elevated in colon and pancreatic cancers, and HRAS activity is upregulated in bladder and kidney cancers. This upregulation of RAS activity within the context of cancer leads to the dysregulation of downstream protein kinase activities.
RAS structures and activations mechanism
RAS activation is triggered by various extracellular stimuli, with the primary mechanism involving the formation of complexes comprising autophosphorylated growth factor receptors, the adapter protein GRB2 and the exchange factor SOS.71 It has been proposed that RAS dimerization plays a critical role in facilitating RAS signal transmission, directing influencing RAF activation.72
In their normal and resting states, RAS proteins exist in an inactive GDP-bound form.73 Guanine nucleotide exchange factors (GEFs), like SOS, are recruited to the plasma membrane following the stimulation of mitogenic growth factors (Fig. 3b). Once GEFs bind to RAS, the stability of nucleotide binding disrupted, leading to the release of GDP from RAS and the transient formation of a nucleotide-free state. This, in turn, activates RAF and other downstream targets recruited by RAS-GTP. The signaling from RAS is terminated by the hydrolysis of GTP, which is mediated by the intrinsic enzymatic activity of RAS. Mammalian cells typically express three GEFs that are recognized as RAS activators: SOS, RASGRF, and RASGRP.74 Some RAS-related malignancies have been associated with GAPs, including NF1, p120GAP/RASA1, SynGAP/RASA5, GAP1 family, DAB2IP, and GAPVD174. The conformational changes that accompany GTP hydrolysis are critical for RAS to function as a molecular switch in signaling pathways.75 There are four isomers of RAS genes, including KRAS4A, KRAS4B, NRAS, and HRAS. These isomers exhibit relatively consistent sequences or structures, encompassing N-terminal G-domains (1–166 a.a.) and C-terminal hypervariable regions (HVR) (166-189 a.a.).76 The G-domain conveys signals to downstream RAS effectors, featuring switch 1 (30–40 a.a.), switch 2 (60–76 a.a.), and a P loop (10–17a.a.) (Fig. 3c).77,78 The C-terminal region (final four amino acids, CAAX) undergoes posttranslational modification like iso-prenylation, proteolysis, and methylation, facilitating RAS localization and attachment to the membrane.77
RAF
RAF proteins, part of serine/threonine kinase family encoded by the RAF gene, serve as the upstream activator of MAPK and direct effector of RAS. In mammalian cells, there exist three distinct RAF proteins; ARAF, BRAF, and CRAF (also known as RAF-1). The initial member of the RAF family, CRAF, was initially characterized as an oncogene. Fusion of the CRAF catalytic domain with the retroviral Gag protein results in constitutive RAF kinase activation.79 Subsequently, two additional RAF family proteins, ARAF and BRAF, were discovered, each demonstrating similar function to that of CRAF.32,80–82 CRAF forms interactions with MEK, a dual-specificity kinase responsible for activating ERK.
RAF structure and activation mechanisms
The MAPK pathway is tightly regulated by several activation steps. The physical interaction between the RAF regulatory domain and membrane-bound RAS results in the attachment of RAF to the membrane, dephosphorylation, a conformation change for the kinase domain, and the subsequent phosphorylation of active sites (Ser338 and Tyr341).83 Many modulators mediate the negative or positive regulation of RAF activity through the formation of signaling complexes, which play critical roles in cancer growth and progression.83,84 To date, approximately 30 RAF-interacting proteins have been identified as putative RAF regulators.83
CRAF activation is a complex process in which Ser338, Tyr340, and Tyr341 are phosphorylated in response to oncogenes and growth factors. Many up- and downstream RAF effectors are associated with cancer transformation. Although the exact mechanisms underlying CRAF regulation remain unclear, phosphorylation of Ser338 and Tyr341 have been identified as crucial regulators of RAF kinase activity.85 In humans, all three RAF proteins are activated by phosphorylation at shared, conserved residues. Direct interaction between RAS and the N-terminal regulatory domain of CRAF is essential for RAF activation, and RAS mutations that cause constitutive RAF activation are detected in more than 30% of all human malignancies.86–88 However, RAS interaction alone is not sufficient to activate CRAF in vitro, indicating that other biochemical activities are necessary for RAF activation (Table 1). Some RAS mutants, including V12Y32F and V12T35S, are incapable of RAF activation but are able to interact with members of the RHO GTPase family.13 Previous studies have shown that p21-activated kinase (PAK) family members serve as molecular linkers, connecting RAS with RHO GTPases, including RAC and CDC42.89 In addition, CK2, JNK2, or SRC may be involved in CRAF activation through either RAS-dependent or RAS-independent mechanisms.90–92
Table 1.
RAF-interacting activator proteins and regulatory mechanisms
| RAF regulators | Regulatory mechanisms | Ref. | |
|---|---|---|---|
| RAS | Direct activation | RAS plays an essential role in the activation of CRAF kinase, which is directly responsible for the activation of the MEK–ERK pathway. | 13 |
| An undamaged CRAF zinc finger is necessary to bind to RAS and activate RAF in situ. | 85 | ||
| Indirect activation | Dominant-negative Rac, Rho, and Cdc42 mutants prevent RAS-dependent transformation, whereas activated mutants work with CRAF to transform cells. | 573–578 | |
| Type I PAKs (PAK1/2/3) | PAK1 | PAK1 acts as a physiological candidate for CRAF phosphorylation on Ser338 during RAF activation. | 579,580 |
| PAK2 | Microtubule integrity regulates RAS-independent activation of CRAF through co-expression of small GTPases, including Rac, Cdc42, and PAK1/2. | 581 | |
| PAK3 | PAK3 regulates CRAF activity by phosphorylating Ser338. | 582 | |
| Type II PAKs (PAK4/5/6) | PAK4 | PAK4 promotes premature senescence through a pathway including p16INK4/p19ARF and MAPK signaling. | 583 |
| PAK5 | PAK5 phosphorylates CRAF at Ser338, directing RAF to the mitochondria and contributing to anti-apoptotic action by phosphorylating BAD. | 582,584 | |
| Other interacting proteins | CK2 | CK2 acts as a component of the KSR1 scaffold complex during C/BRAF activation. | 167,585 |
| Rac/Cdc42 | Rac and Cdc42 act together with RAS and PI3K to achieve CRAF activation. | 586 | |
| Rac and Cdc42 induce CRAF activation with RAS. | 13 | ||
| Src | Activated Src tyrosine kinase stimulates CRAF and MAPK. | 587 | |
| Src activates CRAF via RAS-independent pathways in vivo and in vitro. | 90 | ||
| CNK1 regulates CRAF activation through Src. | 588 | ||
| Hsp90 | The Hsp90 and p50 (cdc37) complex regulates CRAF activity and stability. | 589,590 | |
| Cdc25A | Cdc25A regulates CRAF tyrosine phosphorylation. | 591 | |
| PKCs | Sequential activation of PKC isoforms (α and ε) contributes to CRAF and ERK1/2 activation. | 592 | |
| AKT | AKT physically interacts with BRAF and balances the cross-regulation between the PI3K–AKT and RAS–RAF–MEK signaling cascades. | 593 | |
| AKT3 collaborates with BRAF V600E, reducing activity to levels that favor cell proliferation rather than senescence. | 239 | ||
| JAK | The Hopscotch JAK kinase requires the CRAF pathway to enhance blood cell activation and differentiation. | 594 | |
| JAK2, together with RAS and CRAF, activates ERK and MAPK in response to growth hormones. | 91 | ||
| PP2A | PP2A functions as a CRAF-associated kinase activator involving the dephosphorylation of 14-3-3 binding sites in KSR and CRAF. | 595,596 | |
AKT protein kinase B, BAD BCL2-assocaited agonist of cell death, CNK connector enhancer of KSR, CK2 casein kinase 2, ERK extracellular signal-related kinase, JAK Janus kinase, KSR kinase suppressor of RAS, MAPK mitogen-activated protein kinase, MEK MAPK kinase, PAK p21-activated kinase, PI3K phosphoinositide 3-kinase, PKC protein kinase C, PP2A protein phosphatase 2A
RAF proteins do not possess inherent subcellular localization motifs, and initially, they are within the cytoplasm in an inactive monomeric form.93 The activation of RAF, transitioning it from its autoinhibited, pre-signaling, and inactive state, necessitates a series of regulatory steps. These include the relief of autoinhibition, the formation of dimers or higher-order multimers, and phosphorylation.
Autoinhibition of the pre-signaling, inactive state
All RAF family members contain three conserved regions (CR1, CR2, and CR3) and two functional domains: an N-terminal regulatory domain and a C-terminal catalytic domain. The N-terminal regulatory domain contains both CR1, composed of a RAS-binding domain (RBD) and a cysteine-rich domain (CRD), and CR2, which is enriched in Ser/Thr residues, whereas CR3 is located at the C-terminal domain (Fig. 3d). RAF activation is largely accomplished through the removal of inhibitory enforcement at the RAF catalytic domain. The N-terminal regulatory region interacts with the kinase domain, leading to RAF autoinhibition, a fundamental regulatory mechanism shared by all three RAF proteins (ARAF, BRAF, and CRAF). However, both ARAF and CRAF require additional steps to achieve maximal activity, such as the phosphorylation of activating residues and the dephosphorylation of negative regulatory residues.94
Autoinhibition relief
In the absence of cellular stimuli, RAF proteins exist in a monomeric, autoinhibited, inactive form. Activation of the RAF kinase domain requires the relief of N-terminal autoinhibition, which is accomplished through a series of events, including a change in the subcellular localization, protein–protein interactions, lipid interactions, and regulatory phosphorylation.95,96 RAF activation first requires the translocation of RAF from the cytosol to the plasma membrane, which represents a vital step. Experiments have shown that retaining RAF on the plasma membrane results in the constitutive activation of RAF in a RAS-independent manner.97,98 The RAF RBD interacts with the GTP-bound RAS effector domain by adopting a conserved, ubiquitin-like structure,99,100 and RAS binding with the RAF CRD (zinc-coordinated structure) can relocate RAF to phosphatidylserines in the plasma membrane regardless whether RAS is bound to GTP.101–104 However, both the RBD and the CRD are involved in the full activation of RAF.
Dimerization and activating phosphorylation
RAS engagement on the membrane increases the phosphorylation of the RAF kinase domain and RAF dimerization. Recent work indicates that RAF dimerization is necessary for RAS-dependent RAF kinase activity and correlates with the pathogenic role of disease-associated mutant RAF, which displays strong intrinsic kinase activity.105 The formation of the side-by-side RAF dimer involves a structural association between the N- and C-terminal regions of the kinase domain.106 Following the release of inhibitory domain from the complex, the RAF kinase domain readily forms RAF–RAF homodimers, subsequently leading in kinase activation.107 The RAF-related pseudo-kinase KSR (kinase suppressor of RAS) also participates in forming side-to-side heterodimers with RAF (RAF-KSR heterodimer). Activated RAF kinase phosphorylates target proteins, such as MEK1 and MEK2, leading to the subsequent activation of ERK1 and ERK2 (Fig. 3e). By contrast, inhibitory phosphorylation of the RAF hinge region can disrupt and inactivate dimeric structures. Hyperphosphorylated RAF proteins are recycled to an inactive state, ready to receive a new round of activating signals.108–110
RAF and MEK1 activity can also be regulated by activated ERK via a feedback loop. Phosphorylation regulates the activities of RAF, MEK1, and ERK depending on the phosphorylation site.111–113 Upon signal engagement, active RAS promotes the exchange capacity of son of sevenless (SOS) through a positive feedback loop, eventually activating ERK1/2. By contrast, ERK-dependent SOS phosphorylation and disassociation of the SOS-Grb2 complex prevents RAS activation through a negative feedback loop.114,115 Therefore, the phosphorylation of target proteins in the ERK pathway can regulate associated signaling pathways based on the functional location of target proteins.116,117 Improved understanding of the RAS–RAF axis and RAF dimerization has revealed the roles played by RAF in many cellular conditions. In patients with cancer, RAF homo- and heterodimers likely mediate cellular responses to ATP-competitive inhibitors and cancer progression, suggesting that RAF dimers may represent potential therapeutic targets.
MEK and ERK
MEK represents a family of protein kinases that possess dual-specificity for tyrosine and serine/threonine residues, facilitating the activation of ERK by phosphorylating regulatory Tyr and Thr sites. Upon interaction of the catalytic VIII sub-region of RAF with MEK via its C-terminal catalytic domain, a serine residue becomes phosphorylated, thereby initiating MEK activation. The primary targets for phosphorylation by activated RAF are dual-specificity kinases, such as MEK1 and MEK2, with molecular weights of 44 and 45 kDa, respectively.83 Subsequent to MEK-dependent phosphorylation, ERK is set into action, triggering a range of functional responses in cells in response to growth factors or stressors. These responses are mediated by various cytoplasmic and nuclear substrates, including transcription factors.118–120
ERK (MAPK), a Ser/Thr protein kinase, occupies a crucial position in the cellular signal transduction network, and any aberrations in its activation faults can significantly impact cellular functions. When activated, MEK directly interacts with ERKs via its N-terminal region. In situations where multiple kinases are at work, they catalyze the bispecific phosphorylation of Tyr and Thr residues within the 8 “TEY box” of the sub-functional region of ERK, thereby activating ERK. Activated ERKs subsequently translocate to the nucleus, where they increase the phosphorylation of target proteins in the cytoplasm or regulate the activity of other protein kinases. This occurs before further phosphorylation and dimerization of ERK in response to signals that promote ERK activation.121
ERK downstream signals
Several ERK1/2 target proteins are ubiquitously found in cells.60 Including the cytoplasmic substrates death-associated protein kinase (DAPK), tuberous sclerosis complex 2, RSK, and MNK, and the nuclear transcription factor substrates nuclear factor of activated T cells (NF-AT), Elk-1, myocyte enhancer factor 2 (MEF2), c-Fos, c-Myc, and signal transducer and activator of transcription (STAT3). Some membrane-associated proteins (e.g., CD120a, Syk, and Calnexin) and cytoskeleton proteins (e.g., Neurofilaments and Paxillin) are also directly phosphorylated by ERK1/2.
Other MAPK pathway
As summarized in Fig. 2, other several classical and atypical pathways and their related proteins are regulated by MAPKs.
The p38 signaling pathway
The p38 is reliably activated by a wide range of environmental stressors and inflammation and, in some cell types, by insulin and growth hormones. The p38 pathway is regulated by apoptosis-related receptors and physical sensors, including CDC42, RAC1, and mammalian Ste20-like kinases (MSTs), which also regulate JNK, resulting in the phosphorylation of the activation loop of MEK3/6. In particular, RAC1, a small G protein, controls the activation of p38 MAPK by a retinoic acid–induced beta1 integrin. p38 isoforms phosphorylate a wide range of cytoplasmic (e.g., cPLA2, MNK1/2, MK2/3, HuR, Bax, and Tau) and nuclear proteins (e.g., ATF1/2/6, MEF2, Elk-1, GADD153, Ets1, p53, and MSK1/2).122 p38 signaling is involved in immunological and inflammatory responses,123 cell fate determinants, and other stress responses124. Three anticancer compounds (e.g., SB203580, SB202190, and BIRB0796) specifically inhibit p38 isoforms (p38α and p38β) by competing with ATP in the binding pocket.125
The JNK pathway
The JNK pathway responds strongly to cytokines, growth factor deprivation, intracellular stimuli (e.g., DNA damage, cytoskeletal changes, oxidative, and ER stress), and extracellular stressors (e.g., UV radiation and osmotic stress). The JNK cascade is activated by adapter proteins in the TNF receptor-associated factor (TRAF) family, such as TRAF6, which is involved in the IL-1–induced activation of JNK.126 Two ATP-competitive JNK inhibitors, SP600125 (also known as JNK inhibitor II)127 and AS601245 (JNK inhibitor V),128 have been widely employed in the cancer research, although they exhibit low specificity. Many transcriptional factors (e.g., c-Jun, p53, ATF-2, NF-ATc1, Elk-1, HSF-1, STAT3, c-Myc, and JunB) are also regulated by JNK-directed phosphorylation.129 JNK plays an essential role in cell proliferation by modulating cell cycle genes130 and is involved in the differentiation of hematopoietic populations and the apoptotic response to cellular stressors.131
The ERK5 pathway
The ERK5 Pathway is activated by growth factors (e.g., epidermal growth factor [EGF], nerve growth factor, fibroblast growth factor [FGF]-2, and brain-derived neurotrophic factor); some cytokines, including leukemia inhibitory factor; and stressors, such as osmotic stress and hydrogen peroxide. ERK5 can be activated by several upstream factors, such as c-SRC, RAS, LAD1 adapter protein, and WNK Ser/Thr kinases132 In addition, ERK5 is activated by dual phosphorylation with a unique MAPK/ERK kinase 5 and MEK5,133,134 and activated ERK5 phosphorylates several cellular proteins, including the MEF2 transcription factor family, Sap1a (ETS domain transcription factor), c-MYC, serum and glucocorticoid inducible protein kinase, Connexin 43, and Bcl-2 agonist of cell death (BAD).132 Similar to ERK1/2, ERK5 is involved in cell survival and proliferation, increasing cyclin D1 expression during the G1/S transition.135 ERK5 is also necessary for vascular endothelial growth factor (VEGF)-mediated survival and tubular morphogenesis in primary human microvascular endothelial cells and the MEK inhibitors PD98059 and U0126 effectively inhibit ERK5.136
ERK3/4, ERK 7/8, and NLK
The ERK3/4 and ERK7 pathways are poorly characterized, although these proteins autophosphorylate the activating loop residues in vitro and in vivo.137–139 Serum and hydrogen peroxide stimulate ERK8 phosphorylation via conventional MAPKs. The MAPKAPK MK5 is the only known target of ERK3/4 according to several previous studies.137,140–144 ERK7/8 directly controls a number of proteins (e.g., Myelin basic protein [MBP], c-FOS, and c-MYC) in vitro, although their cellular functions are not clear.139,145–147 Many cytokines, including interleukin (IL)-6, granulocyte colony-stimulating factor, and transforming growth factor-beta (TGF-β), are associated with the activation of NLK, which is a key regulator of cell fate determination. NLK is triggered by Wnt pathway stimulation (Wnt-1 and Wnt-5a) and TGF-β.148 Last, NLK targets T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors and STAT3.148 ERK3, ERK7, and NLK are involved in cell proliferation, cell cycle progression, migration, invasion, apoptosis, and cell differentiation. However, few inhibitors for atypical MAPKs have been validated as anticancer drugs, although GPLG0259, an inhibitor of MK5, is currently under clinical study for use in obesity and diabetes.
Accessory proteins in the RAS/RAF/MAPK cascade
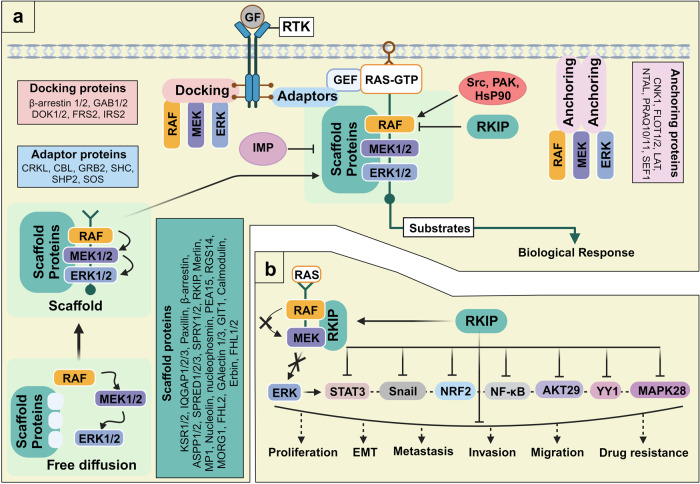
MAPK signaling is linked to various malignancies in humans, and its activation is associated with many extracellular signals and intracellular proteins.149 Therefore, targeting the constituents of this signaling cascade frequently results in severe toxicity, activation of backup mechanisms, and reduced drug efficacy, often associated with an increase in therapy burden. To avoid these undesirable effects, other approaches targeting RAF modulators must be developed. The spatiotemporal characteristics of MAPK pathway constituents may offer an alternative strategy MAPK accessory proteins are spatially assembled to promote cooperation during signaling150 and can be divided into four categories: (1) anchoring proteins, (2) docking proteins, (3) adapter proteins, and (4) scaffold proteins (Fig. 4a).
Fig. 4.
RAF signaling regulation by accessory proteins. a Accessory proteins consist of anchoring proteins, docking proteins, adapter proteins, and scaffold proteins. Anchoring proteins bind to membrane kinases and other effectors, whereas adapter proteins link receptor kinases with guanine exchange factors (GEFs). Docking proteins connect active receptors with multiple effectors. Scaffold proteins offer a signaling platform for the spatial regulation of the mitogen-activated protein kinase (MAPK) pathway. b CRAF kinase inhibitor protein (RKIP) is a tumor suppressor. RKIP, an intrinsic RAF kinase inhibitor, is associated with many malignant features, including metastasis and chemotherapy resistance, through the regulation of oncogenic mediators and signaling axes, such as NF-kB, YY1, MAPK28, STAT3, NRF2, and AKT29. Arrows and bars indicate stimulating and inhibiting signals, respectively. This figure was created with BioRender.com
Anchoring proteins
Anchoring proteins are bound to the membrane and connect and associate with effector proteins, which primarily consist of kinases. For example, the A-kinase-anchoring protein (AKAP-Lbc) and the scaffolding protein (KSR-1) constitute a signaling network that efficiently relays signals from RAF to MEK to ERK1/2.151,152 Many anchoring proteins have been discovered in other signaling pathways, including connector enhancer of KSR (CNK)1,153 Flotillin ½,154 linker for activation of T cells,155 non–T cell activation linker (NTAL),156 progestin and adipoQ receptor family members (PAQR10/11),157 and Sef1.158
RAF docking proteins
Docking proteins play a crucial role in cellular signaling by binding to receptors like RTKs and GPCRs, subsequently recruiting effector molecules. These specialized proteins typically possess PTB domains that enable them to selectively interaction with activated RTKs, as well as PH domains that serve to extend their presence at the cell membrane.159 The FGF receptor substrate 2 (FRS2) was initially identified as a substrate for the FGF receptor160 but was later found to be a key docking protein for various RTKs.161 Other identified docking proteins include docking protein 1/2 and GRB2-associated binding protein GAB1/2.162
RAF adapter proteins
Adapter proteins connect two functional components (e.g., receptor and GEF), providing additional docking sites for signaling proteins and promoting signal transduction from one-way receptors. Phosphorylation at Ser residues generates protein–protein interaction sites mediated by adapter proteins, such as the 14-3-3 family, facilitating associations with various signaling modulators, including CRAF, KSR, B-cell receptor (BCR), and phosphoinositide 3-kinase (PI3K).163 Many adapter proteins play important roles in signaling pathways, including CRK proto-oncogene (CRK and CRKL), Casitas B-lineage lymphoma, GRB2,164 SHC, SH2-containing protein tyrosine phosphatase 2 (SHP2) and SOS.165
RAF scaffolding proteins
Scaffolding proteins bind two or more partners to provide a signaling platform able to regulate the MAPK pathway both spatially and temporally.166 MAPK signaling components that exist as freely diffuse cytosolic forms are unable to effectively transmit signals to corresponding partners. Scaffolding proteins offer a platform at which many components can associate, allowing the efficient propagation of signals. Scaffolds also facilitate tighter control of MAPK signaling.150 KSR is a scaffold protein in the MAPK pathway that assembles B/CRAF, MEK1/2, and ERK1/2.167 In addition, both KSR and MEK partner-1 (MP1) retain ERK proteins in the proximity of critical cellular effectors.168 Several scaffolding proteins are involved in cellular functions169, including IQGAP1/2/3,170 Paxillin,171 β-arrestin,172 apoptosis stimulating proteins of p53 1/2 (ASPP1/2),173 SPRED1/2/3,174 SPRY1/2,175 RAF kinase inhibitory protein (RKIP), Merlin,176 Nucleolin and Nucleophosmin,177,178 PEA15,179 Regulator of G-protein signaling 14 (RGS14),180 MAPK organizer 1 (MORG1),181 Galectin 1/3,182 GIT1,183 Calmodulin,184 Erbin,185 and FHL1/2.186
RKIP is ubiquitously expressed in a broad range of cells and serves as an integral scaffolding protein187 and a negative modulator of the RAF–MEK–ERK signaling pathway188. RKIP directly binds CRAF and inhibits the MEK-dependent phosphorylation of CRAF by interfering with the formation of a kinase–substrate complex between CRAF and MEK.189
RKIP as a tumor suppressor
RKIP belongs to the phosphatidylethanolamine-binding protein family, which functions in lipid metabolism and phospholipid membrane biogenesis.190 RKIP is a highly dynamic protein with a malleable pocket loop that exists in a variety of states, serving as a functional switch. This protein has pleiotropic roles in several signaling pathways involved in physiological processes As a MAPK signaling modulator, RKIP can inhibit the metastatic process by modulating RAF activation and may represent a new avenue for therapeutic intervention (Fig. 4b). RKIP also regulates cancer development and progression, and191 its expression is severely downregulated in many cancer tissues, including breast cancer192, prostate cancer193, gastric cancer,194 lung cancer,195 esophageal squamous cell carcinoma,196 colorectal cancer,197,198 and nasopharyngeal carcinoma.199 Low RKIP expression levels are generally associated with malignant features, such as metastasis and chemotherapy resistance, promoting oncogenic signaling axes, including nuclear factor kappa B (NF-κB),200 YY1,201 MAPK28,202 and AKT29.203 RKIP levels are regulated by STAT3 activation during metastasis in NSCLC cells,204 and RKIP downregulation leads to nuclear factor erythroid 2-related factor 2 (NRF2) hyperactivation, which is responsible for the development of chemotherapeutic resistance in colorectal cancer cells.205 Reduced RKIP levels stimulate invasion, metastasis, and radio-resistance in nasopharyngeal cancer cells.206
Physiological functions of RAS/RAF/MAPK
The RAS/RAF/MAPK pathway involves signal transmission from membrane-based receptors, which interact with mitogens, to various destinations with cells, including the nuclear, cytoplasmic, and cell. These signals play a pivotal role in orchestrating a diverse range of physiological responses, encompassing cell proliferation, tumor invasion and metastasis, cellular metabolism, cell cycle progression, and ultimately cell survival or death.207 Consequently, any disruption or dysregulation of the RAS/RAF/MAPK pathway is closely linked to numerous human disorders, most notably cancer.
Role of ERK/MAPK in tumorigenesis
While tumorigenesis and the metastatic spread of cancer involve multiple cooperative cellular signals, the significance of the ERK/MAPK signaling pathway cannot be overstated when it comes to cancer invasion and metastasis. Notably, a heightened activation of ERK is evident across a spectrum of human cancer types, including ovarian, colon, breast, lung cancer, and others.208 Furthermore, in vitro experiments have revealed that microRNA-508 effectively inhibits EMT, migration, and invasion in ovarian cancer cells by modulating the ERK/MAPK1 signaling system.208 Meanwhile, in vivo studies have demonstrated that blocking the MAP kinase pathway leads to the suppression of growth of colon cancer cells.68 Additionally, Emodin has been found to inhibit the proliferation of non-small-cell lung cancer (NSCLC) cells by inducing PPARs and subsequently reducing Sp1 levels through the activation of ERK and AMPK.209
Cell proliferation and cell apoptosis
The ERK/MAPK signaling pathway primarily plays a role in promoting cell proliferation and exerts an anti-apoptotic influence. Specifically, under hypoxia conditions, it facilitates the survival of nutrient-starved tumor cell by reducing their susceptibility to apoptosis.210 Moreover, in the context of large B-lymphoma cells, microRNA-101 exerts control over cell proliferation and apoptosis by directly modulating MEK1 in the RAS/RAF/MAPK pathway.211 Furthermore, both ERK1 and ERK2 contribute to cell proliferation in a manner that depends on their expression levels, as they integrate signals from RAS, RAF, and MEK.212 The constitutive activation of the RAS/RAF/MAPK pathway contributes to tumorigenesis by inhibiting Caspase-9 through MAPK-dependent phosphorylation at Thr125.213
Cell cycle progression
Numerous downstream effectors of the RAS/RAF/MAPK pathway have a multifaceted impact. They not only drive cell cycle progression by instigating the production of cyclins and cell cycle-dependent protein kinases (CDKs) through the regulation of MYC and E2F but also orchestrate an early G1 cell cycle arrest. This arrest is achieved by influencing the expression of various CDK inhibitor proteins, including p16Ink4a, p15Ink4b, and p21Cip. Furthermore, the RAS/RAF/MAPK pathway is closely linked to the induction of cellular senescence, which is mediated by these CDK inhibitors, resulting in a premature G1 arrest.214
Tumor ECM Degradation and angiogenesis
Furthermore, the RAS/RAF/MAPK pathway plays a vital role in degradation of extracellular matrix proteins, a crucial process for cancer metastasis and angiogenesis. For instance, Mesothelin, a secretory protein, stimulates the production of MMP-7 by activating the MAPK/ERK and JNK signaling pathways in ovarian cancers215. Additionally, p70S6K, a downstream target of the RAS/RAF/MAPK pathway, exerts control over tumor growth and angiogenesis by promoting the activation of HIF-1alpha and the production of VEGF in ovarian cancer cells.216
It is fair to state that as we understand more about the specifics of RAF (ARAF, BRAF, and CRAF), we found that there are more unanswered questions about how they work and how they specifically affect physiological functions.
Cell regulatory pathways mediated by MAPK-independent RAF kinase
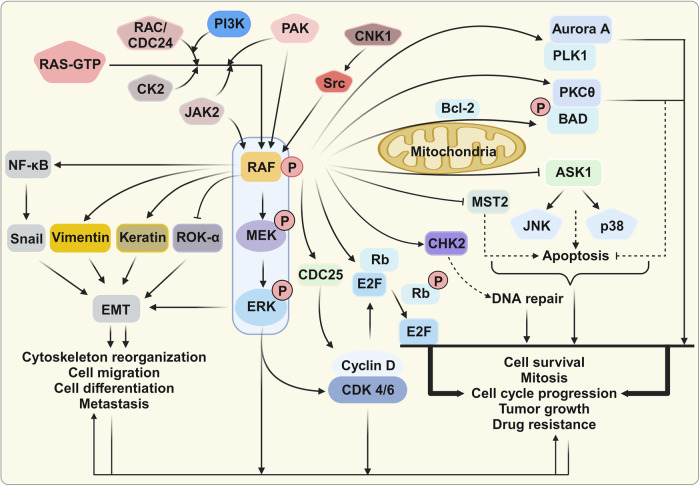
Generally, RAF activation leads to ERK1/2 activation via MEK1/2 phosphorylation217. MEK1/2 was thought to be the only RAF kinase substrate prior to the discovery of MEK1/2-independent RAF functions.218,219 Although only a few MEK-independent RAF targets have been defined, these MEK-independent RAF activities are thought to be important for carcinogenic regulation (Fig. 5).
Fig. 5.
Kinase-independent regulation of RAF-interacting signaling. Three RAF effector proteins, Bcl-2 agonist of cell death (BAD), apoptosis signal-regulating kinase 1 (ASK1), and mammalian Ste20-like kinase 2 (MST2), are kinase-independent negative regulators of apoptosis. RAF can enhance cell cycle progression via the extracellular signal-related kinase (ERK) pathway, and RAF can regulate the cell cycle in a kinase-independent manner. RAF interacts with polo-like kinase 1 (PLK1) and Aurora kinase A (Aurora-A). During cell migration, RAF also functions as a spatial regulator of Rho-associated kinase (ROK)-α, a downstream effector of RHO, in a kinase-independent manner by inhibiting ROK-α activity. Several additional RAF substrates, such as NF-κB, Vimentin, Snail, and Keratin, are associated with cytoskeleton organization. This figure was created with BioRender.com
RAF as an apoptosis regulator
MEK-independent RAF is a negative regulator of apoptosis. Three RAF effector proteins, BAD, apoptosis signal-regulating kinase 1 (ASK1), and MST2, play critical roles in the regulation of apoptosis. Interactions between RAF and these targets occur at the outer mitochondrial membrane, unlike classical RAF signaling, which is localized to the plasma membrane.220
BAD promotes apoptosis by inhibiting the pro-survival effects of BCL2 proteins,221 and RAF increases cell survival through the direct phosphorylation of BAD.222 Raf also acts as an adapter protein that stimulates the binding of BAD with protein kinase C, which phosphorylates BAD and inactivates downstream signals.223
ASK1, also referred to as MAP3K5, is a Ser/Thr kinase able to activate the SAPK, JNK, and p38 pathways to trigger apoptosis under oxidative conditions.217,224 FGF receptor activation increases interactions between RAF and ASK1 in the mitochondria, preventing the activation of the p38 MAPK pathway in an ERK- and PI3K-independent fashion.225,226 RAF modulates ASK1 activity, which is necessary for JNK- and p38-induced apoptosis, and the loss of RAF expression increases ASK1 activity, followed by increased JNK and p38 activation. ASK1 knockout reverses the effects of RAF loss.227
RAF can inhibit the MST2-associated tumor suppression pathway in various cancers. RAF binds MST2 independent of kinase activity, preventing apoptosis in cancer cells via a two-pronged mechanism.228–231 First, RAF binding to MST2 blocks MST2 dimerization, a critical step for MST activation. Second, RAF recruitment of a phosphatase can prevent MST2 autophosphorylation at the Thr180 residue in the activation loop.228,232
RAF as a cell cycle and mitosis checkpoint mediator
CRAF can promote cell cycle progression in a MEK/ERK-independent manner233. CRAF directly interacts with key regulators of mitotic progression, polo-like kinase 1 (PLK1) and Aurora kinase A (Aurora-A).233,234 At the G2/M transition during mitosis, CRAF phosphorylation (Ser338) induces protein localization to centrosomes and mitotic spindle poles, where CRAF interacts with and activates Aurora-A and PLK1, leading to mitosis and tumor growth.
In addition, RAF regulates checkpoint kinase 2 (CHK2) activity during cell cycle progression. Upon DNA damage, PAK1 induces the formation of a RAF–CHK2 complex to stimulate the DNA repair system.235 RAF phosphorylation at Ser338 promotes the RAF–CHK2 interaction, which is associated with radiation resistance and CHK2 activation. In addition, RAF interacts with CDC25 phosphatase, which links mitogenic signaling with cell cycle activation.236,237
RAF as a regulator of the EMT and cytoskeletal organization
Cancer metastasis has been linked to the dysregulation of various signal pathways, including the constitutive activation of MAPK, NF-κB, and PI3K in melanoma.238 RAF-dependent AKT3 inhibition has also been associated with the promotion of cell proliferation, survival, and metastasis and the inhibition of cellular defense mechanisms and cellular senescence.239,240 NF-κB activation promotes metastasis by increasing the expression of many metastatic genes, including cyclooxygenase 2 (COX2), genes encoding metalloproteinases, VEGF, and SNAIL.241,242 Jacqueline’s group demonstrated that RAS can activate NF-κB transcriptional activity via either RAF-dependent or RAF-independent mechanisms, both of which are dependent on SAPKs, such as p38.243 According to another study, RAF-dependent NF-κB activation involves MEKK1 rather than the traditional mitogenic cytoplasmic kinase pathway.244 RAF stimulates the expression of atrial natriuretic factor (ANF) in cardiac myocytes, whereas MEK1/2 inhibits ANF expression.245
RAF-dependent RHO signaling is related to cell migration. During cell migration, RAF functions as a spatial regulator of RHO-associated kinase (ROK)-α, an effector downstream of RHO, in a kinase-independent manner.246 In a conditional knockout study, RAF was found to be necessary for proper wound healing in vivo and for keratinocyte and fibroblast cell migration in vitro. This study also indicated that RAF-mediated ROK-α inhibition is necessary for RAS-dependent carcinogenesis.247 Functionally, the interaction between RAF and ROK-α may be associated with a RAF-induced anti-apoptotic signal, as stimulation of the FAS death receptor increases RAF–ROK-α complex formation.248 In addition, ROK-α kinase activity is inhibited by the RAF regulatory domain.249 However, ROK-α is likely not regulated by RAF in RAS-mutant tumors.250 Moreover, several additional RAF substrates are involved in cytoskeleton organization, including Vimentin251 and Keratin 8.252
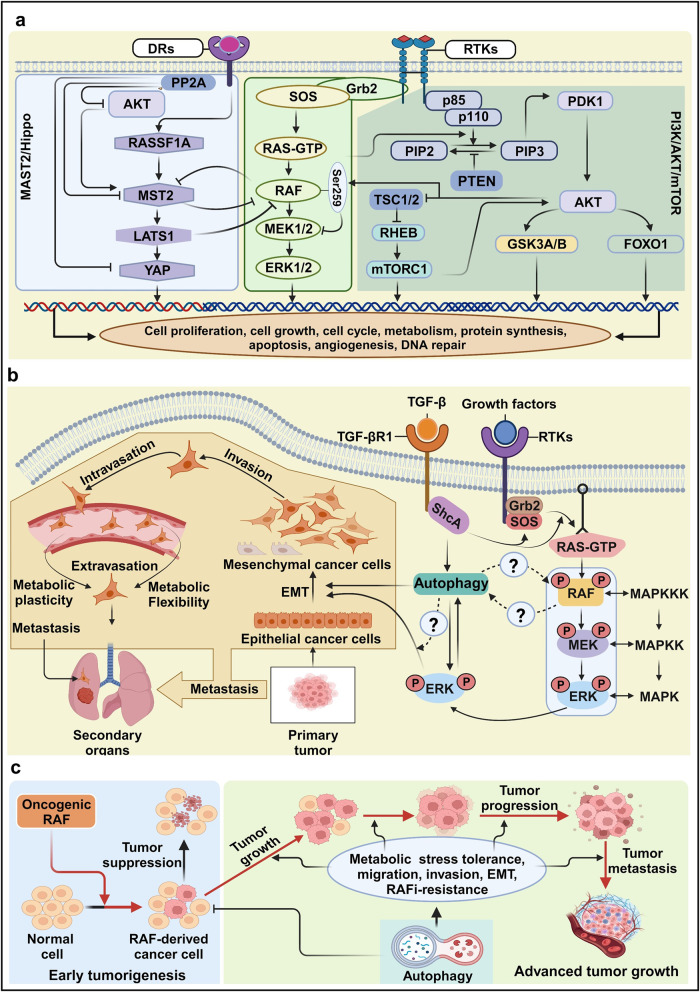
Crosstalk between RAS/RAF/MAPK and PI3K/AKT/mTOR or MST2/Hippo signaling pathways
The PI3K-mTOR (mammalian target of rapamycin) pathway is a crucial mechanism governing cell survival, division, and metabolism. Growth factors can engage the pathway by either directly recruiting PI3K to their receptors or indirectly involving it through docking proteins like IRS (insulin receptor substrate) or GAB (GRB2-associated binder). This activation of PI3K leads to the generation of the secondary messenger phosphatidyl inositol 3,4,5-triphosphate (PIP3), which in turn recruits the protein kinase AKT to the plasma membrane. Subsequent AKT activation, dependent on PDK1, initiates the phosphorylation of numerous factors related to survival, proliferation, motility, and the TSC2 GAP (GTPase Activating Protein). AKT-dependent TSC2 phosphorylation releases TSC1/2 inhibition by the GTPase RHEB (RAS homolog abundant in brain), ultimately activating mTORC1, which regulates cell growth. Both the RAS/RAF/MAPK and PI3K/AKT/mTOR pathways frequently experience dysregulation in many human cancers, often due to genetic alterations in their components or upstream regulators.253 The intricate network of positive feedforward and negative feedback loops in these pathways significantly influences signal dynamics. Notably, the GAB docking proteins, forming the GRB2-SOS complex upon activation of RTKs, are key players in positive loops. This complex, which includes RAS-GAP, SHP2, PI3K, and PIP3 (a protein-tyrosine phosphatase with a Src homology two domain), contributes to RAS activation. Dephosphorylation of RAS-GAP docking sites on GAB1 by SHP2 reduces RAS activation and enhances RAS-ERK signaling. Additionally, GAB2-mediated PI3K recruitment generates local PIP3, further stimulating PI3K signaling. Furthermore, SOS, RAF, and MEK1 can be phosphorylated by ERK, which creates a negative feedback loop by dampening ERK activity (Fig. 6a). Indeed, comprehensive cancer genome analyses have revealed that over-activation or mutations that enhance the MAPK and PI3K pathways are characteristic features of many human cancer types. Importantly, the interplay between RAS/RAF/MAPK and PI3K/AKT pathways is tightly controlled in response to ligands in a dose-dependent manner. For Instance, elevated levels of insulin-like growth factor I (IGF-I) induce a rapid and potent phosphorylation of AKT at the serine 259 residue, effectively restraining RAF kinase activity. Conversely, low concentration of IGH-1 fail to induce such crosstalk, but still exert mitogenic effects254
Fig. 6.
The crosstalk of RAS/RAF/MAPK with other pathways. a Crosstalk between the RAS/RAF/MAPK, PI3K/mTOR or MAT2/Hippo signaling pathways. The RAS/RAF/MAPK signal collaborates within its own cascade and interfaces with the PI3K/mTOR pathway, where its influence is MAPK-dependent. Conversely, the MST-2/Hippo pathway operates independently of MAPK activity but relies on the presence of RAF for its functionality. b Functional interactions between autophagy and mitogen-activated protein kinase (MAPK) signaling during the epithelial-to-mesenchymal transition (EMT). The MAPK pathway can be activated by canonical receptor tyrosine kinases (RTKs) and through Smad-independent activation by transforming growth factor-beta (TGF-β). Both signals activate a typical RAS–RAF–MAPK cascade, stimulating the EMT process. During the metastasis process, cancer cells migrate from a primary site to a secondary site facing many stressors, and therefore metabolic and cellular alterations are necessary to overcome these stressors. c The role of autophagy in tumorigenesis. During early tumorigenesis, autophagy acts as a tumor suppressor. However, autophagy drives tumor growth, progression, and metastasis by enhancing migration, invasion, EMT, and metabolic tolerance during advanced stages of cancer, allowing cancer cells to evade RAFi therapy. Arrows and bars indicate stimulating and inhibiting signals, respectively. This figure was created with BioRender.com
The MST2 protein family, characterized by serine/threonine kinases, becomes activated in response to stress signals in mammalian cells, and their overexpression has been observed to trigger apoptosis.232 The MST2/Hippo pathway is intricately linked to the RAS/RAF/MAPK pathway (Fig. 6a). Specifically, the MST-CRAF complex, induced by mitogenic and apoptotic signals, acts as a safeguard against unchecked cell proliferation.229 A previous study has shed light on the dynamic changes in protein-protein interactions (PPIs) arising from the function association between the kinases MST2 and CRAF kinases, along with the modulation of their respective upstream activators RASSF1A and RAS231. The interaction between CRAF and MST2 inhibits the binding of the scaffold protein RASSF1A to MST2, leading to MST2 dimerization and activation. Interestingly, activated AKT promotes the binding of MST2 to RAF and subsequently preventing MST2 activation.255 Additionally, MST2 can be inhibited by a phosphatase, likely PP2A, associated with RAF.256 Conversely, MST2 interferes with RAS-dependent RAF activation by blocking the RAS-binding domain (RBD) domain in CRAF. RASSF1A can rerelease MST2 from its inhibitory complex with CRAF, activating LATS1. This activation of LATS1, in turn, induces the formation of the YAP1-p73 transcriptional complex, ultimately leading to apoptosis.257 Furthermore, the RAS/RAF/MAPK pathway engages in crosstalk with several other signal pathways, including those involved in DNA repair process or cell cycle regulation, as depicted in Fig. 5.
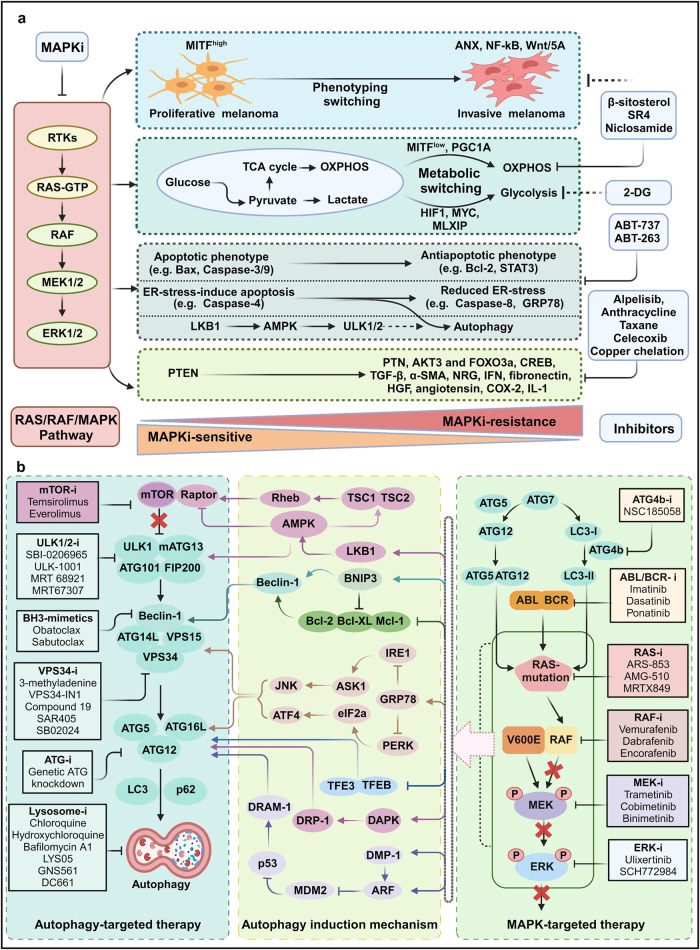
The link between autophagy and RAF signaling in cancer metastasis
Autophagy is an essential process that maintains cellular homeostasis via the lysosome-dependent degradation of cellular components, such as proteins and organelles, allowing cells to recycle macromolecules258,259. Irregular autophagy activation can lead to cellular dysfunction, as observed in many human diseases, including neurodegenerative diseases, heart disease, infectious diseases intervertebral disc degeneration, liver disorders, and cancers.260–262 Autophagy is also involved in various stress responses, developmental processes, and aging.263,264
Although autophagy can be either tumor-suppressive or tumor-promoting, depending on the context, the precise mechanisms that determine the role of autophagy are not well-defined. In general, increased catabolism driven by autophagy promotes cell survival, and autophagic abnormalities induce cell death in cancer cells. Cancer cells exhibit more microenvironmental and metabolic dependencies than normal cells, and targeting the double-edged process of autophagy represents an appealing option for the development of future therapeutic agents.265
The MAPK pathway drives the expression of autophagy-related-8 (Atg8) in cancer cells.266 Studies of RAF mutations suggest a functional association between autophagy and V600E-mutant BRAF. Autophagy accelerates the growth of V600E-mutant melanoma in mice by allowing the bypass of the senescence process.267 The reduced expression of autophagy-related ATG genes in patients with BRAFV600E-mutant melanoma appears to suppress carcinogenesis.268
Cancers can spread to secondary organs through a complicated, efficient, and deadly process called metastasis, which represents a substantial contributor to mortality.269 Metastasis requires coordination between the genetic programs that promote and prevent metastasis and the tumor microenvironment to allow cancer cells to transition from their initial locations and develop in secondary organs. Metastasis suppressor genes prevent metastasis at secondary sites with no impact on the primary tumor.
During metastasis, cancer cells dissociate from the primary sites, travel through the circulation, and deposit in a secondary site, overcoming many obstacles, including nutrient limitations and cellular stress.270 In metastatic cells, cellular metabolism undergoes dynamic alterations that promote a shift toward the metastatic cascade.271 Metabolic plasticity and flexibility in cancer cells play important roles in the metastatic process.272 Metabolic plasticity allows a single metabolite to meet multiple metabolic needs during metastasis, whereas metabolic flexibility allows several metabolites to fulfill the same metabolic need.273,274 The metabolites sapienate, generated by fatty acid desaturase 2, and palmitoleate, generated by stearoyl-CoA desaturase-1, are mono-unsaturated fatty acids involved in the metabolic flexibility of primary tumors.275,276 The presence of these metabolites in the bloodstream is essential for the invasion, migration, and survival of metastatic cells. These metabolites are also associated with the EMT,277 which represents a necessary step through which cancer cells acquire metastatic properties and plays important roles in cancer progression, metastasis, and drug resistance.278
Based on previous studies, MAPK signaling, autophagy, and EMT are physically and functionally interconnected during cancer metastasis (Fig. 6b). EMT is a multidimensional process that involves the remodeling of the cytoskeleton, cell membrane, and cell–cell junctions, resulting in the loss of epithelial characteristics and the acquisition of mesenchymal properties facilitated by MAPK activation.279 Autophagy supplies the energy required for TGF-β–induced EMT and cancer metastasis. TGF-β is involved in many cellular processes, including tissue fibrosis, growth inhibition, and EMT. TGF-β activates Smad-dependent and Smad-independent signaling pathways, including the ERK1/2 pathway, which is involved in cytoskeletal organization, cell growth, survival, migration, and invasion.280 TGF-β–directed MAPK signaling also activates common downstream signaling molecules induced by RTKs. In response to TGF-β, RAS triggers ERK1 and ERK2 activation, leading to the activation of RAF and MEK1/2, as shown in Fig. 2.281,282 Following TGF-β-induced Ser or tyrosine phosphorylation of the type I TGF-β receptor, ShcA recruits GRB2 and SOS to activate ERK1/2 through RAS, RAF, and MEK1/2.281,283 Other signaling molecules, such as integrin, Notch, Wnt, TNF-α, long non-coding (lnc) RNA, and EGF, synergize with TGF-β signaling to promote tumor invasion and metastasis.284–290 Many studies suggest the existence of functional connections between TGF-β and RAS–RAF signaling in tumorigenesis. TGF-β–induced EMT is enhanced by increased RAS–ERK291 signaling, and MEK1/2 pharmacological inhibition prevents TGF-β–induced EMT.292
EMT and autophagy are connected through multiple pathways, although the exact role of autophagy in EMT remains unclear. Autophagy inhibition promotes EMT, invasion, and metastasis in many cancer cells, including gastric, colorectal, melanoma, and pancreatic cancer cells, mouse embryonic fibroblasts, and keratinocytes.293–295 By contrast, TGF-β–induced autophagy promotes cancer cell migration via MAPK–ERK activation in NSCLC and SMAD4-negative pancreatic cancer cells.296,297 However, autophagy inhibits metastasis in HCC298 and prevents EMT in breast and SMAD4-positive pancreatic cancer cells.297,299
Autophagy plays a significant role in RAF-driven tumorigenesis, functioning as a tumor suppressor during early stages and as a tumor promotor during advanced stages (Fig. 6c). BRAF and CRAF activate autophagy to promote tumor cell survival.27 Atg7-knockout mice with RAF-mutant melanoma display reduced tumor growth and significantly increased survival compared to wild-type mice.267 Interestingly, autophagy exhibits both tumor-promoting and tumor-suppressive roles in the same mouse model of BRAFV600E–mutant lung cancer.300 However, autophagy promotes tumor growth and metabolism in BRAFV600E–mutant lung cancer,301 and BRAFV600E–mutant cancers promote autophagy to maintain mitochondrial function and glutamine metabolism.302
The fact that human cancer frequently exhibits dysregulation of the autophagy and RAS/RAF/MAPK pathways makes the components of these signaling cascades intriguing candidates for therapeutic intervention. Recent research has shown the existence of positive and negative feedback loops in these pathways, which activate one pathway when the other signaling cascade is blocked. Therefore, blocking both pathways with a combination of signaling inhibitors may have a stronger antitumor effect than using a single medication.
Targeting the RAS/RAF/MAPK pathway for cancer therapy
Our understanding of the roles played by RAS/RAF/MAPK components in both normal physiology and pathological conditions has significantly progressed. In the realm of therapeutic interventions, RAS/RAF/MAPK inhibitors have emerged as promising targets for addressing BRAF-mutated cancers and other disorders. These inhibitors have gained approval from the FDA and are employed either as standalone treatments or in combination with two or more agents. These FDA-approved RAS/RAF/MAPK-targeted medications, including their most up-to-date information, are represented in Table 2 in the latest medication guide available at www.accessdata.fda.gov.
Table 2.
FDA-approved Ras/Raf/MAPK targeting drugs and their limitations
| Strategy/Targets | Drugs (Trade name) NDA/BLA | Company/Approval date | Indications/Biomarkers | Limitations | |
|---|---|---|---|---|---|
| Common adverse effects (AD)/Incidence (%) | Remarks | ||||
| Single therapy | |||||
| RAS |
Adagrasib (KrazatiI) NDA 216340 |
Mirati Theraps Dec 12, 2022 |
NSCLC/KRasG12C | Nausea, diarrhea, vomiting, fatigue, dyspnea musculoskeletal pain, hepatotoxicity, renal impairment, and decreased appetite. ≥25% | The pharmacokinetics (PK) of adagrasib is constrained by CYP3A and ABCB1 |
|
Cetuximab (Erbitux) BLA 125084 |
Imclone Feb 12, 2004 |
CRC, HNC/KRASWT, EGFR | Fever, sepsis, kidney failure, dehydration, skin drying, fissuring, blepharitis, cheilitis, cellulitis, pulmonary embolus, and cyst. ≥25% | Approved and effective only in KRAS wild-type patients | |
|
Panitumumab (Vectibix) BLA 125147 |
Amgen Inc Sept. 27, 2006 |
CRC/KRASWT, NRASWT | Dermatitis acneiform, pruritus, erythema, rash, skin exfoliation, paronychia, dry skin, and skin fissures. ≥20% | Approved and effective only in KRAS wild-type patients | |
|
Sotorasib (Lumakras) NDA 214665 |
Amgen Inc May 28, 2021 |
NSCLC/KRASG12C | Diarrhea, musculoskeletal pain, nausea, fatigue, hepatotoxicity, and cough. ≥20% | Oral availability and brain accumulation of sotorasib were limited by CYP3A and ABCB1, respectively | |
| RAF |
Vemurafenib (Zelboraf) NDA 202429 |
Hoffmann- La Roche Aug 17, 2011 |
Melanoma/BRaf V600E | Arthralgia, maculopapular rash, alopecia fatigue change in the heart’s electrical activity, and skin growth (papilloma). ≥10% | Zelboraf has not been studied in patients with BRAFV600-negative, it blocks certain enzymes that promote cell growth |
|
Dabrafenib (Tafinlar) NDA 202806 |
Novartis May 29, 2013 |
Melanoma/BRAFV600EBRAFV600K | Hyperkeratosis, headache pyrexia, arthralgia, rash, papilloma, alopecia, palmar-plantar, back pain, erythrodysesthesia, cough, constipation, myalgia, and nasopharyngitis. ≥10% | Tafinlar is not indicated for the treatment of patients with wild-type BRAF melanoma. | |
|
Encorafenib (Braftovi) NDA 210496 |
Array Biopharma Inc Jue 27, 2018 |
Melanoma, CRC/BRAFV600E BRAFV600K |
Hyperkeratosis, alopecia, PPES, fatigue, rash, arthralgia, dry skin, nausea, myalgia, headache, vomiting and pruritus. ≥25% | Braftovi can cause tumor promotion in BRAF wild-type tumors, new primary malignancies, fetal harm, impaired fertility, uveitis, hemorrhage | |
| MEK |
Trametinib (Mekinist) NDA 204114 |
Novartis May 29, 2013 |
Melanoma/BRAFV600E BRAFV600K |
Rash, diarrhea, lymphedema, stomatitis, hypertension, abdominal pain, hemorrhage, dry skin, pruritis, and paronychia. ≥10% | Mekinist is not indicated for the treatment of patients who have received a prior BRAFi therapy |
|
Cobimetinib (Cotellic) NDA 206192 |
Genentech Nov 10, 2015 |
Melanoma/BRAFV600E BRAFV600K |
Diarrhea, sensitivity to ultraviolet (UV) light (photosensitivity reaction), nausea, fever (pyrexia) and vomiting. ≥20% | Coadministration of colic with itraconazole (increased cobimetinib systemic exposure by 6.7-fold | |
| Selumetinib (Koselugo) NDA 213756 |
Astrazeneca April 10, 2020 |
PN/NF1 | Vomiting, rash, abdominal pain, diarrhea, nausea, dry skin, fatigue, musculoskeletal pain, pyrexia, acneiform rash, stomatitis, headache, paronychia, and pruritus. ≥40% | It is not known if Koselugo is safe and effective in children under 2 years of age | |
| Combination therapy | |||||
| RAF and MEK | Vmurafenib+cobimetinibe NDA 206192 |
Genentech Nov 1, 2022 |
Melanoma, histiocytic neoplasm/BRAFV600E BRAF V600K |
Hyperkeratosis, headache, pyrexia, arthralgia, rash, papilloma, alopecia, palmar-plantar, back pain, erythrodysesthesia, cough, constipation, myalgia, and nasopharyngitis.10% | Tafinlar is not indicated for the treatment of patients with wild-type BRAF melanoma |
| Dabrafenib + trametinib |
Novartis Jun 22, 2017 April 30, 2018 June 22, 2022 Mar 16, 2023 |
Solid cancers, NSCLC, melanoma with involvement of lymph node(s), LGG/BRAFV600E BRAFV600K |
Pyrexia, fatigue, nausea, vomiting, diarrhea, dry skin, decreased appetite, edema, rash, chills, hemorrhage, cough headache, dyspnea arthralgia, myalgia, dyspnea, musculoskeletal pain, abdominal pain, dermatitis acneiform, dizziness, upper respiratory tract infection and weight increased. ≥15% | Tumor promotion in wild-type BRAF, resistance to BRAFi, melanoma, hemolytic anemia, hyperglycemia, erythema, serious febrile drug reactions, uveitis, and iritis, RPED, RVO, cardiomyopathy, venous thromboembolic events, hemorrhage, and embryofoetal toxicity | |
| RAF and RAS | Encorafenib+cetuximab |
Array Biopharma Inc April 8, 2020 |
CRC/BRAFV600E | Fatigue, nausea, diarrhea, dermatitis acneiform, abdominal pain, decreased appetite, arthralgia, and rash. ≥25% | Around 10% of patients who received Braftovi in combination with cetuximab had pancreatitis |
| PDL-1, RAF and MEK | Atezolizumab (Tecentriq) +vemurafenib or cobimetinib |
Genentech July 30, 2020 |
Melanoma/BRAFV600E | Rash, musculoskeletal pain, fatigue, hepatotoxicity, pyrexia, nausea, pruritus, edema, stomatitis, hypothyroidism, and photosensitivity reaction. ≥20% | Tecentriq may cause fertility problems in females. The safety and effectiveness in children are unknown |
FDA US food and drug administration, NDA new drug application, BLA biological license application, NSCLC non-small cancer lung cell, CRC colorectal cancer, HNC head and neck cancer, LGG low grade gliomas, PN plexiform neurofibromas, NF1 Nnurofibromatosis type 1, RPED retinal pigment epithelial detachment, PPE palmar-plantar erythrodysesthesia, RVO retinal vein occlusion, UV ultraviolet, ≥ greater or equal, % percentage
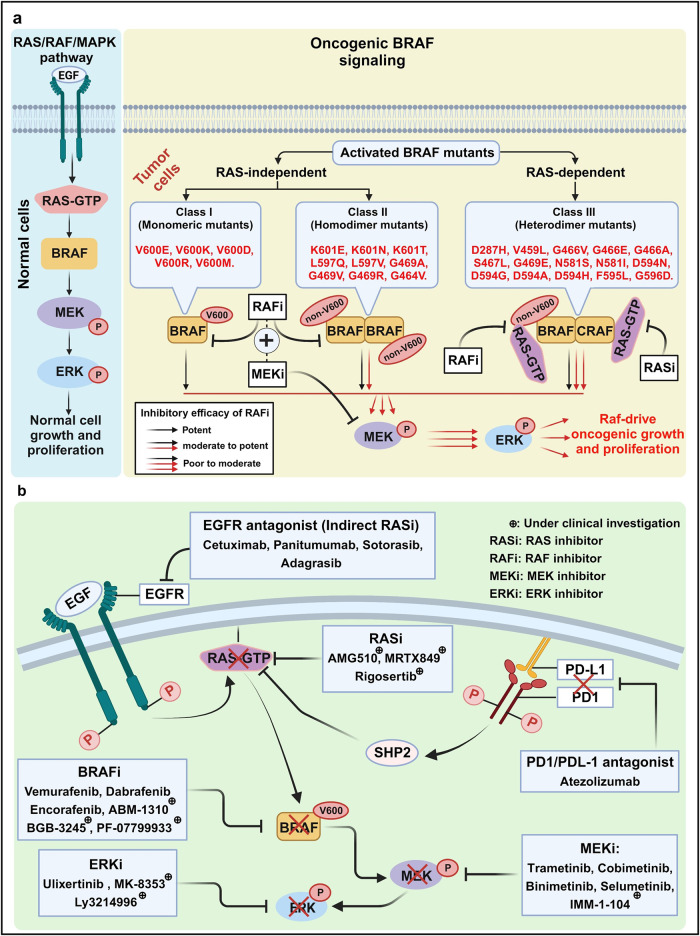
RAS/RAF/MAPK inhibitors
RAS/Raf/MAPK inhibitors represent a category of precision therapies employed in the management of diverse cancers, especially those marked by mutations in the RAS/RAF/MAPK pathway (Fig. 7a). These pathway constituents play a critical role in regulating cell proliferation, differentiation, and survival. However, when mutations disrupt their normal function, they can contribute to the initiation and progression of cancer.
Fig. 7.
RAS/RAF/MAPK-targeted therapy in BRAF-mutated malignancies. a Functional classifications of BRAF mutations. In Class I, BRAF mutants (e.g., V600E) transmit signals via a monomeric form independent of RAS activation, leading to increased extracellular signal-related kinase (ERK) activation. In Class II, BRAF mutants (e.g., K601E) are RAS-independent, forming mutant–mutant BRAF dimers. RAF inhibitors (RAFi), such as Vemurafenib and Dabrafenib, block both Class I and II RAF kinases. In Class III, mutant BRAF (e.g., D287H, V459L) exhibits increased RAS binding and heterodimer formation between mutant BRAF and wild-type CRAF. MEK inhibitors (MEKi), such as Trametinib or Cobimetinib, show an additive effect when combined with RAFi for cancer treatment. b RAS/RAF/MAPK-targeted therapies. Specific Inhibitors targeting the RAS/RAF/MAPK pathway represent as each group of action: EGFR agonists (EGFRi), RAS inhibitors (RASi), RAF inhibitors (RAFi), MEK inhibitors (MEKi), and ERK inhibitors (ERKi). Arrows and bars indicate stimulating and inhibiting signals, respectively. This figure was created with BioRender.com
KRAS-targeted therapy
The proto-oncogene KRAS plays a crucial role in cell signaling pathways governing cell growth and differentiation. In various cancers, mutations in the KRAS gene have been linked to poor prognosis and a limited range of targeted treatment options. In Western nations, lung adenocarcinomas exhibit KRAS mutations in 20–40% of cases, while the prevalence is slightly lower, around, in Asian countries.303 KRAS mutations pose therapeutic challenges as they are often associated with resistance to specific target therapies, such as epidermal growth factor receptor (EGFR) inhibitors in non-small cell lung cancer (NSCLC).304 While the development of the direct RAS inhibitors has been proven challenging due to the nature of RAS protein, ongoing research and clinical trials are exploring strategies and therapies to effectively target KRAS-mutant tumors.
Cetuximab and Panitumumab
Cetuximab and Panitumumab are monoclonal antibody therapies that target the epidermal growth factor receptor (EGFR) and are employed in the treatment of various tumors, including head and neck and colorectal cancer.305 Importantly, these drugs are approved and effective exclusively in KRAS wild-type patients with advanced colorectal cancer.306 While they are not direct RAS-mutant targeted therapeutics, they may occasionally have an indirect impact on RAS signaling pathways.307 For instance, when combined with Cetuximab, LSN3074753 (a pan-RAFi) demonstrates synergistic anticancer efficacy in BRAF or KRAS-mutant CRC PDX models.308 It has been suggested that colorectal cancers with the KRASG13D mutation may respond more favorably to Cetuximab or Panitumumab treatment compared to other more prevalent KRAS mutations.309 In the realm of KRAS-mutant targeted therapeutics, Adagrasib (MRTX849) demonstrated favorable tolerability and exhibited anticancer activity in patients with advanced solid tumors harboring the KRASG12C mutation in a first-in-human phase I/IB clinical trial (KRYSTAL-1) (NCT03785249).310
Sotorasib
The U.S. FDA-approved Sotorasib (LumakrasTM, Amgen) has been employed in the treatment of advanced NSCLC patients with and KRASG12C mutation who have undergone at least one prior systemic therapy.311 The FDA’s rapid approval of Sotorasib serves as a remarkable example of recent expeditious approvals for clinically effective drugs. In phase I clinical trials, Sotorasib demonstrated promising anticancer activity in heavily pretreated patients with advanced solid tumors bearing the KRASG12C mutation (NCT03600883).312 Subsequently, in phase II clinical trials, Sotorasib therapy provided sustained clinical benefit without revealing new safety concerns in previously treated KRASG12C-mutated NSCLC patients (NCT03600883). It is noteworthy that Sotorasib’s oral availability is significantly restricted by CYP3A, while its brain accumulation is robustly constrained by ABCB1.313
Adagrasib
Adagrasib (KRAZATITM, Mirati Therapeutics) is an orally administered, and highly effective small molecule inhibitor, irreversibly covalent binding to KRAS. It have been developed for the treatment of solid tumors harboring the KRASG12C oncogenic driver mutation, including NSCLC and CRC.314 Tian et al. unequivocally demonstrates the promising effectiveness and acceptable safety profile of Adagrasib based on multiple registered interventional clinical trials (e.g., NCT05375994 and NCT05472623) in patients with KRASG12C-mutated NSCLC.315 They have also recommended further research to explore Adagrasib’s potential in various contexts and combination therapies. The US FDA has granted approval for expanded access to Adagrasib (MRTX849) in patients with advanced solid tumors carrying the KRASG12C mutation (NCT05162443), citing data from several interventional clinical trials (e.g., NCT04975256, NCT05853575, NCT03785249, NCT04330664, and NCT05609578). A recent phase I/II study showcased Adagrasib’s antitumor activity in heavily pretreated patients with metastatic CRC bearing mutant KRASG12C, both as oral monotherapy and in combination with Cetuximab (NCT03785249).316 The pharmacokinetics (PK) of Adagrasib is constrained by the CYP3A and ABCB1 activity, and it can be modified by mouse plasma carboxylesterase 1 c.317
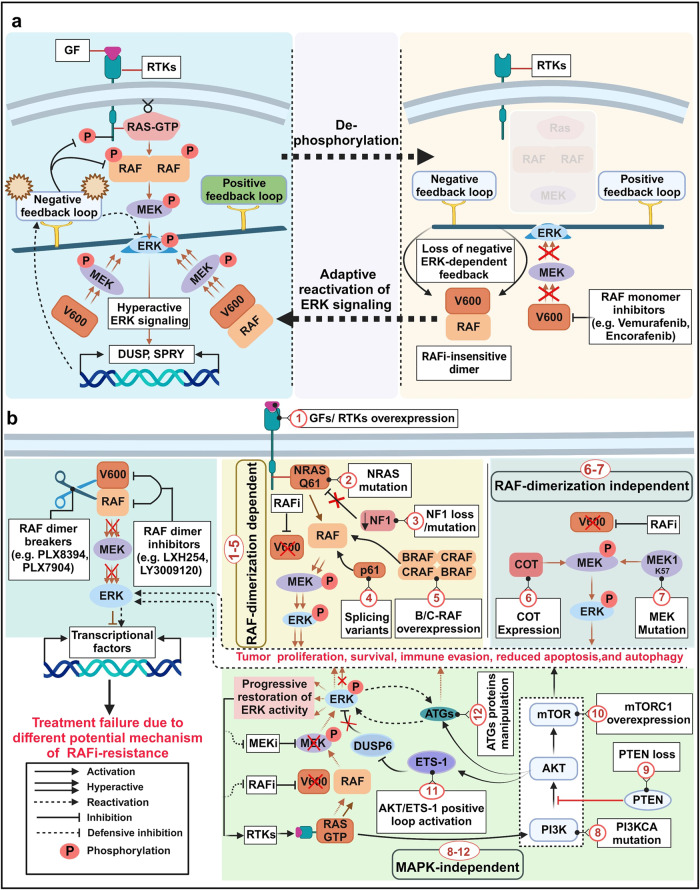
BRAF-mutant targeted therapy
RAF inhibitors (RAFi)
Mutations of RAF are frequently found in melanoma and other cancers. The prevailing mutation of RAF is BRAFV600E. Abnormal activation of the RAS/RAF/MAPK pathway, a hallmark of cancer, often originates from genetic alterations in RAF-encoding genes or RAF upstream genes. Consequently, the RAF kinase family is a promising target for potential cancer therapies. The RAF kinase activity is frequently disrupted in human cancer due to RAF itself or mutations affecting the upstream regulators and downstream effectors proteins as depicted in Fig. 7b.
PLX4720, a potent and selective RAF inhibitor, has demonstrated robust antitumor activity against RAF-mutant melanoma in vitro and in vivo.318 Vemurafenib (PLX4032), an analog of PLX4720, exhibits improved pharmacokinetic properties compared to PLX4720, and has received approval for the treatment of advanced melanomas and other cancers harboring RAF mutations.319,320 Vemurafenib has shown exceptional efficacy with manageable side effects, both as a monotherapy and when combined with MEK inhibitors, for patients with RAF mutant cancer. Dabrafenib is another effective therapeutic option for melanomas bearing the V600E BRAF mutation, achieving tumor shrinkage in over 90% of patients with half experiencing partial to complete responses.321,322 Furthermore, clinical phase III studies have demonstrated that Encorafenib, a BRAF inhibitor, either standalone treatment or in combination with MEK inhibitors, significantly prolongs progression-free survival compared to Vemurafenib.323 In previous studies involving patients with BRAFV600E metastatic melanoma, treatment of Vemurafenib, Dabrafenib, and Encorafenib resulted in dose-dependent tumor suppression and improved overall survival rates relative to other therapeutic approaches.324,325 Notably, patients with BRAF-mutant melanoma have displayed significant responses to BRAF inhibitors such as vemurafenib (PLX4032, RG7204) and Dabrafenib (GSK2118436), while those with BRAF wild-type melanoma have not shown similar responses.326
Despite the encouraging beneficial observed with RAF inhibitors, approximately half of the patients treated with RAFi experience cancer recurrence due to the development of resistance to the RAF inhibitors within 6–7 months of initiating treatment.327
MEK inhibitors (MEKi)
While activated MEK mutations are relatively rare in human tumors, mutations in upstream genes like RAS or RAF are implicated in over 85% of malignancies, often resulting in elevated MEK activity.328 Targeting MEK has thus emerged as a promising and cutting-edge therapeutic approach. Combinations of MEK and BRAF inhibitors have received FDA approval and have proven effective in inhibiting tumor growth in both preclinical models and patients with RAS or RAF mutations.329 One notable MEK inhibitor, Trametinib, was identified progression-free and overall survival in patients with BRAFV600E-mutated metastatic melanoma.330 Furthermore, combinations of Dabrafenib and Trametinib led to improved progression-free survival in patients with metastatic BRAFV600E-mutated melanoma.331 Cobimetinib (GDC-0973) is another highly selective allosteric MEKi that has demonstrated effectiveness in BRAFV600E/K-mutant patients as well as in cell lines with BRAF- or KRAS-mutations.332 The FDA approved Cobimetinib in combination with Vemurafenib for the treatment of BRAFV600E/K-mutant and unresectable melanoma in 2015.333 Binimetinib, also known as MEK162, ARRY-162, or ARRY-438162, is an effective oral MEKi approved for patients with BRAFV600E/K-mutant and unresectable melanoma. The FDA approved the combination therapy of Binimetinib and Encorafenib in 2018334. In 2020, Selumetinib (KOSELUGO, AstraZeneca), a highly specific MEKi, is approved for the treatment of pediatric children with neurofibromatosis type 1 (NP1) who have symptomatic, inoperable plexiform neurofibromas (PN).335 Selumetinib is also under evaluation for use in other cancers such as melanoma, gliomas, and non-small cell lung cancers, where MEK1/2 is overexpressed.336
Ongoing clinical studies on the combination of BRAFi and MEKi
In patients with BRAF mutated cancer, a molecular mechanism analysis has revealed potential target for RAS/RAF/MAPK inhibitors. This analysis has led to a significant shift in the treatment approach for patients with BRAF mutated cancer. It is believed that inhibiting both BRAF and MEK greatly can improve clinical outcomes for melanoma patients by slowing or preventing the development of resistance to BRAFi alone.337 In Table 3, we have compiled an update overview of clinical studies related to combination of BRAFi and MEKi based on information from the ClinicalTrials.go database.
Table 3.
Combinations of BRAFi and MEKi under clinical investigations
| Targeted therapy | Condition (s) | Phase | Participants | Estimated study completion | Current status/Other ID | NTC identifier |
|---|---|---|---|---|---|---|
| Vemurafenib + cobimetinib | BRAFV600E, PCP | II | 36 | August 2026 | Recruiting/NCI-2017-00740 | NCT03224767 |
| HM, melanoma, TC, PON, CRN, NSCLC, Gloma, MM, and ATC | II, III | 30 | September, 2029 | Recruiting/CRUKD/21/004 | NCT05768178 | |
| Melanoma | II | 24 | January 2024 | Active, not recruiting/ML28606 | NCT02036086 | |
| Encorafenib + binimetinib | Advanced BRAF-mutant cancers | I, II | 17 | September, 2023 | Completed, no result posted/18-547 | NCT03843775 |
| Solid tumor | II | 26 | December, 2023 | Recruiting/BEAVER-001 | NCT03839342 | |
| Locally advanced, metastatic, recurrent and stage III/IV PC AJCC v8 | II | 29 | December, 2024 | Active, not recruiting/NCI-2020-02972 | NCT04390243 | |
| NSCLC | II | 55 | March, 2025 | Recruiting/CTR20212962 | NCT05195632 | |
| HCL | II | 45 | July, 2024 | Recruiting/200076 | NCT04324112 | |
| NSCLC | II | 98 | June, 2024 | Active, not recruiting/ARRAY-818-202 | NCT03915951 | |
| Melanoma stage III, in-transit metastasis of CM | II | 28 | January, 2026 | NL77905.058.21 | NCT05767879 | |
| Relapsed or refractory MM with BRAFV600E/K mutation | II | 12 | December, 2022 | Active, not recruiting/CLGX818ADE01T | NCT02834364 | |
| Dabrafenib + trametinib | Melanoma, NSCL, solid tumor, rare cancers, and HGG | IV | 100 | December, 2027 | Recruiting/CDRB436X2X02B | NCT03340506 |
| Stage IIIB/C CM AJCC v7 | II | 66 | April, 2024 | Active, not recruiting/NCI-2014-01969 | NCT02231775 |
PCP papillary craniopharyngioma, HM hematologic malignancy, TC thyroid cancer, PON papillary ovarian neoplasms, CRN colorectal neoplasms, LNC laryngeal neoplasms carcinoma, NSCLC non-small-cell lung, MM multiple myeloma, ECD erdheim-chester disease, ATC anaplastic thyroid carcinoma, PC pancreatic carcinoma, HCL hairy cell leukemia
Vemurafenib and Cobimetinib
Hoffmann-La Roche/Genentech sponsored a Phase III clinical trial that employed a double-blind, placebo-controlled design to compare Vemurafenib alone with a combination of Vemurafenib and Cobimetinib (GDC-0973). This trial focused on patients who had not received prior treatment and were diagnosed with BRAFV600-mutattion-positive, unresectable locally advanced, or metastatic melanoma (NCT01689519). Although there was a slight increase in treatment-related side effects, the addition of Cobimetinib to Vemurafenib led to a significant improvement in progression-free survival (PSR) among patients with BRAFV600-mutated metastatic melanoma.337 Also, a recent clinical study demonstrated that the combination of Vemurafenib and Cobimetinib produced a partial response or better in 15 out of patients with papillary craniopharyngiomas, as part of a single-group study (NCT03224767). The average reduction in tumor size was 91%, and progression-free survival (PFS) was 87% at 12 months (95% CI, 57 to 98) and 58% at 24 months.338 Numerous ongoing studies are currently underway, with participants either receiving intervention or undergoing examination to assess the clinical and pathological responses to Vemurafenib and Cobimetinib in BRAF-positive cancers (NCT05768178, NCT02036086).
Encorafenib and Binimetinib
An observational study has been initiated to investigate the real-world effectiveness, impact on quality of life, safety, and tolerability of Encorafenib in combination with Binimetinib for the treatment of unresectable advanced or metastatic BRAF cancers in Germany, Austria and Switzerland (NCT04045691). Several clinical trials are currently underway, with participants either receiving an intervention or undergoing examination to assess the safety, tolerability and efficacy of combining Encorafenib with Binimetinib in patients with BRAFV600-mutant metastatic cancers, including pancreatic cancer (PC) (NCT04390243), NSCLC (NCT05195632, NCT03915951), hairy cell leukemia (HCL) (NCT04324112), multiple myeloma (MM) (NCT02834364) and melanoma (NCT05767879) Despite the significant improvement in survival seen with combination therapy involving of BRAFi and MEKi for BRAFV600-mutant melanomas, there remain limited options for targeted therapy in cases of BRAFnonV600-mutant melanomas. It has been observed that BRAFV600 mutations often co-occur with NF1 deletion, and that these mutations can signal as monomer and dimers when NF1 loss present.339 Consequently, in BRAFnonV600-mutant melanomas with co-occurring NF1 loss-of-function mutations, the combination of BRAFi targeting both monomeric and dimeric BRAF, along with MEK inhibition, has been shown to significantly reduce cell viability in vitro and tumor growth in vivo. Recently, two ongoing clinical studies are exploring whether patients with BRAFnonV600-mutant melanomas may benefit from the currently FDA-approved combination therapy of Encorafenib (BRAFi) and Bonimetinib (MEKi), which is reserved for BRAFnonV600-patients (NCT03839342, NCT03843775). These results are considered hypothesis-generating and will need further confirmation in larger clinical trials in the future.
Dabrafenib and Trametinib
Recent reports have shown that previously untreated individuals with metastatic melanoma and BRAFV600E/K mutations exhibited superior responses to the combination therapy of Dabrafenib and Trametinib compared to Dabrafenib alone.340 The randomized Phase III study funded by GlaxoSmithKline compared the efficacy of combination therapy involving Dabrafenib and Trametinib to that of administered alongside a placebo (Dabrafenib monotherapy) (NCT01584648). Moreover, the Novartis designed non-interventional study has initiated to evaluate the use of Dabrafenib in combination with Trametinib as adjuvant treatment for Chinese patients with stage III melanoma carrying the BRAFV600E mutation after complete resection (NCT04666272). Additionally, a managed access program (MAP) cohort clinical trial provides guidelines to physician for the treatment and monitoring of Trametinib/Dabrafenib, in eligible patients diagnosed with advanced NSCLC featuring the BRAFV600E/K-activating mutations (CTMT212X2002I, NCT04507919).
Assessment of a dual RAF/MEK inhibitor: VS-6766
A novel RAF/MEK inhibitor CH5126766/RO5126766 was discovered while screening for compounds that induce p27. It exhibits more prolonged inhibition of MAPK signaling compared to PD0325901, a well-known ATP-competitive MEK inhibitor.341 VS-6766, also known as CH5126766, is a first-in-class dual RAF-MEK inhibitor currently under investigation as a potential treatment for multiple myeloma and solid tumors harboring different RAS-RAF mutations. It targets two key nodes in the RAS/RAF/MAPK signaling pathway, demonstrating both safety and promising efficacy.342 Numerous interventional clinical trials are ongoing to evaluate the effectiveness of VS-6766, both as a standalone therapy and in combination regimens (Table 4). Notably, an intermittent dosage regimen of VS-6766 and Defactinib has shown clinical activity in patients with recurrent low-grade serous ovarian cancer (LGSOC). These findings have paved the way for an ongoing registration-directed study investigating combination of VS-6766 and Defactinib in patients with recurrent LGSOC (NCT04625270).
Table 4.
Ongoing interventional clinical trials of VS-6766 (Dual RAF/MEK inhibitor) for cancer therapy
| NTC identifier | Adjunctive agent (s) | Conditions | Phase/Participants | Study status/Estimated completion date |
|---|---|---|---|---|
| NCT00773526 | - | Neoplasms | Phase I/18 | Completed/Sep 2011 |
| NCT05187169 | - | Food effect | Phase I/52 | Completed/April 2022 |
| NCT03875820 | Defactinib | NSCLC, LGSOC, EEC, PC | Phase I/87 | Active, not recruiting/Oct 2023 |
| NCT05512208 | Defactinib | EEC, MOC, HGSOC, CC | Phase II/55 | Recruiting/Dec 2029 |
| NCT05787561 | Defactinib | MGC | Phase II/20 | Recruiting/March 2025 |
| NCT04625270 | Defactinib | OC, LGSOC, adenocarcinoma | Phase II/225 | Recruiting/Dec 2026 |
| NCT04620330 | Defactinib | NSCLC, KRAS mutation | Phase 2/100 | Recruiting/Dec 2025 |
| NCT02407509 | Everolimus | Solid tumors, MM, LC, OC | Phase II/104 | Recruiting/May 2024 |
| NCT04720417 | Defactinib | Metastatic UV | Phase II/18 | Recruiting/Sep 2023 |
| NCT05798507 | Defactinib | GBM | Early phase I/12 | Recruiting/Sep 2024 |
| NCT06007924 | Defactinib | TC | Phase II/30 | Recruiting/Aug 2027 |
| NCT05669482 | Defactinib, gemcitabine, nab-paclitaxel | KRAS mutation, PDAC | Phase I, II/40 | Recruiting/May 2025 |
| NCT05375994 | Adagrasib | NSCLC, KRAS mutation, LC, MND | Phase I, II/58 | Recruiting/July 2024 |
| NCT05608252 | Abemaciclib, fulvestrant | Met HR + /HER- BC | Phase I, II/63 | Recruiting/Dec 2027 |
| NCT05200442 | Cetuximab, Pill Diary | CRC, COADREAD, mCRC | Phase I, II/53 | Recruiting/April 2024 |
| NCT05074810 | Sotorasib | NSCLC, KRAS mutation | Phase I, II/53 | Recruiting/Dec 2023 |
NSCLC non-small cell lung cancer, LGSOC low-grade serous ovarian cancer, PC pancreatic cancer, EEC endometrioid cancer, MOC mucinous ovarian cancer, HGSOC high-grade serous ovarian cancer, CC cervical cancer, MM multiple myeloma, LC lung cancer, OC ovarian cancer, MND malignant neoplastic disease, MGC mesonephric gynecologic cancer, BrC Breast cancer, CRC colorectal cancer, mCRC metastatic colorectal cancer, COADREAD colorectal adenocarcinoma, LC lung cancer, Met HR+/HER− BC metastatic hormone receptor-positive breast cancer, PDAC pancreatic ductal adenocarcinoma, mUV metastatic uveal melanoma, GBM glioblastoma, TC thyroid cancer
Targeted therapy in pediatric BRAF-mutational gliomas
The BRAFV600E mutations gas been associated with reduced responsiveness to conventional treatment in pediatric low-grade gliomas (pLGG).343 Recent findings from the phase II TADPOLE trial suggest that targeted therapy focusing on the RAS/RAF/MAPK signaling is beneficial for pediatric brain tumors with BRAF mutations. In earlier clinical studies, Dabrafenib demonstrated significant clinical efficacy and tolerability, both as monotherapy and in combination with trametinib, for the treatment of pLGG with BRAFV600E mutations (NCT01677741). These results have spurred further exploration of this combination as a first-line therapy (NCT02684058). Notably, Dabrafenib plus Trametinib, when used as a first-line therapy, achieved significantly higher response rates, longer progression-free survival, and improved safety profiles compared to standard monotherapy in pLGG with BRAFV600E mutations.344 In this randomized study, the combination therapy of Dabrafenib and Trametinib showed an overall response rate (ORR) of 47%, while monotherapy yielded an ORR of only 11%. Furthermore, the Dabrafenib and Trametinib combination significantly extended median progression-free survival compared to chemotherapy (20.1 months vs. 7.4 months) and led to clinical improvement in 86% of patients, as opposed to 46% in the monotherapy group. Additionally, the study with BRAFV600E -mutant pediatric high-grade gliomas (pHGG) demonstrated that combining Dabrafenib with Trametinib exhibits an enhanced ORR of 56 %, associated with long-lasting responses and improved duration of response (22.2 months), overall survival (32.8 months), and along with a favorable safety profile, in relapsed/refractory BRAFV600-mutant pHGG patients. In 2023, the FDA granted its approval for the utilization of the Dabrafenib and Trametinib combination as the primary treatment for pLGG in children aged 1 year or older. This milestone marks the successful outcomes of the TADPOLE trials in the pLGG population. Moreover, in 2022, the FDA also sanctioned the use of this combination therapy for patients aged six and above who were dealing with relapsed/refractory BRAFV600 solid tumors. This additional approval serves to reinforce the positive results observed in the pHGG cohort.
Combining BRAF and MEK inhibitors with other targeted therapy
Combining BRAF and MEK inhibitors with other targeted therapies is currently under investigation, aiming to prolong tumor responses.345 These therapies are also gaining traction in neoadjuvant and adjuvant settings (see Table 5).
Table 5.
Combinations of other targeted therapy along with RAF/MAPK inhibitors under clinical investigations
| Targeted therapy | Condition (s) | Phase | Participants | Estimated study completion | Current status/Other ID | NTC identifier |
|---|---|---|---|---|---|---|
| PF-07284890, binimetinib, midazolam | Malignant melanoma carcinoma, NSCLC, brain neoplasms, malignant neoplasms | I | 57 | November 2023 | Active, not recruiting/C4471001 | NCT04543188 |
| PF-07799933, binimetinib, cetuximab | Melanoma, CRC, NSCLC, TC, Glioma | I | 174 | January 2028 | Recruiting/C4761001 | NCT05355701 |
| Encorafenib, PF07799544, PF07284890, PF07799933, | Melanoma, glioma, TC, NSCLC, malignant neoplasms, brain neoplasms, CRC | I | 124 | March 2027 | Recruiting/C4901001 | NCT05538130 |
| Spartalizumab, dabrafenib trametinib | Melanoma | III | 568 | December 2023 | Active, not recruiting/CPDR001F2301 | NCT02967692 |
| Encorafenib, nivolumab, ipilimumab, binimetinib | Melanoma | I, II | 2 | August 2023 | Active, not recruiting/HCC 20-190 | NCT04655157 |
| Dabrafenib, trametinib pembrolizumab | Melanoma | II | 60 | November 2024 | Active, not recruiting/MIA2015/179 | NCT02858921 |
| Dabrafenib, trametinib, Pembrolizumab | TGAC, TGSCC | II | 30 | June 2024 | Recruiting/NCI-2020-09803 | NCT04675710 |
| Encorafenib, Binimetinib, Nivolumab | Melanoma, skin cancer | I | 13 | May 2028 | Active, not recruiting/MCC-19441 | NCT03543969 |
| Dabrafenib trametinib, nilotinib | Metastatic melanoma, BRAF mutation | I | 15 | March 2027 | Recruiting/MCC-20-MEL-11-PMC | NCT04903119 |
| Vemurafenib, HL-085 | Solid tumor | I | 45 | December 2023 | Recruiting/HL-085-102 | NCT03781219 |
| ABM-1310, cobimetinib | Advanced solid tumor, BRAFV600 mutation | I | 112 | January 2025 | Recruiting/ ABM1310X1101 | NCT04190628 |
| Mirdametinib, BGB-3245 | Advanced solid tumor | I, II | 136 | June 2027 | Recruiting/MEKRAF-AST-101 | NCT05580770 |
| XL888, Vemurafenib, Cobimetinib | Melanoma, skin cancer | I | 26 | October 2023 | Active, not recruiting/MCC-18597 | NCT02721459 |
| AT13387, Dabrafenib, trametinib | Clinical and pathological stage III, IIIa/b/c/d CM AJCC v6/7 | I | 22 | March 2019- | Active, not recruiting/NCI-2014-00615 | NCT02097225 |
| Trametinib, CDK4/6 | Melanoma | II | 1000 | December 2028 | Recruiting/MIA2015/174 | NCT02645149 |
| Vemurafenib, tiragolumab atezolizumab, cobimetinib, | Clinical and pathological stage III, IIIa/b/c/d CM AJCC v8 | I | 30 | June 2024 | Recruiting NCI-2018-01018 | NCT03554083 |
| Dabrafenib, trametinib, PDR001 | mCRC | II | 25 | December 2022 | Recruiting/18-144 | NCT03668431 |
| Dabrafenib, Trametinib, HCQ | Brain LGG and HGG with BRAF aberration and brain LGG with NP1 aberration | I, II | 75 | June 2027 | Recruiting/PBTC-055 | NCT04201457 |
| Dabrafenib, tazemetostat, trametinib | Clinical stage IV CM AJCC v8,Metastatic melanoma | I, II | 58 | December 2024 | Recruiting/NCI-2020-07044 | NCT04557956 |
| Dabrafenib, trametinib, radiation | AA, AGG, APXA, GBM, malignant grade 3 glioma | II | 58 | September 2027 | Recruiting/NCI-2019-02289 | NCT03919071 |
CM cutaneous melanoma, AJCC v8 American joint committee on cancer version 8, mCRC metastatic colorectal cancer, LGG low grade glioma, HGG high grade glioma, NP1 neurofibromatosis type 1, AA anaplastic astrocytoma, AGG anaplastic ganglioglioma, GBM glioblastoma, HCQ hydroxychloroquine, mCRC, metastatic colorectal cancer, APXA anaplastic pleomorphic xanthoastrocytoma, CRC colorectal cancer, NSCLC non-small cancer lung cell, TC thyroid cancer, TGAC thyroid gland anaplastic carcinoma, TGSCC thyroid gland squamous cell carcinoma
Patients with BRAFV600-mutant malignancies are benefiting from RAF/MEKi inhibitor therapy. However, long-term efficacy is limited by disease progression in the brain due to inadequate pharmacokinetics (PK) and pharmacodynamics (PD).346,347 Researchers are working on enhancing the intrinsic properties of drugs, such as their PK and PD, to increase the effectiveness of RAS/RAF/MAPK inhibitors. One promising molecule is PF-07284890 (BRAFi), a potent brain-penetrant compound, currently undergoing evaluation in a first-in-human trial for patients (NCT05538130, NCT04543188).
Similar to BRAF/MEKi, combining adjuvant therapy together with specific checkpoint inhibitors, such as anti-PD-1 (Nivolumab and Pembrolizumab) or anti-CTLA-4 (Ipilimumab), has shown promising results. It leads to extended recurrence-free survival and a reduced occurrence of severe adverse events in patients who have undergone resection for stage IIIB, IIIC, or IV melanoma (NCT02388906, NCT04949113, NCT01972347). Additionally, the combination of these immune checkpoint inhibitors with BRAF/MEK-targeted medications appears to be a more viable treatment option due to their immunomodulatory action (NCT02967692, NCT04655157, NCT02858921). Furthermore, a phase I trial study has demonstrated the potential synergy between anti-PD-1 inhibitor and other BRAF/MEK inhibitors in treating patients with BRAF-mutant stage IIIC-IV melanoma (NCT03543969). Meanwhile, a phase II trial is evaluating the impact of Pembrolizumab, Dabrafenib, and Trametinib as a neoadjuvant therapy for patients with BRAFV600E-mutated anaplastic thyroid cancer (ATC) before surgical intervention (NCT04675710). An ongoing neoadjuvant phase II study of Dabrafenib, Trametinib and/or Pembrolizumab in BRAFV600-mutant respectable stage IIIB/C melanoma is investigating the effect of Pembrolizumab, Dabrafenib, and/or Trametinib in reducing tumor sizes before surgery and preventing melanoma recurrence after surgery in patients with BRAFV600-mutant respectable stage IIIB/C melanoma (NCT02858921).
However, a randomized phase II trial, advanced melanoma patients with BRAFV600E/K mutations who received a combination of Dabrafenib (BRAFi), Trametinib (MEKi), and Pembrolizumab (a PD-1-blocking antibody) displayed numerically longer progression-free survival (PFR) and duration of response but experienced with a higher rate of grade 3/4 adverse events compared to those treated with the doublet therapy of Dabrafenib and Trametinib along with a placebo (NCT02130466).
In addition to the established interventions using BRAF and MEK inhibitors, HSP90 inhibitors for patients with the BRAFV600-mutant melanoma are under investigation due to their manageable side-effect profiles when combined with BRAFi.348 HSP90 inhibitors, such as AT13387 or XL888, offer a promising strategy to combat drug resistance resulting from BRAF and MEK inhibition in melanomas.348 Clinical trials investigating the combination of BRAF/MEKi with XL888 or AT13387 are currently underway (NCT02721459, NCT02097225). Moreover, targeting the CDK4 pathway has emerged as a therapeutic approach for patients with BRAF-mutant metastatic melanomas.349 A patient-centered translational study has shown improved clinical outcomes for individuals with stage III or stage IV metastatic BRAF and NRAS wild-type melanoma who do not respond to standard therapy, typically immunotherapy. Lastly, there are ongoing clinical trials exploring various combinations with BRAF/MEKi, which information can be found in Table 5, provided by ClinicalTrials.gov.
RAFi resistance
The discovery of small-molecule BRAF inhibitors was the start of a revolution in the treatment of advanced melanoma.324,350 Some BRAF inhibitors have demonstrated promising results for the treatment of patients with melanoma featuring the BRAFV600E mutation. Despite the exceedingly positive effects of these inhibitors, the development of drug resistance is a widely occurring phenomenon in patients with cancer.16,351 RAFi resistance can be characterized as either intrinsic acquired or adaptive.14
Intrinsic RAFi resistance
Although cancers featuring activating BRAF mutations respond well to RAFi, a considerable fraction of melanoma cells harboring the BRAF V600E mutation show inherent resistance to RAFi.8 BRAFV600E–mutant tumors do not sensitize to either Vemurafenib or Trametinib due to high cyclin D1 and YAP1 expression.8,352–354 Activation of phosphatase and tensin homolog (PTEN), forkhead box O3, AKT, or insulin-like growth factor 1 (IGF-1) generally leads to the development of intrinsic RAFi resistance due to the suppression of apoptosis mediated by BCL2-interacting mediator of cell death.355–358 Intrinsic RAFi resistance represents an adaptation mediated by transcriptional and epigenetic rewiring.359 Relief from the human EGFR 3 feedback inhibition mediated by RAFis and MEK inhibitors reduces the anticancer effects of these treatments against BRAF-mutant thyroid carcinomas.360 Human growth factor (HGF) released by stromal cells activates the HGF receptor (MET), reactivating the MAPK and PI3K–AKT signaling pathways, resulting in the rapid development of RAFi resistance.361
Acquired RAFi resistance
Acquired RAFi resistance generally develops due to the competitive burden introduced by drug treatment, which results in diverse genetic abnormalities or the acquisition of therapy-induced de novo cellular epigenetic and transcriptional reprogramming events.362 In nearly all cases, acquired drug resistance is linked to secondary mutations in RAF, leading to the activation of parallel signaling pathways (such as RTKs) and the “on-target” accumulation of mutations in the RAF pathway.363–368 These secondary mutations often occur at “gatekeeper” residues within a RAF ATP-binding site, preventing inhibitors from binding to the existing hydrophobic pocket of the ATP-binding domain. A BRAF mutation at Thr259 confers resistance to some anticancer reagents (e.g., SB590885 and PLX4720), although intrinsic kinase activity is maintained.363 Acquired resistance to Vemurafenib is related to the increased expression of several RTKs, especially PDGF receptor. In addition, CRAF overexpression leads to acquired resistance to BRAF inhibition mediated by a RAF kinase switch in melanoma.368,369 MAP3K8 (COT) reactivates ERK in some melanoma samples that are resistant to RAFis and MEK inhibitors.370,371
Adaptive RAFi resistance
Adaptive resistance to RAFi arises from internal compensating mechanisms, often termed adaptive responses or feedback loops. These mechanisms enhance the survival and proliferation potential of a subset of the initial tumor population, ultimately promoting tumor growth. Such adaptive RAFi resistances can manifest in two main forms: non-transcriptional adaptive response, which regulate post-translational modifications of upstream kinases, and transcriptionally mediated adaptive responses. Many of these inherent resistance mechanisms are primarily underscored by the reactivation of the MAPK signaling pathway, consequently, reduces patient responses. Notably, the reactivation of MAPK via paradoxical signaling (as described in section 4.3) accounts for a significant portion of adaptive mechanisms, contributing to the diminished sensitivity to pathway-targeted drugs and overall patient responses.372
RAFi adaptation to RAF dimerization: feedback inhibition and paradoxical activation in RAF-mutant cancers
Aberrant ERK1/2 signaling can lead to oncogenicity, depending on the extent of ERK1/2 activation373. ERK dysregulation is associated with cell death and senescence,374,375 and the feedback mechanisms in ERK signaling are closely connected to cellular dysfunction.376 The negative feedback of ERK signaling is regulated directly via ERK pathway phosphorylation376 or indirectly through de novo gene expression, such as those encoding dual-specificity phosphatases (DUSPs) and Sprouty (SPRY).377,378 ERK-mediated feedback regulation directly targets RAF proteins, reducing dimerization.379 RAFi resistance reactivates ERK signaling and induces RAFi insensitivity via mechanisms that bypass RAF-dependent signaling.322 ERK reactivation is very subtlety regulated at multiple levels via “intra-pathway” feedback loops and mediated by phosphorylation, intracellular localization, and complex formation (Fig. 8a).380. Under normal conditions, RAS-activated RAF dimerizes and phosphorylates the downstream effectors MEK and ERK. Interestingly, ERK also activates other genes (e.g., DUSPs and SPRY) that inhibit the ERK pathway via a negative feedback loop, resulting in RAS dephosphorylation, which weakens the signal.14,15,378,381–383 The network of negative feedback loops limits ERK activation driven by the direct phosphorylation of components in the MAPK cascade.384–386 In BRAF-mutant cancers, the MEK–ERK axis is constitutively active, independent of RAS-mediated RAF dimerization, and low levels of basal RAS activation combined with direct phosphorylation result in the strong activation of negative feedback regulators (SPRY and DUSP), inhibiting signaling intermediates, such as EGFR and SOS. As a result, signaling is hampered downstream of activated receptors in BRAFV600E–mutant cells. Treatment with Vemurafenib, a first‐generation ATP-competitive RAFi, results in a temporary reduction in ERK activity,324 which rapidly recovers in BRAF-mutant cancer cells,387 possibly due to the inhibition of ERK-dependent negative feedback.15,388 The restoration of RTK signaling increases RAS-GTP levels, followed by an increase in the homo- or heterodimerization of RAF, which paradoxically reactivates ERK signaling in the presence of RAFis.389 This paradoxical activation of RAFi-insensitive BRAFV600E–mutant dimers result in the formation of a new steady-state signaling network, even in the presence of RAFis.389,390 Thus, RAFi are ineffective against these tumors due to the development of constitutive BRAF-mutant dimers or the transactivation of BRAF–CRAF heterodimers.389,391
Fig. 8.
Mechanistic basis of RAF inhibitor (RAFi) resistance. a RAFi resistance in RAF-mutant cancer. BRAFV600E–mutant cancers are characterized by hyperactive extracellular signal-related kinase (ERK) signaling during early disease, resulting in negative feedback inhibition of upstream signaling. RAS-GTP expression is minimal, but monomeric BRAFV600E responds to signals. Under these conditions, RAFi inhibits ERK signaling. Subsequent treatment with Vemurafenib suppresses ERK-dependent negative feedback, restoring receptor tyrosine kinase (RTK) signaling. Despite the presence of RAFi, RTK signal restoration elevates RAS-GTP levels, drives the formation of RAFi-insensitive RAF dimers, and reactivates ERK signaling. Over time, negative feedback pathways are partially restored, and a new steady-state condition featuring reactivated ERK signaling develops. b RAFi resistance mechanisms. RAFi resistance promotes significant RAF dimerization through growth factors or RTK activation, NRAS mutation (NRAS Q61), NF1 loss, expression of RAF splice variants (p61), or overexpression of BRAF or CRAF. Reactivation of ERK signaling and RAFi resistance can also develop in a RAF dimerization–independent manner involving MEK mutations or RAF bypass activation by COT (an ERK kinase kinase). This figure was created with BioRender.com
New RAFi families, such as pan-RAF dimer inhibitors and paradox breakers (dimer breakers), appear to be more effective therapeutic options than previous monomeric-targeting RAFis in tumors with RAF dimer–induced signals (Fig. 8b).392–394 pan-RAF dimer inhibitors (e.g., LXH254 and LY3009120) bind directly with RAF dimeric domains and inhibit RAF dimerization in BRAFV600E–mutant tumors. However, RAS mutation or amplification, BRAFV600E amplification, and intragenic deletion or BRAF splice variants can lead to RAFi resistance in patients with BRAFV600E–mutant tumors.390,395,396 Importantly, these inhibitors target both active RAF dimers and monomers to block ERK signaling in cancer cells.397 Most pan-RAF inhibitors (e.g., AZ628, Belvarafenib, CCT196969, CCT241161, LY3009120, and TAK-580 [MLN2480]) are successful when tested using in vitro cellular assays.398,399 However, the use of these inhibitors in patients has been limited by a lack of selectivity for mutant BRAF.400 Although LY3009120 exhibits antitumor effects in patients with mutant RAF dimers, persistent treatment with this drug causes severe resistance due to BRAFV600E dimerization.390 A limited dose escalation has been examined to determine the maximally tolerated dose in patients with RAS-mutant cancer, indicating the potential of this drug as an anticancer therapy.400 LXH254, an inhibitor of RAF dimerization, shows specific paralog selectivity, preventing dimeric formation of BRAF and CRAF but not ARAF.397 These drugs are unable to suppress MAPK signaling in normal cells because they inhibit mutant RAF dimers and monomers at great potency compared with their effects on wild-type RAF dimers. As a result, V600E–mutant BRAF that dimerizes with other RAF mutants or with wild-type BRAF or CRAF exhibit drug resistance due to the unintended paradoxical activation of ERK,322,389,393 which may also be responsible for the elevated toxicity and disruption in RAF signaling observed in non-cancer cells. As a result, these medications prove ineffectual when confronted with tumors carrying BRAFnonV600E-mutations. Additionally, an increase in the expression of BRAFV600E -dimers can lead to acquire resistance, primarily because of unintentional paradoxical activation of ERK.390 This paradoxical activation might also be accountable for heightened toxicity and disturbances in RAF signaling, which are observable in non-cancer cells.401
In addition to pan-RAF dimer inhibitors, paradox breakers (e.g., PLX8394 and PLX7904) disrupt homo- and heterodimers that contain BRAF, resulting in reduced paradoxical ERK activation in cells expressing both mutant and wild-type RAF.394,402 These inhibitors selectively bind BRAF to suppress ERK1/2 in BRAFV600E–mutant cells without inducing the paradoxical activation of ERK1/2 observed in RAS-mutant cells. PLX8394 specifically inhibits BRAF in colon adenocarcinoma cells, preventing the paradoxical activation of the MAPK pathway403 with greater potency than Encorafenib, a selective inhibitor of active BRAF monomers, in BRAF-mutant colorectal cancer.404 However, PLX8394 deteriorates ERK signaling via the specific dissociation of BRAF–containing dimers (BRAF homodimers and BRAF–CRAF heterodimers) but has no effect on CRAF homodimers or dimers containing ARAF due to differences in the amino acid residues within the N-terminal region of the kinase domain among RAF isoforms.402 In addition, PLX8394 activates RAF signaling via BRAF dimerization in several melanoma cells.405 RTK-induced mammalian target of rapamycin (mTOR) activity and other bypass signaling may be able to compensate for decreased p-ERK levels when using paradox breakers.406
The ability of RAF-mutant tumors to adapt to RAFi is primarily due to an increase in RAF dimerization (Fig. 8b, RAF dimerization–dependent).106 Mutant RAF dimerization is often associated with RAFi resistance. An inhibitory bypass mechanism can also result in resistance to RAFi, via RAF-independent ERK reactivation (Fig. 8b, RAF dimerization–independent). For example, mutations in the MEK kinase COT (MAP3K8) and MEK1 (encoded by MAP2K1) are closely related to inhibitory bypass resistance that develops in response to RAFi.371,407 RAFi-resistant tumor cells show the rapid recovery of MAPK pathway activation, allowing escape from RAFi therapy; therefore, the total blockade of the entire pathway is essential for stimulating apoptosis in RAF-mutant cancers.408 The combination of RAFi together with other downstream inhibitors (e.g., MEK inhibitors) can maximize MAPK pathway inhibition and minimize cancer resistance. Compared with typical single-agent chemotherapy, Trametinib, a MEK inhibitor, enhances progression-free and overall survival rates in patients with metastatic melanoma who harbor a BRAFV600E/V600K mutations.19 However, BRAFV600E–mutant tumors still can develop resistance to combination RAFi and MEK inhibitor therapy over the course of months to years.409 Most patients who are treated with RAFi and MEK inhibitor combination therapy develop mild (Grade 1–2) toxicity in response to these drugs.22,410,411 Therefore, a solution to the underlying toxicity induced by RAFi is necessary to develop effective therapies for RAF-mutant malignancies.
Strategies to overcome chemoresistance related to targeting the RAS/RAF/MAPK pathway
The hyperactivation of the RAS/RAF/MAPK signaling pathway plays a significant role in the development of numerous human malignancies. While current targeted therapies like BRAFi and MEKi show varying levels of effectiveness across different cancer types, s substantial number of patients develop resistance relatively quickly.412 Ongoing cancer research is now actively addressing the challenge of countering both intrinsic and acquired drug resistance to small-molecule RAF inhibitors. Strategies involving combination therapies are been exploring as potential approaches to address RAF inhibitor resistance.413
Utilizing existing combination therapeutic strategies against RAFi resistance
The RAS/RAF/MAPK inhibitors currently in use have been through clinical development as combination therapies. They can either vertically target specific proteins within the MAPK pathway or horizontally inhibit multiple signaling pathways.414
Vertical pathway inhibition
BRAF-targeted single-agent therapy frequently results in the development of drug resistance due to MAPK pathway reactivation. The vertical blockade of MAPK signaling mediated by RAFi may be supplemented by other agents targeting the intermittent pathway (RTK–RAF–MEK–ERK).415–417 Combination therapy has become an important therapeutic goal in class-specific polytherapy.417,418 A vertical blockade combining the RAFi Dabrafenib with the EGFR inhibitor Panitumumab and the MEK inhibitor Trametinib is able to greatly suppress MAPK signaling, demonstrating improved therapeutic efficacy in metastatic colon cancer compared with other therapies.419,420 Although several therapeutic approaches using various medication doses and regimens can extend therapeutic responses, these strategies can also accelerate the development of resistance mechanisms. Continuous drug-related toxicity is also a risk, and the intermittent targeting of mutant BRAF can lead to the development of new drug-resistant tumors.418,421–423 As Vemurafenib-resistant cancer cells are reliant on the presence of the drug, withdrawal of drug treatment could be sufficient to induce tumor regression.418,422 Although the development of resistance to multiple inhibitors targeting the MAPK pathway remains to be addressed, this drug combination approach improves patient outcomes.424,425
Horizontal pathway inhibition
Targeting multiple signaling pathways simultaneously, known as horizontal pathway inhibition, can be an effective strategy to counteract potential compensatory or escape mechanisms that selective pathway inhibition might trigger. Cancer cells develop heterogenous resistance mechanisms to evade various drugs, but most of these mechanisms involve the reactivation of MEK–ERK signaling, combined with enhanced signaling output via the PI3K–AKT–mTOR and other pathways.426 Therefore, understanding the MAPK-related mechanisms underlying the development of resistance to RAFi and combination therapies may improve their potential therapeutic effects.427,428 Reactivation of ERK signaling can also occur via MAPK-independent pathways, including the bypassing of the paradoxical MAPK arm through PI3KC mutations,429,430 activation of an AKT–ETS-1–mediated positive feedback loop,431 loss of PTEN activity432 overexpression of mTOR,433 or manipulation of autophagy proteins (ATGs)434,435 (Fig. 8b, MAPK-independent). The combined inhibition of both the RAF–MEK and PI3K–AKT–mTOR pathways has a synergistic pro-apoptotic effect. For example, the addition of an AKT or mTOR inhibitor to Vemurafenib therapy or MEK inhibitor therapy results in synergistic effects.436 SHP2 inhibitors efficiently bypass the paradoxical MAPK signaling arm in RAS-mutant tumors treated with RAFi.437,438
The sequential or intermittent dosing of BRAF/MEK inhibitors
Patients with metastatic melanoma harboring activating BRAF mutations have demonstrated improved survival when receiving combination therapy with both BRAFi and MEKi.439 Unfortunately, the therapeutic effectiveness of such treatments often proves transient, with resistance emerging within a few months.440 Intermittent therapy has been explored as a strategy to halt or delay resistance to BRAF inhibition.422 In pursuit of curation approach for melanoma patients with BRAF mutations, researchers have proposed that modifying dosing patterns could prevent the development of lethal drug resistance and extend the durability of BRAFi responses.441 The optional implementation of this strategy in treating advanced BRAFV600E-mutant melanoma patients is currently under investigation in randomized phase II and III clinical trials comparing intermittent and continuous dosing schedules of BRAFi in combination with MEKi (NCT02224781, NCT02196181). However, one completed trial did not need its primary objective of improving progression-free survival, indicating that the experimental intermittent schedule of both Vemurafenib and Cobimetinib did not demonstrate superior anticancer activity compared to the regular continuous schedule (NCT02583516).
Potential alternative strategies for further optimizing the use of RAF/MEKi
The potential of different combination therapies is being explored to delay the emergence of resistance to MAPKi or to precisely target BRAF/MEKi-resistance. Recent preclinical research has identified alternative strategies for combating resistance, including bolstering apoptosis, modulating autophagy, and directing attention on mitochondrial metabolism and phenotyping switch (Fig. 9a).
Fig. 9.
The possible therapeutic strategies for overcome resistance to RAS/RAF/MAPK inhibitors. a Alternative approaches to overcome MAPK inhibitor (RAFi) resistance. b Targeting autophagy. In cancer cells, autophagy-related genes are functionally and physically associated with mitogen-activated protein kinase (MAPK)-targeted therapy and cancer resistance induced by MAPK signaling inhibitors (e.g., RAFi, MEKi). The regulatory mechanisms involved in autophagy induction and the mediators that regulate autophagy are described in each rectangle. Arrows and bars indicate stimulating and inhibiting signals, respectively. This figure was created with BioRender.com
Targeting mitochondrial and energy metabolism
Metabolic disorders have long been associated with cancers, characterized by metabolic reprogramming, which involves alterations in metabolic pathways enabling cancer cells to proliferate rapidly, survive in hypoxia and nutrient-deprived conditions, and evade the immune system.442 Furthermore, metabolic reprogramming events play a crucial role in tumorigenesis, drug resistance, and metastases in melanomas.443 Research on chemotherapy-resistant, slow-cycling BRAFV600E melanomas has shown increased level of oxidative phosphorylation (OXPHOS) enzymes and blocking them resulted in cell death. Additionally, the pivotal role of the microphthalmia-associated transcription factor (MITF) and PGC1A in regulating phenotyping plasticity is indispensable for steering cellular metabolism towards OXPHOS. A two-tiered strategy involves combining anticancer drugs that target rapidly proliferating melanoma cells with drugs that suppress the slow-cycling, drug-resistant subpopulation. Several drugs targeting mitochondrial respiration have been investigated in preclinical settings with promising results.444 For instances, β-sitosterol, due to its favorable tolerability, excellent bioavailability, and ability to inhibit mitochondrial respiration, serves as a viable adjuvant to BRAFi therapy for individuals with or at risk for melanoma brain metastases. Combining Vemurafenib with either β-sitosterol or a functional knockdown of mitochondrial complex I completely eliminated BRAFi resistance.444 Also, uncouplers like SR4 and Niclosamide that disrupt mitochondrial OXPHOS may be effective as first-line adjuvant treatments for melanoma patients who are not responding to MAPK inhibitors.445
Cancer starvation therapy, which relies on glucose restriction to induce oxidative stress and slow tumor growth has proven effective in curbing the rapid multiplication of cancer cells.446 The glucose analog 2-Deoxy-D-glucose (2-DG), when used in combination with Cisplatin or Erlotinib, enhances the cytotoxicity of head and neck squamous cell carcinoma (HNSCC) cells through metabolic oxidative stress. While 2-DG treatment alone may not cause significant cell death in most cancer cells, it does sensitize them to oxidative stress induced by radiotherapy or chemotherapy. Using 2-DG as an alternative therapeutic approach for the treatment of radioresistant and highly glycolytic cervical malignancies involves inhibiting intracellular redox metabolism and glycolysis.447 Also, some transcriptional factors, including hypoxia-inducible factor-1 (HIF1), MYC, and MONDOA (MLXIP), involve the regulation of glycolysis by inhibiting BRAF in melanomas, and further the combined inhibition of BRAF and glycolysis triggers cell death in BRAFi-resistant melanoma cells.448 Although there is currently no clinical evidence of this, targeting glucose metabolism could potentially be adapted therapeutically to overcome BRAFi resistance, particularly in pancreatic cancer driven by KRAS-dependent resistance to MAPK inhibition.449
Therapeutic modulation of metabolic difference in lipid metabolism can be a promising strategy to overcome BRAFi resistance. AMP-activated protein kinase (AMPK), a nutritional sensor present in both healthy and malignant cells, regulates lipid breakdown, phosphorylated, and inactivates acetyl-CoA carboxylase, preventing fatty acid production, and promotes lipid utilization through beta-oxidation in mitochondria.450 AMPK-dependent phosphorylation of BRAF at serine 729 reduces MAPK signal in BRAF wild-type cells by Inhibiting BRAF interaction with CRAF and KSR1.451 A study elucidated how 14-3-3 controls dimerization-driven RAF activation and mitigate the negative side effects of RAF inhibitors in cancer therapy.452 Additionally, the combination of Phenformin and PLX4720 resulted in tumor regression in some mice models. This study suggests that combining AMPK activators, such as Phenformin, with BRAF inhibitors may offer significant therapeutic advantages in melanoma treatment.
Targeting phenotyping switching
Phenotypic switching and metabolic reprogramming play distinct roles in the development of resistance in therapy-resistant clones of BRAFV600-mutated melanoma cells.453 The significance of MITF, a primary transcription factor in melanocyte differentiation/dedifferentiation, has been emphasized in relation to phenotypic switching. It has been observed that drug-sensitive cell lines and patient biopsies typically exhibit high MITF levels, whereas inherently resistant cell lines and patient biopsies demonstrated low MITF expression but elevated levels of NF-kB signaling and the receptor tyrosine kinase AXL.454 These MITF-low/NF-kB-high melanomas display resistance to ERK, RAF, and MEK inhibition in vitro. Remarkably, MITF expression show an inverse correlation with AXL expression, suggesting that cell lines can be categorized based on their metastatic potential regardless of whether they carry the BRAF or NRAS mutations.455 Additionally, Wnt5a signaling directly influences the motility and invasion of metastatic melanoma cells with low MITF levels.456 The phenomenon of EMT observed in cancer cells bears a striking resemblance to phenotypic switching.457 Exploring phenotypic switching may be essential in developing novel therapeutics capable of restoring sensitivity to RAFi or MEKi, providing a critical avenue for overcoming resistance.
Targeting the PI3K/AKT, tumor microenvironment and inflammatory responses
Resistance commonly develops in the majority of advancing melanomas through the reactivation of MAPK signaling, often driven by alterations in BRAF, NRAS, and MEK.458 While the activation of the PI3K/AKT pathway does not limit patient responses to BRAF/MEK inhibition, a small subset of resistant melanoma relies on the compensatory PI3K/AKT signaling cascade.459 Initial clinical trials with PI3K inhibitors in combination with Vemurafenib have shown promising results.460 In challenging-to-treat BRAF-mutant mCRC patients, a combination therapy comprising Alpelisib (a PI3K inhibitor), Encorafenib, and Cetuximab (an anti-EGFR) has demonstrated effectiveness.461 Notably, in BRAFV600E-mutated CRC cancers, activation of the PI3K/AKT pathway serves as a mechanism for both innate and acquired resistance to BRAF inhibitors, with proposed combination approaches to improve outcomes in this challenging patient population.462 PTEN, a major antagonist PI3K, is frequently mutated in various cancer tissues and is implicated in 17% of melanoma cases, 10% of colorectal cancer cases, and 4% of lung adenocarcinoma cases.463 Mutations in PTEN, resulting loss of function, are associated with a higher proportion of BRAF-mutant alleles, and are linked to lower progression-free survival (PFS), overall survival (OS), and response rates in melanoma patients. PTEN loss induces BRAFi-resistance in melanoma cells by inhibiting BIM expression.355 The involvement of AKT3 and the activation of FOXO3a play a role in enhancing apoptosis when PLX4720 and a PI3K inhibitor are combined to treat PTEN-negative cells.355 Importantly, HSP90 inhibition appears to be more effective in restoring BIM expression and downregulating Mcl-1 expression compared to the combined MEK/PI3K inhibitor therapy, making it a potentially highly effective strategy for managing the diverse array of resistance mechanism observed in BRAF resistance.464 AEBP1 (adipocyte enhancer-binding protein 1) is significantly upregulated in melanoma cells resistant to PLX4032 due to over-activation of the PI3K/AKT/CREB signal pathway. Blockage of this signaling pathway effectively restores the PLX4032-resistant phenotype of melanoma cells.
The development of resistance to BRAFi resistance in melanoma is a well-recognized challenge, stemming not only from genomic or epigenetic aberrations but also from the crucial role played by the tumor microenvironment. In response to BRAF inhibition, melanoma cells and fibroblasts undergo microenvironmental changes that enhance PI3K/AKT survival signaling, allowing tumor cells to evade treatment.465 When treated with Emurafenib, melanoma cells release TGF-β, triggering fibroblasts to increase expression levels of α-smooth muscle actin (α-SMA), neuregulin (NRG), and fibronectin. Paradoxically, Vemurafenib’s off-target effects lead to the secretion of hepatocyte growth factor (HGF) by fibroblasts. Furthermore, tumor-associated macrophages (TAMs) contribute to the development of a pro-tumorigenic microenvironment that fosters resistance to MAPKi therapy by providing an abundance of oxygen, nutrients (resulting in hypoxia and metabolic stress), and extracellular matrix proteins.466 Subsequently, TAMs secrete angiotensin, COX-2, IFN, and IL-1, further promoting the growth and metastasis of melanoma.467 Thus, to enhance the effectiveness of combination therapy for melanoma patients, targeting spatiotemporal interactions within tumor microenvironments holds significant promise.
Inflammatory signals establish a novel epigenetic program that silences a specific set of genes contributing to inflammation-induced cellular transformation in tumor cells.468 Tumor-induced inflammatory responses have adverse effects on the adaptive immune system and open up possibilities for new therapeutic strategies address the immunological dysfunctions caused by tumors.469 A novel independent predictor of Anthracycline/Taxane neoadjuvant chemotherapy response in breast cancer is the presence of tumor-associated lymphocytes, aiding healthcare professionals in identifying patients who will benefit most from this treatment.470 Pharmacological targeting of key factors derived from tumor-associated inflammation presents a unique strategy to eliminate therapy-resistant tumors. Celecoxib, a COX-2 inhibitor, significantly reduced tumor burden by 90% and notably delay tumor growth induced by the BRAFi inhibitor PLX7420.471 Copper chelation emerges as a potential treatment strategy for a specific subset of tumors characterized by activating BRAFV600E mutations. Copper chelators, commonly used to treat Wilson’s disease, inhibit tumor formation in human or mouse cells harboring BRAFV600E mutations or engineered to be resistant to BRAFi.472
Targeting apoptosis
Tumor cells often develop resistance to chemotherapy and radiation by disrupting the regulation of apoptosis-related mediators, especially by increasing the levels of anti-apoptotic BCL-2 family proteins.473 BH3 mimics have gained popularity as a means to induce cell death effectively by targeting the primary anti-apoptotic proteins without necessitating significant involvement of pro-apoptotic proteins.474 ABT-263, a BH3 mimic, when used in combination with selective BRAFi (PLX4720), enhances both the extent and speed of responses in treatment-naïve patients with BRAFV600E-mutated melanoma but it is less effective in individuals who have developed resistance to these drugs.475 Surprisingly, ABT-737 significantly increases the sensitivity of melanoma cell lines to conventional chemotherapeutics, leading to BIM-mediated apoptosis.476 Inhibiting anti-apoptotic BCL-2 proteins can improve primary PLX-4032 responses and reduces the development of resistance to both targeted and standard therapies.477
In addition to BCL-2 family proteins, STAT3 is a promising target for anti-apoptotic drug development against BRAFi resistance. Inhibiting the STAT3 pathway demonstrates significantly higher cytotoxicity compared to the currently used therapeutic drug PLX-4032. This approach targets both BRAF-mutant and WT melanoma cells without selectivity.478 Combing Vemurafenib with STAT3 silencing or miR-579-3p overexpression proves effective in overcoming Vemurafenib resistance in cancer cells.479 Furthermore, activation of the RAS-RAF-MAPK pathway is associated with the prevention of Caspase-3 activation, cell protection against apoptosis, and direct phosphorylation of caspase-9.213 Caspase-3 inhibits MEK1 through proteolytic means, leading to reduced pro-survival ERK signaling and increased susceptibility of cell to apoptosis.480
Endoplasmic reticulum (ER) stress can potentially activate various apoptotic signaling pathways in melanoma cells in a context-dependent manner. The MEK/ERK signaling pathway plays a pivotal role in preventing caspase-4 activation induced by ER stress. Additionally, the apoptosis repressor with caspase recruitment domain (ARC) proteins appears essential in preventing Caspase-8 activation in melanoma cells under ER stress.481 Inhibiting the MEK/ERK pathway makes melanoma cells more susceptible to apoptosis induced by ER stress, partially through caspase-4 activation, and is also associated with the inhibition of ER chaperon glucose-regulated protein 78 (GRP78) production.482
Expanding current strategies using high-throughput technology
High-throughput techniques for profiling tumor-associated gene expression, including miRNAs, have a great prospect for clinical applications. A recent advancement involves a high-throughput quantitative method based on RCA, enhancing the sensitivity of detecting miRNAs associated with Triple-negative breast cancer (TNBC) using fluorescence-encoded microspheres.483
Genome editing
The translation of gene editing from theory to clinical practice has been accelerated by recent advancements in programmable nucleases, including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR/Cas9-associated nucleases.484 One of the pioneering pooled CRISPR screens involved a genome-scale knockout library of melanoma cells treated with the BRAF inhibitor, Vemurafenib, revealing genes responsible for treatment resistance.485 Another study found that CRISPR/Cas9 editing of RAS and MEK mutant cells led to resistance to BRAF and MEK inhibitors, but MEK1 Q56P restored sensitivity to the MEK/BRAF inhibitor combination, and KRAS G13D increased sensitivity to immunotherapy.486
Cancer vaccines
Cancer vaccines aim to activate latent or unresponsive tumor-specific T cells, bolstering the innate immune defense against cancer.487 Various cancer immunotherapies have been developed to trigger tumor-specific immune response in tumor patients. Clinical trials have demonstrated the effectiveness of cytosine-phosphorothioate-guanine oligodeoxynucleotides (CpG ODN) as a cancer vaccine adjuvant. The GO-PEI-OVA-PEG-CpG nano-vaccine exhibited favorable safety profiles and significant anticancer efficacy, extending mouse survival, and limiting tumor growth in vivo. When combined with NLG918 (antitumor immunotherapy), the vaccine displayed even greater therapeutic.488 Peptide-based drug delivery system, like cancer vaccines, offer an additional promising avenue to enhance existing BRAFi and MEKi strategies. An early-phase I clinical trial is therapy with the administration of six melanoma helper peptides (6MHP). The trial also assesses the impact of peptide vaccination, BRAF inhibitors, and MEK inhibitors on the immune system using participant blood and tumor samples. Initial results indicate that 77% of patients exhibited antibody responses to 6MHP by week 7, peaking 6 weeks after the last vaccine and persisting for 6 months (NCT02382549).
Leveraging online resources for compound screening
Resources for studying the RAF-centered signaling network are expanding, facilitating the discovery of protein sets that physically interact with MAPK network through high-throughput techniques, such as protein-interaction based screening approaches.489 The development of the NanoBRET screening platform employs live-cell bioluminescence resonance energy transfer (BRET) to identify substances that modify the binding between activated KRAS and CRAF kinase.489 Additionally, a subtractive forward two-hybrid method is employed to identify small molecule that disrupt the interaction between RAS and RAF.490 Furthermore, numerous online databases, including Cell Circuits, NCBI GEO, DIP, BIND, KEGG, NetPath, LINCS, BioGrid, and expert systems, enable individual researcher to construct and query protein interaction maps tailored to their gene of interest.
Targeting autophagy: a new therapeutic avenue for RAS/RAF/MAPK inhibitor-resistance
Autophagy is generally affected by a wide range of anticancer medications, including DNA-damaging agents, microtubule-targeted therapies, antimetabolites, death receptor agonists, hormonal agents, antiangiogenic agents, proteasome inhibitors, histone deacetylase inhibitors, and kinase inhibitors.262 Both chemotherapeutic agents and ionizing radiation stimulate autophagy by inducing autophagosome formation.262,491,492 Some agents promote autophagosome formation but prevent lysosome fusion, consequently blocking autophagic flux.493 For example, mTOR inhibitors, such as Rapamycin and Temsirolimus, are well-known anticancer drugs that directly promote autophagy by disrupting the activity of the negative autophagy regulator mTOR complex 1 (mTORC1).494 ABT-737, a Bcl-2 inhibitor, directly targets the core autophagy machinery.495,496
Some autophagy inhibitors can attenuate tumor growth. Autophagy inhibition increases Icariin-induced cytotoxicity in colorectal cancer cells.497 In addition, several PI3K inhibitors, including 3-Methyladenine, Wortmannin, and LY294002, used as anticancer therapeutics, directly inhibit autophagy.498,499 Anticancer therapies that activate autophagy frequently induce a pro-survival response that may contribute to the development of drug resistance and refractory cancer.500–503 Therefore, drugs that target autophagy should be considered for combination therapy strategies together with other anticancer drugs. The antimalarial drugs Chloroquine (CQ) and Hydroxychloroquine (HCQ) are currently being studied as potential components of combination therapies together with standard treatments in multiple tumor types.504–506
Autophagy-associated RAFi resistance
Two recent studies have delved into the potential therapeutic benefits of combining autophagy inhibitors with targeted therapy.507,508 Based on a wealth of preclinical research, clinical trials, and recent literature, we have compiled a summary of autophagy-related RAFi-resistance and potential therapeutic interventions. MAPK inhibitors are used as conventional therapy in cancer patients with activated RAF mutations. However, their antitumorigenic effects and clinical benefits are temporary due to the eventual development of drug resistance and relapse. Drug resistance can occur when upstream factors activate these signaling pathways, bypassing RAF inhibition (Fig. 9b). The MAPK pathway is often aberrantly stimulated by BCR-ABL oncoproteins through direct binding with RAS activators, and aberrant MAPK activation plays significant roles in the onset and progression of leukemia.509 In addition, MAPK phosphorylation is frequently associated with autophagic structures, suggesting that microtubule-associated protein 1 A/1B-light chain 3 (LC3)-II–positive membranes and ATG5- and ATG12-positive pre-autophagosomes may serve as scaffolds or cellular signaling platforms for the RAF–MEK–ERK cascade, facilitating ERK pathway activation510. The ATG7–ATG5–ATG12–LC3-II cascade modulates MAPK phosphorylation in an unconventional manner (Fig. 9b). Atg4b, an LC3-specific protease, catalyzes the conversion of the LC3 precursor into its active forms and deconjugates modifiers from phospholipids.511,512
AMPK plays a crucial in promoting resistance to RAS-RAF-MAPK pathway inhibitors triggering autophagy.513 The oncogenic BRAF negatively regulates the tumor suppressor LKB1, fostering the proliferation of melanoma cells. Consequently, the RAS/RAF/MAPK pathway initiates autophagy activation through the LKB1-AMPK-ULK1 signaling axis. To induce mTORC1 inhibition and cell cycle arrest in response to energy stress, AMPK must phosphorylate Raptor, a part of mTORC1.514 The oncogenic BRAF leads to heightened basal autophagy and increased resistance to apoptosis in cutaneous melanomas, also causing chronic ER stress.515 ER stress-induced autophagy is diminished in cells lacking IRE1 or treated with a JNK inhibitor, highlighting the essential role of the IRE1-JNK pathway in autophagy activation following ER stress.516 Activation of the IRE1/ASK1/JNK and TRB3 pathways is induced by p38 activation, which is driven by BRAFV600E.515
BCL-2 family members (BCL-2, BCL-xL, and MCL-1) inhibit autophagy, whereas BNIP3 stimulates autophagy by releasing Beclin1 from the BCL2/Beclin1 complex.496 p53 family isoforms contribute to acquired resistance to targeted MAPK inhibitors in melanoma cells,517 and p53-dependent activation of damage-regulated autophagy modulator-1 (DRAM-1) triggers autophagy.518 RAF-mutant–induced chronic ER stress stimulates basal autophagy, and RAF-mediated p38 activation augments the IRE1–ASK1–JNK and Tribbles 3 (TRB3) pathways.519 DMP1 also acts as a critical molecule connecting oncogenic RAS/RAF/MEK/ERK signaling with the tumor-suppressive ARF–MDM2–p53 pathway.520 In conclusion, RAFi resistance in cancer is often associated with increased autophagy, driven by multiple interconnected pathways and molecular events, suggesting that understanding these mechanism may have implications for developing more effective treatment strategies in cancer.
Targeting autophagy to overcome RAFi resistance as a therapeutic strategy
BRAFV600E-mutant MAPK activation in cancer highlights the potential therapeutic benefits of using MAPK inhibitors, such as Vemurafenib, which have demonstrated beneficial outcomes when used to treat patients with late-stage melanoma.324 In addition, pharmacological inhibitors of BRAFV600E-mutant and MEK have been used to treat metastatic carcinoma.331,337,521 However, the rapid development of drug resistance has limited the use of these drugs in cancer therapy, resulting in temporary benefits lasting from several months to less than 2 years.522 When combined with immune checkpoint inhibitors targeting programmed death protein 1 or ipilimumab targeting cytotoxic T lymphocyte–associated protein 4, these medications greatly enhance life expectancy up to 4 years in 50% of patients with metastatic melanoma.523,524 However, combination therapy using RAFis or MEK inhibitors together with immune checkpoint inhibitors remains restricted to patients with certain types of cancer.525–528 To enhance therapeutic efficacy, a wide range of adaptive responses must be studied, including therapy resistance mechanisms and autophagy, which enable drug sequestration and cell survival.
Inhibition of adaptive protective autophagy induced by MAPK re-sensitization in RAFi resistance
Numerous avenues that lead to acquired resistance to RAFis generally utilize multiple methods to target the same or parallel pathways.366 Vertical inhibition of the MAPK signaling pathway using combinations of RAFi together with MEK inhibitors has received FDA approval as a first-line treatment strategy for patients with advanced RAF-mutant melanoma, NSCLC, and thyroid cancer. Suppression of RAF-mutant signaling promotes autophagy in cancer cells,25,26 suggesting that targeting autophagy in cancer cells may be desirable when using pathway-targeted inhibitors in RAF-mutant cancer.529 Many tumors harboring RAS and RAF mutations develop an “addiction” to autophagy, which is required to maintain cellular homeostasis; thus, the inhibition of protective autophagy induced by MAPK pathway activation may represent a therapeutic option in RAS- and RAF-mutant malignancies.435,530–533 The genetic suppression of autophagy genes may also represent a novel approach for the treatment of lung cancers harboring mutant RAS534 or BRAFV600E.535 Pharmacologic RAF inhibition dramatically increases autophagy in RAFi-resistant melanoma cells,26 and the genetic or pharmacological inhibition of autophagy counters RAFi resistance in brain tumors.434 Additionally, inhibiting autophagy in RAFi-resistant tumor cells in vitro and in patients reverses drug resistance.434 Combining autophagy inhibition with MEK inhibition increases apoptotic cell death in a RAF-mutant colorectal cancer cell line.536 The combined inhibition of autophagy and MEK significantly accelerates regression in RAF-mutant patient-derived colorectal cancer cells xenografted onto an animal model.537 Similarly, RAF-mutant thyroid cancer cells are sensitive to autophagy targeting in the presence of vemurafenib.320,538
Therapeutics success beyond autophagy targeted RAFi therapy
In cancer, autophagy plays adaptive and protective roles when MAPK signaling is hampered.539 Accordingly, combined treatment using both MAPK and autophagy inhibitors represents one of the best options for treating various carcinomas.26,531 However, tumor responses to autophagy inhibitors are not clinically consistent due to non-uniform permeability across tumor tissues and the potential toxicity of combination treatments using multiple chemotherapeutics.540
Because autophagy can act as a double-edged sword during tumorigenesis, autophagy-targeted treatment outcomes are highly dependent on cancer types, contexts, and stages.541,542 In addition, the inhibition of autophagy initiation and late fusion steps results in differential responses. For example, early-stage autophagy inhibition using ULK1/2 inhibitors (e.g., MRT 68921) decreases tumor cell death, whereas inhibitors (HCQ) that target the later fusion stage greatly increase cell death in mesothelioma cells.543 CQ and its variants, such as HCQ, are currently the most frequently tested autophagy inhibitors in cancer clinical trials. These chemicals impede the late fusion stage between autophagosomes and lysosomes, causing deacidification and impairing enzymatic function.540,544,545 Interestingly, these inhibitors lack selectivity, affecting total lysosomal function.546 However, CQ largely improved the sensitivity of Vemurafenib in an ex vivo primary cell culture derived from patients harboring the BRAFV600E mutation435. Autophagy-independent vascular normalization of CQ inhibits tumor invasion and metastasis by enhancing chemotherapeutic effects.547,548 The autophagy inhibitor HCQ has been more widely used in clinical trials over CQ because HCQ is less toxic at optimum doses.504,549–551 HCQ shows beneficial effects and synergistic effects when combined with the mTOR inhibitor, Temsirolimus, in melanoma552 and breast cancer.553,554 Importantly, HCQ affects chemotherapy and radiation sensitivity in non-selective cancers.555 Lys05, a novel lysosomal autophagy inhibitor, has also been suggested as a potential therapeutic agent for cancer.556,557
CQ and HCQ are non-selective autophagy inhibitors, and specific small-molecule inhibitors that target earlier stages of autophagy are preferred for cancer therapy.547,558–560 ULK1, a key autophagy regulator, can be specifically targeted by cellular energy status regulators, such as mTORC1 and AMPK.561 AMPK, a low-energy sensor, activates ULK1 to induce autophagy.562 ATG13, an autophagy protein in the ULK1 complex, is a static autophagy marker that corresponds closely with autophagic flux in mesothelioma.563 In addition, multiple ATP-competitive ULK1 kinase inhibitors have demonstrated compelling evidence to support clinical use,564,565 such as the successful suppression of NSCLC growth by SBI-0206965.565,566 Vacuolar protein sorting 34 (VPS34), a component of the Beclin1 complex, also plays an important early role in autophagosome formation and is considered a potential therapeutic target.567,568 Several VPS34 inhibitors (e.g., SAR405) have demonstrated anticancer effects in kidney carcinoma cells,569 including VPS34-IN1, a particularly powerful and selective VPS34 inhibitor with cancer therapeutic potential.570 Furthermore, transitional small molecules tend to form prolonged associations with target proteins, leading to the inhibition of their enzymatic activity. This extended binding duration poses challenges in devising small molecule structures and countering drug resistance resulting from target mutations. In contrast, the proteolysis-targeting chimera (PROTAC) exhibits the ability to specifically eliminate target proteins, thereby intensifying the cytotoxic impact on mutant tumors. In the realm of both fundamental research and pharmaceutical advancement, the autophagy-targeting chimera (AUTOTAC) provides a versatile platform for precise proteolysis.571 Also, high-throughput technology presents a promising approach for identifying selective autophagy inhibitors that are both effective and less cytotoxic when used in combination therapy. Arzonol, for instance, emerged as a distinct chemotherapeutic candidate through high-throughput screening of natural compounds aimed at modulating autophagy.572 Overall, targeting autophagy initiation, together with improved investigations of the functional mechanisms underlying existing chemotherapies, could be crucial for developing novel cancer therapies.
Conclusion and future directions
RAF inhibitors exhibit exceptional clinical efficacy in patients with RAF-mutant carcinoma, although their therapeutic effects are restricted due to the development of drug resistance. Recent in vitro and in vivo translational studies have elucidated the molecular basis underlying the development of RAFi resistance in cancer. Obtaining additional biological insights into the process of drug resistance represents a necessary, long-term goal for the development of new RAFi and combination treatments able to slow or prevent the development of drug resistance in cancer. The combined regulation of MAPK signaling and RAFi-induced autophagy may represent a potential strategy for overcoming drug resistance and inform the development of novel cancer therapies. Because autophagy can act as either a pro-death or a pro-survival factor in tumorigenesis, targeting autophagy is highly dependent on cancer type and stage. Autophagy activation can promote tumorigenesis, and autophagy inhibition can improve the therapeutic efficacy of RAFi. A better understanding of the molecular mechanisms and precise targets underlying the complex process of autophagy modulation could lead to the development of agonists or blockers that could serve as potential therapeutic agents for future use in RAF-mutant cancers.
Pharmacological autophagy inhibitors hold significant potential in combating resistance to RAF inhibitors in cancer patients with RAF mutations. Recent research highlights the efficacy of compounds like 3-Methyladenine and hydroxychloroquine (HCQ) in suppressing autophagy when combined with anti-EGFR monoclonal antibodies (mAbs) and checkpoint inhibitors. The BAMM trial (BRAF Autophagy and MEK Inhibition in Melanoma), which combined Dabrafenib, Trametinib, and HCQ, demonstrated these combinations are effective and safe in patients with BRAFV600-mutant melanoma. Beyond classical autophagy inhibitors, emerging indirect inhibitors like ABT-737, natural products such as Resveratrol, and synthetic compounds like Quinacrine may warrant clinical investigation. The AUTOTAC chemical biology platform has garnered significant attention within the area of targeted protein degradation. It employs bifunctional molecules that consist of ligand binding to specific targets linked with ligands targeting the autophagy processes. Furthermore, recent advancements in autophagy-based degraders and molecular adhesives, such as autophagosome-tethering compounds (AATEC) or lysosome-targeting chimera (LYTAC), have enabled the successful degradation of numerous target proteins by routing them to the lysosome.
Also, the genetic inhibition of autophagy genes via RNA interference and CRISPR/Cas9 targeting holds promising. Evaluating the safety of combing BRAF and MEK inhibitor therapy with melanoma helper peptides (6MHP) administration assesses the impact on the immune system. In addition, cancer vaccines aim to activate tumor-specific T cells, strengthening the immune defenses. Future research emphasizes high-throughput technology for preclinical evaluations of cytotoxicity and combination therapy between RAS/RAF/MAPK inhibitors and other compounds.
Acknowledgements
We thank all the lab members for their thoughtful feedback on this study. All figures were created with BioRender.com.
Author contributions
E.M.B. conceptualized, wrote the manuscript, and created all original figures. H.J.K. collected data for clinical trials, and edited the manuscript during revision. D.R.K. conceptualized, provided financial support, edited all figures, and contributed to writing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants from the Basic Science Research Program through the National Research Foundation of Korea (RS-2023-00219399, RS-2023-00238051) and by the Commercializations Promotion Agency for R&D Outcomes (COMPA) grant funded by the Korea government (MSIT) (1711173796).
Competing interests
The authors declare no competing interests.
References
- 1.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 2.Gysin S, Salt M, Young A, McCormick F. Therapeutic strategies for targeting ras proteins. Genes Cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W, Fang G, Rudolph J. Ras inhibition via direct Ras binding–is there a path forward? Bioorg. Med. Chem. Lett. 2012;22:5766–5776. doi: 10.1016/j.bmcl.2012.07.082. [DOI] [PubMed] [Google Scholar]
- 5.Spiegel J, et al. Small-molecule modulation of Ras signaling. Nat. Chem. Biol. 2014;10:613–622. doi: 10.1038/nchembio.1560. [DOI] [PubMed] [Google Scholar]
- 6.Sayyed-Ahmad A, Gorfe AA. How to make an undruggable enzyme druggable: lessons from ras proteins. Adv. Protein Chem. Struct. Biol. 2020;122:181–202. doi: 10.1016/bs.apcsb.2020.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Cuadrado A, et al. H-ras and raf-1 cooperate in transformation of NIH3T3 fibroblasts. Oncogene. 1993;8:2443–2448. [PubMed] [Google Scholar]
- 8.Fedorenko IV, Paraiso KH, Smalley KS. Acquired and intrinsic BRAF inhibitor resistance in BRAF V600E mutant melanoma. Biochem. Pharmacol. 2011;82:201–209. doi: 10.1016/j.bcp.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Downward J. Targeting RAS signaling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 10.Leicht DT, et al. Raf kinases: function, regulation and role in human cancer. Biochim. Biophys. Acta. 2007;1773:1196–1212. doi: 10.1016/j.bbamcr.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dillon, M. et al. Progress on Ras/MAPK signaling research and targeting in blood and solid cancers. Cancers13, 5059 (2021). [DOI] [PMC free article] [PubMed]
- 12.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Chong H, Guan KL. Function of the Rho family GTPases in Ras-stimulated Raf activation. J. Biol. Chem. 2001;276:34728–34737. doi: 10.1074/jbc.M103496200. [DOI] [PubMed] [Google Scholar]
- 14.Holderfield M, Deuker MM, McCormick F, McMahon M, Targeting RAF. kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaeger R, Corcoran RB. Targeting alterations in the RAF-MEK pathway. Cancer Discov. 2019;9:329–341. doi: 10.1158/2159-8290.CD-18-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haarberg HE, Smalley KS. Resistance to Raf inhibition in cancer. Drug Discov. Today Technol. 2014;11:27–32. doi: 10.1016/j.ddtec.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oddo D, et al. Molecular landscape of acquired resistance to targeted therapy combinations in BRAF-mutant colorectal cancer. Cancer Res. 2016;76:4504–4515. doi: 10.1158/0008-5472.CAN-16-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatzivassiliou G, et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature. 2013;501:232–236. doi: 10.1038/nature12441. [DOI] [PubMed] [Google Scholar]
- 19.Flaherty KT, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 20.Michielin O, et al. Cutaneous melanoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-updagger. Ann. Oncol. 2019;30:1884–1901. doi: 10.1093/annonc/mdz411. [DOI] [PubMed] [Google Scholar]
- 21.Adamopoulos C, et al. Exploiting allosteric properties of RAF and MEK inhibitors to target therapy-resistant tumors driven by oncogenic BRAF signaling. Cancer Discov. 2021;11:1716–1735. doi: 10.1158/2159-8290.CD-20-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsh SJ, Corrie PG. Management of BRAF and MEK inhibitor toxicities in patients with metastatic melanoma. Ther. Adv. Med. Oncol. 2015;7:122–136. doi: 10.1177/1758834014566428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinha R, et al. Cutaneous adverse events associated with vemurafenib in patients with metastatic melanoma: practical advice on diagnosis, prevention and management of the main treatment-related skin toxicities. Br. J. Dermatol. 2012;167:987–994. doi: 10.1111/bjd.12010. [DOI] [PubMed] [Google Scholar]
- 24.Yun, C. W. & Lee, S. H. The roles of autophagy in cancer. Int. J. Mol. Sci. 19, 3466 (2018). [DOI] [PMC free article] [PubMed]
- 25.Goulielmaki M, et al. BRAF associated autophagy exploitation: BRAF and autophagy inhibitors synergise to efficiently overcome resistance of BRAF mutant colorectal cancer cells. Oncotarget. 2016;7:9188–9221. doi: 10.18632/oncotarget.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma XH, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J. Clin. Invest. 2014;124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee CS, et al. MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc. Natl Acad. Sci. USA. 2019;116:4508–4517. doi: 10.1073/pnas.1817494116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J. Thorac. Oncol. 2006;1:7–9. doi: 10.1016/S1556-0864(15)31506-9. [DOI] [PubMed] [Google Scholar]
- 29.Harvey JJ. An unidentified virus which causes the rapid production of tumours in mice. Nature. 1964;204:1104–1105. doi: 10.1038/2041104b0. [DOI] [PubMed] [Google Scholar]
- 30.Tur’ianov, M. et al. [Present-day characteristics of etiology, epidemiology and clinical aspects of diphtheria in adults]. Sov. Med.1, 75–7 (1991). [PubMed]
- 31.Scolnick EM, Rands E, Williams D, Parks WP. Studies on the nucleic acid sequences of Kirsten sarcoma virus: a model for formation of a mammalian RNA-containing sarcoma virus. J. Virol. 1973;12:458–463. doi: 10.1128/jvi.12.3.458-463.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapp UR, et al. Structure and biological activity of v-raf, a unique oncogene transduced by a retrovirus. Proc. Natl Acad. Sci. USA. 1983;80:4218–4222. doi: 10.1073/pnas.80.14.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moelling K, et al. Serine- and threonine-specific protein kinase activities of purified gag-mil and gag-raf proteins. Nature. 1984;312:558–561. doi: 10.1038/312558a0. [DOI] [PubMed] [Google Scholar]
- 34.Bonner TI, et al. Structure and biological activity of human homologs of the raf/mil oncogene. Mol. Cell Biol. 1985;5:1400–1407. doi: 10.1128/mcb.5.6.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huleihel M, et al. Characterization of murine A-raf, a new oncogene related to the v-raf oncogene. Mol. Cell Biol. 1986;6:2655–2662. doi: 10.1128/mcb.6.7.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sturgill TW, Ray LB, Erikson E, Maller JL. Insulin-stimulated MAP-2 kinase phosphorylates and activates ribosomal protein S6 kinase II. Nature. 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- 37.Courchesne WE, Kunisawa R, Thorner J. A putative protein kinase overcomes pheromone-induced arrest of cell cycling in S. cerevisiae. Cell. 1989;58:1107–1119. doi: 10.1016/0092-8674(89)90509-6. [DOI] [PubMed] [Google Scholar]
- 38.Ahn NG, Weiel JE, Chan CP, Krebs EG. Identification of multiple epidermal growth factor-stimulated protein serine/threonine kinases from Swiss 3T3 cells. J. Biol. Chem. 1990;265:11487–11494. doi: 10.1016/S0021-9258(19)38423-6. [DOI] [PubMed] [Google Scholar]
- 39.Kyriakis JM, et al. Raf-1 activates MAP kinase-kinase. Nature. 1992;358:417–421. doi: 10.1038/358417a0. [DOI] [PubMed] [Google Scholar]
- 40.Zhang XF, et al. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature. 1993;364:308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 41.Troppmair J, Hartkamp J, Rapp UR. Activation of NF-kappa B by oncogenic Raf in HEK 293 cells occurs through autocrine recruitment of the stress kinase cascade. Oncogene. 1998;17:685–690. doi: 10.1038/sj.onc.1201981. [DOI] [PubMed] [Google Scholar]
- 42.Kane RC, et al. Sorafenib for the treatment of advanced renal cell carcinoma. Clin. Cancer Res. 2006;12:7271–7278. doi: 10.1158/1078-0432.CCR-06-1249. [DOI] [PubMed] [Google Scholar]
- 43.Ambrosini G, et al. Sorafenib inhibits growth and mitogen-activated protein kinase signaling in malignant peripheral nerve sheath cells. Mol. Cancer Ther. 2008;7:890–896. doi: 10.1158/1535-7163.MCT-07-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bollag G, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat. Rev. Drug Discov. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 45.Wright CJ, McCormack PL. Trametinib: first global approval. Drugs. 2013;73:1245–1254. doi: 10.1007/s40265-013-0096-1. [DOI] [PubMed] [Google Scholar]
- 46.Menzies AM, Long GV. Dabrafenib and trametinib, alone and in combination for BRAF-mutant metastatic melanoma. Clin. Cancer Res. 2014;20:2035–2043. doi: 10.1158/1078-0432.CCR-13-2054. [DOI] [PubMed] [Google Scholar]
- 47.New Medical Devices. P&T.40, 792–806 (2015). [PMC free article] [PubMed]
- 48.Braicu, C. et al. A comprehensive review on MAPK: a promising therapeutic target in cancer. Cancers11, 1618 (2019). [DOI] [PMC free article] [PubMed]
- 49.Krall, E. B. et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife6, e18970 (2017). [DOI] [PMC free article] [PubMed]
- 50.Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta. 2010;1802:396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 51.Karin M. Mitogen activated protein kinases as targets for development of novel anti-inflammatory drugs. Ann. Rheum. Dis. 2004;63(Suppl. 2):ii62–ii64. doi: 10.1136/ard.2004.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coskun M, Olsen J, Seidelin JB, Nielsen OH. MAP kinases in inflammatory bowel disease. Clin. Chim. Acta. 2011;412:513–520. doi: 10.1016/j.cca.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 53.Yu W, Song X, Liu Y. TRB3 regulates pulmonary interstitial fibrosis through the MAPK signaling pathway. Int. J. Clin. Exp. Pathol. 2019;12:3247–3257. [PMC free article] [PubMed] [Google Scholar]
- 54.Pearson G, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 55.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 56.Qi M, Elion EA. MAP kinase pathways. J. Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 57.Chen Z, et al. MAP kinases. Chem. Rev. 2001;101:2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 58.Coulombe P, Meloche S. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta. 2007;1773:1376–1387. doi: 10.1016/j.bbamcr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 59.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 60.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramos JW. The regulation of extracellular signal-regulated kinase (ERK) in mammalian cells. Int. J. Biochem. Cell Biol. 2008;40:2707–2719. doi: 10.1016/j.biocel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 62.Levitzki A. Protein kinase inhibitors as a therapeutic modality. Acc. Chem. Res. 2003;36:462–469. doi: 10.1021/ar0201207. [DOI] [PubMed] [Google Scholar]
- 63.Faivre S, Djelloul S, Raymond E. New paradigms in anticancer therapy: targeting multiple signaling pathways with kinase inhibitors. Semin. Oncol. 2006;33:407–420. doi: 10.1053/j.seminoncol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 64.Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer. 2018;17:58. doi: 10.1186/s12943-018-0782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edin ML, Juliano RL. Raf-1 serine 338 phosphorylation plays a key role in adhesion-dependent activation of extracellular signal-regulated kinase by epidermal growth factor. Mol. Cell Biol. 2005;25:4466–4475. doi: 10.1128/MCB.25.11.4466-4475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamden KE, et al. Raf and VEGF: emerging therapeutic targets in Kaposi’s sarcoma-associated herpesvirus infection and angiogenesis in hematopoietic and nonhematopoietic tumors. Leukemia. 2005;19:18–26. doi: 10.1038/sj.leu.2403532. [DOI] [PubMed] [Google Scholar]
- 67.Dibb NJ, Dilworth SM, Mol CD. Switching on kinases: oncogenic activation of BRAF and the PDGFR family. Nat. Rev. Cancer. 2004;4:718–727. doi: 10.1038/nrc1434. [DOI] [PubMed] [Google Scholar]
- 68.Sebolt-Leopold JS, et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 69.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 70.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J. Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 71.Downward J. Control of ras activation. Cancer Surv. 1996;27:87–100. [PubMed] [Google Scholar]
- 72.Herrero, A. & Crespo, P. RAS dimers: the novice couple at the RAS-ERK pathway ball. Genes12, 1556 (2021). [DOI] [PMC free article] [PubMed]
- 73.Scolnick EM, Papageorge AG, Shih TY. Guanine nucleotide-binding activity as an assay for src protein of rat-derived murine sarcoma viruses. Proc. Natl Acad. Sci. USA. 1979;76:5355–5359. doi: 10.1073/pnas.76.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hennig A, et al. Ras activation revisited: role of GEF and GAP systems. Biol. Chem. 2015;396:831–848. doi: 10.1515/hsz-2014-0257. [DOI] [PubMed] [Google Scholar]
- 75.Hall BE, Bar-Sagi D, Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc. Natl Acad. Sci. USA. 2002;99:12138–12142. doi: 10.1073/pnas.192453199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen K, Zhang Y, Qian L, Wang P. Emerging strategies to target RAS signaling in human cancer therapy. J. Hematol. Oncol. 2021;14:116. doi: 10.1186/s13045-021-01127-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 78.Schopel M, et al. The small GTPases Ras and Rheb studied by multidimensional NMR spectroscopy: structure and function. Biol. Chem. 2017;398:577–588. doi: 10.1515/hsz-2016-0276. [DOI] [PubMed] [Google Scholar]
- 79.Heidecker G, et al. Mutational activation of c-raf-1 and definition of the minimal transforming sequence. Mol. Cell Biol. 1990;10:2503–2512. doi: 10.1128/mcb.10.6.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marx M, et al. A novel oncogene related to c-mil is transduced in chicken neuroretina cells induced to proliferate by infection with an avian lymphomatosis virus. EMBO J. 1988;7:3369–3373. doi: 10.1002/j.1460-2075.1988.tb03209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 82.Niault TS, Baccarini M. Targets of Raf in tumorigenesis. Carcinogenesis. 2010;31:1165–1174. doi: 10.1093/carcin/bgp337. [DOI] [PubMed] [Google Scholar]
- 83.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 2000;351(Pt 2):289–305. doi: 10.1042/bj3510289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trakul N, Rosner MR. Modulation of the MAP kinase signaling cascade by Raf kinase inhibitory protein. Cell Res. 2005;15:19–23. doi: 10.1038/sj.cr.7290258. [DOI] [PubMed] [Google Scholar]
- 85.Carey KD, Watson RT, Pessin JE, Stork PJ. The requirement of specific membrane domains for Raf-1 phosphorylation and activation. J. Biol. Chem. 2003;278:3185–3196. doi: 10.1074/jbc.M207014200. [DOI] [PubMed] [Google Scholar]
- 86.Goitre L, Trapani E, Trabalzini L, Retta SF. The Ras superfamily of small GTPases: the unlocked secrets. Methods Mol. Biol. 2014;1120:1–18. doi: 10.1007/978-1-62703-791-4_1. [DOI] [PubMed] [Google Scholar]
- 87.Hancock JF. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 88.Cox AD, Der CJ. Ras history: the saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dharmawardhane S, et al. Regulation of macropinocytosis by p21-activated kinase-1. Mol. Biol. Cell. 2000;11:3341–3352. doi: 10.1091/mbc.11.10.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stokoe D, McCormick F. Activation of c-Raf-1 by Ras and Src through different mechanisms: activation in vivo and in vitro. EMBO J. 1997;16:2384–2396. doi: 10.1093/emboj/16.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Winston LA, Hunter T. JAK2, Ras, and Raf are required for activation of extracellular signal-regulated kinase/mitogen-activated protein kinase by growth hormone. J. Biol. Chem. 1995;270:30837–30840. doi: 10.1074/jbc.270.52.30837. [DOI] [PubMed] [Google Scholar]
- 92.Firnau, M. B. & Brieger, A. CK2 and the hallmarks of cancer. Biomedicines10, 1987 (2022). [DOI] [PMC free article] [PubMed]
- 93.Nan X, et al. Single-molecule superresolution imaging allows quantitative analysis of RAF multimer formation and signaling. Proc. Natl Acad. Sci. USA. 2013;110:18519–18524. doi: 10.1073/pnas.1318188110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maurer G, Tarkowski B, Baccarini M. Raf kinases in cancer-roles and therapeutic opportunities. Oncogene. 2011;30:3477–3488. doi: 10.1038/onc.2011.160. [DOI] [PubMed] [Google Scholar]
- 95.Tran TH, et al. KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat. Commun. 2021;12:1176. doi: 10.1038/s41467-021-21422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chao FA, et al. Insights into the cross talk between effector and allosteric lobes of KRAS from methyl conformational dynamics. J. Am. Chem. Soc. 2022;144:4196–4205. doi: 10.1021/jacs.2c00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stokoe D, et al. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 98.Leevers SJ, Paterson HF, Marshall CJ. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 99.Herrmann C, Martin GA, Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J. Biol. Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 100.Nassar N, et al. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375:554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- 101.Nguyen K, et al. Exploring CRD mobility during RAS/RAF engagement at the membrane. Biophys. J. 2022;121:3630–3650. doi: 10.1016/j.bpj.2022.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Williams JG, et al. Elucidation of binding determinants and functional consequences of Ras/Raf-cysteine-rich domain interactions. J. Biol. Chem. 2000;275:22172–22179. doi: 10.1074/jbc.M000397200. [DOI] [PubMed] [Google Scholar]
- 103.Mott HR, et al. The solution structure of the Raf-1 cysteine-rich domain: a novel ras and phospholipid binding site. Proc Natl Acad Sci USA. 1996;93:8312–8317. doi: 10.1073/pnas.93.16.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hekman M, et al. Associations of B- and C-Raf with cholesterol, phosphatidylserine, and lipid second messengers: preferential binding of Raf to artificial lipid rafts. J. Biol. Chem. 2002;277:24090–24102. doi: 10.1074/jbc.M200576200. [DOI] [PubMed] [Google Scholar]
- 105.Freeman AK, Ritt DA, Morrison DK. The importance of Raf dimerization in cell signaling. Small GTPases. 2013;4:180–185. doi: 10.4161/sgtp.26117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rajakulendran T, et al. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 107.Matallanas D, et al. Raf family kinases: old dogs have learned new tricks. Genes Cancer. 2011;2:232–260. doi: 10.1177/1947601911407323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ritt DA, Monson DM, Specht SI, Morrison DK. Impact of feedback phosphorylation and Raf heterodimerization on normal and mutant B-Raf signaling. Mol Cell Biol. 2010;30:806–819. doi: 10.1128/MCB.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dougherty MK, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell. 2005;17:215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 110.Brummer T, Naegele H, Reth M, Misawa Y. Identification of novel ERK-mediated feedback phosphorylation sites at the C-terminus of B-Raf. Oncogene. 2003;22:8823–8834. doi: 10.1038/sj.onc.1207185. [DOI] [PubMed] [Google Scholar]
- 111.Yang S, Liu G. Targeting the Ras/Raf/MEK/ERK pathway in hepatocellular carcinoma. Oncol. Lett. 2017;13:1041–1047. doi: 10.3892/ol.2017.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shin SY, et al. Positive- and negative-feedback regulations coordinate the dynamic behavior of the Ras-Raf-MEK-ERK signal transduction pathway. J. Cell Sci. 2009;122:425–435. doi: 10.1242/jcs.036319. [DOI] [PubMed] [Google Scholar]
- 113.Sturm OE, et al. The mammalian MAPK/ERK pathway exhibits properties of a negative feedback amplifier. Sci. Signal. 2010;3:ra90. doi: 10.1126/scisignal.2001212. [DOI] [PubMed] [Google Scholar]
- 114.Kamioka Y, et al. Multiple decisive phosphorylation sites for the negative feedback regulation of SOS1 via ERK. J. Biol. Chem. 2010;285:33540–33548. doi: 10.1074/jbc.M110.135517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Waters SB, et al. Desensitization of Ras activation by a feedback disassociation of the SOS-Grb2 complex. J. Biol. Chem. 1995;270:20883–20886. doi: 10.1074/jbc.270.36.20883. [DOI] [PubMed] [Google Scholar]
- 116.Trujillo JI. MEK inhibitors: a patent review 2008–2010. Expert Opin. Ther. Pat. 2011;21:1045–1069. doi: 10.1517/13543776.2011.577068. [DOI] [PubMed] [Google Scholar]
- 117.Chung E, Kondo M. Role of Ras/Raf/MEK/ERK signaling in physiological hematopoiesis and leukemia development. Immunol. Res. 2011;49:248–268. doi: 10.1007/s12026-010-8187-5. [DOI] [PubMed] [Google Scholar]
- 118.Schulze A, et al. The transcriptional response to Raf activation is almost completely dependent on Mitogen-activated Protein Kinase Kinase activity and shows a major autocrine component. Mol. Biol. Cell. 2004;15:3450–3463. doi: 10.1091/mbc.e03-11-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mansour SJ, et al. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 120.Cowley S, Paterson H, Kemp P, Marshall CJ. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 121.Avruch J, et al. Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog. Horm. Res. 2001;56:127–155. doi: 10.1210/rp.56.1.127. [DOI] [PubMed] [Google Scholar]
- 122.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 2010;429:403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 123.Kim C, et al. The kinase p38 alpha serves cell type-specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat. Immunol. 2008;9:1019–1027. doi: 10.1038/ni.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nebreda AR, Porras A. p38 MAP kinases: beyond the stress response. Trends Biochem. Sci. 2000;25:257–260. doi: 10.1016/S0968-0004(00)01595-4. [DOI] [PubMed] [Google Scholar]
- 125.Regan J, et al. Pyrazole urea-based inhibitors of p38 MAP kinase: from lead compound to clinical candidate. J. Med. Chem. 2002;45:2994–3008. doi: 10.1021/jm020057r. [DOI] [PubMed] [Google Scholar]
- 126.Li M, et al. TRAF6-p38/JNK-ATF2 axis promotes microglial inflammatory activation. Exp. Cell Res. 2019;376:133–148. doi: 10.1016/j.yexcr.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 127.Bennett BL, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl Acad. Sci. USA. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Carboni, S. et al. AS601245 (1, 3-benzothiazol-2-yl (2-[[2-(3-pyridinyl) ethyl] amino]-4 pyrimidinyl) acetonitrile): a c-Jun NH2-terminal protein kinase inhibitor with neuroprotective properties. J. Pharmacol. Exp. Ther. 310, 25–32 (2004). [DOI] [PubMed]
- 129.Bogoyevitch MA, et al. c-Jun N-terminal kinase (JNK) signaling: recent advances and challenges. Biochim. Biophys. Acta. 2010;1804:463–475. doi: 10.1016/j.bbapap.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 130.Sabapathy K, et al. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol. Cell. 2004;15:713–725. doi: 10.1016/j.molcel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 131.Tournier C, et al. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 132.Hoang VT, et al. Oncogenic signaling of MEK5-ERK5. Cancer Lett. 2017;392:51–59. doi: 10.1016/j.canlet.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang X, et al. Activation of extracellular signal-regulated protein kinase 5 downregulates FasL upon osmotic stress. Cell Death Differ. 2006;13:2099–2108. doi: 10.1038/sj.cdd.4401969. [DOI] [PubMed] [Google Scholar]
- 134.Zehorai E, Yao Z, Plotnikov A, Seger R. The subcellular localization of MEK and ERK–a novel nuclear translocation signal (NTS) paves a way to the nucleus. Mol. Cell Endocrinol. 2010;314:213–220. doi: 10.1016/j.mce.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 135.Mulloy R, Salinas S, Philips A, Hipskind RA. Activation of cyclin D1 expression by the ERK5 cascade. Oncogene. 2003;22:5387–5398. doi: 10.1038/sj.onc.1206839. [DOI] [PubMed] [Google Scholar]
- 136.Roberts OL, et al. ERK5 is required for VEGF-mediated survival and tubular morphogenesis of primary human microvascular endothelial cells. J. Cell Sci. 2010;123:3189–3200. doi: 10.1242/jcs.072801. [DOI] [PubMed] [Google Scholar]
- 137.Deleris P, et al. Activation loop phosphorylation of the atypical MAP kinases ERK3 and ERK4 is required for binding, activation and cytoplasmic relocalization of MK5. J. Cell Physiol. 2008;217:778–788. doi: 10.1002/jcp.21560. [DOI] [PubMed] [Google Scholar]
- 138.Abe MK, et al. ERK8, a new member of the mitogen-activated protein kinase family. J. Biol. Chem. 2002;277:16733–16743. doi: 10.1074/jbc.M112483200. [DOI] [PubMed] [Google Scholar]
- 139.Klevernic IV, et al. Characterization of the reversible phosphorylation and activation of ERK8. Biochem. J. 2006;394:365–373. doi: 10.1042/BJ20051288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kant S, et al. Characterization of the atypical MAPK ERK4 and its activation of the MAPK-activated protein kinase MK5. J. Biol. Chem. 2006;281:35511–35519. doi: 10.1074/jbc.M606693200. [DOI] [PubMed] [Google Scholar]
- 141.Aberg E, et al. Docking of PRAK/MK5 to the atypical MAPKs ERK3 and ERK4 defines a novel MAPK interaction motif. J. Biol. Chem. 2009;284:19392–19401. doi: 10.1074/jbc.M109.023283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schumacher S, et al. Scaffolding by ERK3 regulates MK5 in development. EMBO J. 2004;23:4770–4779. doi: 10.1038/sj.emboj.7600467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aberg E, et al. Regulation of MAPK-activated protein kinase 5 activity and subcellular localization by the atypical MAPK ERK4/MAPK4. J. Biol. Chem. 2006;281:35499–35510. doi: 10.1074/jbc.M606225200. [DOI] [PubMed] [Google Scholar]
- 144.Seternes OM, et al. Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. EMBO J. 2004;23:4780–4791. doi: 10.1038/sj.emboj.7600489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Abe MK, Kuo WL, Hershenson MB, Rosner MR. Extracellular signal-regulated kinase 7 (ERK7), a novel ERK with a C-terminal domain that regulates its activity, its cellular localization, and cell growth. Mol. Cell Biol. 1999;19:1301–1312. doi: 10.1128/MCB.19.2.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Saelzler MP, et al. ERK8 down-regulates transactivation of the glucocorticoid receptor through Hic-5. J. Biol. Chem. 2006;281:16821–16832. doi: 10.1074/jbc.M512418200. [DOI] [PubMed] [Google Scholar]
- 147.Henrich LM, et al. Extracellular signal-regulated kinase 7, a regulator of hormone-dependent estrogen receptor destruction. Mol. Cell Biol. 2003;23:5979–5988. doi: 10.1128/MCB.23.17.5979-5988.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kojima H, et al. STAT3 regulates Nemo-like kinase by mediating its interaction with IL-6-stimulated TGFbeta-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc. Natl Acad. Sci. USA. 2005;102:4524–4529. doi: 10.1073/pnas.0500679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nakhaei-Rad S, et al. Structural fingerprints, interactions, and signaling networks of RAS family proteins beyond RAS isoforms. Crit. Rev. Biochem. Mol. Biol. 2018;53:130–156. doi: 10.1080/10409238.2018.1431605. [DOI] [PubMed] [Google Scholar]
- 150.Kolch W. Coordinating ERK/MAPK signaling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 151.Smith FD, et al. AKAP-Lbc enhances cyclic AMP control of the ERK1/2 cascade. Nat. Cell Biol. 2010;12:1242–1249. doi: 10.1038/ncb2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dumaz N, Marais R. Integrating signals between cAMP and the RAS/RAF/MEK/ERK signalling pathways. Based on the anniversary prize of the Gesellschaft fur Biochemie und Molekularbiologie Lecture delivered on 5 July 2003 at the Special FEBS Meeting in Brussels. FEBS J. 2005;272:3491–3504. doi: 10.1111/j.1742-4658.2005.04763.x. [DOI] [PubMed] [Google Scholar]
- 153.Fischer A, Muhlhauser WWD, Warscheid B, Radziwill G. Membrane localization of acetylated CNK1 mediates a positive feedback on RAF/ERK signaling. Sci. Adv. 2017;3:e1700475. doi: 10.1126/sciadv.1700475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Amaddii M, et al. Flotillin-1/reggie-2 protein plays dual role in activation of receptor-tyrosine kinase/mitogen-activated protein kinase signaling. J. Biol. Chem. 2012;287:7265–7278. doi: 10.1074/jbc.M111.287599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Harder T, Kuhn M. Selective accumulation of raft-associated membrane protein LAT in T cell receptor signaling assemblies. J. Cell Biol. 2000;151:199–208. doi: 10.1083/jcb.151.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Thome CH, et al. NTAL is associated with treatment outcome, cell proliferation and differentiation in acute promyelocytic leukemia. Sci. Rep. 2020;10:10315. doi: 10.1038/s41598-020-66223-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jin T, et al. PAQR10 and PAQR11 mediate Ras signaling in the Golgi apparatus. Cell Res. 2012;22:661–676. doi: 10.1038/cr.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen C, Noble SM. Post-transcriptional regulation of the Sef1 transcription factor controls the virulence of Candida albicans in its mammalian host. PLoS Pathog. 2012;8:e1002956. doi: 10.1371/journal.ppat.1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kouhara H, et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/S0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 161.Hadari YR, et al. Critical role for the docking-protein FRS2 alpha in FGF receptor-mediated signal transduction pathways. Proc. Natl Acad. Sci. USA. 2001;98:8578–8583. doi: 10.1073/pnas.161259898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lamothe B, et al. The docking protein Gab1 is an essential component of an indirect mechanism for fibroblast growth factor stimulation of the phosphatidylinositol 3-kinase/Akt antiapoptotic pathway. Mol. Cell Biol. 2004;24:5657–5666. doi: 10.1128/MCB.24.13.5657-5666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xiao B, et al. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376:188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- 164.Levy-Apter E, et al. Adaptor protein GRB2 promotes Src tyrosine kinase activation and podosomal organization by protein-tyrosine phosphatase ϵ in osteoclasts. J. Biol. Chem. 2014;289:36048–36058. doi: 10.1074/jbc.M114.603548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Marie-Cardine A, et al. SHP2-interacting transmembrane adaptor protein (SIT), a novel disulfide-linked dimer regulating human T cell activation. J. Exp. Med. 1999;189:1181–1194. doi: 10.1084/jem.189.8.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 167.Ritt DA, et al. CK2 Is a component of the KSR1 scaffold complex that contributes to Raf kinase activation. Curr. Biol. 2007;17:179–184. doi: 10.1016/j.cub.2006.11.061. [DOI] [PubMed] [Google Scholar]
- 168.Schaeffer HJ, et al. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- 169.Pudewell, S. & Ahmadian, M. R. Spotlight on accessory proteins: RTK-RAS-MAPK modulators as new therapeutic targets. Biomolecules11, 895 (2021). [DOI] [PMC free article] [PubMed]
- 170.McNulty DE, et al. MAPK scaffold IQGAP1 binds the EGF receptor and modulates its activation. J. Biol. Chem. 2011;286:15010–15021. doi: 10.1074/jbc.M111.227694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ishibe S, Joly D, Liu ZX, Cantley LG. Paxillin serves as an ERK-regulated scaffold for coordinating FAK and Rac activation in epithelial morphogenesis. Mol. Cell. 2004;16:257–267. doi: 10.1016/j.molcel.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 172.DeFea KA, et al. beta-arrestin-dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J. Cell Biol. 2000;148:1267–1281. doi: 10.1083/jcb.148.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Posada IM, et al. ASPP2 is a novel pan-ras nanocluster scaffold. PLoS One. 2016;11:e0159677. doi: 10.1371/journal.pone.0159677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li M, et al. Mechanistic insights into the long-range allosteric regulation of KRAS Via neurofibromatosis Type 1 (NF1) scaffold upon SPRED1 loading. J. Mol. Biol. 2022;434:167730. doi: 10.1016/j.jmb.2022.167730. [DOI] [PubMed] [Google Scholar]
- 175.Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16:45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 176.Zhou L, Hanemann CO. Merlin, a multi-suppressor from cell membrane to the nucleus. FEBS Lett. 2012;586:1403–1408. doi: 10.1016/j.febslet.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 177.Inder KL, Hill MM, Hancock JF. Nucleophosmin and nucleolin regulate K-Ras signaling. Commun. Integr Biol. 2010;3:188–190. doi: 10.4161/cib.3.2.10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Inder KL, et al. Nucleophosmin and nucleolin regulate K-Ras plasma membrane interactions and MAPK signal transduction. J. Biol. Chem. 2009;284:28410–28419. doi: 10.1074/jbc.M109.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ikedife J, He J, Wei Y. PEA-15 engages in allosteric interactions using a common scaffold in a phosphorylation-dependent manner. Sci. Rep. 2022;12:116. doi: 10.1038/s41598-021-04099-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lee SE, et al. RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc. Natl Acad. Sci. USA. 2010;107:16994–16998. doi: 10.1073/pnas.1005362107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Vomastek T, et al. Modular construction of a signaling scaffold: MORG1 interacts with components of the ERK cascade and links ERK signaling to specific agonists. Proc. Natl Acad. Sci. USA. 2004;101:6981–6986. doi: 10.1073/pnas.0305894101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Blazevits O, et al. Galectin-1 dimers can scaffold Raf-effectors to increase H-ras nanoclustering. Sci. Rep. 2016;6:24165. doi: 10.1038/srep24165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yin G, Haendeler J, Yan C, Berk BC. GIT1 functions as a scaffold for MEK1-extracellular signal-regulated kinase 1 and 2 activation by angiotensin II and epidermal growth factor. Mol. Cell Biol. 2004;24:875–885. doi: 10.1128/MCB.24.2.875-885.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Villalobo A, Ishida H, Vogel HJ, Berchtold MW. Calmodulin as a protein linker and a regulator of adaptor/scaffold proteins. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:507–521. doi: 10.1016/j.bbamcr.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 185.Jang H, Stevens P, Gao T, Galperin E. The leucine-rich repeat signaling scaffolds Shoc2 and Erbin: cellular mechanism and role in disease. FEBS J. 2021;288:721–739. doi: 10.1111/febs.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Park J, et al. Deficiency in the LIM-only protein FHL2 impairs assembly of extracellular matrix proteins. FASEB J. 2008;22:2508–2520. doi: 10.1096/fj.07-095521. [DOI] [PubMed] [Google Scholar]
- 187.Chivers SB, Brackley AD, Jeske NA. Raf kinase inhibitory protein reduces bradykinin receptor desensitization. J. Neurochem. 2022;162:156–165. doi: 10.1111/jnc.15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dautel G, Merle M. Pronator quadratus free muscle flap for treatment of palmar defects. J. Hand. Surg. Br. 1993;18:576–578. doi: 10.1016/0266-7681(93)90006-2. [DOI] [PubMed] [Google Scholar]
- 189.Rath O, et al. The RKIP (Raf-1 Kinase Inhibitor Protein) conserved pocket binds to the phosphorylated N-region of Raf-1 and inhibits the Raf-1-mediated activated phosphorylation of MEK. Cell Signal. 2008;20:935–941. doi: 10.1016/j.cellsig.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 190.Granovsky AE, Rosner MR. Raf kinase inhibitory protein: a signal transduction modulator and metastasis suppressor. Cell Res. 2008;18:452–457. doi: 10.1038/cr.2008.43. [DOI] [PubMed] [Google Scholar]
- 191.Escara-Wilke J, Yeung K, Keller ET. Raf kinase inhibitor protein (RKIP) in cancer. Cancer Metastasis Rev. 2012;31:615–620. doi: 10.1007/s10555-012-9365-9. [DOI] [PubMed] [Google Scholar]
- 192.Zou Q, et al. RKIP suppresses the proliferation and metastasis of breast cancer cell lines through up-regulation of miR-185 targeting HMGA2. Arch Biochem. Biophys. 2016;610:25–32. doi: 10.1016/j.abb.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 193.Du Y, et al. MiR-543 promotes proliferation and epithelial-mesenchymal transition in prostate cancer via targeting RKIP. Cell Physiol. Biochem. 2017;41:1135–1146. doi: 10.1159/000464120. [DOI] [PubMed] [Google Scholar]
- 194.Li Y, et al. Bmi-1-induced miR-27a and miR-155 promote tumor metastasis and chemoresistance by targeting RKIP in gastric cancer. Mol. Cancer. 2020;19:109. doi: 10.1186/s12943-020-01229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Raquel-Cunha, A., Cardoso-Carneiro, D., Reis, R. M. & Martinho, O. Current status of Raf Kinase Inhibitor Protein (RKIP) in lung cancer: Behind RTK Signal. Cells8, 442 (2019). [DOI] [PMC free article] [PubMed]
- 196.Kim HS, et al. Reduced expression of Raf-1 kinase inhibitory protein predicts regional lymph node metastasis and shorter survival in esophageal squamous cell carcinoma. Pathol. Res. Pract. 2012;208:292–299. doi: 10.1016/j.prp.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 197.Koelzer VH, et al. Geographic analysis of RKIP expression and its clinical relevance in colorectal cancer. Br. J. Cancer. 2013;108:2088–2096. doi: 10.1038/bjc.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Minoo P, et al. Loss of raf-1 kinase inhibitor protein expression is associated with tumor progression and metastasis in colorectal cancer. Am. J. Clin. Pathol. 2007;127:820–827. doi: 10.1309/5D7MM22DAVGDT1R8. [DOI] [PubMed] [Google Scholar]
- 199.Huang W, et al. Downregulation of RKIP promotes radioresistance of nasopharyngeal carcinoma by activating NRF2/NQO1 axis via downregulating miR-450b-5p. Cell Death Dis. 2020;11:504. doi: 10.1038/s41419-020-2695-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 200.Jing, S. H., Gao, X., Yu, B. & Qiao, H. Raf Kinase Inhibitor Protein (RKIP) Inhibits Tumor Necrosis Factor-alpha (TNF-alpha) Induced Adhesion Molecules Expression in Vascular Smooth Muscle Bells by Suppressing (Nuclear Transcription Factor-kappaB (NF-kappaB) Pathway. Med. Sci. Monit. 23, 4789–4797 (2017).. [DOI] [PMC free article] [PubMed]
- 201.Wottrich S, et al. Inverse correlation between the metastasis suppressor RKIP and the metastasis inducer YY1: Contrasting roles in the regulation of chemo/immuno-resistance in cancer. Drug Resist. Updat. 2017;30:28–38. doi: 10.1016/j.drup.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 202.Yang K, et al. KRAS promotes tumor metastasis and chemoresistance by repressing RKIP via the MAPK-ERK pathway in pancreatic cancer. Int. J. Cancer. 2018;142:2323–2334. doi: 10.1002/ijc.31248. [DOI] [PubMed] [Google Scholar]
- 203.Yuan L, et al. Reduced RKIP enhances nasopharyngeal carcinoma radioresistance by increasing ERK and AKT activity. Oncotarget. 2016;7:11463–11477. doi: 10.18632/oncotarget.7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wang A, et al. Reduced RKIP expression levels are associated with frequent non-small cell lung cancer metastasis and STAT3 phosphorylation and activation. Oncol. Lett. 2017;13:3039–3045. doi: 10.3892/ol.2017.5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Al-Mulla F, et al. A new model for raf kinase inhibitory protein induced chemotherapeutic resistance. PLoS One. 2012;7:e29532. doi: 10.1371/journal.pone.0029532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.He QY, et al. Reduction of RKIP expression promotes nasopharyngeal carcinoma invasion and metastasis by activating Stat3 signaling. Oncotarget. 2015;6:16422–16436. doi: 10.18632/oncotarget.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Baccarini M. Second nature: biological functions of the Raf-1 “kinase”. FEBS Lett. 2005;579:3271–3277. doi: 10.1016/j.febslet.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 208.Hong L, Wang Y, Chen W, Yang S. MicroRNA-508 suppresses epithelial-mesenchymal transition, migration, and invasion of ovarian cancer cells through the MAPK1/ERK signaling pathway. J. Cell Biochem. 2018;119:7431–7440. doi: 10.1002/jcb.27052. [DOI] [PubMed] [Google Scholar]
- 209.Tang, Q., Wu, J., Zheng, F., Hann, S.S. & Chen, Y. Emodin Increases Expression of Insulin-Like Growth Factor Binding Protein 1 through Activation of MEK/ERK/AMPKα and Interaction of PPARγ and Sp1 in Lung Cancer. Cell Physiol. Biochem.41, 339–357 (2017). [DOI] [PubMed]
- 210.Baek JH, et al. Hypoxia-induced VEGF enhances tumor survivability via suppression of serum deprivation-induced apoptosis. Oncogene. 2000;19:4621–4631. doi: 10.1038/sj.onc.1203814. [DOI] [PubMed] [Google Scholar]
- 211.Huang Y, et al. miR‑101 regulates the cell proliferation and apoptosis in diffuse large B‑cell lymphoma by targeting MEK1 via regulation of the ERK/MAPK signaling pathway. Oncol. Rep. 2019;41:377–386. doi: 10.3892/or.2018.6821. [DOI] [PubMed] [Google Scholar]
- 212.Lefloch R, Pouyssegur J, Lenormand P. Total ERK1/2 activity regulates cell proliferation. Cell Cycle. 2009;8:705–711. doi: 10.4161/cc.8.5.7734. [DOI] [PubMed] [Google Scholar]
- 213.Allan LA, et al. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat. Cell Biol. 2003;5:647–654. doi: 10.1038/ncb1005. [DOI] [PubMed] [Google Scholar]
- 214.Malumbres M, et al. Cellular response to oncogenic ras involves induction of the Cdk4 and Cdk6 inhibitor p15(INK4b) Mol. Cell Biol. 2000;20:2915–2925. doi: 10.1128/MCB.20.8.2915-2925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chang MC, et al. Mesothelin enhances invasion of ovarian cancer by inducing MMP-7 through MAPK/ERK and JNK pathways. Biochem. J. 2012;442:293–302. doi: 10.1042/BJ20110282. [DOI] [PubMed] [Google Scholar]
- 216.Bian CX, et al. P70S6K 1 regulation of angiogenesis through VEGF and HIF-1alpha expression. Biochem. Biophys. Res. Commun. 2010;398:395–399. doi: 10.1016/j.bbrc.2010.06.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chen J, et al. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK-ERK independent mechanism. Proc. Natl Acad. Sci. USA. 2001;98:7783–7788. doi: 10.1073/pnas.141224398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Nolan, A. A., Aboud, N. K., Kolch, W. & Matallanas, D. Hidden targets in RAF signaling pathways to block oncogenic RAS signaling. Genes12, 553 (2021). [DOI] [PMC free article] [PubMed]
- 219.Huser M, et al. MEK kinase activity is not necessary for Raf-1 function. EMBO J. 2001;20:1940–1951. doi: 10.1093/emboj/20.8.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Galabova-Kovacs G, et al. ERK and beyond: insights from B-Raf and Raf-1 conditional knockouts. Cell Cycle. 2006;5:1514–1518. doi: 10.4161/cc.5.14.2981. [DOI] [PubMed] [Google Scholar]
- 221.Bui NL, et al. Bad phosphorylation as a target of inhibition in oncology. Cancer Lett. 2018;415:177–186. doi: 10.1016/j.canlet.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 222.Wang HG, Rapp UR, Reed JC. Bcl-2 targets the protein kinase Raf-1 to mitochondria. Cell. 1996;87:629–638. doi: 10.1016/S0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- 223.Marliss EB, Caron D, Albisser AM, Zinman B. Present and future expectations regarding insulin infusion systems. Diabetes Care. 1981;4:325–327. doi: 10.2337/diacare.4.2.325. [DOI] [PubMed] [Google Scholar]
- 224.Matsuzawa A, et al. Physiological roles of ASK1-mediated signal transduction in oxidative stress- and endoplasmic reticulum stress-induced apoptosis: advanced findings from ASK1 knockout mice. Antioxid. Redox. Signal. 2002;4:415–425. doi: 10.1089/15230860260196218. [DOI] [PubMed] [Google Scholar]
- 225.Alavi AS, Acevedo L, Min W, Cheresh DA. Chemoresistance of endothelial cells induced by basic fibroblast growth factor depends on Raf-1-mediated inhibition of the proapoptotic kinase, ASK1. Cancer Res. 2007;67:2766–2772. doi: 10.1158/0008-5472.CAN-06-3648. [DOI] [PubMed] [Google Scholar]
- 226.Du J, Cai SH, Shi Z, Nagase F. Binding activity of H-Ras is necessary for in vivo inhibition of ASK1 activity. Cell Res. 2004;14:148–154. doi: 10.1038/sj.cr.7290214. [DOI] [PubMed] [Google Scholar]
- 227.Yamaguchi O, et al. Cardiac-specific disruption of the c-raf-1 gene induces cardiac dysfunction and apoptosis. J. Clin. Invest. 2004;114:937–943. doi: 10.1172/JCI200420317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.O’Neill E, Rushworth L, Baccarini M, Kolch W. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science. 2004;306:2267–2270. doi: 10.1126/science.1103233. [DOI] [PubMed] [Google Scholar]
- 229.O’Neill E, Kolch W. Taming the Hippo: Raf-1 controls apoptosis by suppressing MST2/Hippo. Cell Cycle. 2005;4:365–367. doi: 10.4161/cc.4.3.1531. [DOI] [PubMed] [Google Scholar]
- 230.Kilili GK, Kyriakis JM. Mammalian Ste20-like kinase (Mst2) indirectly supports Raf-1/ERK pathway activity via maintenance of protein phosphatase-2A catalytic subunit levels and consequent suppression of inhibitory Raf-1 phosphorylation. J. Biol. Chem. 2010;285:15076–15087. doi: 10.1074/jbc.M109.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rauch J, et al. Heterogeneous nuclear ribonucleoprotein H blocks MST2-mediated apoptosis in cancer cells by regulating A-Raf transcription. Cancer Res. 2010;70:1679–1688. doi: 10.1158/0008-5472.CAN-09-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Galan JA, Avruch J. MST1/MST2 protein kinases: regulation and physiologic roles. Biochemistry. 2016;55:5507–5519. doi: 10.1021/acs.biochem.6b00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Mielgo A, et al. A MEK-independent role for CRAF in mitosis and tumor progression. Nat. Med. 2011;17:1641–1645. doi: 10.1038/nm.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Joukov, V. & De Nicolo, A. Aurora-PLK1 cascades as key signaling modules in the regulation of mitosis. Sci. Signal. 11, eaar4195 (2018). [DOI] [PubMed]
- 235.Advani SJ, et al. Kinase-independent role for CRAF-driving tumour radioresistance via CHK2. Nat. Commun. 2015;6:8154. doi: 10.1038/ncomms9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Khazak V, Astsaturov I, Serebriiskii IG, Golemis EA. Selective Raf inhibition in cancer therapy. Expert Opin. Ther. Targets. 2007;11:1587–1609. doi: 10.1517/14728222.11.12.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Galaktionov K, Jessus C, Beach D. Raf1 interaction with Cdc25 phosphatase ties mitogenic signal transduction to cell cycle activation. Genes Dev. 1995;9:1046–1058. doi: 10.1101/gad.9.9.1046. [DOI] [PubMed] [Google Scholar]
- 238.Dhawan P, Richmond A. A novel NF-kappa B-inducing kinase-MAPK signaling pathway up-regulates NF-kappa B activity in melanoma cells. J. Biol. Chem. 2002;277:7920–7928. doi: 10.1074/jbc.M112210200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cheung M, Sharma A, Madhunapantula SV, Robertson GP. Akt3 and mutant V600E B-Raf cooperate to promote early melanoma development. Cancer Res. 2008;68:3429–3439. doi: 10.1158/0008-5472.CAN-07-5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Palmieri G, et al. Main roads to melanoma. J. Transl. Med. 2009;7:86. doi: 10.1186/1479-5876-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Amiri KI, Richmond A. Role of nuclear factor-kappa B in melanoma. Cancer Metastasis Rev. 2005;24:301–313. doi: 10.1007/s10555-005-1579-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Templer DI, et al. Exploration of head injury without medical attention. Percept. Mot. Skills. 1992;75:195–202. doi: 10.2466/pms.1992.75.1.195. [DOI] [PubMed] [Google Scholar]
- 243.Choux M, et al. Hydatid cyst of the fourth ventricle in a six-year-old child. Mod. Probl. Paediatr. 1976;18:273–275. [PubMed] [Google Scholar]
- 244.Baumann B, et al. Raf induces NF-kappaB by membrane shuttle kinase MEKK1, a signaling pathway critical for transformation. Proc. Natl Acad. Sci. USA. 2000;97:4615–4620. doi: 10.1073/pnas.080583397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Jette C, Thorburn A. A Raf-induced, MEK-independent signaling pathway regulates atrial natriuretic factor gene expression in cardiac muscle cells. FEBS Lett. 2000;467:1–6. doi: 10.1016/S0014-5793(00)01114-5. [DOI] [PubMed] [Google Scholar]
- 246.Ehrenreiter K, et al. Raf-1 regulates Rho signaling and cell migration. J. Cell Biol. 2005;168:955–964. doi: 10.1083/jcb.200409162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ehrenreiter K, et al. Raf-1 addiction in Ras-induced skin carcinogenesis. Cancer Cell. 2009;16:149–160. doi: 10.1016/j.ccr.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 248.Piazzolla D, et al. Raf-1 sets the threshold of Fas sensitivity by modulating Rok-alpha signaling. J. Cell Biol. 2005;171:1013–1022. doi: 10.1083/jcb.200504137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Niault T, et al. From autoinhibition to inhibition in trans: the Raf-1 regulatory domain inhibits Rok-alpha kinase activity. J. Cell Biol. 2009;187:335–342. doi: 10.1083/jcb.200906178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kern F, et al. Essential, non-redundant roles of B-Raf and Raf-1 in Ras-driven skin tumorigenesis. Oncogene. 2013;32:2483–2492. doi: 10.1038/onc.2012.254. [DOI] [PubMed] [Google Scholar]
- 251.Janosch P, et al. The Raf-1 kinase associates with vimentin kinases and regulates the structure of vimentin filaments. FASEB J. 2000;14:2008–2021. doi: 10.1096/fj.99-0883com. [DOI] [PubMed] [Google Scholar]
- 252.Ku NO, Fu H, Omary MB. Raf-1 activation disrupts its binding to keratins during cell stress. J. Cell Biol. 2004;166:479–485. doi: 10.1083/jcb.200402051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.De Luca A, et al. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin. Ther. Targets. 2012;16(Suppl 2):S17–S27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 254.Moelling K, et al. Regulation of Raf-Akt Cross-talk. J. Biol. Chem. 2002;277:31099–31106. doi: 10.1074/jbc.M111974200. [DOI] [PubMed] [Google Scholar]
- 255.Nguyen LK, et al. Competing to coordinate cell fate decisions: the MST2-Raf-1 signaling device. Cell Cycle. 2015;14:189–199. doi: 10.4161/15384101.2014.973743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Romano D, et al. Proapoptotic kinase MST2 coordinates signaling crosstalk between RASSF1A, Raf-1, and Akt. Cancer Res. 2010;70:1195–1203. doi: 10.1158/0008-5472.CAN-09-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Matallanas D, et al. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol. Cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 259.Ravikumar B, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 260.Khandia, R. et al. A comprehensive review of autophagy and its various roles in infectious, non-infectious, and lifestyle diseases: current knowledge and prospects for disease prevention, novel drug design, and therapy. Cells8, 674 (2019). [DOI] [PMC free article] [PubMed]
- 261.Mulcahy Levy JM, Thorburn A. Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 2020;27:843–857. doi: 10.1038/s41418-019-0474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Mol. Pharmacol. 2014;85:830–838. doi: 10.1124/mol.114.091850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 265.Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307–3318. doi: 10.1002/cncr.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kim JH, et al. Raf/MEK/ERK can regulate cellular levels of LC3B and SQSTM1/p62 at expression levels. Exp. Cell Res. 2014;327:340–352. doi: 10.1016/j.yexcr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Xie X, et al. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov. 2015;5:410–423. doi: 10.1158/2159-8290.CD-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Liu H, He Z, Simon HU. Autophagy suppresses melanoma tumorigenesis by inducing senescence. Autophagy. 2014;10:372–373. doi: 10.4161/auto.27163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wu Y, Zhang T, Zhang X, Gao Q. Decoding the complexity of metastasis. Cancer Biol. Med. 2022;19:284–288. doi: 10.20892/j.issn.2095-3941.2022.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Massague J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Bergers G, Fendt SM. The metabolism of cancer cells during metastasis. Nat. Rev. Cancer. 2021;21:162–180. doi: 10.1038/s41568-020-00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Faubert, B., Solmonson, A. & DeBerardinis, R. J. Metabolic reprogramming and cancer progression. Science368, eaaw5473 (2020). [DOI] [PMC free article] [PubMed]
- 273.Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes. 2000;49:677–683. doi: 10.2337/diabetes.49.5.677. [DOI] [PubMed] [Google Scholar]
- 274.Randle PJ, Garland PB, Hales CN, Newsholme EA. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1:785–789. doi: 10.1016/S0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- 275.Vriens K, et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature. 2019;566:403–406. doi: 10.1038/s41586-019-0904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Triki M, et al. mTOR Signaling and SREBP activity increase FADS2 expression and can activate sapienate biosynthesis. Cell Rep. 2020;31:107806. doi: 10.1016/j.celrep.2020.107806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Elia I, Doglioni G, Fendt SM. Metabolic hallmarks of metastasis formation. Trends Cell Biol. 2018;28:673–684. doi: 10.1016/j.tcb.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 278.Krebs AM, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 279.Pastushenko I, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 280.Hao, Y., Baker, D. & Ten Dijke, P. TGF-beta-mediated epithelial-mesenchymal transition and cancer metastasis. Int. J. Mol. Sci. 20, 2767 (2019). [DOI] [PMC free article] [PubMed]
- 281.Lee MK, et al. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 2007;26:3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Mulder KM, Morris SL. Activation of p21ras by transforming growth factor beta in epithelial cells. J. Biol Chem. 1992;267:5029–5031. doi: 10.1016/S0021-9258(18)42722-6. [DOI] [PubMed] [Google Scholar]
- 283.Wong A, et al. FRS2 alpha attenuates FGF receptor signaling by Grb2-mediated recruitment of the ubiquitin ligase Cbl. Proc. Natl Acad. Sci. USA. 2002;99:6684–6689. doi: 10.1073/pnas.052138899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Yuan X, et al. Notch signaling and EMT in non-small cell lung cancer: biological significance and therapeutic application. J. Hematol. Oncol. 2014;7:87. doi: 10.1186/s13045-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Cai J, et al. Simultaneous overactivation of Wnt/beta-catenin and TGFbeta signalling by miR-128-3p confers chemoresistance-associated metastasis in NSCLC. Nat Commun. 2017;8:15870. doi: 10.1038/ncomms15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zhang, N. et al. The interaction of the senescent and adjacent breast cancer cells promotes the metastasis of heterogeneous breast cancer cells through notch signaling. Int. J. Mol. Sci. 22, 849 (2021). [DOI] [PMC free article] [PubMed]
- 287.Kolbe N, et al. Data-based stochastic modeling reveals sources of activity bursts in single-cell TGF-beta signaling. PLoS Comput. Biol. 2022;18:e1010266. doi: 10.1371/journal.pcbi.1010266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98:1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 290.Ko, C. C., Hsieh, Y. Y. & Yang, P. M. Long non-coding RNA MIR31HG promotes the transforming growth factor beta-induced epithelial-mesenchymal transition in pancreatic ductal adenocarcinoma cells. Int. J. Mol. Sci.23, 6559 (2022). [DOI] [PMC free article] [PubMed]
- 291.Janda E, et al. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J. Cell Biol. 2002;156:299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Xie L, et al. Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 2004;6:603–610. doi: 10.1593/neo.04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wang Y, et al. Autophagy inhibition specifically promotes epithelial-mesenchymal transition and invasion in RAS-mutated cancer cells. Autophagy. 2019;15:886–899. doi: 10.1080/15548627.2019.1569912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Qin W, et al. Inhibition of autophagy promotes metastasis and glycolysis by inducing ROS in gastric cancer cells. Oncotarget. 2015;6:39839–39854. doi: 10.18632/oncotarget.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Qiang L, He YY. Autophagy deficiency stabilizes TWIST1 to promote epithelial-mesenchymal transition. Autophagy. 2014;10:1864–1865. doi: 10.4161/auto.32171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Alizadeh J, et al. Autophagy modulates transforming growth factor beta 1 induced epithelial to mesenchymal transition in non-small cell lung cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:749–768. doi: 10.1016/j.bbamcr.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 297.Liang C, et al. TGFB1-induced autophagy affects the pattern of pancreatic cancer progression in distinct ways depending on SMAD4 status. Autophagy. 2020;16:486–500. doi: 10.1080/15548627.2019.1628540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Peng YF, et al. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy. 2013;9:2056–2068. doi: 10.4161/auto.26398. [DOI] [PubMed] [Google Scholar]
- 299.Lv Q, Hua F, Hu ZW. DEDD, a novel tumor repressor, reverses epithelial-mesenchymal transition by activating selective autophagy. Autophagy. 2012;8:1675–1676. doi: 10.4161/auto.21438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Chen S, Guan JL. Tumor-promoting and -suppressive roles of autophagy in the same mouse model of BrafV600E-driven lung cancer. Cancer Discov. 2013;3:1225–1227. doi: 10.1158/2159-8290.CD-13-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Karsli-Uzunbas G, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Strohecker AM, White E. Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy. 2014;10:384–385. doi: 10.4161/auto.27320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Yuan M, et al. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal. Transduct. Target Ther. 2019;4:61. doi: 10.1038/s41392-019-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Massarelli E, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin. Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 305.Bou-Assaly W, Mukherji S. Cetuximab (erbitux) AJNR Am. J. Neuroradiol. 2010;31:626–627. doi: 10.3174/ajnr.A2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Petrelli F, et al. Cetuximab and panitumumab in KRAS wild-type colorectal cancer: a meta-analysis. Int. J. Colorectal. Dis. 2011;26:823–833. doi: 10.1007/s00384-011-1149-0. [DOI] [PubMed] [Google Scholar]
- 307.Tejpar S, et al. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J. Clin. Oncol. 2012;30:3570–3577. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 308.Yao YM, et al. Mouse PDX trial suggests synergy of concurrent inhibition of RAF and EGFR in colorectal cancer with BRAF or KRAS mutations. Clin Cancer Res. 2017;23:5547–5560. doi: 10.1158/1078-0432.CCR-16-3250. [DOI] [PubMed] [Google Scholar]
- 309.Schirripa M, et al. Phase II study of single-agent cetuximab in KRAS G13D mutant metastatic colorectal cancer. Ann. Oncol. 2015;26:2503. doi: 10.1093/annonc/mdv385. [DOI] [PubMed] [Google Scholar]
- 310.Ou SI, et al. First-in-human phase I/IB dose-finding study of adagrasib (MRTX849) in patients with advanced KRAS(G12C) solid tumors (KRYSTAL-1) J. Clin. Oncol. 2022;40:2530–2538. doi: 10.1200/JCO.21.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Nakajima EC, et al. FDA Approval Summary: Sotorasib for KRAS G12C-Mutated Metastatic NSCLC. Clin. Cancer Res. 2022;28:1482–1486. doi: 10.1158/1078-0432.CCR-21-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Skoulidis F, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N. Engl. J. Med. 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Loos NHC, et al. ABCB1 limits brain exposure of the KRAS(G12C) inhibitor sotorasib, whereas ABCB1, CYP3A, and possibly OATP1a/1b restrict its oral availability. Pharmacol. Res. 2022;178:106137. doi: 10.1016/j.phrs.2022.106137. [DOI] [PubMed] [Google Scholar]
- 314.De, S. K. First approval of adagrasib for the treatment of non-small cell lung cancer harboring a KRASG12C mutation. Curr. Med. Chem.31, 266–272 (2024). [DOI] [PubMed]
- 315.Tian H, Yang Z, He J. Adagrasib: a landmark in the KRAS(G12C)-mutated NSCLC. MedComm (2020) 2022;3:e190. doi: 10.1002/mco2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Yaeger R, et al. Adagrasib with or without cetuximab in colorectal cancer with mutated KRAS G12C. N. Engl. J. Med. 2023;388:44–54. doi: 10.1056/NEJMoa2212419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Loos NHC, et al. Pharmacokinetics of the KRAS(G12C) inhibitor adagrasib is limited by CYP3A and ABCB1, and influenced by binding to mouse plasma carboxylesterase 1c. Biomed. Pharmacother. 2023;166:115304. doi: 10.1016/j.biopha.2023.115304. [DOI] [PubMed] [Google Scholar]
- 318.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl Acad. Sci. USA. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Hyman DM, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N. Engl. J. Med. 2015;373:726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Ballantyne AD, Garnock-Jones KP. Dabrafenib: first global approval. Drugs. 2013;73:1367–1376. doi: 10.1007/s40265-013-0095-2. [DOI] [PubMed] [Google Scholar]
- 322.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat. Med. 2013;19:1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 323.Koelblinger P, Thuerigen O, Dummer R. Development of encorafenib for BRAF-mutated advanced melanoma. Curr. Opin. Oncol. 2018;30:125–133. doi: 10.1097/CCO.0000000000000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Delord JP, et al. Phase I dose-escalation and -expansion study of the BRAF inhibitor encorafenib (LGX818) in metastatic BRAF-mutant melanoma. Clin. Cancer Res. 2017;23:5339–5348. doi: 10.1158/1078-0432.CCR-16-2923. [DOI] [PubMed] [Google Scholar]
- 326.Sosman JA, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Johnson DB, et al. Acquired BRAF inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur. J. Cancer. 2015;51:2792–2799. doi: 10.1016/j.ejca.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Song Y, et al. Targeting RAS-RAF-MEK-ERK signaling pathway in human cancer: current status in clinical trials. Genes Dis. 2023;10:76–88. doi: 10.1016/j.gendis.2022.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kirchberger MC, et al. MEK inhibition may increase survival of NRAS-mutated melanoma patients treated with checkpoint blockade: Results of a retrospective multicentre analysis of 364 patients. Eur. J. Cancer. 2018;98:10–16. doi: 10.1016/j.ejca.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 330.Yamaguchi T, et al. Antitumor activities of JTP-74057 (GSK1120212), a novel MEK1/2 inhibitor, on colorectal cancer cell lines in vitro and in vivo. Int. J. Oncol. 2011;39:23–31. doi: 10.3892/ijo.2011.1015. [DOI] [PubMed] [Google Scholar]
- 331.Flaherty KT, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Sarkisian S, Davar D. MEK inhibitors for the treatment of NRAS mutant melanoma. Drug Des. Dev. Ther. 2018;12:2553–2565. doi: 10.2147/DDDT.S131721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Andrlova H, Zeiser R, Meiss F. Cobimetinib (GDC-0973, XL518) Recent Results Cancer Res. 2018;211:177–186. doi: 10.1007/978-3-319-91442-8_12. [DOI] [PubMed] [Google Scholar]
- 334.Shirley M. Encorafenib and Binimetinib: first global approvals. Drugs. 2018;78:1277–1284. doi: 10.1007/s40265-018-0963-x. [DOI] [PubMed] [Google Scholar]
- 335.Casey D, et al. FDA approval summary: selumetinib for plexiform neurofibroma. Clin. Cancer Res. 2021;27:4142–4146. doi: 10.1158/1078-0432.CCR-20-5032. [DOI] [PubMed] [Google Scholar]
- 336.Khan S, et al. Intermittent MEK inhibition for the treatment of metastatic uveal melanoma. Front. Oncol. 2022;12:975643. doi: 10.3389/fonc.2022.975643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Larkin J, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 338.Brastianos PK, et al. BRAF-MEK inhibition in newly diagnosed papillary craniopharyngiomas. N. Engl. J. Med. 2023;389:118–126. doi: 10.1056/NEJMoa2213329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Rajkumar S, et al. Melanomas with concurrent BRAF non-p.V600 and NF1 loss-of-function mutations are targetable by BRAF/MEK inhibitor combination therapy. Cell Rep. 2022;39:110634. doi: 10.1016/j.celrep.2022.110634. [DOI] [PubMed] [Google Scholar]
- 340.Long GV, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 341.Ishii N, et al. Enhanced inhibition of ERK signaling by a novel allosteric MEK inhibitor, CH5126766, that suppresses feedback reactivation of RAF activity. Cancer Res. 2013;73:4050–4060. doi: 10.1158/0008-5472.CAN-12-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Guo C, et al. Intermittent schedules of the oral RAF-MEK inhibitor CH5126766/VS-6766 in patients with RAS/RAF-mutant solid tumours and multiple myeloma: a single-centre, open-label, phase 1 dose-escalation and basket dose-expansion study. Lancet Oncol. 2020;21:1478–1488. doi: 10.1016/S1470-2045(20)30464-2. [DOI] [PubMed] [Google Scholar]
- 343.Nobre, L. et al. Outcomes of BRAF V600E pediatric gliomas treated with targeted BRAF inhibition. JCO Precis. Oncol. 4, PO.19.00298 (2020). [DOI] [PMC free article] [PubMed]
- 344.Bouffet E, et al. Dabrafenib plus Trametinib in Pediatric Glioma with BRAF V600 Mutations. N. Engl. J. Med. 2023;389:1108–1120. doi: 10.1056/NEJMoa2303815. [DOI] [PubMed] [Google Scholar]
- 345.Eroglu Z, Ozgun A. Updates and challenges on treatment with BRAF/MEK-inhibitors in melanoma. Expert Opin. Orphan Drugs. 2018;6:545–551. doi: 10.1080/21678707.2018.1512402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Selvasaravanan KD, et al. The limitations of targeting MEK signalling in Glioblastoma therapy. Sci. Rep. 2020;10:7401. doi: 10.1038/s41598-020-64289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Sakji-Dupre L, et al. Cerebrospinal fluid concentrations of vemurafenib in patients treated for brain metastatic BRAF-V600 mutated melanoma. Melanoma Res. 2015;25:302–305. doi: 10.1097/CMR.0000000000000162. [DOI] [PubMed] [Google Scholar]
- 348.Eroglu Z, et al. Combined BRAF and HSP90 Inhibition in Patients with Unresectable BRAF (V600E)-Mutant Melanoma. Clin. Cancer Res. 2018;24:5516–5524. doi: 10.1158/1078-0432.CCR-18-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Nassar KW, et al. Targeting CDK4/6 represents a therapeutic vulnerability in acquired BRAF/MEK inhibitor-resistant melanoma. Mol. Cancer Ther. 2021;20:2049–2060. doi: 10.1158/1535-7163.MCT-20-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Hauschild A, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomized controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 351.Spagnolo F, et al. BRAF-mutant melanoma: treatment approaches, resistance mechanisms, and diagnostic strategies. Onco. Targets Ther. 2015;8:157–168. doi: 10.2147/OTT.S39096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Smalley KS, et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol. Cancer Ther. 2008;7:2876–2883. doi: 10.1158/1535-7163.MCT-08-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Lin L, Bivona TG. The Hippo effector YAP regulates the response of cancer cells to MAPK pathway inhibitors. Mol. Cell Oncol. 2016;3:e1021441. doi: 10.1080/23723556.2015.1021441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Lin L, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 2015;47:250–256. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Paraiso KH, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Yang JY, et al. Activation of FOXO3a is sufficient to reverse mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor chemoresistance in human cancer. Cancer Res. 2010;70:4709–4718. doi: 10.1158/0008-5472.CAN-09-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Shao Y, Aplin AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer Res. 2010;70:6670–6681. doi: 10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.De Bruyne E, et al. IGF-1 suppresses Bim expression in multiple myeloma via epigenetic and posttranslational mechanisms. Blood. 2010;115:2430–2440. doi: 10.1182/blood-2009-07-232801. [DOI] [PubMed] [Google Scholar]
- 359.Neel, D. S. & Bivona, T. G. Resistance is futile: overcoming resistance to targeted therapies in lung adenocarcinoma. NPJ Precis. Oncol. 1, 3 (2017). [DOI] [PMC free article] [PubMed]
- 360.Montero-Conde C, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 2013;3:520–533. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Straussman R, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Zaman, A., Wu, W. & Bivona, T. G. Targeting oncogenic BRAF: past, present, and future. Cancers11, 1197 (2019). [DOI] [PMC free article] [PubMed]
- 363.Whittaker S, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci. Transl. Med. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 364.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Lin L, et al. Mapping the molecular determinants of BRAF oncogene dependence in human lung cancer. Proc. Natl Acad. Sci. USA. 2014;111:E748–E757. doi: 10.1073/pnas.1320956111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Rizos H, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin. Cancer Res. 2014;20:1965–1977. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 367.Garraway LA, Hahn WC. On or off target: mutations, models, and predictions. Sci. Transl. Med. 2010;2:35ps28. doi: 10.1126/scitranslmed.3001263. [DOI] [PubMed] [Google Scholar]
- 368.Montagut C, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Sharma, V. et al. Registered report: COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Elife5, e11414 (2016). [DOI] [PMC free article] [PubMed]
- 371.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Saei, A. & Eichhorn, P. J. A. Adaptive responses as mechanisms of resistance to BRAF inhibitors in melanoma. Cancers11, 1176 (2019). [DOI] [PMC free article] [PubMed]
- 373.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 374.Unni, A. M. et al. Hyperactivation of ERK by multiple mechanisms is toxic to RTK-RAS mutation-driven lung adenocarcinoma cells. Elife7, e33718 (2018). [DOI] [PMC free article] [PubMed]
- 375.Leung GP, et al. Hyperactivation of MAPK signaling is deleterious to RAS/RAF-mutant melanoma. Mol. Cancer Res. 2019;17:199–211. doi: 10.1158/1541-7786.MCR-18-0327. [DOI] [PubMed] [Google Scholar]
- 376.Lake D, Correa SA, Muller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol. Life Sci. 2016;73:4397–4413. doi: 10.1007/s00018-016-2297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Kidger AM, Keyse SM. The regulation of oncogenic Ras/ERK signalling by dual-specificity mitogen activated protein kinase phosphatases (MKPs) Semin. Cell Dev. Biol. 2016;50:125–132. doi: 10.1016/j.semcdb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signaling pathway. Nat. Cell Biol. 2002;4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- 379.Ueki K, et al. Feedback regulation of mitogen-activated protein kinase kinase kinase activity of c-Raf-1 by insulin and phorbol ester stimulation. J. Biol. Chem. 1994;269:15756–15761. doi: 10.1016/S0021-9258(17)40745-9. [DOI] [PubMed] [Google Scholar]
- 380.Friday BB, et al. BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase and Raf proteins. Cancer Res. 2008;68:6145–6153. doi: 10.1158/0008-5472.CAN-08-1430. [DOI] [PubMed] [Google Scholar]
- 381.Brondello JM, Brunet A, Pouyssegur J, McKenzie FR. The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J. Biol. Chem. 1997;272:1368–1376. doi: 10.1074/jbc.272.2.1368. [DOI] [PubMed] [Google Scholar]
- 382.Amit I, et al. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 383.Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: a developing story. Nat. Rev. Mol. Cell Biol. 2004;5:441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- 384.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat. Rev. Mol. Cell Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 385.Li X, Huang Y, Jiang J, Frank SJ. ERK-dependent threonine phosphorylation of EGF receptor modulates receptor downregulation and signaling. Cell Signal. 2008;20:2145–2155. doi: 10.1016/j.cellsig.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl Acad. Sci. USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Joseph EW, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc. Natl Acad. Sci. USA. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Wan PT, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 389.Poulikakos PI, et al. RAF inhibitors transactivate RAF dimers and ERK signaling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Yao Z, et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell. 2015;28:370–383. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Karoulia Z, Gavathiotis E, Poulikakos PI. New perspectives for targeting RAF kinase in human cancer. Nat. Rev. Cancer. 2017;17:676–691. doi: 10.1038/nrc.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 394.Zhang C, et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature. 2015;526:583–586. doi: 10.1038/nature14982. [DOI] [PubMed] [Google Scholar]
- 395.Koumaki K, et al. BRAF paradox breakers PLX8394, PLX7904 are more effective against BRAFV600Epsilon CRC cells compared with the BRAF inhibitor PLX4720 and shown by detailed pathway analysis. Biochim. Biophys. Acta Mol. Basis Dis. 2021;1867:166061. doi: 10.1016/j.bbadis.2020.166061. [DOI] [PubMed] [Google Scholar]
- 396.Karoulia Z, et al. An integrated model of RAF inhibitor action predicts inhibitor activity against oncogenic BRAF signaling. Cancer Cell. 2016;30:485–498. doi: 10.1016/j.ccell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Monaco KA, et al. LXH254, a Potent and Selective ARAF-Sparing Inhibitor of BRAF and CRAF for the Treatment of MAPK-Driven Tumors. Clin. Cancer Res. 2021;27:2061–2073. doi: 10.1158/1078-0432.CCR-20-2563. [DOI] [PubMed] [Google Scholar]
- 398.Sun Y, et al. A brain-penetrant RAF dimer antagonist for the noncanonical BRAF oncoprotein of pediatric low-grade astrocytomas. Neuro. Oncol. 2017;19:774–785. doi: 10.1093/neuonc/now261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Girotti MR, et al. Paradox-breaking RAF inhibitors that also target SRC are effective in drug-resistant BRAF mutant melanoma. Cancer Cell. 2015;27:85–96. doi: 10.1016/j.ccell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Sullivan RJ, et al. A phase I study of LY3009120, a Pan-RAF inhibitor, in patients with advanced or metastatic cancer. Mol. Cancer Ther. 2020;19:460–467. doi: 10.1158/1535-7163.MCT-19-0681. [DOI] [PubMed] [Google Scholar]
- 401.Noeparast A, et al. Type II RAF inhibitor causes superior ERK pathway suppression compared to type I RAF inhibitor in cells expressing different BRAF mutant types recurrently found in lung cancer. Oncotarget. 2018;9:16110–16123. doi: 10.18632/oncotarget.24576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Yao Z, et al. RAF inhibitor PLX8394 selectively disrupts BRAF dimers and RAS-independent BRAF-mutant-driven signaling. Nat. Med. 2019;25:284–291. doi: 10.1038/s41591-018-0274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Tutuka CSA, et al. PLX8394, a new generation BRAF inhibitor, selectively inhibits BRAF in colonic adenocarcinoma cells and prevents paradoxical MAPK pathway activation. Mol. Cancer. 2017;16:112. doi: 10.1186/s12943-017-0684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Pickles OJ, et al. Paradox breaker BRAF inhibitors have comparable potency and MAPK pathway reactivation to encorafenib in BRAF mutant colorectal cancer. Oncotarget. 2020;11:3188–3197. doi: 10.18632/oncotarget.27681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Botton T, et al. Genetic heterogeneity of BRAF fusion kinases in melanoma affects drug responses. Cell Rep. 2019;29:573–588.e577. doi: 10.1016/j.celrep.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Jain P, et al. Overcoming resistance to single-agent therapy for oncogenic BRAF gene fusions via combinatorial targeting of MAPK and PI3K/mTOR signaling pathways. Oncotarget. 2017;8:84697–84713. doi: 10.18632/oncotarget.20949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Shi H, et al. Preexisting MEK1 exon 3 mutations in V600E/KBRAF melanomas do not confer resistance to BRAF inhibitors. Cancer Discov. 2012;2:414–424. doi: 10.1158/2159-8290.CD-12-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Paraiso KH, et al. Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. Br. J. Cancer. 2010;102:1724–1730. doi: 10.1038/sj.bjc.6605714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Frohlich F, Gerosa L, Muhlich J, Sorger PK. Mechanistic model of MAPK signaling reveals how allostery and rewiring contribute to drug resistance. Mol. Syst. Biol. 2023;19:e10988. doi: 10.15252/msb.202210988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Schreck KC, et al. RAF and MEK inhibitor therapy in adult patients with brain tumors: a case-based overview and practical management of adverse events. Neurooncol. Pract. 2020;7:369–375. doi: 10.1093/nop/npaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Daud A, Tsai K. Management of treatment-related adverse events with agents targeting the MAPK pathway in patients with metastatic melanoma. Oncologist. 2017;22:823–833. doi: 10.1634/theoncologist.2016-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Carlino MS, et al. Antiproliferative effects of continued mitogen-activated protein kinase pathway inhibition following acquired resistance to BRAF and/or MEK inhibition in melanoma. Mol. Cancer Ther. 2013;12:1332–1342. doi: 10.1158/1535-7163.MCT-13-0011. [DOI] [PubMed] [Google Scholar]
- 413.Smalley KS, Flaherty KT. Integrating BRAF/MEK inhibitors into combination therapy for melanoma. Br. J. Cancer. 2009;100:431–435. doi: 10.1038/sj.bjc.6604891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Hendrikse CSE, et al. The potential of RAS/RAF/MEK/ERK (MAPK) signaling pathway inhibitors in ovarian cancer: a systematic review and meta-analysis. Gynecol. Oncol. 2023;171:83–94. doi: 10.1016/j.ygyno.2023.01.038. [DOI] [PubMed] [Google Scholar]
- 415.Whittaker SR, et al. Combined Pan-RAF and MEK inhibition overcomes multiple resistance mechanisms to selective RAF inhibitors. Mol. Cancer Ther. 2015;14:2700–2711. doi: 10.1158/1535-7163.MCT-15-0136-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Del Curatolo A, et al. Therapeutic potential of combined BRAF/MEK blockade in BRAF-wild type preclinical tumor models. J. Exp. Clin. Cancer Res. 2018;37:140. doi: 10.1186/s13046-018-0820-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Gao Y, et al. Allele-specific mechanisms of activation of MEK1 mutants determine their properties. Cancer Discov. 2018;8:648–661. doi: 10.1158/2159-8290.CD-17-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Abdel-Wahab O, et al. Efficacy of intermittent combined RAF and MEK inhibition in a patient with concurrent BRAF- and NRAS-mutant malignancies. Cancer Discov. 2014;4:538–545. doi: 10.1158/2159-8290.CD-13-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Corcoran RB, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF(V600E)-mutant colorectal cancer. Cancer Discov. 2018;8:428–443. doi: 10.1158/2159-8290.CD-17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 422.Das Thakur M, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Kong X, et al. Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature. 2017;550:270–274. doi: 10.1038/nature24037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Welsh SJ, Rizos H, Scolyer RA, Long GV. Resistance to combination BRAF and MEK inhibition in metastatic melanoma: Where to next? Eur. J. Cancer. 2016;62:76–85. doi: 10.1016/j.ejca.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 425.Xue Y, et al. An approach to suppress the evolution of resistance in BRAF(V600E)-mutant cancer. Nat. Med. 2017;23:929–937. doi: 10.1038/nm.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Rittler, D. et al. Horizontal combination of MEK and PI3K/mTOR inhibition in BRAF mutant tumor cells with or without concomitant PI3K pathway mutations. Int. J. Mol. Sci. 21, 7649 (2020). [DOI] [PMC free article] [PubMed]
- 427.Corcoran RB. New therapeutic strategies for BRAF mutant colorectal cancers. J. Gastrointest. Oncol. 2015;6:650–659. doi: 10.3978/j.issn.2078-6891.2015.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Dankner M, et al. Classifying BRAF alterations in cancer: new rational therapeutic strategies for actionable mutations. Oncogene. 2018;37:3183–3199. doi: 10.1038/s41388-018-0171-x. [DOI] [PubMed] [Google Scholar]
- 429.Candido, S. et al. The PIK3CA H1047R mutation confers resistance to BRAF and MEK inhibitors in A375 melanoma cells through the cross-activation of MAPK and PI3K-Akt pathways. Pharmaceutics14, (2022). [DOI] [PMC free article] [PubMed]
- 430.Bonazzoli E, et al. PI3K oncogenic mutations mediate resistance to afatinib in HER2/neu overexpressing gynecological cancers. Gynecol. Oncol. 2019;153:158–164. doi: 10.1016/j.ygyno.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Phuchareon J, McCormick F, Eisele DW, Tetsu O. EGFR inhibition evokes innate drug resistance in lung cancer cells by preventing Akt activity and thus inactivating Ets-1 function. Proc. Natl Acad. Sci. USA. 2015;112:E3855–E3863. doi: 10.1073/pnas.1510733112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Schreck KC, et al. Deconvoluting mechanisms of acquired resistance to RAF inhibitors in BRAF(V600E)-Mutant Human Glioma. Clin. Cancer Res. 2021;27:6197–6208. doi: 10.1158/1078-0432.CCR-21-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008;27(Suppl 2):S43–S51. doi: 10.1038/onc.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Mulcahy Levy, J. M. et al. Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. Elife6, e19671 (2017). [DOI] [PMC free article] [PubMed]
- 435.Levy JM, et al. Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 2014;4:773–780. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Atefi M, et al. Reversing melanoma cross-resistance to BRAF and MEK inhibitors by co-targeting the AKT/mTOR pathway. PLoS One. 2011;6:e28973. doi: 10.1371/journal.pone.0028973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Nichols RJ, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat. Cell Biol. 2018;20:1064–1073. doi: 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Liu C, et al. Combinations with allosteric SHP2 inhibitor TNO155 to block receptor tyrosine kinase signaling. Clin Cancer Res. 2021;27:342–354. doi: 10.1158/1078-0432.CCR-20-2718. [DOI] [PubMed] [Google Scholar]
- 439.Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther. Adv. Med. Oncol. 2016;8:48–56. doi: 10.1177/1758834015616934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Das Thakur M, Stuart DD. The evolution of melanoma resistance reveals therapeutic opportunities. Cancer Res. 2013;73:6106–6110. doi: 10.1158/0008-5472.CAN-13-1633. [DOI] [PubMed] [Google Scholar]
- 441.Schreuer M, et al. Combination of dabrafenib plus trametinib for BRAF and MEK inhibitor pretreated patients with advanced BRAF(V600)-mutant melanoma: an open-label, single arm, dual-centre, phase 2 clinical trial. Lancet Oncol. 2017;18:464–472. doi: 10.1016/S1470-2045(17)30171-7. [DOI] [PubMed] [Google Scholar]
- 442.Fernandez LP, Gomez de Cedron M, Ramirez de Molina A. Alterations of lipid metabolism in cancer: implications in prognosis and treatment. Front. Oncol. 2020;10:577420. doi: 10.3389/fonc.2020.577420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Abildgaard C, Guldberg P. Molecular drivers of cellular metabolic reprogramming in melanoma. Trends Mol. Med. 2015;21:164–171. doi: 10.1016/j.molmed.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 444.Sundstrom T, et al. Inhibition of mitochondrial respiration prevents BRAF-mutant melanoma brain metastasis. Acta Neuropathol. Commun. 2019;7:55. doi: 10.1186/s40478-019-0712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Figarola JL, et al. Bioenergetic modulation with the mitochondria uncouplers SR4 and niclosamide prevents proliferation and growth of treatment-naive and vemurafenib-resistant melanomas. Oncotarget. 2018;9:36945–36965. doi: 10.18632/oncotarget.26421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Yu J, et al. Advanced cancer starvation therapy by simultaneous deprivation of lactate and glucose using a MOF nanoplatform. Adv. Sci. 2021;8:e2101467. doi: 10.1002/advs.202101467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Simons AL, et al. 2-Deoxy-D-glucose combined with cisplatin enhances cytotoxicity via metabolic oxidative stress in human head and neck cancer cells. Cancer Res. 2007;67:3364–3370. doi: 10.1158/0008-5472.CAN-06-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Parmenter TJ, et al. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov. 2014;4:423–433. doi: 10.1158/2159-8290.CD-13-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Rashmi R, et al. Radioresistant cervical cancers are sensitive to inhibition of glycolysis and redox metabolism. Cancer Res. 2018;78:1392–1403. doi: 10.1158/0008-5472.CAN-17-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Zhang M, et al. Adipocyte-derived lipids mediate melanoma progression via FATP proteins. Cancer Discov. 2018;8:1006–1025. doi: 10.1158/2159-8290.CD-17-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Yuan J, et al. The AMPK inhibitor overcomes the paradoxical effect of RAF inhibitors through blocking phospho-Ser-621 in the C terminus of CRAF. J Biol. Chem. 2018;293:14276–14284. doi: 10.1074/jbc.RA118.004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Yuan P, et al. Phenformin enhances the therapeutic benefit of BRAF(V600E) inhibition in melanoma. Proc. Natl Acad. Sci. USA. 2013;110:18226–18231. doi: 10.1073/pnas.1317577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Han J, et al. Elucidating molecular mechanisms of acquired resistance to BRAF inhibitors in melanoma using a microfluidic device and deep sequencing. Genom. Inform. 2021;19:e2. doi: 10.5808/gi.20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Konieczkowski DJ, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4:816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Hoek KS, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 456.Weeraratna AT, et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/S1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 457.Kemper K, de Goeje PL, Peeper DS, van Amerongen R. Phenotype switching: tumor cell plasticity as a resistance mechanism and target for therapy. Cancer Res. 2014;74:5937–5941. doi: 10.1158/0008-5472.CAN-14-1174. [DOI] [PubMed] [Google Scholar]
- 458.Long GV, et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 459.Irvine M, et al. Oncogenic PI3K/AKT promotes the step-wise evolution of combination BRAF/MEK inhibitor resistance in melanoma. Oncogenesis. 2018;7:72. doi: 10.1038/s41389-018-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.van Geel R, et al. A phase Ib dose-escalation study of encorafenib and Cetuximab with or without Alpelisib in Metastatic BRAF-mutant colorectal cancer. Cancer Discov. 2017;7:610–619. doi: 10.1158/2159-8290.CD-16-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Mao M, et al. Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin. Cancer Res. 2013;19:657–667. doi: 10.1158/1078-0432.CCR-11-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Forbes SA, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Catalanotti, F. et al. PTEN Loss-of-function alterations are associated with intrinsic resistance to BRAF inhibitors in metastatic melanoma. JCO Precis Oncol. 1, (2017). [DOI] [PMC free article] [PubMed]
- 464.Paraiso KH, et al. The HSP90 inhibitor XL888 overcomes BRAF inhibitor resistance mediated through diverse mechanisms. Clin. Cancer Res. 2012;18:2502–2514. doi: 10.1158/1078-0432.CCR-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Fedorenko IV, et al. BRAF inhibition generates a host-tumor niche that mediates therapeutic escape. J. Invest. Dermatol. 2015;135:3115–3124. doi: 10.1038/jid.2015.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Almeida FV, Douglass SM, Fane ME, Weeraratna AT. Bad company: microenvironmentally mediated resistance to targeted therapy in melanoma. Pigment Cell Melanoma Res. 2019;32:237–247. doi: 10.1111/pcmr.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Wang H, et al. Pro-tumor activities of macrophages in the progression of melanoma. Hum. Vaccine Immunother. 2017;13:1556–1562. doi: 10.1080/21645515.2017.1312043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Abu-Remaileh M, et al. Chronic inflammation induces a novel epigenetic program that is conserved in intestinal adenomas and in colorectal cancer. Cancer Res. 2015;75:2120–2130. doi: 10.1158/0008-5472.CAN-14-3295. [DOI] [PubMed] [Google Scholar]
- 469.Gallina G, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J. Clin. Invest. 2006;116:2777–2790. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Denkert C, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 471.Escuin-Ordinas H, et al. COX-2 inhibition prevents the appearance of cutaneous squamous cell carcinomas accelerated by BRAF inhibitors. Mol. Oncol. 2014;8:250–260. doi: 10.1016/j.molonc.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Brady DC, et al. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature. 2014;509:492–496. doi: 10.1038/nature13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Dogan, E., Kara, H. G., Kosova, B. & Cetintas, V. B. in Metastasis (ed C. M. Sergi) (2022). [PubMed]
- 474.Mukherjee N, et al. BH3 mimetics induce apoptosis independent of DRP-1 in melanoma. Cell Death Dis. 2018;9:907. doi: 10.1038/s41419-018-0932-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Wroblewski D, et al. The BH3-mimetic ABT-737 sensitizes human melanoma cells to apoptosis induced by selective BRAF inhibitors but does not reverse acquired resistance. Carcinogenesis. 2013;34:237–247. doi: 10.1093/carcin/bgs330. [DOI] [PubMed] [Google Scholar]
- 476.Anvekar RA, et al. Sensitization to the mitochondrial pathway of apoptosis augments melanoma tumor cell responses to conventional chemotherapeutic regimens. Cell Death Dis. 2012;3:e420. doi: 10.1038/cddis.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Serasinghe MN, et al. Anti-apoptotic BCL-2 proteins govern cellular outcome following B-RAF(V600E) inhibition and can be targeted to reduce resistance. Oncogene. 2015;34:857–867. doi: 10.1038/onc.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Alcolea, V. et al. Identification of a novel quinoxaline-isoselenourea targeting the STAT3 pathway as a potential melanoma therapeutic. Int. J. Mol. Sci. 20, 512 (2019). [DOI] [PMC free article] [PubMed]
- 479.Su Y, et al. Targeting STAT3 restores BRAF inhibitor sensitivity through miR-759-3p in human cutaneous melanoma cells. Int. J. Clin. Exp. Pathol. 2018;11:2550–2560. [PMC free article] [PubMed] [Google Scholar]
- 480.Moriizumi H, et al. Caspase 3-specific cleavage of MEK1 suppresses ERK signaling and sensitizes cells to stress-induced apoptosis. FEBS Open Bio. 2023;13:684–700. doi: 10.1002/2211-5463.13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Chen LH, et al. Inhibition of endoplasmic reticulum stress-induced apoptosis of melanoma cells by the ARC protein. Cancer Res. 2008;68:834–842. doi: 10.1158/0008-5472.CAN-07-5056. [DOI] [PubMed] [Google Scholar]
- 482.Jiang CC, et al. Inhibition of MEK sensitizes human melanoma cells to endoplasmic reticulum stress-induced apoptosis. Cancer Res. 2007;67:9750–9761. doi: 10.1158/0008-5472.CAN-07-2047. [DOI] [PubMed] [Google Scholar]
- 483.Liu J, et al. High-throughput quantitative detection of triple-negative breast cancer-associated expressed miRNAs by rolling circle amplification on fluorescence-encoded microspheres. Chin. Chem. Lett. 2023;34:108141. doi: 10.1016/j.cclet.2023.108141. [DOI] [Google Scholar]
- 484.Li H, et al. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct. Target Ther. 2020;5:1. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Olivieri I, Vitali C, Gemignani G, Pasero G. Concomitant ankylosing spondylitis and DISH. J. Rheumatol. 1989;16:1170–1172. [PubMed] [Google Scholar]
- 486.Turner, E. et al. CRISPR/Cas9 Edited RAS & MEK Mutant Cells Acquire BRAF and MEK Inhibitor Resistance with MEK1 Q56P Restoring Sensitivity to MEK/BRAF Inhibitor Combo and KRAS G13D Gaining Sensitivity to Immunotherapy. Cancers14, 5449 (2022). [DOI] [PMC free article] [PubMed]
- 487.Yao M, et al. Research progress of nanovaccine in anti-tumor immunotherapy. Front. Oncol. 2023;13:1211262. doi: 10.3389/fonc.2023.1211262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Zhang L, et al. A novel therapeutic vaccine based on graphene oxide nanocomposite for tumor immunotherapy. Chin. Chem. Lett. 2022;33:4089–4095. doi: 10.1016/j.cclet.2022.01.071. [DOI] [Google Scholar]
- 489.Durrant DE, et al. Development of a high-throughput NanoBRET screening platform to identify modulators of the RAS/RAF interaction. Mol. Cancer Ther. 2021;20:1743–1754. doi: 10.1158/1535-7163.MCT-21-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Khazak V, et al. A two-hybrid approach to identify inhibitors of the RAS-RAF interaction. Enzymes. 2013;33(Pt A):213–248. doi: 10.1016/B978-0-12-416749-0.00010-5. [DOI] [PubMed] [Google Scholar]
- 491.Levy JM, Thorburn A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol. Ther. 2011;131:130–141. doi: 10.1016/j.pharmthera.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Shen S, et al. Association and dissociation of autophagy, apoptosis and necrosis by systematic chemical study. Oncogene. 2011;30:4544–4556. doi: 10.1038/onc.2011.168. [DOI] [PubMed] [Google Scholar]
- 493.Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- 494.Hua H, et al. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019;12:71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 496.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 497.Zhou C, et al. AMPK-autophagy inhibition sensitizes icaritin-induced anti-colorectal cancer cell activity. Oncotarget. 2017;8:14736–14747. doi: 10.18632/oncotarget.14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Blommaart EF, et al. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 499.Wu YT, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Buchser WJ, et al. Cell-mediated autophagy promotes cancer cell survival. Cancer Res. 2012;72:2970–2979. doi: 10.1158/0008-5472.CAN-11-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Bahar, E., Han, S. Y., Kim, J. Y. & Yoon, H. Chemotherapy resistance: role of mitochondrial and autophagic components. Cancers14, 1462 (2022). [DOI] [PMC free article] [PubMed]
- 502.Al-Ejeh F, et al. Harnessing the complexity of DNA-damage response pathways to improve cancer treatment outcomes. Oncogene. 2010;29:6085–6098. doi: 10.1038/onc.2010.407. [DOI] [PubMed] [Google Scholar]
- 503.John S, et al. Regulation of estrogenic effects by beclin 1 in breast cancer cells. Cancer Res. 2008;68:7855–7863. doi: 10.1158/0008-5472.CAN-07-5875. [DOI] [PubMed] [Google Scholar]
- 504.Manic G, et al. Chloroquine and hydroxychloroquine for cancer therapy. Mol. Cell Oncol. 2014;1:e29911. doi: 10.4161/mco.29911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Chen N, Karantza V. Autophagy as a therapeutic target in cancer. Cancer Biol. Ther. 2011;11:157–168. doi: 10.4161/cbt.11.2.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Amaravadi RK, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin. Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Fratta E, et al. Autophagy in BRAF-mutant cutaneous melanoma: recent advances and therapeutic perspective. Cell Death Discov. 2023;9:202. doi: 10.1038/s41420-023-01496-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Foth, M. & McMahon, M. Autophagy inhibition in BRAF-driven cancers. Cancers13, 3498 (2021). [DOI] [PMC free article] [PubMed]
- 509.Ma L, et al. Matrine inhibits BCR/ABL mediated ERK/MAPK pathway in human leukemia cells. Oncotarget. 2017;8:108880–108889. doi: 10.18632/oncotarget.22353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Martinez-Lopez N, et al. Autophagy proteins regulate ERK phosphorylation. Nat. Commun. 2013;4:2799. doi: 10.1038/ncomms3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Kabeya Y, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 512.Tanida I, et al. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 513.Sanduja S, et al. AMPK promotes tolerance to Ras pathway inhibition by activating autophagy. Oncogene. 2016;35:5295–5303. doi: 10.1038/onc.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Corazzari M, et al. Oncogenic BRAF induces chronic ER stress condition resulting in increased basal autophagy and apoptotic resistance of cutaneous melanoma. Cell Death Differ. 2015;22:946–958. doi: 10.1038/cdd.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Ogata M, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Vlasic, I., Horvat, A., Tadijan, A. & Slade, N. p53 family in resistance to targeted therapy of melanoma. Int. J. Mol. Sci. 24, 65 (2022). [DOI] [PMC free article] [PubMed]
- 518.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 519.Sreeramaneni R, et al. Ras-Raf-Arf signaling critically depends on the Dmp1 transcription factor. Mol. Cell Biol. 2005;25:220–232. doi: 10.1128/MCB.25.1.220-232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Inbal B, et al. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J. Cell Biol. 2002;157:455–468. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Ascierto PA, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14:249–256. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 522.Gibney GT, et al. Paradoxical oncogenesis–the long-term effects of BRAF inhibition in melanoma. Nat. Rev. Clin. Oncol. 2013;10:390–399. doi: 10.1038/nrclinonc.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Yun S, et al. Targeting immune checkpoints in unresectable metastatic cutaneous melanoma: a systematic review and meta-analysis of anti-CTLA-4 and anti-PD-1 agents trials. Cancer Med. 2016;5:1481–1491. doi: 10.1002/cam4.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Weiss SA, Wolchok JD, Sznol M. Immunotherapy of melanoma: facts and hopes. Clin. Cancer Res. 2019;25:5191–5201. doi: 10.1158/1078-0432.CCR-18-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Huynh, S. et al. Combined therapy with Anti-PD1 and BRAF and/or MEK inhibitor for advanced melanoma: a multicenter cohort study. Cancers12, 1666 (2020). [DOI] [PMC free article] [PubMed]
- 526.Hu-Lieskovan S, et al. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAF(V600E) melanoma. Sci. Transl. Med. 2015;7:279ra241. doi: 10.1126/scitranslmed.aaa4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Dummer R, et al. Rationale for immune checkpoint inhibitors plus targeted therapy in metastatic melanoma: a review. JAMA Oncol. 2020;6:1957–1966. doi: 10.1001/jamaoncol.2020.4401. [DOI] [PubMed] [Google Scholar]
- 528.Sanlorenzo M, et al. BRAF and MEK Inhibitors increase PD-1-positive melanoma cells leading to a potential lymphocyte-independent synergism with anti-PD-1 antibody. Clin. Cancer Res. 2018;24:3377–3385. doi: 10.1158/1078-0432.CCR-17-1914. [DOI] [PubMed] [Google Scholar]
- 529.Li S, et al. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat. Commun. 2019;10:1693. doi: 10.1038/s41467-019-09634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Kinsey CG, et al. Protective autophagy elicited by RAF–>MEK–>ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 2019;25:620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Bryant KL, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 2019;25:628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Goodall ML, et al. Development of potent autophagy inhibitors that sensitize oncogenic BRAF V600E mutant melanoma tumor cells to vemurafenib. Autophagy. 2014;10:1120–1136. doi: 10.4161/auto.28594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Seton-Rogers S. Eliminating protective autophagy in KRAS-mutant cancers. Nat. Rev. Cancer. 2019;19:247. doi: 10.1038/s41568-019-0137-5. [DOI] [PubMed] [Google Scholar]
- 534.Guo JY, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Strohecker AM, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Koustas E, Papavassiliou AG, Karamouzis MV. The role of autophagy in the treatment of BRAF mutant colorectal carcinomas differs based on microsatellite instability status. PLoS One. 2018;13:e0207227. doi: 10.1371/journal.pone.0207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Zhao H, Zheng B. Dual Targeting of Autophagy and MEK in KRAS Mutant Cancer. Trends Cancer. 2019;5:327–329. doi: 10.1016/j.trecan.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 538.Wang W, et al. Targeting Autophagy Sensitizes BRAF-Mutant Thyroid Cancer to Vemurafenib. J. Clin. Endocrinol. Metab. 2017;102:634–643. doi: 10.1210/jc.2016-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Chude, C. I. & Amaravadi, R. K. Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int. J. Mol. Sci. 18, 1279 (2017). [DOI] [PMC free article] [PubMed]
- 541.Li C, et al. Impact of autophagy inhibition at different stages on cytotoxic effect of autophagy inducer in glioblastoma cells. Cell Physiol. Biochem. 2015;35:1303–1316. doi: 10.1159/000373952. [DOI] [PubMed] [Google Scholar]
- 542.Palmeira dos Santos C, et al. Comparative study of autophagy inhibition by 3MA and CQ on Cytarabine‑induced death of leukaemia cells. J. Cancer Res. Clin. Oncol. 2014;140:909–920. doi: 10.1007/s00432-014-1640-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Follo C, et al. Inhibition of autophagy initiation potentiates chemosensitivity in mesothelioma. Mol. Carcinog. 2018;57:319–332. doi: 10.1002/mc.22757. [DOI] [PubMed] [Google Scholar]
- 544.Jain V, Singh MP, Amaravadi RK. Recent advances in targeting autophagy in cancer. Trends Pharmacol. Sci. 2023;44:290–302. doi: 10.1016/j.tips.2023.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Shi TT, Yu XX, Yan LJ, Xiao HT. Research progress of hydroxychloroquine and autophagy inhibitors on cancer. Cancer Chemother. Pharmacol. 2017;79:287–294. doi: 10.1007/s00280-016-3197-1. [DOI] [PubMed] [Google Scholar]
- 546.Piao S, Amaravadi RK. Targeting the lysosome in cancer. Ann. N. Y. Acad. Sci. 2016;1371:45–54. doi: 10.1111/nyas.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Maes H, Kuchnio A, Carmeliet P, Agostinis P. How to teach an old dog new tricks: autophagy-independent action of chloroquine on the tumor vasculature. Autophagy. 2014;10:2082–2084. doi: 10.4161/auto.36259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Maes H, et al. Tumor vessel normalization by chloroquine independent of autophagy. Cancer Cell. 2014;26:190–206. doi: 10.1016/j.ccr.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 549.Barnard RA, et al. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy. 2014;10:1415–1425. doi: 10.4161/auto.29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Warhurst DC, et al. Hydroxychloroquine is much less active than chloroquine against chloroquine-resistant Plasmodium falciparum, in agreement with its physicochemical properties. J. Antimicrob. Chemother. 2003;52:188–193. doi: 10.1093/jac/dkg319. [DOI] [PubMed] [Google Scholar]
- 551.Finbloom DS, Silver K, Newsome DA, Gunkel R. Comparison of hydroxychloroquine and chloroquine use and the development of retinal toxicity. J. Rheumatol. 1985;12:692–694. [PubMed] [Google Scholar]
- 552.Xie X, White EP, Mehnert JM. Coordinate autophagy and mTOR pathway inhibition enhances cell death in melanoma. PLoS One. 2013;8:e55096. doi: 10.1371/journal.pone.0055096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Dragowska WH, et al. Induction of autophagy is an early response to gefitinib and a potential therapeutic target in breast cancer. PLoS One. 2013;8:e76503. doi: 10.1371/journal.pone.0076503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Cook KL, et al. Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clin. Cancer Res. 2014;20:3222–3232. doi: 10.1158/1078-0432.CCR-13-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.McAfee Q, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc. Natl Acad. Sci. USA. 2012;109:8253–8258. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Zhou W, Guo Y, Zhang X, Jiang Z. Lys05 induces lysosomal membrane permeabilization and increases radiosensitivity in glioblastoma. J. Cell Biochem. 2020;121:2027–2037. doi: 10.1002/jcb.29437. [DOI] [PubMed] [Google Scholar]
- 557.Amaravadi RK, Winkler JD. Lys05: a new lysosomal autophagy inhibitor. Autophagy. 2012;8:1383–1384. doi: 10.4161/auto.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Maycotte P, et al. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8:200–212. doi: 10.4161/auto.8.2.18554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Eng CH, et al. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc. Natl Acad. Sci. USA. 2016;113:182–187. doi: 10.1073/pnas.1515617113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Zahedi S, et al. Effect of early-stage autophagy inhibition in BRAF(V600E) autophagy-dependent brain tumor cells. Cell Death Dis. 2019;10:679. doi: 10.1038/s41419-019-1880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Roach PJ. AMPK -> ULK1 -> autophagy. Mol. Cell Biol. 2011;31:3082–3084. doi: 10.1128/MCB.05565-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Follo C, et al. Autophagy initiation correlates with the autophagic flux in 3D models of mesothelioma and with patient outcome. Autophagy. 2016;12:1180–1194. doi: 10.1080/15548627.2016.1173799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Petherick KJ, et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 2015;290:11376–11383. doi: 10.1074/jbc.C114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Egan DF, et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Tang F, et al. SBI0206965, a novel inhibitor of Ulk1, suppresses non-small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol. Rep. 2017;37:3449–3458. doi: 10.3892/or.2017.5635. [DOI] [PubMed] [Google Scholar]
- 567.Dowdle WE, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 2014;16:1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 568.Bilanges B, Vanhaesebroeck B. Cinderella finds her shoe: the first Vps34 inhibitor uncovers a new PI3K-AGC protein kinase connection. Biochem. J. 2014;464:e7–e10. doi: 10.1042/BJ20141218. [DOI] [PubMed] [Google Scholar]
- 569.Ronan B, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 570.Bago R, et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 2014;463:413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Ji CH, et al. The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system. Nat. Commun. 2022;13:904. doi: 10.1038/s41467-022-28520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Deitersen J, et al. High-throughput screening for natural compound-based autophagy modulators reveals novel chemotherapeutic mode of action for arzanol. Cell Death Dis. 2021;12:560. doi: 10.1038/s41419-021-03830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Qiu RG, Chen J, McCormick F, Symons M. A role for Rho in Ras transformation. Proc. Natl Acad. Sci. USA. 1995;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Qiu RG, Abo A, McCormick F, Symons M. Cdc42 regulates anchorage-independent growth and is necessary for Ras transformation. Mol. Cell Biol. 1997;17:3449–3458. doi: 10.1128/MCB.17.6.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Qiu RG, et al. An essential role for Rac in Ras transformation. Nature. 1995;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 576.Soriano, O., Alcon-Perez, M., Vicente-Manzanares, M. & Castellano, E. The crossroads between RAS and RHO signaling pathways in cellular transformation, motility and contraction. Genes12, 819 (2021). [DOI] [PMC free article] [PubMed]
- 577.Tang Y, Yu J, Field J. Signals from the Ras, Rac, and Rho GTPases converge on the Pak protein kinase in Rat-1 fibroblasts. Mol. Cell Biol. 1999;19:1881–1891. doi: 10.1128/MCB.19.3.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Wang Y, Senoo H, Sesaki H, Iijima M. Rho GTPases orient directional sensing in chemotaxis. Proc. Natl Acad. Sci. USA. 2013;110:E4723–E4732. doi: 10.1073/pnas.1312540110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Zang M, Hayne C, Luo Z. Interaction between active Pak1 and Raf-1 is necessary for phosphorylation and activation of Raf-1. J. Biol. Chem. 2002;277:4395–4405. doi: 10.1074/jbc.M110000200. [DOI] [PubMed] [Google Scholar]
- 580.Tran NH, Frost JA. Phosphorylation of Raf-1 by p21-activated kinase 1 and Src regulates Raf-1 autoinhibition. J. Biol. Chem. 2003;278:11221–11226. doi: 10.1074/jbc.M210318200. [DOI] [PubMed] [Google Scholar]
- 581.Zang M, et al. Microtubule integrity regulates Pak leading to Ras-independent activation of Raf-1. insights into mechanisms of Raf-1 activation. J. Biol. Chem. 2001;276:25157–25165. doi: 10.1074/jbc.M100152200. [DOI] [PubMed] [Google Scholar]
- 582.King AJ, et al. The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature. 1998;396:180–183. doi: 10.1038/24184. [DOI] [PubMed] [Google Scholar]
- 583.Cammarano MS, Nekrasova T, Noel B, Minden A. Pak4 induces premature senescence via a pathway requiring p16INK4/p19ARF and mitogen-activated protein kinase signaling. Mol. Cell Biol. 2005;25:9532–9542. doi: 10.1128/MCB.25.21.9532-9542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Cotteret S, Jaffer ZM, Beeser A, Chernoff J. p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol. Cell Biol. 2003;23:5526–5539. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 586.Sun H, King AJ, Diaz HB, Marshall MS. Regulation of the protein kinase Raf-1 by oncogenic Ras through phosphatidylinositol 3-kinase, Cdc42/Rac and Pak. Curr. Biol. 2000;10:281–284. doi: 10.1016/S0960-9822(00)00359-6. [DOI] [PubMed] [Google Scholar]
- 587.Chao TS, et al. Src tyrosine kinase mediates stimulation of Raf-1 and mitogen-activated protein kinase by the tumor promoter thapsigargin. Cancer Res. 1997;57:3168–3173. [PubMed] [Google Scholar]
- 588.Ziogas A, Moelling K, Radziwill G. CNK1 is a scaffold protein that regulates Src-mediated Raf-1 activation. J. Biol. Chem. 2005;280:24205–24211. doi: 10.1074/jbc.M413327200. [DOI] [PubMed] [Google Scholar]
- 589.Grammatikakis N, et al. p50(cdc37) acting in concert with Hsp90 is required for Raf-1 function. Mol. Cell Biol. 1999;19:1661–1672. doi: 10.1128/MCB.19.3.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Wallace AG, von Witt R. Modifast and weight reduction. Med. J. Aust. 1984;140:598. doi: 10.5694/j.1326-5377.1984.tb108429.x. [DOI] [PubMed] [Google Scholar]
- 591.Xia K, et al. Tyrosine phosphorylation of the proto-oncoprotein Raf-1 is regulated by Raf-1 itself and the phosphatase Cdc25A. Mol. Cell Biol. 1999;19:4819–4824. doi: 10.1128/MCB.19.7.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Cheng JJ, Wung BS, Chao YJ, Wang DL. Sequential activation of protein kinase C (PKC)-alpha and PKC-epsilon contributes to sustained Raf/ERK1/2 activation in endothelial cells under mechanical strain. J. Biol. Chem. 2001;276:31368–31375. doi: 10.1074/jbc.M011317200. [DOI] [PubMed] [Google Scholar]
- 593.Reusch HP, et al. Regulation of Raf by Akt controls growth and differentiation in vascular smooth muscle cells. J. Biol. Chem. 2001;276:33630–33637. doi: 10.1074/jbc.M105322200. [DOI] [PubMed] [Google Scholar]
- 594.Luo H, Rose PE, Roberts TM, Dearolf CR. The Hopscotch Jak kinase requires the Raf pathway to promote blood cell activation and differentiation in Drosophila. Mol. Genet. Genom. 2002;267:57–63. doi: 10.1007/s00438-001-0632-7. [DOI] [PubMed] [Google Scholar]
- 595.Abraham D, et al. Raf-1-associated protein phosphatase 2A as a positive regulator of kinase activation. J. Biol. Chem. 2000;275:22300–22304. doi: 10.1074/jbc.M003259200. [DOI] [PubMed] [Google Scholar]
- 596.Raabe T, Rapp UR. Ras signaling: PP2A puts Ksr and Raf in the right place. Curr. Biol. 2003;13:R635–R637. doi: 10.1016/S0960-9822(03)00568-2. [DOI] [PubMed] [Google Scholar]