Extended Data Fig. 4. RBMY1 gene similarity and architecture.
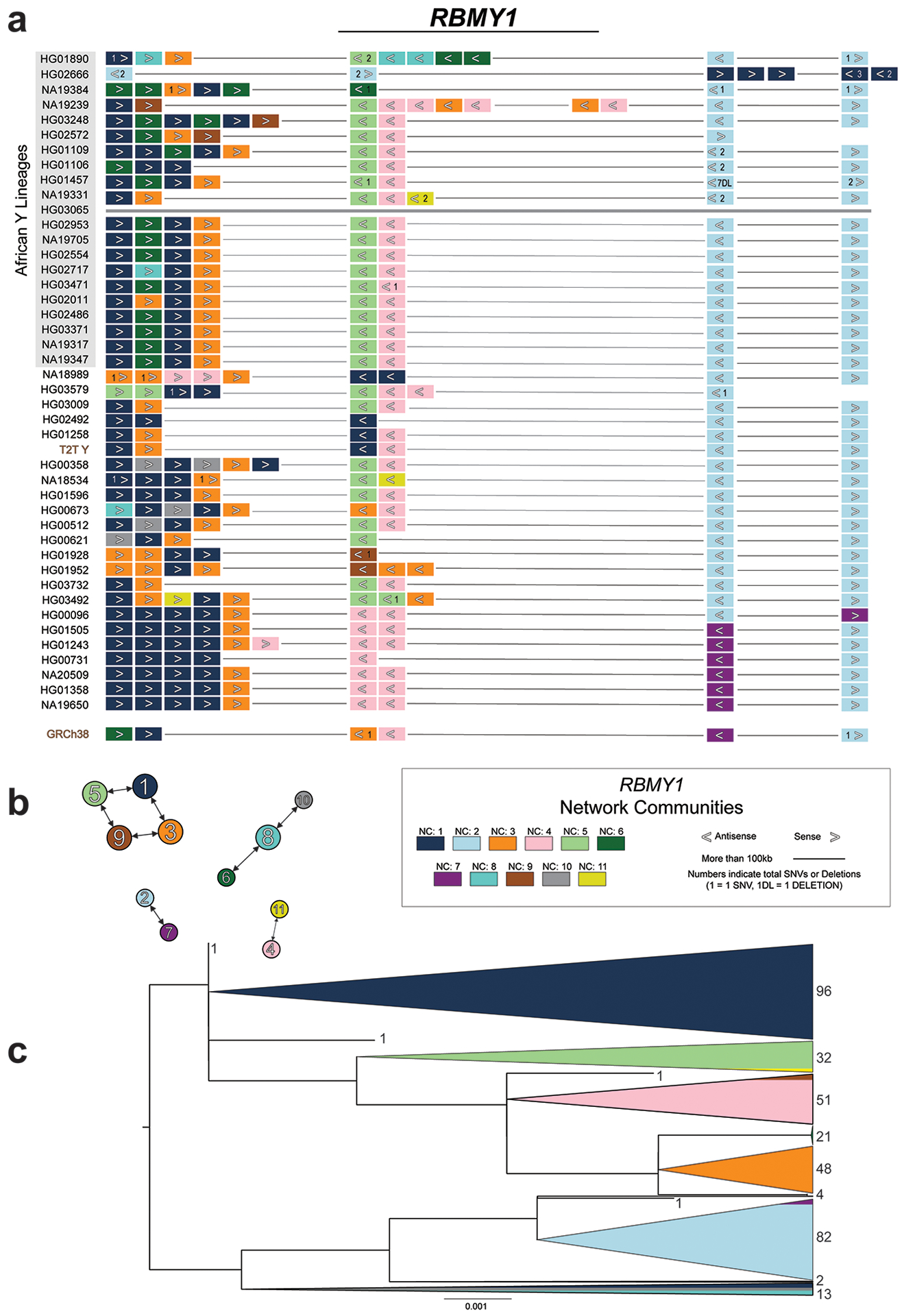
a. A schematic distribution of individual RBMY1 gene copies (filled rectangles) within analysed Y chromosome assemblies (42 + T2T + GRCh38). The RBMY1 gene copies are located in four primary regions (NA19239 carries a partial duplication of gene region 2 and the composition of HG02666 suggests at least one inversion within the RBMY regions). Fill colours refer to the assigned network community (NC) and indicates a similar sequence (Methods). Assembly of this region was not contiguous in HG03065 (brown line) and was not included in the analysis.
b. A secondary directed network showing connections between NCs with the most similar consensus sequences. An edge pointing from one node to a second node indicates that the second node was the first’s closest match (i.e., most similar sequence; ties are allowed and shown as multiple edges stemming from a node). The width of the edge represents the sequence similarity between two nodes (i.e., NC consensus sequence similarity; thicker means fewer SNVs). The node size is representative of the total edges pointing to the node.
c. RBMY1 phylogenetic analysis of exonic nucleotide sequences. Shown is the unrooted phylogenetic tree of RBMY1 genes constructed using a maximum likelihood approach (Methods). This tree is rooted at the midpoint with the total count of RBMY1 copies shown on the right. The scale bar represents the average number of substitutions per site. RBMY1 copies located in regions 1 and 2 (primarily dark blue, orange, dark/light green, and pink) distinguish themselves from those located downstream in regions 3 and 4 (primarily light blue and purple copies).
