Abstract
Background and purpose
Accurate CT numbers in Cone Beam CT (CBCT) are crucial for precise dose calculations in adaptive radiotherapy (ART). This study aimed to generate synthetic CT (sCT) from CBCT using deep learning (DL) models in head and neck (HN) radiotherapy.
Materials and methods
A novel DL model, the 'self-attention-residual-UNet' (ResUNet), was developed for accurate sCT generation. ResUNet incorporates a self-attention mechanism in its long skip connections to enhance information transfer between the encoder and decoder. Data from 93 HN patients, each with planning CT (pCT) and first-day CBCT images were used. Model performance was evaluated using two DL approaches (non-adversarial and adversarial training) and two model types (2D axial only vs. 2.5D axial, sagittal, and coronal). ResUNet was compared with the traditional UNet through image quality assessment (Mean Absolute Error (MAE), Peak-Signal-to-Noise Ratio (PSNR), Structural Similarity Index (SSIM)) and dose calculation accuracy evaluation (DVH deviation and gamma evaluation (1 %/1mm)).
Results
Image similarity evaluation results for the 2.5D-ResUNet and 2.5D-UNet models were: MAE: 46±7 HU vs. 51±9 HU, PSNR: 66.6±2.0 dB vs. 65.8±1.8 dB, and SSIM: 0.81±0.04 vs. 0.79±0.05. There were no significant differences in dose calculation accuracy between DL models. Both models demonstrated DVH deviation below 0.5 % and a gamma-pass-rate (1 %/1mm) exceeding 97 %.
Conclusions
ResUNet enhanced CT number accuracy and image quality of sCT and outperformed UNet in sCT generation from CBCT. This method holds promise for generating precise sCT for HN ART.
Keywords: CBCT, Synthetic CT, Deep-learning, Attention, UNet, Adaptive Radiotherapy
1. Introduction
Accurate patient positioning is important for the successful outcome of radiotherapy treatment. Image-guided radiation therapy (IGRT) uses advanced imaging technologies to provide critical insights into a patient's internal anatomy, thereby enhancing the accuracy of patient positioning. However, IGRT approaches encounter significant hurdles when dealing with anatomical variations that may occur in head and neck (HN) cancer patients undergoing radiotherapy, such as target shrinkage or weight loss [1], [2]. In addressing this challenge, the adoption of in-room Cone Beam Computed Tomography (CBCT) imaging emerges as a necessary tool. CBCT's ability to capture three-dimensional anatomical data allows the creation of personalized adaptive treatment (ART) strategies, effectively accommodating the dynamic anatomical changes encountered in each treatment session [2], [3]. Nevertheless, despite the many advantages offered by CBCT, it also presents several challenges. These include compromised image quality attributed to increased cone beam scattering, limited field-of-view, and inaccuracies in CT numbers [4].
Accurate CT numbers in CBCT are crucial for precise dose calculations in ART. Various methods are proposed to correct the CBCT for ART calculations: CT number conversion curve, density assignment, deformable image registration (DIR), and machine learning. The CT number conversion curve involves a detailed calibration process of CBCT images with standard density calibration phantoms, but it is sensitive to artifacts and scattering conditions [5]. The density assignment method assigns specific densities to CBCT contours, requiring tissue class segmentation [6], [7], but it is limited in covering tissue types. DIR between planning CT (pCT) and CBCT provides density information, but its effectiveness can be compromised when the pCT does not anatomically correspond to the CBCT, and the poor image quality of CBCT can affect DIR accuracy [8], [9]. Alternatively, recent works [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24] have explored deep learning (DL) methods for synthetic CT (sCT) generation, producing sCT images with accurate CT numbers closely resembling pCT. Comprehensive insights into various DL methods can be found in recent reviews by Spadea et al. [25] and Rusanov et al. [26].
Chen et al. [11] employed a UNet-based network, utilizing CBCT and registered pCT images of HN patients as input to generate sCT. Likewise, Eckl et al. [14] utilized cycle generative adversarial networks (GAN) to design site-specific DL models customized for various body regions, including HN. Additionally, Barateau et al. [10] used GAN-based DL models for sCT generation in HN patients. These studies collectively demonstrated the successful production of sCT images using DL models, closely resembling pCT scans with precise CT numbers.
While there has been extensive research on the generation of sCT from CBCT images, there remains a need for accurate DL models capable of handling limited training data effectively. In light of this gap, our study aimed to develop a robust sCT generation model tailored specifically for HN patients. Our approach centered on the creation of a novel DL network architecture designed to address the challenges posed by smaller training datasets.
2. Materials and methods
2.1. Patient data
A total of 93 previously treated HN patients with a pair of pCT and first-day CBCT images were included. This retrospective study was approved (MRC-01–21-119) by our Institutional Review Board. CT images were acquired with the patient supine position on a multi-detector-row spiral CT scanner (Somatom Definition AS, Siemens Healthineers, Forchheim, Germany). CBCT images were acquired using the On-Board-Imager kV system attached to Clinac series linear accelerator (Varian Medical Systems, Palo Alto, CA).
The pCT and CBCT axial images were re-sampled to 1x1x3 mm3 voxel size. To minimize the anatomical difference, CBCT images were aligned with respective pCT images using rigid registration followed by intensity-based DIR using MIM MaestroTM v6.4.5 (MIM Software, Cleveland, OH). In addition, the 3D-CBCT and CT volumes were sliced along sagittal and coronal planes for multiplane model (2.5D) training.
An external body contour mask (CBCTbody) on CBCT was created using threshold and morphological operations, and the CBCT and pCT images were cropped to exclude the background regions. The intensities of these images were clipped to the range of [-1000 3500] HU and divided by 2000 before being used for model training.
2.2. Self-attention residual UNet (ResUNet)
In this work, a novel 2D convolutional neural network called ‘self-attention-residual-UNet’ (ResUNet) was designed to generate the sCT. The architecture, as shown in the supplementary fig. SF1(a), was inspired by the combination of traditional UNet [27] and residual neural networks [28]. The encoder section performs downsampling three times on the input image, while the decoder section upsamples three times. To learn the important information between the encoder and decoder, long skip connections were included along with a special self-attention mechanism as illustrated in the supplementary fig. SF1(b). The central part of the model architecture consisted of five residual blocks, as shown in the supplementary fig. SF1(c). Overall, the ResUNet architecture leverages the strengths of UNet, residual networks, and self-attention mechanisms to effectively generate sCT images.
The ResUNet performance was compared against standard UNet (supplementary fig. SF2) for two different DL approaches (non-adversarial training and adversarial training [29]) and DL models (2D (axial only) vs. 2.5D (axial, sagittal and coronal)). The multiplane 2.5D approach was implemented to enhance the predictive accuracy of the DL models (see supplementary fig. SF4), following a previously reported method [25]. Further details about these training methodologies can be found in the supplementary material.
2.3. Training details
Patient images were randomly split into training (58), validation (10), and testing (25) datasets. The testing data were exclusively used for model evaluation, not for network optimization, and validation data was used to monitor the training process and prevent overfitting.
The training was performed in MATLAB using an NVIDIA GeForce RTX-3090 24 GB GPU. To ensure a fair comparison, fixed hyper-parameters were used for all the network training. The optimal hyper-parameters such as learning rate (0.2e-4), L2-regularization (1e-7), and batch size (4) were determined using a simple grid search on a small training dataset. The ADAM optimizer was used for network optimization [30]. The maximum epoch was set to 20 and the network training stopped automatically before the completion of 20 epochs when the model performance on the validation data stopped improving.
Training times varied from 1.4 to 6.5 h for UNet and ResUNet with non-adversarial training, extending to 4.4 to 14 h for adversarial training. All models maintained quick prediction times per patient, staying under 20 s.
The final model, taking axial CBCT slices as input, generates the corresponding sCT slice. Areas outside the CBCT mask (CBCTbody) were replaced with corresponding regions from pCT to address truncation issues due to CBCT's limited FOV and ensure that the new sCT has a complete image covering the entire anatomical regions necessary for adaptive planning.
2.4. Evaluation
The mean absolute error (MAE) and mean error (ME) were calculated within the external body contour of CBCT (excluding the stitched parts) to measure the CT number differences between the pCT and sCT images. To evaluate the image similarity between pCT and sCT images, structural similarity index (SSIM) and peak-signal-to-noise ratio (PSNR) were used (equation SE5 and SE8, supplementary material).
Dose volume histograms (DVH) and gamma evaluations were used in dose calculation accuracy evaluation. The model-generated sCT images were imported into the EclipseTM (v16.1.0) planning system (Varian Medical Systems, Palo Alto, CA). The targets and organs at risk (OARs) were mapped onto both the sCT and CBCT images from their corresponding pCT images using rigid registration. Clinical plans were devised for dose prescriptions of 70 Gy, 60 Gy, and 54 Gy delivered in 35 fractions to the primary and elective planning target volumes (PTVs), respectively, utilizing a 6 MV volumetric modulated arc (VMAT) technique. The original plans, initially calculated on the pCT, were transferred onto both the sCT and CBCT. The dose distribution was then recalculated using the Acuros dose calculation algorithm. DVH metrics were extracted for OARs (spinal cord-Dmax, parotids-Dmean, brainstem-Dmax, optic nerve-Dmax, optic chiasm-Dmax, eyes-Dmean, larynx-Dmean, oral cavity-Dmean) and PTVs (D95). Gamma evaluation (global-gamma) [31] was conducted using VeriSoft (PTW Dosimetry, Freiburg, Germany) to compare dose distributions on pCT and recalculated sCT and CBCT, employing 1 %/1mm and 2 %/2mm criteria with a 10 % threshold. The statistical significance of disparities in image quality results for UNet vs. ResUNet and 2D vs. 2.5D models was evaluated using an independent two-sample Student’s t-test with equal variances. This study also included an assessment of image quality and dose comparison assessment for CBCT, comparing it with pCT.
This study also evaluated the robustness and generalizability of DL models across different data sizes. Initially, models trained with 100 % data were used as references. Subsequently, training data sizes were reduced to 50 % and 25 %, and models were trained accordingly. The performance of models trained with smaller datasets (50 % and 25 %) was compared to reference models trained with 100 % data.
3. Results
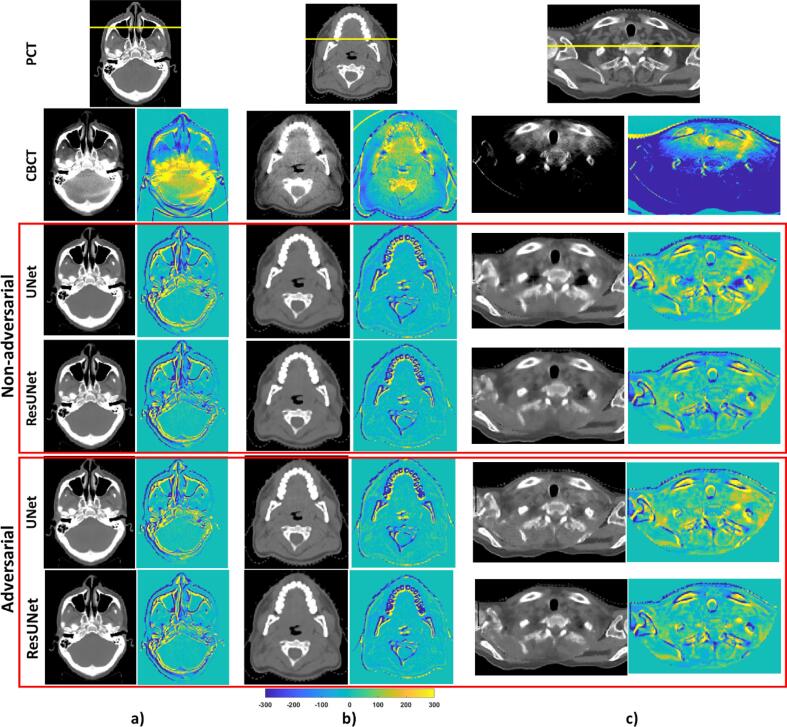
The image quality assessment results are summarized in Table 1, and Fig. 1 provides a visual representation of the accuracy of sCT images in the axial view, with supplementary fig. SF5 showing the sagittal view. Supplementary figs. SF6 and SF7 further compare the line profiles through the images.
Table 1.
Results (mean ± SD) of image quality evaluation and dose calculation accuracy.
| Training | Model | MAE (HU) | ME (HU) | PSNR (dB) | SSIM | DVH deviation (%) | Gamma-pass-rate (%) |
|
|---|---|---|---|---|---|---|---|---|
| 1 %/1mm | 2 %/2mm | |||||||
| CBCT | 163±29 | 73±35 | 60.1±1.8 | 0.6±0.1 | 2.4±5.4 | 58.9±14.5 | 76.6±12.8 | |
| Non-adversarial | 2D-UNet | 55±9 | 1±11 | 64.8±1.7 | 0.8±0.1 | 0.2±0.4 | 97.4±2.0 | 99.7±0.2 |
| 2D-ResUNet | 48±7 | 0.1±10 | 66.0±1.9 | 0.8±0.0 | 0.2±0.3 | 97.7±2.0 | 99.8±0.2 | |
| 2.5D-UNet | 51±9 | 5±10 | 65.8±1.8 | 0.8±0.1 | 0.2±0.3 | 97.6±2.0 | 99.8±0.2 | |
| 2.5D-ResUNet | 46±7 | 3±10 | 66.6±2.0 | 0.8±0.0 | 0.2±0.3 | 97.7±1.9 | 99.8±0.2 | |
| Adversarial | 2D-UNet | 51±8 | 3±11 | 65.5±2.0 | 0.8±0.0 | 0.2±0.3 | 97.5±2.1 | 99.6±0.5 |
| 2D-ResUNet | 49±8 | 4±9 | 65.6±1.8 | 0.8±0.0 | 0.2±0.4 | 97.4±2.0 | 99.7±0.3 | |
| 2.5D-UNet | 50±9 | −9±10 | 65.8±1.9 | 0.8±0.0 | 0.2±0.3 | 97.7±1.9 | 99.8±0.2 | |
| 2.5D-ResUNet | 46±8 | 7±10 | 66.4±2.0 | 0.8±0.1 | 0.2±0.4 | 97.4±2.4 | 99.8±0.3 | |
MAE: Mean Absolute Error, ME: Mean error, PSNR: Peak Signal-to-Noise Ratio, SSIM: Structural Similarity Index.
Fig. 1.
Comparative analysis of planning CT (pCT), CBCT, synthetic CTs generated by UNet and ResUNet using non-adversarial and adversarial training, and the corresponding difference images relative to pCT. Figures a), b), and c) represent the best, median, and worst prediction results based on the image quality assessment. All images are displayed within the window of (-300, 500) HU. The yellow horizontal lines on the pCT indicate the locations from which the CT number line profiles were extracted (refer to supplementary figures SF6 and SF7 for details).
For the non-adversarial training approach, notable differences were seen between the performance of the 2D-UNet and 2D-ResUNet models. Specifically, 2D-ResUNet's MAE exhibited a significant 11 % (p < 0.005) average improvement over 2D-UNet. The integration of a multiplane 2.5D model further increased accuracy, with 2.5D-ResUNet models displaying a superior performance (p < 0.0005) over 2D models. In the adversarial training, while ResUNet demonstrated a benefit over UNet, the improvement was relatively modest, with an average MAE enhancement of 4 % (p < 0.002) and 8 % (p < 0.0005) observed in the 2D and 2.5D models respectively.
Supplementary Fig. SF8 shows the comparison of dose distribution on sCT images and corresponding gamma evaluation results in the sagittal view and the detailed DVH deviations can be found in Supplementary Table ST1. While DL models notably improved sCT image quality, this enhancement was not uniformly reflected in the dose comparison. For both training methods, average deviations of DVH parameters consistently remained below 0.5 %, in contrast to CBCT, where these deviations were slightly higher at 3 %. The gamma pass rate for all DL models was more than 97 % and 99 % for 1 %/1mm and 2 %/2mm criteria, respectively. Although some structures on CBCT exhibited smaller DVH deviations due to their distal location, the average gamma-pass-rate was particularly lower for CBCT, at 59 % (ranging from 36 % to 86 %) and 77 % (ranging from 59 % to 95 %) for 1 %/1mm and 2 %/2mm, respectively.
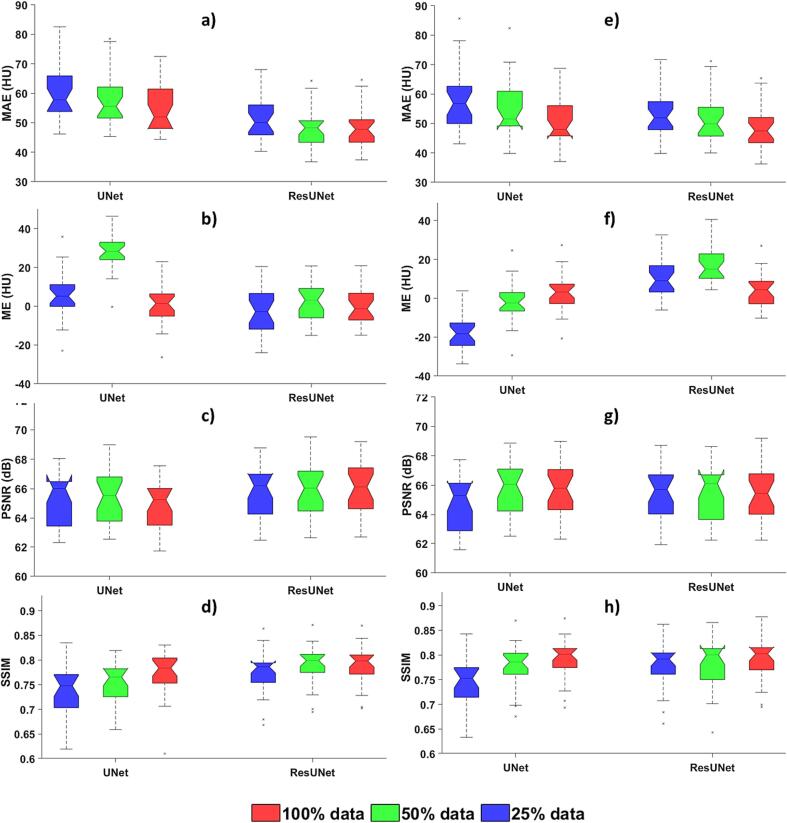
The ResUNet maintained sCT generation accuracy even with reduced training data, as seen in Fig. 2. For instance, when comparing non-adversarial training with 100 % and 50 % of the data, average MAE values were 55 HU and 58 HU for the UNet, 48 HU, and 49 HU for the ResUNet. Likewise, in the adversarial training, the average MAE values were 51 HU and 56 HU for the UNet, 49 HU, and 52 HU for the ResUNet.
Fig. 2.
Impact of training data size on UNet and ResUNet. Box and whisker plot of image quality assessment results with 2D models. The upper, middle, and lower box lines represent the 75th, 50th (median), and 25th percentiles, respectively. Figures (a-d): non-adversarial training and Figures (e-h): adversarial training.
4. Discussions
In this study, we introduced a novel network architecture, ResUNet, and compared its performance to the conventional UNet architecture in the context of sCT generation from CBCT images for HN radiotherapy. Our image quality assessment showed significant differences in performance between various model configurations. Particularly, the ResUNet model exhibited superior image quality compared to UNet, with an 8–11 % improvement in metrics such as MAE, PSNR, and SSIM.
Furthermore, we explored the impact of training strategies, specifically adversarial versus non-adversarial training, and the dimensionality of models (2D versus 2.5D). These investigations unveiled that the benefits of ResUNet were most pronounced when employed with non-adversarial training alongside 2.5D models. The enhanced performance of the ResUNet model can be attributed to its utilization of a larger number of trainable parameters. While the UNet model consists of 7.6 million trainable parameters, the ResUNet model incorporates 21.2 million parameters. Consequently, the training duration for the ResUNet model was marginally longer than that of the UNet model, owing to its larger parameter size. It is important to note that this slightly extended training time is a one-time operation and does not pose significant constraints in the context of practical clinical deployment.
One striking advantage of ResUNet was its ability to maintain performance even with smaller training datasets. This robustness holds a unique significance in radiotherapy, given the logistical complexities of acquiring extensive training data. Our results demonstrated that ResUNet's performance remained consistent when trained with a reduced dataset, highlighting its practical utility in real-world clinical settings.
However, it is important to note that while image quality assessments revealed substantial differences, dose comparison did not exhibit the same level of variation, aligning with the findings from a prior study [17]. Specifically, when examining dose calculation accuracy, both ResUNet and UNet consistently showed DVH deviations below 0.5 % and gamma pass rates (1 %/1mm) exceeding 97 %. These findings indicate that the marked improvement in image quality did not automatically translate into dose calculation accuracy. This was in contrast to CBCT, where the gamma pass rate was notably lower. In addition to its dose calculation implications, improved image quality can have a profound impact on other critical aspects of radiotherapy. It can significantly improve precision in patient set-up during IGRT and enhance the accuracy of OAR/tumor contouring during ART. Moreover, superior image quality may emerge as a crucial requirement in the context of particle beam radiotherapy.
Numerous studies focus on sCT generation using CBCT [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22] and supplementary table ST2 provides a brief comparison of our work with other studies. Among these studies, research focused on employing DL models for sCT generation from HN CBCT has reported mean MAEs calculated within the body contour ranging from 43 to 141 HU. In contrast, our 2.5D-ResUNet model achieved MAEs within the range of 33 to 63 HU, signifying a substantial 72 % enhancement in image quality compared to CBCT images. Nonetheless, it is important to note that drawing direct comparisons between these studies can be intricate due to variations in network architectures, pre-processing techniques, imaging protocols, and patient cohorts.
Yuan et al. [21] employed a modified UNet++ to generate sCT from ultra-low-dose CBCT images of HN patients, resulting in a 74 % improvement in CBCT image quality, as indicated by a reduction in MAEs from 166 to 43 HU. Maspero et al. [17] employed cycle-GAN to create sCTs from CBCT images of HN, lung, and breast patients. They assessed the performance of a single network trained for all three sites versus networks trained individually for specific anatomical sites. Their results showed a 74 % improvement (MAEs) in CBCT image quality. Notably, the single network exhibited superior generalization for HN cases, with MAE values of 53±12 HU (37 HU-77 HU) compared to 51±12 HU (35 HU-74 HU) for the individual networks. A recent work by Deng et al. [13] also reported the use of respath-cycleGAN to generate sCT images from HN CBCT images and demonstrated a notable improvement of 29 % in CBCT image quality, as indicated by a reduction in MAEs from 198 to 141 HU.
Our DL model, trained on paired CT and CBCT data, encountered challenges due to anatomical discrepancies despite rigorous registration, particularly around the spine and oral cavity-pharynx areas. These disparities arise from inherent differences in a patient's anatomy between the pCT and subsequent CBCT scans (see supplementary fig. SF5(c)). This mismatch introduces inaccuracies into our DL models, affecting image quality and dose calculation assessments.
Several limitations warrant consideration when interpreting our study's results. Firstly, we primarily focused on 2D and 2.5D DL models, while acknowledging that a 3D model could harness additional advantages by integrating spatial information. Secondly, our dose calculation evaluation exclusively employed photon beams, while some recent studies incorporated particle beams for a more comprehensive assessment [18]. Finally, our current evaluation was confined to an internal dataset and future work should evaluate ResUNet's performance on external datasets.
In this study, we introduced a novel DL model to generate sCT images from daily CBCT scans for HN patients. Our proposed ResUNet architecture was compared to the conventional UNet architecture, resulting in a notable improvement in the accuracy of CT numbers and overall image quality of the sCT. The ResUNet demonstrated a remarkable 8–11 % enhancement in the quality of the sCT images, outperforming the UNet model. This superiority was especially pronounced when dealing with limited training data. There was no significant difference in dose calculation accuracy among the models. Both models demonstrated DVH deviation below 0.5 % and a gamma-pass-rate (1 %/1mm) exceeding 97 %. Our approach holds significant promise in the domain of HN ART planning, presenting the opportunity to create highly accurate sCT images.
CRediT authorship contribution statement
S.A. Yoganathan: Conceptualization, Methodology, Software, Validation, Writing – original draft. Souha Aouadi: Writing – original draft. Sharib Ahmed: Writing – original draft. Satheesh Paloor: Writing – original draft. Tarraf Torfeh: Writing – original draft. Noora Al-Hammadi: Writing – original draft. Rabih Hammoud: Writing – original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Article processing charge is provided by Medical Research Center, Hamad Medical Corporation, Doha, Qatar.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2023.100512.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Brouwer C.L., Steenbakkers R.J., Langendijk J.A., Sijtsema N.M. Identifying patients who may benefit from adaptive radiotherapy: Does the literature on anatomic and dosimetric changes in head and neck organs at risk during radiotherapy provide information to help? Radiother and Oncol. 2015;115:285–294. doi: 10.1016/j.radonc.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Sonke J.J., Aznar M., Rasch C. Adaptive Radiotherapy for Anatomical Changes. Semin Radiat Oncol. 2019;29:245–257. doi: 10.1016/j.semradonc.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Lim-Reinders S., Keller B.M., Al-Ward S., Sahgal A., Kim A. Online Adaptive Radiation Therapy. Int J Radiat Oncol Biol Phys. 2017;99:994–1003. doi: 10.1016/j.ijrobp.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 4.Kida S., Kaji S., Nawa K., Imae T., Nakamoto T., Ozaki S., et al. Visual enhancement of Cone-beam CT by use of CycleGAN. Med Phys. 2020;47:998–1010. doi: 10.1002/mp.13963. [DOI] [PubMed] [Google Scholar]
- 5.Richter A., Hu Q., Steglich D., Baier K., Wilbert J., Guckenberger M., et al. Investigation of the usability of conebeam CT data sets for dose calculation. Radiat Oncol. 2008;3:42. doi: 10.1186/1748-717X-3-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barateau A., Perichon N., Castelli J., Schick U., Henry O., Chajon E., et al. A density assignment method for dose monitoring in head-and-neck radiotherapy. Strahlenther Onkol. 2019;195:175–185. doi: 10.1007/s00066-018-1379-y. [DOI] [PubMed] [Google Scholar]
- 7.Giacometti V., King R.B., Agnew C.E., Irvine D.M., Jain S., Hounsell A.R., et al. An evaluation of techniques for dose calculation on cone beam computed tomography. Br J Radiol. 2019;92:20180383. doi: 10.1259/bjr.20180383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang T., Chi Y., Meldolesi E., Yan D. Automatic delineation of on-line head-and-neck computed tomography images: toward on-line adaptive radiotherapy. Int J Radiat Oncol Biol Phys. 2007;68:522–530. doi: 10.1016/j.ijrobp.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 9.Nithiananthan S., Brock K.K., Daly M.J., Chan H., Irish J.C., Siewerdsen J.H. Demons deformable registration for CBCT-guided procedures in the head and neck: convergence and accuracy. Med Phys. 2009;36:4755–4764. doi: 10.1118/1.3223631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barateau A., De Crevoisier R., Largent A., Mylona E., Perichon N., Castelli J., et al. Comparison of CBCT-based dose calculation methods in head and neck cancer radiotherapy: from Hounsfield unit to density calibration curve to deep learning. Med Phys. 2020;47:4683–4693. doi: 10.1002/mp.14387. [DOI] [PubMed] [Google Scholar]
- 11.Chen L., Liang X., Shen C., Jiang S., Wang J. Synthetic CT generation from CBCT images via deep learning. Med Phys. 2020;47:1115–1125. doi: 10.1002/mp.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L., Liang X., Shen C., Nguyen D., Jiang S., Wang J. Synthetic CT generation from CBCT images via unsupervised deep learning. Phys Med Biol. 2021;66 doi: 10.1088/1361-6560/ac01b6. [DOI] [PubMed] [Google Scholar]
- 13.Deng L., Hu J., Wang J., Huang S., Yang X. Synthetic CT generation based on CBCT using respath-cycleGAN. Med Phys. 2022;49:5317–5329. doi: 10.1002/mp.15684. [DOI] [PubMed] [Google Scholar]
- 14.Eckl M., Hoppen L., Sarria G.R., Boda-Heggemann J., Simeonova-Chergou A., Steil V., et al. Evaluation of a cycle-generative adversarial network-based cone-beam CT to synthetic CT conversion algorithm for adaptive radiation therapy. Phys Med Biol. 2020;80:308–316. doi: 10.1016/j.ejmp.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Harms J., Lei Y., Wang T., Zhang R., Zhou J., Tang X., et al. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Med Phys. 2019;46:3998–4009. doi: 10.1002/mp.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang X., Chen L., Nguyen D., Zhou Z., Gu X., Yang M., et al. Generating synthesized computed tomography (CT) from cone-beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy. Phys Med Biol. 2019;64 doi: 10.1088/1361-6560/ab22f9. [DOI] [PubMed] [Google Scholar]
- 17.Maspero M., Houweling A.C., Savenije M.H., van Heijst T.C., Verhoeff J.J., Kotte A.N., et al. A single neural network for cone-beam computed tomography-based radiotherapy of head-and-neck, lung and breast cancer. Phys Imaging Radiat Oncol. 2020;14:24–31. doi: 10.1016/j.phro.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thummerer A., Zaffino P., Meijers A., Marmitt G.G., Seco J., Steenbakkers R.J., et al. Comparison of CBCT based synthetic CT methods suitable for proton dose calculations in adaptive proton therapy. Phys Med Biol. 2020;65 doi: 10.1088/1361-6560/ab7d54. [DOI] [PubMed] [Google Scholar]
- 19.Xue X., Ding Y., Shi J., Hao X., Li X., Li D., et al. Cone Beam CT (CBCT) Based Synthetic CT Generation Using Deep Learning Methods for Dose Calculation of Nasopharyngeal Carcinoma Radiotherapy. Technol Cancer Res Treat. 2021;20 doi: 10.1177/15330338211062415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan N., Dyer B., Rao S., Chen Q., Benedict S., Shang L., et al. Convolutional neural network enhancement of fast-scan low-dose cone-beam CT images for head and neck radiotherapy. Phys Med Biol. 2020;65 doi: 10.1088/1361-6560/ab6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan N., Rao S., Chen Q., Sensoy L., Qi J., Rong Y. Head and neck synthetic CT generated from ultra-low-dose cone-beam CT following Image Gently Protocol using deep neural network. Med Phys. 2022;49:3263–3277. doi: 10.1002/mp.15585. [DOI] [PubMed] [Google Scholar]
- 22.O'Hara C.J., Bird D., Al-Qaisieh B., Speight R. Assessment of CBCT-based synthetic CT generation accuracy for adaptive radiotherapy planning. J Appl Clin Med Phys. 2022;23:e13737. doi: 10.1002/acm2.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Zhu J., Liu Z., Teng J., Xie Q., Zhang L., et al. A preliminary study of using a deep convolution neural network to generate synthesized CT images based on CBCT for adaptive radiotherapy of nasopharyngeal carcinoma. Phys Med Biol. 2019;64 doi: 10.1088/1361-6560/ab2770. [DOI] [PubMed] [Google Scholar]
- 24.de Hond Y.J.M., Kerckhaert C.E.M., van Eijnatten M., van Haaren P.M.A., Hurkmans C.W., Tijssen R.H.N. Anatomical evaluation of deep-learning synthetic computed tomography images generated from male pelvis cone-beam computed tomography. Phys Imaging Radiat Oncol. 2023;25 doi: 10.1016/j.phro.2023.100416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spadea M.F., Maspero M., Zaffino P., Seco J. Deep learning based synthetic-CT generation in radiotherapy and PET: A review. Med Phys. 2021;48:6537–6566. doi: 10.1002/mp.15150. [DOI] [PubMed] [Google Scholar]
- 26.Rusanov B., Hassan G.M., Reynolds M., Sabet M., Kendrick J., Rowshanfarzad P., et al. Deep learning methods for enhancing cone-beam CT image quality toward adaptive radiation therapy: A systematic review. Med Phys. 2022;49:6019–6054. doi: 10.1002/mp.15840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18: Springer; 2015. p. 234-41.
- 28.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition2016. p. 770-8.
- 29.Isola P, Zhu J-Y, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. Proceedings of the IEEE conference on computer vision and pattern recognition2017. p. 1125-34.
- 30.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014.
- 31.Low D.A., Dempsey J.F. Evaluation of the gamma dose distribution comparison method. Med Phys. 2003;30:2455–2464. doi: 10.1118/1.1598711. [DOI] [PubMed] [Google Scholar]
- 32.Taigman Y, Polyak A, Wolf L. Unsupervised cross-domain image generation. arXiv preprint arXiv:161102200. 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.