Abstract
Human serum albumin (HSA) is a multi-domain macromolecule with diverse ligand binding capability because of its ability to allow allosteric modulation despite being a monomeric protein. Physiologically, HSA act as the primary carrier for various exogenous and endogenous compounds and fatty acids, and alter the pharmacokinetic properties of several drugs. It has antioxidant properties and is utilized therapeutically to improve the drug delivery of pharmacological agents for the treatment of several disorders. The flexibility of albumin in holding various types of drugs coupled with a variety of modifications makes this protein a versatile drug carrier with incalculable potential in therapeutics. This review provides a brief outline of the different structural properties of HSA, and its various binding sites, moreover, an overview of the genetic, biomedical, and allosteric modulation of drugs and drug delivery aspects of HSA is also included, which may be helpful in guiding advanced clinical applications and further research on the therapeutic potential of this extraordinary protein.
Keywords: Human serum albumin, Allosteric modulation, Drug delivery, Drug specificity, Albumin fusion proteins
Graphical abstract

Highlights
-
•
Human Serum Albumin (HSA) excels in drug accommodation, making it a versatile drug carrier with vast therapeutic potential.
-
•
This review summarizes HSA's structural diversity and explores genetic, biomedical, and allosteric aspects in drug delivery.
-
•
Additionally, it covers in-silico studies on HSA's drug binding affinities and its impact on SARS-CoV-2.
-
•
Understanding HSA's binding sites and mechanisms aids in designing drug molecules for enhanced delivery and treatment.
1. Introduction
Human serum albumin belongs to the homologous protein family, having a unique structure with diverse ligand binding ability. It is a soluble and monomeric, multi-domain macromolecule, abundantly present in the human blood plasma, responsible for buffering of pH and maintaining the oncotic pressure of plasma (Peters, 1995). Moreover, the proper distribution of fluids among different body compartments is also the role of this dominating plasma protein (Evans, 2002; Mendez et al., 2005). Intracellular and extracellular transportation of various endogenous and exogenous substances is also possible by HSA which serves as a transporter and depository protein for them. HSA binding to a drug molecule reduces its toxicity by holding the toxic compound hostage till its excretion, increases plasma solubility, provides protection against oxidation and prolongs its half-life (Carter and Ho, 1994; Kragh-Hansen, 1981; Kragh-Hansen et al., 2002). Besides this, pharmacokinetics and pharmacodynamics profiling of drugs is also characterized via drug binding to HSA, as it exhibits the capability to attach to nearly all recognized pharmaceuticals, along with numerous nutraceuticals and harmful substances, significantly influencing their pharmacokinetics and toxicokinetics. (Kragh-Hansen et al., 2002; Herv et al., 1994; Vorum, 1999; Belinskaia et al., 2021). Furthermore, uniform and buffered distribution of hydrophobic drugs through hydrophilic medium become favorable only by HSA. Hydrophobic molecules binding to HSA, not only solubilize them for transportation but also prolong their biological lifetime by avoiding them from being metabolized (Rang et al., 1999). In clinical diagnosis, major plasma protein acts as a useful biomarker of various diseases, including ischemia, cancer, rheumatoid arthritis, severe acute graft-versus-host disease, post-menopausal obesity as well as for conditions that require glycemic control monitoring (Gupta and Lis, 2010). Furthermore, HSA is extensively used, therapeutically, to treat surgical blood loss, burns, shock, hypovolemia, trauma, hemorrhage, cardiopulmonary bypass, acute respiratory distress syndrome, acute and chronic liver disease, hemodialysis, nutrition support, resuscitation and hypoalbuminemia (Alexander et al., 1982; Erstad et al., 1991; Koga et al., 2010; Sbarouni et al., 2011; Tullis, 1977).
2. Transcription of HSA
HSA transcription is carried out by a gene located on the chromosome 4 (Harper and Dugaiczyk, 1983; Mikkelsen et al., 1977; Minghetti et al., 1986; Nishio and Dugaiczyk, 1996), containing 16,961 nucleotides from the putative ‘cap’ site to the first poly (A) site. This gene consists of 15 exons and is divided into 14 introns, situated inside the three domains which are supposed to have evolved via triplication of a single primordial domain (Fig. 1). Mutations in this gene can result in abnormal proteins that are responsible for its altered binding and carrying capability, affect the pharmacological effect of the molecule. In humans, a copy of ALB (albumin) gene must be functional, constitutively to produce circulating levels of human serum albumin. Consistently responsive ALB genes have thirty processing signals, an active promoter, and splicing signals which are efficiently recognized via the splicing machinery of the cell for activity (Minghetti et al., 1986). HSA can also undergo post-translational modifications, under some diseased and other such states which can influence its drug-interaction capabilities. Among all the amino acids, cysteine 34 in domain I appears to be the primary target for such modifications (Lee and Wu, 2015). Otagiri et al. have highlighted the importance of post-translational modifications, such as oxidation, glycation, and S-nitrosylation in albumin mutant analogues for pharmaceutical purposes (Otagiri and Chuang, 2009). Nakajou et al. have studied the effect of glycation on the structure, function and biological fate of recombinant human serum albumin (Nakajou et al., 2003).
Fig. 1.
ALB gene and mRNA organization. The ALB gene is present on the long arm of chromosome 4, near the centromere at position q11-22, consisting of 22,306 base pairs (bp) further subdivided into 15 exons and 14 introns. The numbering of exons and introns is based on the GenBank reference sequence NC_000004.11. The ALB gene transcript consists of 2250 bp; the numbering of exon bp is based on the GenBank reference sequence NM_000477.3 (Fanali et al., 2012).
3. Biosynthesis of HSA
HSA is synthesized in the hepatocytes as pre-pro-albumin, followed by its modification into pro-albumin within the endoplasmic reticulum. Pre-pro albumin has an N-terminal oligopeptide containing six amino acids, further cleaved by furin enzyme in the Golgi vesicles to produce the mature serum albumin, containing 585 residues (single chain) and molecular weight of 66,438 kDa (Peters, 1995). HSA may undergo various modifications during its long lifespan (28–30 days) that affect the anti-oxidant and ligand binding properties (Roche et al., 2008).
4. Structure of HSA
HSA is rich in Cysteine, Leucine, Glutamine, and Lysine residues with few Methionine, Glycine, and Isoleucine residues and only one Tryptophan residue located at 214 positions. The net charge of the HSA protein is very high due to the abundance of ionized residues, which promotes its solubility. Additionally, this net charge is negative with a value of 15 at pH 7.0 because the polypeptide chain contains more acidic residues than basic ones (Peters, 1995). The secondary structure of HSA contains 68% α-helices with no β-sheets. HSA is a globular protein organized into three structurally similar domains known as Domain I, II and III (Fig. 2), each of which consists of two subdomains, generally designated as subdomain IA, IB, IIA, IIB, IIIA, and IIIB (Curry, 2002, 2009; Fasano et al., 2005). Moreover, three-dimensional structural stability and biological activity are possible by 17 disulfide bridges, established by 34 Cysteine residues except one free Cysteine residue situated at position 34, unable to make a pairing (Sugio et al., 1999).
Fig. 2.
Organization of HSA Modular domain. The upper panel demonstrates the SA sequence architecture. The lower panel illustrates the 3D structure of HSA with the sub domains presented with different colors (domain IA, in pink; domain IIA; in sandy brown; domain IIIA, in green; domain IB, in purple; domain IIB, in blue; domain IIIB, in rosy brown). Atomic coordinates were retrieved from the Protein Data Bank PDB ID 1AO6 (Fanali et al., 2012). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
All the three domains of HSA are assembled asymmetrically; with approximate dimensions of 80 × 80 × 30 Å (Sugio et al., 1999). All the domains consist of ten α-helices and are further divided into subdomain A and subdomain B. These domains are linked together with a long extending loop, which comprises of six and four α-helices, correspondingly (Fig. 2). The disulfide bond topology and polypeptide chain folding are comparable for all subdomains. Subdomain IIA is linked via hydrogen and hydrophobic bonding between IA and IB subdomains interface area. As a result, domains I and II appear in a T-shaped arrangement, approximately perpendicular to each other. On the other hand, domain III protrudes from subdomain IIB forming a Y-shaped assembly for domains II and III, whereas domain III interacts with subdomain IIB only. Few contacts link domains I and III and are divided by a large channel formed by subdomains IB, IIIA, and IIIB. Terminal regions of domains forming inter-domains, nine turn long α-helices, responsible for connecting IIB to IIIA and IB to IIA domains. All domains show interaction with their neighbouring domains in a different manner, despite the similar structure. Consequently, the positioning of domains establishes an extremely asymmetric environment where several binding sites for ligands are situated (Sugio et al., 1999; Ascenzi et al., 2009a; Curry et al., 1998).
5. Albumin binding sites
5.1. Drug site I
Pioneering studies by Sudlow and their co-workers revealed the existence of two distinct potential drug-binding sites on HSA (Sudlow et al., 1975, 1976). Bos et al. used fragments of albumin which are derived from the digestion of pepsin and trypsin enzymes and suggested that drug sites I and II are located in domains II and III, correspondingly (Bailey and Dickinson, 2003; Bos et al., 1988a, 1988b). Recent crystallographic investigations also suggest that the drug sites I and II are the predominant drug binding sites (Watanabe et al., 2000). In subdomain IIA, entrance to the site I face subdomain IIIA. Additionally, drug site I have an extended binding region because of the presence of residues from subdomains IIB and IIIA (Curry, 2009; Ghuman et al., 2005).
After the pivotal work by Sudlow, various research studies were focused towards describing the environment of drug site I. Primarily, Fehske et al. stated that drug site I contain a binding area for warfarin–azapropazone, which contains two overlapping binding sites for azapropazone and warfarin (Fehske et al., 1980, 1981, 1982). Correspondingly, Yamasaki and their co-workers suggested that within site I, three binding regions are located known as subsite IA, IB, and IC (Yamasaki et al., 1996). Sudlow's site I is recognized for its affinity for dicarboxylic acids and large heterocyclic molecules possessing a negative charge, such as warfarin. In contrast, Sudlow's site II is characterized by its binding to aromatic carboxylic acids containing a single negatively charged acid group separated by a hydrophobic region, as seen in diazepam and ibuprofen, for example. (Table 1) (Kragh-Hansen et al., 2002; Curry, 2009; OTAGIRI, 2005). Kragh-Hansen defines the site I as “a large and flexible region” on the basis of various ligands' binding ability or to accommodate many ligands at the same time (Kragh-Hansen, 1988). Some studies helped to explore that drug site I is more prominent than drug site II, however, drugs that are responsive to the site I, while binding, cover various parts of the subdomain IIA and IIB (adjacent to the interface) (Ghuman et al., 2005; Zhu et al., 2008). Moreover, a site I consist of two massive polar clusters; including a pair of centrally-located polar features, formed by the tyrosine (Tyr150), histidine (His242) and arginine (Arg257) residues side chain, located near the end of the site. While lysine (Lys195 &199) and arginine (Arg218 & 222) residues result in another polar centre on an outer cluster at the entrance of the binding site (Fig. 3a) (Ghuman et al., 2005; OTAGIRI, 2005).
Table 1.
The table illustrates drugs that have been suggested to bind to the site I with higher affinity (association constant: K1).
| Drugs (Association Constants) | Structures | Drugs (Association Constants) | Structures |
|---|---|---|---|
| Warfarin (3.4 × 105 M−1) | 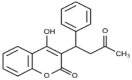 |
Iodipamide (9.9 × 106 M−1) | 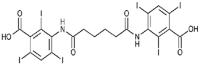 |
| Phenylbutazone (1.5 × 106 M−1) | 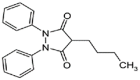 |
Iophenoxic acid (7.7 × 107 M−1) | 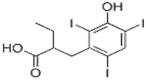 |
| Azapropazone (2.8 × 105 M−1) | 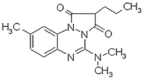 |
Furosemide (2.0 × 105 M−1) |  |
| Indomethacin (1.4 × 106 M−1) | 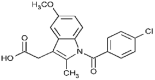 |
Bucolome (1.5 × 106 M−1) | 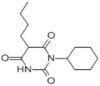 |
| Tolbutamide (4.0 × 104 M−1)a[10] | 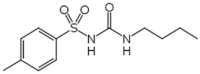 |
Sulfisoxazole (1.8 × 105 M−1)a[55] | 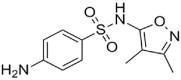 |
Fig. 3.
HSA Crystal structure complexed with warfarin (a) and diazepam (b). Amino acid residues and Drugs mentioned were presented in stick, and color-coded by atom-type), orange (diazepam) or gray (residues), carbon; yellow (warfarin), oxygen; red, nitrogen; blue, chlorine; green. Atomic coordinates were retrieved from the Protein Data Bank I.Ds 2BXD and 2BXF for warfarin and diazepam, respectively (Fanali et al., 2012). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
5.2. Drug site II
Additionally, exhaustive research has also been performed to understand the environment within drug site II such as a quantitative structure-activity relationship (QSAR) study conducted by Wanwimorluck et al. stated that the site II consists of the wide and broader hydrophobic cleft with a cationic moiety situated adjacent to the surface. On the structural basis of HSA, site II is considered principally a polar cavity with one polar patch present close to the opening of the site, centralized on tyrosine positioned at 411 and arginine positioned at 410 (Fig. 3b) (Curry, 2009). These apolar and polar features arrangement is constant with the drugs of site II, which are negatively charged aromatic carboxylic acids moiety at one side of the molecule and divided by a hydrophobic centre (Table 2) (Kragh-Hansen et al., 2002; OTAGIRI, 2005). The drugs of site II (e.g. Diazepam, ibuprofen, and diflunisal) interact with the hydroxyl group of tyrosine, arginine, and serine residues involved in salt-bridge and hydrogen bonding (Itoh et al., 1997). Drug site II was suggested to be narrow than drug site I because it hardly binds to larger molecules (Kragh-Hansen et al., 2002; Curry, 2009; OTAGIRI, 2005). To date, no study has been reported for separating this site into different sub-binding regions as observed in the drug site I. Drug binding site II is less flexible, often shows stereoselectivity and is influenced by the modification of ligand structure with a comparatively small group (Curry, 2009; OTAGIRI, 2005). Ibuprofen in its (R) form binds with 2.3 times higher affinity to site II than the (S)-enantiomer (Itoh et al., 1997). Additionally, diazepam binds to the site II, but flunitrazepam (fluorinated analogue) cannot bind to site II (Chuang and Otagiri, 2001). Therefore, the site II can bind with a variety of ligands, but it is more selective than site I. Bhattacharya et al. suggests that two molecules of long-chain fatty acid can accommodate in site II at a time due to its flexibility (Bhattacharya et al., 2000a).
Table 2.
The table illustrates drugs that have been suggested to bind to site II with higher affinity (association constant: K1).
| Drugs (association constants) | Structures | Drugs (association constants) | Structures |
|---|---|---|---|
| Diazepam (1.3 × 106 M−1) | 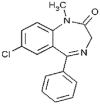 |
6-MNAe (1.2 × 106 M−1) | 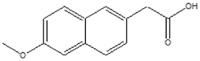 |
| Diflunisal (5.3 × 105 M−1) | 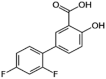 |
Diclofenac (3.8 × 106 M−1) | 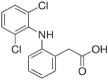 |
| Ibuprofen (3.5 × 106 M−1) | 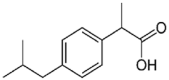 |
Etodolac (2.0 × 105 M−1) | 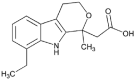 |
| Ketoprofen (2.5 × 106 M−1) | 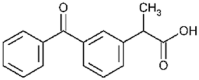 |
Clofibrate (7.6 × 105 M−1) |  |
| Naproxen (1.2 × 106 M−1) | 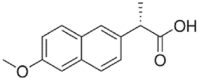 |
Iopanoic acid (6.7 × 106 M−1) |  |
5.3. Drug site III
A third binding site has also been discovered within subdomain IB as the primary binding pocket of a photoisomer of bilirubin, hemin, the steroid antibiotic such as fusidic acid, and sulphonamide derivatives (Zsila, 2013; Zunszain et al., 2008). Some crystallographic studies reported that the prominent cleft of subdomain IB has secondary binding sites for some other compounds as well (Table 3) (Hall et al., 2013). Carter et al. have suggested that site IB is another main drug-binding site of human serum albumin (Carter and Ho, 1994). Kragh-Hansen proposed more than two drug sites for digitoxin, warfarin, phenol red, and diazepam on human serum albumin (Kragh-Hansen, 1983). As reported by some crystallographic studies hemin and heme are enclosed inside the hydrophobic D-shaped cavity of subdomain IB of HSA.
Table 3.
List of Ligands that binds with HSA Binding Site in Subdomain IB.
| Site IB Ligands | PDB code |
|---|---|
| Bilirubinb | 2VUE |
| Compound 3 | 3LU8 |
| Fusidic Acid | 2VUF |
| Hemin | 1N5U, 1O9X |
| Lidocaine | 3JQZ |
| Arachidonic Acid | 1GNJ |
| Azapropazone | 2BXI, 2BX8 |
| Azidothymidine | 3B9L |
| Capric Acid | 1E7E |
| Dansyl-L-Arginine | 2XVW |
| Dansyl-L-Asparagine | 2XVV |
| Dansyl-L-Glutamate | 2XSI |
| Indomethacin | 2BXQ, 2BXM |
| Iophenoxic Acid | 2YDF |
| Lauric Acid | 1E7F |
| Naproxen | 2VDB |
| Myristic Acid | 1BJ5 |
| Palmitic Acid | 1E7H |
| Salicylic Acid | 2I30, 3B9M |
| Stearic Acid | 1E7I |
| Tri-Iodobenzoic Acid | 1BKE |
| Warfarin | 2BXD, 1HA2, 1H9Z |
6. Fatty acid (FA) binding sites
Since it was first reported in 1941 (Kendall, 1941), many studies have been reported for better characterization of FA binding potential of HSA, which is capable of binding nine long-chain FAs, at a time, signify the principal biological ligands at different binding sites (Curry et al., 1998). These sites are asymmetrically distributed all over the human serum albumin protein with different affinities (Bhattacharya et al., 2000b; Simard et al., 2006). Among other FA-binding sites; FA-binding sites 4 and 5 showed the highest affinity for fatty acids, whereas, FA2 showed moderate affinity. FA2 is entirely present inside the N-terminal and found at the interface between IA and IIA subdomain, while fatty acid binding sites 4 and 5 are present inside the domain III. These fatty acid binding sites provide the most enclosed binding environment on HSA, which allows the binding of FA methylene tail linearly, whereas, carboxylate moieties of fatty acid form a salt bridge with side chain of one basic residue (Simard et al., 2006). FA-binding sites 8 and nine are generally considered as additional sites, while, they show occupancy of ligand only, when short chain (i.e., FA8) and saturating (i.e., FA9) fatty acids are present, respectively (Bhattacharya et al., 2000b). Numerous endogenous and exogenous drugs also can bind with FAs sites of HSA, showing its significance in drug transportation to the target areas. Drug affinity with FAs binding sites increases the drug half-life and plasma solubility but reduces the free active concentration of drugs (Ascenzi et al., 2006).
6.1. Fatty acid-binding site 1 (FA1, heme pocket)
FA-binding site 1 is found in the D-shaped cavity of subdomain IB, which is also reported as the potential binding site for a number of active molecules (Fig. 4). If any compound does not bind at the heme binding pocket, the pocket is closed by the stacking of Tyr138 with Tyr161. FA-binding site 1 partly corresponds to the heme binding site (Fig. 5a). Binding of Heme–Fe (III) produce massive conformational changes, resulting in the rearrangement of tyrosine residues (Heys et al., 1998; Kouzuma et al., 2004) that are involved in p–p interaction with porphyrin and tyrosine (Tyr161) residue provide donor oxygen atom for the Penta-coordinated heme–Fe-atom. Long IA–IB connecting loop protects Heme–Fe (III) which fits in the opening of the cleft. Heme–Fe (III) propionates directed towards the interface between domains I and III and are supported via salt bridge formation with arginine (Arg114), histidine (His146), and lysine (lys190) amino acids as shown in Fig. 5b (Curry et al., 1998).
Fig. 4.
Showing different ligand binding to drug site IB (Fanali et al., 2012).
Fig. 5.
3D structure of HSA complexed with prototypical ligands. Panel A shows ribbon representation of the HSA structure complexed with heme–Fe (III) in FA1 (i.e., heme site), warfarin in FA7 (i.e., Sudlow's site I), and ibuprofen in FA3–FA4 (i.e., Sudlow's site II). Warfarin, Heme and ibuprofen, are represented as as sticks (black). Atomic coordinates were retrieved from PDB ID 1O9X. Panel B shows Heme–Fe (III) binding to the FA1 site. Heme–Fe (III) (red) is bound in the D-shaped cavity in the centre of the four-helix bundle of subdomain IB. Arg114, Tyr138, His146, Tyr161, and Lys190 residues are shown. Atomic coordinates were retrieved from the PDB ID 1O9X (Fanali et al., 2012). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
6.2. Fatty acid-binding site 2
FA-binding site 2 is situated between IA and IIA sub-domains. Numerous pieces of evidence specified that the binding of FA to this site stabilizes. B-conformation of HSA and cause ligand-induced conformational changes. Domain I require rotation to accommodate FA for the formation of adjacent pocket, as opposed to domain II. Thus, ligand binding to FA2 is stabilizing by this rearrangement (Ascenzi and Fasano, 2010; Fanali et al., 2005, 2007). The carboxylate moiety of the fatty acids is buried in subdomain II via hydrogen bonds with the side chain of tyrosine (Tyr150), arginine (Arg257), and serine (Ser287) residues (Bhattacharya et al., 2000b). However, a few reports suggested that FA2 has been considered as a third binding site for ibuprofen (Ascenzi et al., 2009a, 2009b; Nicoletti et al., 2008).
6.3. Fatty acid-binding sites 3 and 4 (Sudlow's site II)
FA-binding sites 3 and 4 are situated in subdomain IIIA and consist of six helices that composed of Sudlow's site II a primary drug-binding site (Carter and Ho, 1994; Sudlow et al., 1975). In FA-binding site 3, the head moieties of fatty acids involved in hydrogen bonding interaction with Ser and Arg positioned at 342 and 348 respectively of the subdomain IIB, and with the Arg at position 485 in subdomain IIIA. In FA-binding site 4 (Curry, 2009), the carboxyl groups of fatty acids form hydrogen bonding interaction with arginine (Arg410), tyrosine (Tyr411), and serine (Ser489) residues of subdomain IIIA.
Indeed, site II contains an apolar region, attached to fatty acid methylene tail bound to binding site 3 (Curry, 2009; Ghuman et al., 2005; Bhattacharya et al., 2000b) and the polar region showing interaction with the carboxyl group of fatty acid bound to binding site 4. To date, no drug has been reported for long hydrophobic tunnel of fatty acid binding site 4. On the other hand, the polar region of Tyr411 residue does not interact with the fatty acids that are bound to binding site 3. Drugs (e.g., ibuprofen) that bind with drug site II, show interaction with hydroxyl moiety of Ty, Arg, and Ser positioned at 411, 410 and 489 respectively and stabilized by the formation of salt-bridge and hydrogen-bonds (Curry, 2009; Ghuman et al., 2005). Both Tyr411 and Arg410 are present nearby, therefore, the nucleophilic nature of the Tyr411 is mostly controlled through electrostatically by Arg410.
6.4. Fatty acid-binding site 5
FA-binding site 5 is situated in subdomain IIIB and formed by a hydrophobic channel. The carboxylate moiety of FA shows interaction with the tyrosine (Tyr401) and lysine (Lys525) residues side chain, while the fatty acid methylene tail extends into the tunnel (Bhattacharya et al., 2000b). Complexes of HSA with arachidonic acid in FA-binding site 5 give a comprehensive view of the unique structure of this site (Petitpas et al., 2001).
6.5. Fatty acid-binding site 6
FA-binding site 6 lies in at the interface of subdomains IIA & IIB. This site is engaged by long and medium-chain fatty acids. The cavity of FA-binding site 6 is different from other fatty acids sites; because of the absence of the cluster of amino acids that sterically stabilizes the carboxylate moiety of fatty acid. The FA carboxylate moiety temporarily recognizes by the arginine (Arg209), lysine (Lys351), and Serine (Ser480) residues side chain, while the fatty acid methylene tail is buried via the formation of a salt-bridge from arginine (Arg209) to aspartate (Asp324) and glutamic acid (Glu354) residues. In FA6, capric acid shows different binding modes only, because two molecules showed a tail-to-tail linear conformation. Additionally, the ligands binding affinity with this site is relatively low (Bhattacharya et al., 2000b; Simard et al., 2006).
6.6. Fatty acid-binding site 7 (Drug site I)
The subdomain IIA is a hydrophobic cavity that contains the seventh FA binding site (drug site I). This site preferred the binding of bulky heterocyclic anions (Curry, 2002, 2009; Sudlow et al., 1975, 1976). However, FA-binding site 7 is small than other binding sites in subdomain IIIA (Curry, 2009; Ghuman et al., 2005). Moreover, FAs-FA7 complexation resulted in a curved configuration indicating low affinity of the binding site towards FA molecules. The arginine residue (Arg257) stabilizes the carboxylate moiety of fatty acid by polar interaction(s) and provides a link between FA-binding site 2 and FA-binding 7 sites. Basic residues of this site do not involve directly in the binding of fatty acid (Bhattacharya et al., 2000a; Ascenzi et al., 2009b). However, Lys199 secured its position at the entrance of the Sudlow's site I where Lys199-dependent esterase activity is mostly inhibited by the ligand bindings. The cleavage of a few substrates like; penicillin, acetylsalicylic acids and trinitrobenzene-sulfonates, into two products, take place at Lys199, which either released or form bonds with Lys199 residue. The stability of the FA7 is directly linked to inter-hydrogen bonding between the basic form of the Lys199 and the acidic form of the neighbouring Lys195 residue through bridging water molecules.
6.7. Fatty acid binding site 8
FA-binding site 8 is found at the base of the gap between IA–IB–IIA and IIB–IIIA–IIIB subdomains. This binding site only allows small-chain FA molecules, like capric acid, to accommodate in it. FA8's hydrophobic end is formed by the contribution of FA6 and capric acid methylene tail. The ligand polar head is stabilized by the polar residues of the cavity such as lysine (Lys195 &199), arginine (Arg218), aspartate (Asp451), and serine (Ser454) residues (Bhattacharya et al., 2000a).
6.8. Fatty acid binding site 9
FA-binding site 9 is present in the uppermost area of the cavity between IA–IB–IIA and IIB-IIIA-IIIB subdomains, consequently, providing an open binding environment. The binding of ligand to the FA9 site is stabilized by salt-bridge formation between the residues of domain I (Glu187) and domain III (Lys432). Remarkably, the FA9 binding site is formed by fatty acid-induced conformational changes, therefore an extra binding pocket in fatty acid-saturated human serum albumin (Bhattacharya et al., 2000a; Petitpas et al., 2001).
6.9. Current research on HSA binding based on FA interplay
In a study conducted by Wu et al. it was observed that the addition of molar quanities of non-esterified fatty acid palmitate reduced the Co2+-binding affinity with HSA (Wu et al., 2023). In another study conducted by Sekula et al. differences were observed in the binding mode of lysophospholipids (LPLs) analogues called cyclic phosphatidic acids (cPAs) with human and equine serum albumin, despite the structural homology between them (Sekula et al., 2016). Accoording to another study conducted by Czub et al. studying the carrying mechanism of testosterone by HSA, It was observed that testosterone binds two sites, neither of which overlaps with the previously suggested Sudlow site I, however, the site remains the same in case of both the human an equine serum albumin (Czub et al., 2019). Other comparative studies of albumin complexes with hormones, drugs, and other biologically relevant compounds strongly suggests interference between the compounds present in blood and the behavior of HSA.
7. Minor binding sites
7.1. Thyroxin binding site
FA-free HSA binds to T4 at four different sites, namely thyroxine-1 to thyroxine-4. These four sites are situated in subdomain IIA, IIIA, and IIIB, sequentially (Fig. 6). The Thyroxin binding crevices partly overlap the FA-binding site 3–4, FA5 and FA7 binding sites, which display double site occupancy (Petitpas et al., 2001). Tryptophan (Trp214) and arginine (Arg218 & 222) side chains do not get affected by the binding of T4 to Tr-1. Specific hydrogen bonding interactions are observed amongst the T4 phenolic hydroxyl group and arginine (Arg257) and tyrosine (Tyr150) residues of the binding site. The carboxylate group of T4 involve in salt-bridge formation with two lysine residues (Lys199 and Lys195) might involve in an electrostatic interaction as well, whereas, the amino group is positioned at the opening of site 7. Structurally, it has been suggested that Tr-1 is the highest affinity T4 binding site rather than Tr-2 and Tr-3 sites (De Chateau et al., 1994).
Fig. 6.
3D structure of HSA complexed with T4 in the absence of myristate (PDB ID: 1HK1).
Myristate (magenta) is represented as space-filled. T4 (olive) is represented as sticks (Fanali et al., 2012).
7.2. Binding site for bacterial protein
It has been stated that HSA binds to bacterial surfaces by PAB protein (De Chateau et al., 1994). PAB protein contains a protein-G related albumin binding module (GA) domain having 60% similarity of sequence with G protein (a bacterial protein which attaches to the Immunoglobulin Fc region) (Björck and Kronvall, 1984). HSA interacts with the GA domain and takes place between h2 of protein-G related albumin binding module including two loops adjacent to it, and the domain II of human serum albumin is surrounded by helix 2–3 and helix 7–8 prior to helix7. A small part of the surface area of the GA domain interacts with human serum albumin, with the contact area composed of a hydrophobic centre surrounded by a pair of hydrogen bonds. The hydrophobic centre on the other hand is flanked by phenylaniline, alanine, valine, and methionine positioned at 228 &326, 229 & 322, 325, and 329 from human serum albumin and phenylaniline, alanine, leucine, and isoleucine positioned at 27, 31, 44, and 48 respectively from Protein-G related albumin binding module (Lejon et al., 2004). In the IIB domain of HSA and the loop preceding the h2 in the protein-G related albumin binding module, a hydrogen bonding interaction is formed between helix 7. The two hydrogen bonds stabilize the HSA-GA binding, mediating from Glu321 of HSA towards the nitrogen of threonine (Thr24) and serine (Ser25) residues of the Protein-G related albumin binding module.
7.3. Binding sites for metals
Human serum albumin shows several binding sites for numerous metal ions, such as Magnesium (II), Aluminum (III), Calcium (II), Manganese (II), Cobalt (II/III), Nickel (II), Copper (I/II), Zinc (II), Cadmium (II), Platinum (II), Gold(I/II), Mercury (II), and Terbium (III) (Peters, 1995; Deng et al., 2010; Duff and Kumar, 2009; Sokoł et al., 2009, 2010). Thus, three major binding pockets of HSA were observed with proper matching of residues for the different geometries of metals. The primary metal binding site is situated at the NTS (N-terminal binding site), where some metal ions such as Cobalt, Nickel and Copper, are coordinated through donor atoms of nitrogen from aspartate (Asp1), alanine (Ala2), and histidine (His3) residues (Sadler et al., 1994). The secondary site is shown by the thiol group of free cysteine residue (Cys34) which binds to gold, platinum, and mercury ions. The third metal binding site is the main Cadmium Site A (MBS-A) which contains some residues such as His67, Asn99, His247, and Asp249 for binding. Finally, further metal ions bind to undefined regions or un-specifically (Shaw, 1989).
Studies are still on-going to gain an in-depth insight into the biochemical processes through which HSA is able to bind and mobilize these metal ions. A study conducted by Gautam et al. in 2023 suggests that HSA mobilizes mercury more effectively compared to Cadmium, also including the reasons for the mentioned observation (Gautam and Gailer, 2023). In 2013, Bal et al. studied in detail, the four metal-binding sites on HSA, their localization and metal preferences, binding of transition metal ions to albumin, their sites, affinities and rates (Bal et al., 2013). The interaction of the organometallic complexes of Ru(η6-p-cymene) and Rh(η5-C5Me5) with human serum albumin (HSA) was studied in detail by Dömötör et al. using a combination of various methods such as ultrafiltration, capillary electrophoresis, 1H NMR spectroscopy, fluorometry and UV–visible spectrophotometry. The mentioned organometallic ions were found bound to HSA via a coordination bond to a high extent. The release of these bound metal ions were therefore kinetically hindered and was not affected by the denaturation of the protein (Dömötö et al., 2019). Al- Harthi et al. investigated zinc interactions and the effect of various FAs, including lipoic acid (LA), on the protein structure, stability, and zinc(II) binding. Upon the binding of FAs to HSA, we observed a range of behaviors in terms of zinc(II) affinity, depending on the type of FA (Al-Harthi et al., 2022).
7.4. Bilirubin binding site
Bilirubin, is an insoluble pigment with yellow-orange colour formed by the catabolism of heme–Fe. It binds to HSA near to the heme site in subdomain IB. However, bilirubin accommodates in the pre-generated L-shaped pocket because of steric hindrance of the exposed tetrapyrrole ring indicatively situated at the FA1 cavity opening. The pigment of bilirubin is held in its place by extension of peptides connecting the residues (Glu110–Glu119) of subdomains IA and IB, and through salt-bridges via arginine positioned at 117 &186 near binding cleft opening (Zunszain et al., 2008).
8. Allosteric modulation of the drugs bound to HSA
The conformational changes in HSA involve instant vicinity generation of the binding site(s) (Peters, 1995; Kragh-Hansen, 1981; Ghuman et al., 2005; Ascenzi et al., 2005). Remarkably, chloroform, halothane, NO (nitric oxide) and N2O bind to HSA, to drug site I, and appear allosterically and spectroscopically linked to Cysteine residue at position 34. It is notable that without nitrosylation of the Cys34 residue nitric oxide can still bring about variations in the structure of HSA. These findings proposed that anaesthetic binding to HSA may change by the nitric oxide and support the fact that anaesthetic-like noncovalent bonding to proteins results from a few physiological effects of nitric oxide (Ascenzi et al., 2001; Sampath et al., 2001; Stamler, 2004). The drug site I and heme binding crevices are associated functionally to lower down the second-order rate constant and binding affinity of the drug. While drug and heme dissociation kinetics from human serum albumin complexes is unchanged by third components. It shows that the Sudlow's site I drug such as warfarin behaves as an allosteric modulator for the connotation of heme and vice versa. On the other hand, the drug site II and heme-binding crevices are functionally disassociated upon binding of the drug to Sudlow's site II such as ibuprofen, does not alter the heme association and vice versa (Ascenzi et al., 2005; Fasano et al., 2001). HSA bind to benzodiazepines at numerous allosterically and functionally linked clefts, depending on the substitution and optical confirmation of benzodiazepines. It has been reported that allosteric interaction affects stereo-selective binding equilibria between warfarin, ibuprofen, lorazepam and HSA. HSA binding either with warfarin and ibuprofen enantiomers affects the binding affinity of lorazepam (Ascenzi et al., 2006; Fitos et al., 1999).
Fatty acids allosterically regulate the binding properties of albumin, mainly drug site I. The HSA binding properties are regulating by myristate are very complicated, including allosteric and competitive both the mechanisms. Initially, ligands of drug site I such as warfarin shift fatty acid linked to binding site 7 and drug site II ligands like Ibuprofen, shift fatty acid bound to sites FA-binding sites 3 & 4. Consequently, HSA binding with heme shifts fatty acid linked to the binding site 1. With respect to conformational changes due to the allosteric modulation of binding properties, the binding of fatty acid leads to the rearrangement of the domain at the interfaces of domains I-II and II-III. Short fatty acids such as octanoate do not show this type of rearrangement, it binds to drug site II and shifts the other ligands like ibuprofen without inducing allosteric rearrangement(s) of albumin. It shows that the hydrophobic interactions between the elongated polymethylene tail of fatty acid and human serum albumin lead to allosteric rearrangement (Curry et al., 1998; OTAGIRI, 2005; Zunszain et al., 2008; Bhattacharya et al., 2000b; Petitpas et al., 2001). Massive conformational changes take place upon binding of myristate with HSA (Simard et al., 2006). It is reported that the addition of three long-chain fatty acids equivalents improved the affinity of the drug site I (fatty acid binding site 7) ligands. Additionally, the binding of myristate to subdomain IA (FA-binding site 2) was recommended to be functionally associated with drug site I. It must be considered that binding of more than three myristates (FAs) equivalents lowers the warfarin affinity for drug site I, similarly as observed in Ferric (III) heme. Based on structure, myristate binding greatly affects the conformation of subdomain IA, while unaffected by the binding of other ligands to HSA with myristate. The interdomain of the IA-IB interface hosted the heme binding site and described allosteric binding interactions between fatty acids and heme. Moreover, in the myristate-HSA-heme complex, subdomain IIB conformation is like the ligand-free conformation of HSA with or without warfarin. Furthermore, subdomain IIB is connected to IIIA by a long α-helix that greatly affects both stereo-selectivity and geometry of the drug site (Kragh-Hansen, 1981; Curry et al., 1998; Bhattacharya et al., 2000a, 2000b; Zunszain et al., 2008; Petitpas et al., 2001; Reichlin et al., 2009; Vorum et al., 1996; Wilding et al., 1977).
Remarkably, Human serum albumin undergoes reversible conformational isomerization due to allosteric and pH-dependent effectors. At acidic pH, HSA exists in a fast (F) form, because of an increase in viscosity, lower solubility, and a significant loss of α-helical content. HSA exists in normal (N) form without the presence of allosteric effectors and shows a structure resembling the shape of a heart at pH 4.3 and 8.0. At the basic pH, HSA exists in the basic (B) form with the complete loss of α-helix content and an improved affinity for some ligands like warfarin (Yamasaki et al., 1996; Fasano et al., 2001, 2004; Wilding et al., 1977).
9. Antioxidant activity of human serum albumin
The antioxidant property of HSA owing to the free sulfhydryl group (–SH), makes it responsible for most antioxidant properties of plasma. These functions make SA a major player in and a mirror of overall health status, ageing, and neurodegeneration.2 The HSA antioxidant property is due to its metal-binding and redox properties of cysteine residue (Cys34). Therefore, Fe (II/III) and Cu (I/II) are very quick to generate reactive oxygen species after reaction with oxygen (O2). Free ions of Copper (I) and Ferric (II) react with hydrogen peroxide (H2O2) leading to the production of the toxic hydroxyl radical (OH−) via the Fenton reaction. The binding of HSA with Copper (I) and Ferric (II) changes their oxidation state to Copper (II) and Ferric (III), because of this oxidation, the participation of these ions is hindered in the Fenton reaction. Human serum albumin traps Ferric (III) and heme–Ferric (III), and plays a critical anti-oxidative function in heme–Ferric (III) and Ferric (III)-overload diseases (Ascenzi et al., 2006; Ascenzi and Fasano, 2009; Cha and Kim, 1996; Walter et al., 2006). The (Asp-Ala-His)–Cu (II) complexation displays a superoxide dismutase-like activity, which prohibits the production of reactive oxygen species. HSA prevents LDL (low-density lipoproteins) peroxidation by trapping Cu(II) ions (Bar-Or et al., 2001). (Sakata et al., 2002) HSA also shows an indirect antioxidant activity by binding with numerous compounds, including, bilirubin, homocysteine and oxysterols. The Oxidation of cholesterol either in vivo and in vitro leads to the formation of biologically active derivatives of oxysterols (van Reyk et al., 2006). Surprisingly, oxysterols are released by HSA more slowly than cholesterol in cells hence, restricting their harmful effects (Roche et al., 2008). Binding of bilirubin to HSA enables tetrapyrrole to act as a lipid peroxidation inhibitor, protecting it from peroxyl radicals. The complex formed between bilirubin and human serum albumin (HSA) also enhances the survival rate of human ventricular myocytes against oxidation in their natural environment (Roche et al., 2008; Neuž et al., 1993; Wu et al., 1991). Moreover, HSA protects from atherosclerosis by trapping of homocysteine because the increased level of homocysteine in the plasma is the main risk factor for atherosclerosis (Papatheodorou and Weiss, 2007). Reduced amount of SA has been linked to cognitive decline in older adults and poor outcomes in MS and ALS. SA has the ability to attach to amyloid-beta (Aβ), the precursor of AD, and prevent its harmful effects outside of the brain. Research is being conducted to investigate the potential use of SA in treating AD. SA therapy may have the potential to rejuvenate the brain (Shojai et al., 2022).
During a long lifespan (28–36 days) of human serum albumin, its molecule makes approximately 15,000 passes throughout the circulation incurring a few damages which affect ligand and anti-oxidant binding properties (Peters, 1995; Roche et al., 2008). Furthermore, the same properties are impaired in diabetes mellitus due to HSA glycation (Bourdon et al., 1999; Van Campenhout et al., 2006). Since the binding capacity of non-glycated protein with Copper (II) is more than that of the glycated HSA protein. Copper (II) induced oxidation of LDL is worsened through glycated HSA due to the production of superoxide (Bourdon et al., 1999; Sakata et al., 2002). Furthermore, the Fe(III)-binding HSA antioxidant capability is distinctly decreased in diabetes. Finally, tryptophan binding and transport are decreased in glycated HSA (Barzegar et al., 2007). Numerous AGEs (advanced glycation end products) receptors begin the signalling mechanism of the cell and increase reactive oxygen species (ROS) production in cells during binding and recognition of glycated macromolecules including human serum albumin (Mera et al., 2005; Goldin et al., 2006). AGEs including glycated serum albumin damage vascular endothelial NO synthesis activity and are also involved in the neurodegeneration (Xue et al., 2004).
10. The significance of human serum albumin in disease diagnosis and prognosis
10.1. Hypoalbuminemia
Hypoalbuminemia is associated with inflammation. Despite being addressed repeatedly in the literature, there remains uncertainty surrounding its origin and clinical importance. Inflammation increases capillary permeability and escape of serum albumin, resulting in interstitial space expansion and an increase in the albumin distribution volume. Even in the presence of high fractional synthesis in plasma, the half-life of albumin is known to be shortened, leading to a decrease in the overall quantity of albumin. This reduction in albumin levels, known as hypoalbuminemia, is primarily attributed to inflammation, which hinders appropriate reactions to conditions like surgery or chemotherapy. Hypoalbuminemia is associated with a lower quality of life and a shortened lifespan. Raising or lowering levels of serum albumin can serve as a reliable indicator of an improvement or worsening in the clinical condition, respectively. The presence and progression of infectious diseases are closely linked to hypoalbuminemia, as adaptive and innate immune responses rely on it. The oxidation and degradation of albumin impact its binding with lipid mediators, which play crucial roles in defence against infections and tissue repair. HSA levels hold prognostic value for complications arising from infectious as well as for non-infectious complexities stemming from chronic situations (Soeters et al., 2019).
In cases of SARS-COv2, hypoalbuminemia is correlated with the infectious load and the extent of lung damage. The decreased levels of HSA are associated with insufficient handling of the infection. The administration of HSA can be effective for antimicrobial therapy (Wiedermann, 2021). Hypoalbuminemia is observed in various conditions, including protein-losing enteropathy, hepatic synthetic failure, malnutrition, inflammatory states, and renal disease. Furthermore, it serves as an indicator of unfavourable outcomes in severely ill children and individuals with other medical interventions (Chen et al., 2021).
10.2. Familial dysalbuminemic hypertriiodothyroninemia and hyperthyroxinemia syndromes (FDH)
FDH are autosomal dominant disorders (Hennemann et al., 1979; Lee et al., 1979; Yabu et al., 1987), characterized by an upsurge of an entire plasma TT4 concentration primarily in Caucasians (Croxson et al., 1985). FDH is categorized by increasing of TT4 concentration, while total reverse T3 (TrT3) is up to a lesser extent (Weiss et al., 1995). Some earlier studies showed the association of the TT4 abnormality to the HSA variant. HSA mutants show a slighter increase in affinity towards T3 (Lee et al., 1979; Lalloz et al., 1983; SILVERBERG and Premachandra, 1982; STOCKIGT et al., 1981). Structural analysis of HSA mutants (Arg218Pro and Arg218His) demonstrates that greater affinity for TT4 occurs due to the replacement of the Arg218 amino acid, which interacts with the hormone linked in IIA domain, responsible for changing of its conformation which helps in overcoming steric limitations preventing the T4 binding (Petitpas et al., 2001; Petersen et al., 1996; Sunthornthepvarakul et al., 1994).
10.3. Familial hyperzincemia
Zn2+ is mainly transported by HSA in blood. This metal ion is necessary for biological processes and induction by a variety of drugs and toxins. It plays a crucial role in the body, being essential for the immune system, as well as for the transfer of genetic information. Remarkably, Zn2+ has structural and catalytic roles in large varieties of protein (Lipscomb et al., 1996). In vitro, experiments have shown that complexes of Zn2+ and HSA are transported through endothelium via receptor-mediated vesicular system (Tibaduiza and Bobilya, 1996). Erythrocytes take up Zn2+, facilitated by HSA, where it binds with glutathione (Rabenstein and Isab, 1980) and haemoglobin (Simons, 1991) which increases its affinity for O2 (Oelshlegel et al., 1974). Familial hyperzincemia is a “non-disease” condition produced by the strong binding of Zn2+ metal with human serum albumin. The specific biochemical mechanism underlying this atypical binding is still under investigation, however, the presence of a genetic variant in human serum albumin is assumed to be potentially responsible for this condition (Krebs et al., 1985).
10.4. Oxidation of HSA
Oxidation of HSA at different sites with a number of agents produced numerous effects on its physiological functions. HSA function can be altered by oxidative stress, nephropathy, diabetes mellitus, and liver diseases are a few examples of this stress. This alteration of HSA, including disulfide (-S-S-) bond formation, glycation, and carbonylation, may be responsible for changing its binding properties by affecting ligand affinity. HSA redox state can influence the allosteric ligand binding properties. Oxidatively modified HSA levels differ over a broad variety in evaluating the clinical significance of modified binding properties of ligands to oxidize HSA species in a variety of diseases (Kawakami et al., 2006; Narazaki and Otagiri, 1997; Oettl and Stauber, 2007). These HSA levels are also linked with the advanced liver diseases like liver failure. The severity of liver failure (acute and chronic) parallel to HSA oxidation, is the indication of oxidation of Cys34 and increase of carbonyl content (Oettl and Stauber, 2007). There is a direct correlation between oxidative stress and the degree of hepatic dysfunction in chronic liver disease (Terawaki et al., 2004). HSA undergo enormous oxidation in primary nephrotic syndrome (Musante et al., 2006). HSA shows low antioxidant activity in hemodialysis patients (Himmelfarb and McMonagle, 2001). Oxidative stress is made worse in Diabetic conditions as it increases the oxidized levels of HSA (Mera et al., 2005). HSA administration can be made more effective by the reduction of damaged proteins and by raising the thiol-dependent anti-oxidation levels of plasma (Quinlan et al., 2004).
11. Albumin as an unconventional chaperone
According to previous reports, Human Serum Albumin (HSA) has the ability to prevent protein aggregation and amyloid formation and acts as a chaperone for misfolded proteins. Recently, Pomier et al. conducted a study examining how HSA interacts with a broad range of client proteins, including intrinsically disordered proteins (IDPs) like Aβ, islet amyloid peptide (IAPP), alpha-synuclein, and stressed globular proteins such as insulin. Although HSA is not the sole extracellular chaperone, its abundance suggests that HSA is responsible for a significant portion of the chaperoning effects in plasma, providing new therapeutic possibilities (Pomier et al., 2022).
12. HSA as a biomarker
HSA is found to be an invaluable biomarker for numerous diseases including cancer, obesity due to menopause, severe acute graft-versus-host reaction, ischemia, rheumatoid arthritis and that which require control of glycemic index. Macromolecules having the potential to act as a biomarker have been discovered in concentrations that effectively interact with HSA. Thus, the albuminome is a vital tool in the discovery of biomarkers. Consequently, it is observed that changes in albuminome with the disease could be a potential biomarker (Gundry et al., 2007).
13. HSA and solid tumours
The HSA level determination in blood is one of the approaches to assessing the nutritional status, the disease progression, disease severity, and the prognosis, mainly in cancer patients. HSA also has been considered as an independent prognosis indicator of continued existence in a variety of cancers, like lung (Subramanian et al., 2007), gastric (Onate-Ocana et al., 2007), pancreatic (Siddiqui et al., 2007), colorectal (Boonpipattanapong and Chewatanakornkul, 2006; Cengiz et al., 2006; Heys et al., 1998) and breast cancer (Lis et al., 2003). A low level of HSA also acts as an independent indicator of cancer patients during prognosis with unfamiliar primaries (Seve et al., 2006). In current years, the malnutrition as a prognosticator in cancer existence has gained significant attention. A contrary correlation is found between HSA synthesis and the body mass index of cancer patients, which makes the option of a compensatory increase in the synthesis of HSA (Gupta and Lis, 2010). Numerous studies have been reported, in which HSA was either conjugated with dyes or radio-labelled and observed that 3%–25% of the applied dose was absorbed in a tumour (Kratz and Beyer, 1998; Kratz, 2008). The highest turnover of HSA in tumours suggests that HSA is the major source of nutrition and energy necessary for tumour development (Kratz, 2008; Stehle et al., 1997). This is a significant factor in the development of cachexia, a condition characterized by weight loss and muscle reduction, induced by various diseases, including malignancies (Kratz, 2008). In the later stages of the disease, however, inflammation and malnutrition reduce the synthesis of HSA and lower the concentration of HSA in the blood due to cytokines production (Ballmer et al., 1994; Barber et al., 1999; Yeun and Kaysen, 1998).
The assessment of HSA levels prior to treatment holds significant potential as a predictive factor in cancer patients, offering substantial benefits in terms of its strength, reliability, and feasibility (Gupta and Lis, 2010; Barber et al., 1999; Dixon et al., 2003; Sun et al., 2009). The main disadvantage of interpreting the levels of HSA is often complicated as factors, such as the progression of the disease can cause ambiguity in the degree of real nutrient deficiency (Lien et al., 2004). Additionally, HSA has a prolonged half-life, therefore, variations in the HSA levels status for a short period of time are often not easy to assess (Gupta and Lis, 2010).
14. Rheumatoid arthritis
Hypoalbuminemia is commonly observed in rheumatoid arthritis patients, mainly caused by the high uptake of HSA at inflammation sites (Kratz, 2008; Ballantyne et al., 1971; Niwa et al., 1990; WILKINSON et al., 1965). The metabolism of synovial cells is significantly enhanced, and their high HSA uptake serves as a crucial means for the fulfilment of nitrogen and energy requirements. Patients suffering from rheumatoid arthritis exhibit a noticeable increase in the permeability of the joint blood barrier, which enables the excessive transportation of HSA to the affected region Methotrexate, a drug used for rheumatoid arthritis showed potent efficacy in this rodent model of collagen-induced arthritis. HSA, therefore, may act as a promising drug carrier to deliver doses to arthritis patients (Kratz, 2008). Preclinical models demonstrated the accumulation of SA in mice with collagen-induced arthritis (Wunder et al., 2003).
15. Ischemia
Reduced level of HSA is linked to a high risk of the coronary heart disease as well as mortality (Chan et al., 1995). The exposure to ischemic tissue modifies the N-terminus of HSA, which decreases its metal binding capability and leads to the formation of ischemia-modified albumin (Bar-Or et al., 2001). In patients suffering from acidosis, oxidative stress, hypoxia, membrane disruption, and exposure to free metals like copper and iron, ischemia-modified HSA has been found present (Bar-Or et al., 2001; Roy et al., 2006). The production of ischemia-modified Albumin is strongly dependent on the degree of free radical injury (Sbarouni et al., 2011).
Nevertheless, the ischemia-modified albumin does not constitute a specific marker for ischemia. So, the high levels of ischemia-modified albumin have also been observed in intracranial hemorrhage, subarachnoid, cerebrovascular-ischemic stroke, acute infections, systemic sclerosis, intrauterine disorders, malignancies, advanced liver cirrhosis and prostatic diseases. Additionally, the modified ischemic albumin does not rise in non-ischemic cardiac diseases or gastrointestinal and immune system disorders (Sbarouni et al., 2011). Furthermore, ischemia-modified albumin is a valuable biomarker currently in the finding of myocardial ischemia without necrosis (Reichlin et al., 2009). The mechanism of action of ischemia-modified HSA still remains unclear, including the kinetics of HSA during early hours following acute coronary syndrome, the cardio-specificity of albumin, and determining the optimal cut-off point for validation in a clinical setting.
16. HSA as a glycemic control biomarker
Diabetic patients show increased levels of protein glycation, further leading to chronic diabetic complications (Reichlin et al., 2009; Cohen, 1988). Glycated HbA1C is frequently used as the biomarker for glucose control in blood (Bunn et al., 1978; Koenig et al., 1976). Increased glycation of serum albumin occurs earlier than that of HbA1C when there is a deterioration in short-term glycemic control, revealing a worsening status at an earlier point (Koga and Kasayama, 2010).
Furthermore, due to the increase in plasma glucose, the elevation of HbA1C is more significant than that of HbA1C at the diagnosis of fulminant type 1 diabetes mellitus (Koga and Kasayama, 2010). As a consequence, the glycated HSA to HbA1C ratio is relatively increased in fulminant type 1 diabetes mellitus patients and therefore acts as a biomarker in its diagnosis. However, in type 2 diabetes, glycated serum albumin is not influenced by erythrocytes' life-span and is not affected by HSA concentration (Kosecki et al., 2005; Kouzuma et al., 2004). In the circulation, HSA becomes non-enzymatically glycated by reducing sugars, and the reference range in healthy humans is 6–10% of glycated HSA. This proportion increases typically about 20%–30% in hyperglycemic patients (Bourdon et al., 1999). Hence, glycated HSA serves as a valuable indicator for assessing short-term glycemic index (within a range of 2–4 weeks) in diabetic patients (Peters, 1995). Notably, glycated HSA remains unaffected by Hb variants and anaemia but can be influenced by dysfunctions related to HSA metabolism (Schleicher et al., 1993).
17. Glycated HSA and anaemia
Low level of HbA1C has been observed in patients with hemolytic anaemia because the erythrocytes’ lifespan is shortened in these patients (Panzer et al., 1982). In the meantime, patients with iron deficiency anaemia reversely show higher HbA1C values relative to plasma glucose levels (Coban et al., 2004; Kim et al., 2010). Patients undergoing treatment with iron supplements for iron-deficiency anaemia, show a temporary decrease in HbA1C levels as a result of a shorter erythrocyte lifespan (Koga et al., 2010; GRAM‐ et al., 1990). In comparison, glycated serum albumin is not influenced by these changes and therefore acts as a biomarker of glucose index in iron-deficiency anaemia (Koga et al., 2009).
18. Glycated HSA and chronic liver diseases
Liver is a crucial organ responsible for plasma glycemic control, dysfunction in glucose metabolism is common in patients with chronic liver ailments. About 30%–60% of patients with diabetes mellitus and about 70%–90% with chronic liver diseases are diagnosed with impaired glucose tolerance (Kingston et al., 1984). Maintaining blood glucose levels is essential because patients with chronic liver disease show weak glycemic control offering a poor prognosis (Bianchi et al., 1994). In chronic liver disease, the glycated HSA to HbA1C ratio is elevated and therefore acts as an indicator of hepatic health regardless of plasma glycemic index (Koga and Kasayama, 2010; Bando et al., 2009).
19. COVID-19
Albumin has been shown to have a down-regulating effect on the countenance of ACE2, the receptor that binds to SARS-CoV-2. The binding domains of SARS-CoV-2 spike glycoprotein receptors, RBDs (Receptor Binding Domain), have a high affinity for ACE2 surface receptors, which enhances the pathogenicity of the virus and allows for viral entry into host cells. Hypoalbuminemia is linked to COVID-19 and is a predictor of outcomes independent of age and morbidity. In a recent study of COVID-19 patients, hypoalbuminemia was found to be an independent risk factor for death, with a 6.394 times higher rate of mortality in those with hypoalbuminemia compared to those with normal albumin. Additionally, with each incremental rise in serum albumin levels, there was a corresponding 14% reduction in the likelihood of mortality. The inflammatory-induced rise in capillary permeability has been demonstrated to result in the leakage of HSA into the interstitial space, consequently leading to an expansion in the volume of HSA. Given the absence of a targeted treatment for general inflammation in COVID cases, utilizing albumin therapy with minimal negative effects can be a promising approach (Huang et al., 2020).
20. HSA as drug delivery carrier
Albumin serves as a multipurpose carrier protein for targeted drug delivery and is also instrumental in improving the pharmacokinetic profiles of protein and peptide medications. HSA is a useful carrier for agents, mainly responsible for the diagnoses and treatment of carcinomas, hyperglycemia, infectious diseases and rheumatoid arthritis. In addition to passive accumulation through enhanced permeability and retention, albumin can actively transport into tumour or inflamed tissues through receptor-mediated mechanisms, making it a potential candidate for drug targeting. The hydrophobic pockets, thiol residue, and surface-exposed termini of albumin make it suitable for acting as a carrier for a range of peptidyl and non-peptidyl drugs. HSA is gaining attention as a versatile drug carrier that can enhance the pharmacokinetics, targetability, solubility, and stability of various therapeutic agents (Rahimizadeh et al., 2020).
Primarily, the four most important ways in which HSA can be used as a drug carrier are shown in Fig. 7. HSA can form covalent or physical bonds with prodrugs, drugs, and polypeptides through ligand or protein-binding interactions. Many complex systems involve multiple targeting ligands and prodrugs attached to the surface of the protein, as well as nanobodies or bispecific antibodies that get attached to HSA, substituting the Fc fragment of IgG. Therapeutic systems based on HSA are typically categorized into four groups: HSA-drug nanoparticles, HSA-drug conjugates, HSA-binding prodrugs, and HSA-based recombinant fusion proteins (Tao et al., 2021).
Fig. 7.
Human Serum Albumin, a versatile drug carrier for developing peptide, drug or antibody based drugs as conjugates, complexes or nanoparticles (Kianfar, 2021).
21. Pharmacokinetic characteristics of therapeutic peptides and proteins
Peptides and proteins with significant therapeutic effects have turned out to be game changers for managing of autoimmune, malignant, and infectious diseases. The progress and effective utilization of therapeutic proteins or peptides often encounter multiple challenges, including inadequate stability and shelf life, potential immunogenicity and allergenicity, production cost, as well as limited sensitivity and bioavailability. Three well-designed technologies have been reported to combat these problems based on albumin's role as a drug carrier:
-
●
The Albumin Fusion Technology developed by Human Genome Sciences (Subramanian et al., 2007)
-
●
The Drug Affinity Complex (DAC™)
-
●
Preformed Conjugate-Drug Affinity Complex (PC-DAC™) technology (Giannoukakis, 2003)
-
●
Development of peptide fatty acid derivatives that bind physically to circulating albumin (Kurtzhals, 2007)
22. Albumin Fusion Technology
Fusion of human albumin gene with the therapeutically active protein drug-gene gives a reformed kind of the protein. Initially applied to cytokines like interleukins and interferons, Albumin Fusion Technology operates in a similar manner to protein pegylation (Haag and Kratz, 2006). It enhances the molecular weight of the protein, thereby prolonging its half-life and therefore requiring less frequent administration of the therapeutic protein. Moreover, the albumin molecule serves as a shield, protecting the protein from proteases and reducing its immunogenicity. In order to enhance the pharmacokinetics of peptides with low molecular weights, which typically have a shorter lifespan, ConjuChem, Incorporated, has developed a novel approach that involves binding albumin to therapeutic peptides-making constructs, comprising of a drug, a chemical group accounting for the permanent binding of the construct with target proteins and a connector molecule. Commonly used chemical groups include salicylate group, isocyanates, maleimide or N-hydroxysuccinimide esters. To ensure that the drug maintains a substantial portion of its original activity when bound to the protein, the drug attachment and connector length are optimized for each construct. In the case of most peptides, maleimide selectively and swiftly reacts with the Cys34 positions of HSA. This technology mainly targets this unique residue on the exogenous or endogenous surface of albumin. Additionally, these peptides efficiently adopt the useful albumin pharmacokinetics properties. The advanced peptide drug is CJC-1134-PC, which is an albumin conjugate of Exendin-4, a glucagon-like Peptide-1 (GLP-1) homolog used for the treatment of type 2 diabetes mellitus (Carter and Ho, 1994; Carter et al., 1989; Peters, 2012).
23. Albumin conjugates and prodrugs
A methotrexate-albumin conjugate (MTX-HSA) is formed by the coupling of Lys residues of HSA and the drug. However, the clinical evaluation of this conjugate is still under work. Some weaknesses reported for MTX-HSA conjugate include:
-
(1)
The underlying chemical mechanism of this conjugate is still not clear as variable quantities of MTX-HSA produce an average of ∼1.3 molecules for each albumin molecule
-
(2)
The precise cleavage rate and resulting products of the prodrug MTX-HSA remain under investigation. Kratz et al. directed their attention towards bioconjugation methods that would improve the coupling techniques of drug analogues in order to obtain pure conjugates with minimal modification of the protein, and a determined breaking point.
Commercially available albumin is a mix of non-mercaptalbumin and mercaptalbumin containing free sulfhydryl compounds. As a result, Cys-34 is obstructed by sulfhydryl groups including cysteine, homocysteine, and glutathione. Cleland's reagent has been developed for reducing HSA selectively and determining the presence of sulfhydryl groups in albumin. Next, derivatives of doxorubicin maleimide (4-maleimidophenylacetyl hydrazone) abbreviated as DOXO-HYD are paired with reduced albumin. At an equitoxic dose, the A-DOXO-HYD conjugate exhibited significantly superior efficacy compared to free doxorubicin in murine renal carcinoma (Kratz, 2008).
Compared to ex vivo synthesized albumin drug conjugates, utilizing albumin as a drug carrier offers several advantages:
-
(1)
Commercially available and possibly pathogenic albumin use should be avoided.
-
(2)
Drugs which bind to albumin have defined chemical properties based on basic organic chemistry.
-
(3)
These drugs are economical to manufacture and relatively simple.
-
(4)
A wide range of drugs can be used for this purpose.
The recent advancement in in situ albumin advancements involve the development of novel prodrugs that exhibit both active and passive targeting capabilities giving double benefit (Fig. 8). Firstly, a ligand is introduced in the construct of prodrugs such as integrins, folate receptors, asialoglycoprotein receptor, which are overexpressed in a variety of solid tumours (e.g. endothelium and liver tumours). By in situ binding of the endogenous or exogenous albumin to the ligand-based albumin-binding prodrugs, a conjugate drug is formed that potentially interacts with tumour-related antigens or receptors and enhance tumour targeting. Another approach involves (Fig. 9) two anticancer agents binding to albumin for cellular combination therapy or dual-acting prodrug tactic. These prodrugs aim to prevent chemoresistance in solid tumours.
Fig. 8.
Concepts of novel prodrug based on albumin as a drug carrier: receptor targeting and dual acting prodrugs (Kratz, 2008).
Fig. 9.
Top: Transmission electron micrograph and model of nab-paclitaxel, a 130 nm albumin paclitaxel nanoparticle (Kratz and Elsadek, 2012).
24. Albumin conjugates for hepatic targeting
Other applications of albumin in the drug delivery system is targeting the liver by using conjugate consisting of HSA and galactose residues. The conjugates of neoglycoprotein move into the hepatocytes selectively via binding to the asialoglycoprotein receptor resulting in internalization and degradation of the transporter in lysosomes, therefore circumventing extrahepatic side effects for instance, neurotoxicity of antiviral derivatives of the nucleoside in the chronic viral hepatitis management. The validation of this therapeutic approach has been established in a clinical study in which adenine arabinoside monophosphate conjugated with lactose aminated albumin employed an antiviral effect as compared to the free drug with no harmful effects containing critical neurotoxicity of free ara-AMP (Fiume et al., 1997). The receptor of asialoglycoprotein acts as a carrier for the delivery of the selective drug to malignant liver tumours (Di Stefano et al., 2003, 2004; Fiume et al., 2005). The utilization of DAC technology has primarily garnered attention due to its ability to improve the effectiveness and pharmacokinetic characteristics of therapeutic peptides and thrombin inhibitors possessing anti-nociceptive, anti-diabetic, hypotensive, or antiviral properties (Kratz, 2002).
25. Albumin fusion proteins
The application of Albumin Fusion Technology involves the creation of modified albumin conjugates through genetic interweaving, where genes from two molecules are fused and expressed in yeast strains. This technology, developed by Human Genome Sciences, has been predominantly utilized with cytokines, particularly interferon α-2b, interleukin-2, granulocyte colony-stimulating factor, and therapeutic peptides (Subramanian et al., 2007). Albuferon- α; a fusion of interferon α-2b and albumin which is undergoing clinical trials for hepatitis C, is the most promising. In early clinical trials, numerous albumin fusion proteins are being investigated, including fusion proteins involving low-molecular-weight peptides like glucagon-like peptide 1 and β-natriuretic peptide (Chemmanur and Wu, 2006). An albumin fusion protein Albuleukin is a target of interest for the oncologist as it possesses recombinant interleukin-2 with antitumor potential (Melder et al., 2005).
26. Albumin microspheres
Typically, the formation of albumin microspheres is achieved through chemical cross-linking or by adding organic solvents and stabilizing them at elevated temperatures. The size range of albumin microspheres is generally 1–100 μm is the crucial aspect for its biodistribution. Smaller microspheres (1–3 μm) are consumed by the reticuloendothelial cells and gathered in the hepatic cells, spleen and also in solid tumours. A larger microsphere will efficiently target the capillary region of the lungs. For the diagnostic purpose, 99mTc macro aggregated albumin shows promise in the detection of breast cancer (Rink et al., 2001) and other solid tumours (Bedrosian et al., 1999; Dhabuwala et al., 2005) such as lymphoscintigraphy (Nishiyama et al., 1998), leg oedema (Suga et al., 2001), protein-losing enteropathy (Chiu et al., 2001) and Rheumatoid arthritis (Adams et al., 2001).
27. Albumin nanoparticle technology (Nab-technology)
Coating therapeutic agents into nanoparticles formed by exogenous HSA is considered an extensive research area for carcinomas, as it takes full advantage of its long half-life and tumor homing characteristics. HSA-based nanoparticles can be prepared using various methods, including desolvation, emulsification, self-assembly, thermal gelation, nano-spray drying, and Nab™ technologies. Nab™ technology offers a simple method and uses non-toxic reagents to encapsulate hydrophobic agents into exogenous HSA (Tao et al., 2021; Karimi et al., 2016). A new technology of albumin-based nanoparticle has been developed by American Bioscience, which is used for encapsulation of lipophilic drugs into nanoparticles. A model and electron micrograph of an albumin nanoparticle with paclitaxel (nab-paclitaxel) is shown in Fig. 9 nab-paclitaxel has a diameter of 130 nm, extensively investigated in preclinical and clinical studies, which was accepted for the treatment of breast carcinomas (Desai et al., 2006).
Pre-clinical data suggests that Abraxane (nab-paclitaxel) is the most promising candidate for drug development. Moreover, in Abraxane, cremophor is not used due to its very low solubility in water, formulation of paclitaxel in polyethoxylated castor oil and ethanol. Paclitaxel in combination with cremophor can lead to neutropenia, prolonged irreversible peripheral neuropathy, mostly linked to axonal degeneration (Gelderblom et al., 2001; Goble and Bear, 2003). Abraxane has further been evaluated by first-line breast cancer treatment, adjuvant, and neoadjuvant as well as for other carcinomas including ovarian cancer, non-small cell cancer and pancreas cancer. Moreover, numerous future pipeline nab-technology-based products are under development, such as with rapamycin and docetaxel, and have entered into advanced clinical trials. Abraxane, a clinically used cancer therapeutic prepared with Nab™, was deserved to be viewed as a milestone for HSA-based nanoparticles (Elzoghby et al., 2012; Loureiro et al., 2016). Apart from their use in the cancer treatment, Albumin nanoparticles have also been employed in the diagnosis of cancer. Colloidal albumin aggregates with radiolabelling are being employed to spot sentinel lymph nodes present in tumour disorders (Zeeshan et al., 2021).
28. An overview of in-silico studies on drug binding affinities of HSA
Several studies employing Molecular Docking and Molecular Dynamic Simulation methods for exploring the interaction mechanism of serum albumin with different drugs such as Neelam et al. studied the binding of CT (N-trans-p-coumaroyltyramine) using molecular docking (Neelam et al., 2010). This study showed that the CT molecule was found deep inside the hydrophobic pocket of subdomain IIA (Fig. 5a) Therefore, the interaction of HSA with CT is principally hydrophobic involving Trp-214 and hydrogen bonding interactions with Arg, Val, Ala located at positions 222, 293, 291, and 298. Aurkie et al. reported a docking study., to predict the ‘superficial binding’ of NSC (Nickel (II)-Schiff base) at domain IIB of HSA. Various techniques including UV–Vis photometry, spectrofluorometry, and 3D modelling were employed to examine the interface between these compounds and serum albumin. The aim was to understand the distribution of protonated and deprotonated forms of the compounds at physiological pH. The observed changes in emission spectra of the coumarin derivatives when bound to serum albumin supported the hypothesis that the binding site is located at site I of HSA. Furthermore, molecular dynamics (MD) simulation calculations indicated the significant role of phenolates in facilitating electrostatic binding between molecular inhibitors and the protein. Various quantitative structure-activity relationship (QSAR) models are regularly employed to calculate binding affinity to albumin, which is valid for the entire domain of chemical-medicinal compounds. This can assist in speeding up the process to help in designing novel composites with suitable binding affinities, leading to pharmacokinetics evaluation. These models also used to predict different significant biopharmaceutical properties, for instance, oral availability, intestinal absorption, blood-brain barrier transportation metabolism, skin and corneal permeability and toxicity.
Few reports that describe HSA binding affinity are as follows:
-
(a)
Gonzalo Colmenarejo et al. utilized genetic function approximation (GFA) to construct projecting models for HSA interaction. These models were developed using Jurs and topological descriptors, and ClogP (Colmenarejo et al., 2001).
-
(b)
Hall et al. made use of multiple linear regression (MLR) to develop QSPR models to predict SA interactions by topological descriptors (Hall et al., 2003).
-
(c)
Hou et al. described analytical models for serum albumin by heuristic regression procedures utilizing seven descriptors. Models for Blood Brain Barrier permeability. Variable Selection and Model Building Using Prediction’ (VSMP) to give the optimal resolution determined by evaluating all potential groupings of the input variables. Thus it has some limitations of being in silico exhaustive, mainly when the number of variables is higher than 3 (Gunturi et al., 2006).
A few models were developed for 1, 4-benzodiazepines, indolocarbazole, and aryl carboxylic acids derivatives. These models contain hydrophobicity parameters and the electronic descriptors. Therefore, the binding of these compounds is taken place by hydrophobic and steric interactions, all authors recommended that hydrophobic interactions are favorable for the binding of the drugs, and also, hydrogen bonding and electrostatic interactions help in retention for the elution of the second enantiomer. Another study reported a structure-based model of SA-binding. They studied that combination of the docking score and predicted log P most precisely differentiate between binders and non-binders. The developed model was effectively used to predict SA binding in an extensive data set of therapeutic drugs which had experimental binding data.
29. Conclusion
The flexibility of albumin in holding various types of drugs coupled with a variety of modifications makes this protein a versatile drug carrier with incalculable potential in therapeutics. HSA shows unusual ligand binding properties making it a multimeric proteins. Clinically, HSA is extensively used to cure diseases and also act as a valuable biomarker of ischemia, cancer, and diabetes mellitus etc. HSA's exceptional ability to bind a wide range of small ligands compared to other proteins can be attributed to its unique structural and biochemical features. This versatility arises from the distinct properties of HSA, such as its large binding pocket, which can accommodate a variety of ligands of different sizes and shapes. Additionally, HSA's conformational flexibility allows it to adapt to different ligands, making it a promiscuous binder. While other proteins may have more specific binding sites tailored to particular ligands, HSA's broad binding capabilities make it a crucial player in various physiological processes, including drug transport and metabolism. As a whole, the complex mechanism modulating ligand binding to HSA represents one of the most important structure-function correlations ever reported.
Additionally, the binding of drugs with HSA form a “nano-drug”, which increases the drug's bioavailability. Consequently, HSA-based delivery systems are being used to enhance the targeting of anti-carcinogenic drugs and reduce their negative effects. The structural investigation of HSA complexes can provide guidance in the rational design of HSA protein by modifying its drug binding ability using numerous strategies including, the modification of compounds' structure, or the regulation of drugs to each other, or by mutations of specific amino acids depicting the serum albumin transporter better for targeted drug delivery. With the increasing demand for safe and effective treatments for various diseases, the future of HSA looks promising. Researchers are exploring new ways to produce and modify HSA to enhance its therapeutic potential and extend its shelf life, making it a vital component in the development of new medical treatments.
CRediT authorship contribution statement
Sajda Ashraf: Writing – original draft. Hina Qaiser: gathered the research, Data curation. Sumayya Tariq: reviewed, revised, and updated the, Writing – original draft. Asaad Khalid: Writing – review & editing, the, Writing – original draft. Hafiz A. Makeen: Writing – review & editing, the, Writing – original draft. Hassan A. Alhazmi: Writing – review & editing, the, Writing – original draft. Zaheer Ul-Haq: Conceived the idea, Writing – review & editing, provided, Supervision, and facilities.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors extend their appreciation to the Deputyship for Research and innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (ISP23-81).
Handling Editor: Dr A Wlodawer
Data availability
Data will be made available on request.
References
- Adams B., Al Attia H., Khadim R., Al Haider Z. 99Tcm nanocolloid scintigraphy: a reliable way to detect active joint disease in patients with peripheral joint pain. Nucl. Med. Commun. 2001;22(3):315–318. doi: 10.1097/00006231-200103000-00008. [DOI] [PubMed] [Google Scholar]
- Al-Harthi S., Chandra K., Jaremko Ł. Lipoic Acid restores binding of Zinc ions to human serum albumin. Front. Chem. 2022;10 doi: 10.3389/fchem.2022.942585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M.R., Alexander B., Mustion A.L., Spector R., Wright C.B. Therapeutic use of albumin: 2. JAMA. 1982;247(6):831–833. [PubMed] [Google Scholar]
- Ascenzi P., Fasano M. Serum heme‐albumin: an allosteric protein. IUBMB Life. 2009;61(12):1118–1122. doi: 10.1002/iub.263. [DOI] [PubMed] [Google Scholar]
- Ascenzi P., Fasano M. Allostery in a monomeric protein: the case of human serum albumin. Biophys. Chem. 2010;148(1):16–22. doi: 10.1016/j.bpc.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Ascenzi P., Salvati L., Bolognesi M., Colasanti M., Polticelli F., Venturini G. Inhibition of cysteine protease activity by NO-donors. Curr. Protein Pept. Sci. 2001;2(2):137–153. doi: 10.2174/1389203013381170. [DOI] [PubMed] [Google Scholar]
- Ascenzi P., Bocedi A., Bolli A., Fasano M., Notari S., Polticelli F. Allosteric modulation of monomeric proteins. Biochem. Mol. Biol. Educ. 2005;33(3):169–176. doi: 10.1002/bmb.2005.494033032470. [DOI] [PubMed] [Google Scholar]
- Ascenzi P., Bocedi A., Notari S., Fanali G., Fesce R., Fasano M. Allosteric modulation of drug binding to human serum albumin. Mini Rev. Med. Chem. 2006;6(4):483–489. doi: 10.2174/138955706776361448. [DOI] [PubMed] [Google Scholar]
- Ascenzi P., di Masi A., Coletta M., Ciaccio C., Fanali G., Nicoletti F.P., et al. Ibuprofen impairs allosterically peroxynitrite isomerization by ferric human serum heme-albumin. J. Biol. Chem. 2009;284(45):31006–31017. doi: 10.1074/jbc.M109.010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi P., di Masi A., De Sanctis G., Coletta M., Fasano M. Ibuprofen modulates allosterically NO dissociation from ferrous nitrosylated human serum heme-albumin by binding to three sites. Biochem. Biophys. Res. Commun. 2009;387(1):83–86. doi: 10.1016/j.bbrc.2009.06.117. [DOI] [PubMed] [Google Scholar]
- Bailey M.J., Dickinson R.G. Acyl glucuronide reactivity in perspective: biological consequences. Chem. Biol. Interact. 2003;145(2):117–137. doi: 10.1016/s0009-2797(03)00020-6. [DOI] [PubMed] [Google Scholar]
- Bal W., Sokołowska M., Kurowska E., Faller P. Binding of transition metal ions to albumin: sites, affinities and rates. Biochim. Biophys. Acta Gen. Subj. 2013;1830(12):5444–5455. doi: 10.1016/j.bbagen.2013.06.018. [DOI] [PubMed] [Google Scholar]
- Ballantyne F.C., Fleck A., Dick W.C. Albumin metabolism in rheumatoid arthritis. Ann. Rheum. Dis. 1971;30(3):265. doi: 10.1136/ard.30.3.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballmer P.E., Ochsenbein A.F., Schütz-Hofmann S. Transcapillary escape rate of albumin positively correlates with plasma albumin concentration in acute but not in chronic inflammatory disease. Metabolism. 1994;43(6):697–705. doi: 10.1016/0026-0495(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Bando Y., Kanehara H., Toya D., Tanaka N., Kasayama S., Koga M. Association of serum glycated albumin to haemoglobin A1C ratio with hepatic function tests in patients with chronic liver disease. Ann. Clin. Biochem. 2009;46(5):368–372. doi: 10.1258/acb.2009.008231. [DOI] [PubMed] [Google Scholar]
- Bar-Or D., Rael L.T., Lau E.P., Rao N.K., Thomas G.W., Winkler J.V., et al. An analog of the human albumin N-terminus (Asp-Ala-His-Lys) prevents formation of copper-induced reactive oxygen species. Biochem. Biophys. Res. Commun. 2001;284(3):856–862. doi: 10.1006/bbrc.2001.5042. [DOI] [PubMed] [Google Scholar]
- Barber M.D., Ross J.A., Fearon K.C. Changes in nutritional, functional, and inflammatory markers in advanced pancreatic cancer. Nutr. Cancer. 1999;35(2):106–110. doi: 10.1207/S15327914NC352_2. [DOI] [PubMed] [Google Scholar]
- Barzegar A., Moosavi-Movahedi A.A., Sattarahmady N., Hosseinpour-Faizi M.A., Aminbakhsh M., Ahmad F., et al. Spectroscopic studies of the effects of glycation of human serum albumin on L-Trp binding. Protein Pept. Lett. 2007;14(1):13–18. doi: 10.2174/092986607779117191. [DOI] [PubMed] [Google Scholar]
- Bedrosian I., Scheff A.M., Mick R., Callans L.S., Bucky L.P., Spitz F.R., et al. ^ 9^ 9^ mTc-human serum albumin: an effective radiotracer for identifying sentinel lymph nodes in melanoma. J. Nucl. Med. 1999;40:1143–1148. [PubMed] [Google Scholar]
- Belinskaia D.A., Voronina P.A., Shmurak V.I., Jenkins R.O., Goncharov N.V. Serum albumin in health and disease: esterase, antioxidant, transporting and signaling properties. Int. J. Mol. Sci. 2021;22(19) doi: 10.3390/ijms221910318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A.A., Grüne T., Curry S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J. Mol. Biol. 2000;303(5):721–732. doi: 10.1006/jmbi.2000.4158. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A.A., Curry S., Franks N.P. Binding of the general anesthetics propofol and halothane to human serum albumin high resolution crystal structures. J. Biol. Chem. 2000;275(49):38731–38738. doi: 10.1074/jbc.M005460200. [DOI] [PubMed] [Google Scholar]
- Bianchi G., Marchesini G., Zoli M., Bugianesi E., Fabbri A., Pisi E. Prognostic significance of diabetes in patients with cirrhosis. Hepatology. 1994;20(1):119–125. doi: 10.1016/0270-9139(94)90143-0. [DOI] [PubMed] [Google Scholar]
- Björck L., Kronvall G. Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. J. Immunol. 1984;133(2):969–974. [PubMed] [Google Scholar]
- Boonpipattanapong T., Chewatanakornkul S. Preoperative carcinoembryonic antigen and albumin in predicting survival in patients with colon and rectal carcinomas. J. Clin. Gastroenterol. 2006;40(7):592–595. doi: 10.1097/00004836-200608000-00006. [DOI] [PubMed] [Google Scholar]
- Bos O.J., Fischer M.J., Wilting J., Janssen L.H. Drug-binding and other physicochemical properties of a large tryptic and a large peptic fragment of human serum albumin. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1988;953:37–47. doi: 10.1016/0167-4838(88)90007-6. [DOI] [PubMed] [Google Scholar]
- Bos O.J., Remijn J.P., Fischer M.J., Wilting J., Janssen L.H. Location and characterization of the warfarin binding site of human serum albumin: a comparative study of two large fragments. Biochem. Pharmacol. 1988;37(20):3905–3909. doi: 10.1016/0006-2952(88)90072-x. [DOI] [PubMed] [Google Scholar]
- Bourdon E., Loreau N., Blache D. Glucose and free radicals impair the antioxidant properties of serum albumin. Faseb. J. 1999;13(2):233–244. doi: 10.1096/fasebj.13.2.233. [DOI] [PubMed] [Google Scholar]
- Bunn H.F., Gabbay K.H., Gallop P.M. The glycosylation of hemoglobin: relevance to diabetes mellitus. Science. 1978;200(4337):21–27. doi: 10.1126/science.635569. [DOI] [PubMed] [Google Scholar]
- Carter D.C., Ho J.X. Structure of serum albumin. Adv. Protein Chem. 1994;45(45):153–203. doi: 10.1016/s0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- Carter D.C., He X.-M., Munson S.H., Twigg P.D., Gernert K.M., Broom M.B., et al. Three-dimensional structure of human serum albumin. Science. 1989;244(4909):1195–1198. doi: 10.1126/science.2727704. [DOI] [PubMed] [Google Scholar]
- Cengiz O., Kocer B., Sürmeli S., Santicky M.-J., Soran A. Are pretreatment serum albumin and cholesterol levels prognostic tools in patients with colorectal carcinoma? Med. Sci. Monitor Basic Res. 2006;12(6):CR240–C247. [PubMed] [Google Scholar]
- Cha M.-K., Kim I.-H. Glutathione-linked thiol peroxidase activity of human serum albumin: a possible antioxidant role of serum albumin in blood plasma. Biochem. Biophys. Res. Commun. 1996;222(2):619–625. doi: 10.1006/bbrc.1996.0793. [DOI] [PubMed] [Google Scholar]
- Chan B., Dodsworth N., Woodrow J., Tucker A., Harris R. Site‐specific N‐terminal auto‐degradation of human serum albumin. Eur. J. Biochem. 1995;227(1‐2):524–528. doi: 10.1111/j.1432-1033.1995.tb20419.x. [DOI] [PubMed] [Google Scholar]
- Chemmanur A.T., Wu G.Y. Drug evaluation: albuferon-alpha--an antiviral interferon-alpha/albumin fusion protein. Curr. Opin. Invest. Drugs. 2006;7(8):750–758. [PubMed] [Google Scholar]
- Chen C.B., Hammo B., Barry J., Radhakrishnan K. Overview of albumin physiology and its role in pediatric diseases. Curr. Gastroenterol. Rep. 2021;23(8):11. doi: 10.1007/s11894-021-00813-6. [DOI] [PubMed] [Google Scholar]
- Chiu N.-T., Lee B.-F., Hwang S.-J., Chang J.-M., Liu G.-C., Yu H.-S. Protein-losing enteropathy: diagnosis with 99mTc-labeled human serum albumin scintigraphy 1. Radiology. 2001;219(1):86–90. doi: 10.1148/radiology.219.1.r01ap2986. [DOI] [PubMed] [Google Scholar]
- Chuang V.T.G., Otagiri M. Flunitrazepam, a 7-nitro-1, 4-benzodiazepine that is unable to bind to the indole-benzodiazepine site of human serum albumin. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2001;1546(2):337–345. doi: 10.1016/s0167-4838(01)00151-0. [DOI] [PubMed] [Google Scholar]
- Coban E., Ozdogan M., Timuragaoglu A. Effect of iron deficiency anemia on the levels of hemoglobin A1c in nondiabetic patients. Acta Haematol. 2004;112(3):126–128. doi: 10.1159/000079722. [DOI] [PubMed] [Google Scholar]
- Cohen M.P. Nonenzymatic glycation: a central mechanism in diabetic microvasculopathy? J. Diabet. Complicat. 1988;2(4):214–217. doi: 10.1016/s0891-6632(88)80012-6. [DOI] [PubMed] [Google Scholar]
- Colmenarejo G., Alvarez-Pedraglio A., Lavandera J.-L. Cheminformatic models to predict binding affinities to human serum albumin. J. Med. Chem. 2001;44(25):4370–4378. doi: 10.1021/jm010960b. [DOI] [PubMed] [Google Scholar]
- Croxson M., Palmer B., Holdaway I., Frengley P., Evans M. Detection of familial dysalbuminaemic hyperthyroxinaemia. BMJ. 1985;290(6475):1099–1102. doi: 10.1136/bmj.290.6475.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry S. Beyond expansion: structural studies on the transport roles of human serum albumin. Vox Sang. 2002;83(s1):315–319. doi: 10.1111/j.1423-0410.2002.tb05326.x. [DOI] [PubMed] [Google Scholar]
- Curry S. Lessons from the crystallographic analysis of small molecule binding to human serum albumin. Drug Metabol. Pharmacokinet. 2009;24(4):342–357. doi: 10.2133/dmpk.24.342. [DOI] [PubMed] [Google Scholar]
- Curry S., Mandelkow H., Brick P., Franks N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Mol. Biol. 1998;5(9):827–835. doi: 10.1038/1869. [DOI] [PubMed] [Google Scholar]
- Czub M.P., Venkataramany B.S., Majorek K.A., Handing K.B., Porebski P.J., Beeram S.R., et al. Testosterone meets albumin–the molecular mechanism of sex hormone transport by serum albumins. Chem. Sci. 2019;10(6):1607–1618. doi: 10.1039/c8sc04397c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chateau M., Björck L. Protein PAB, a mosaic albumin-binding bacterial protein representing the first contemporary example of module shuffling. J. Biol. Chem. 1994;269(16):12147–12151. [PubMed] [Google Scholar]
- Deng B., Wang Y., Zhu P., Xu X., Ning X. Study of the binding equilibrium between Zn (II) and HSA by capillary electrophoresis–inductively coupled plasma optical emission spectrometry. Anal. Chim. Acta. 2010;683(1):58–62. doi: 10.1016/j.aca.2010.10.018. [DOI] [PubMed] [Google Scholar]
- Desai N., Trieu V., Yao Z., Louie L., Ci S., Yang A., et al. Increased antitumor activity, intratumor paclitaxel concentrations, and endothelial cell transport of cremophor-free, albumin-bound paclitaxel, ABI-007, compared with cremophor-based paclitaxel. Clin. Cancer Res. 2006;12(4):1317–1324. doi: 10.1158/1078-0432.CCR-05-1634. [DOI] [PubMed] [Google Scholar]
- Dhabuwala A., Lamerton P., Stubbs R.S. Relationship of 99mtechnetium labelled macroaggregated albumin (99mTc-MAA) uptake by colorectal liver metastases to response following Selective Internal Radiation Therapy (SIRT) BMC Med. Phys. 2005;5(1):7. doi: 10.1186/1471-2385-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano G., Kratz F., Lanza M., Fiume L. Doxorubicin coupled to lactosaminated human albumin remains confined within mouse liver cells after the intracellular release from the carrier. Dig. Liver Dis. 2003;35(6):428–433. doi: 10.1016/s1590-8658(03)00212-3. [DOI] [PubMed] [Google Scholar]
- Di Stefano G., Derenzini M., Kratz F., Lanza M., Fiume L. Liver‐targeted doxorubicin: effects on rat regenerating hepatocytes. Liver Int. 2004;24(3):246–252. doi: 10.1111/j.1478-3231.2004.0916.x. [DOI] [PubMed] [Google Scholar]
- Dixon M.R., Haukoos J.S., Udani S.M., Naghi J.J., Arnell T.D., Kumar R.R., et al. Carcinoembryonic antigen and albumin predict survival in patients with advanced colon and rectal cancer. Arch. Surg. 2003;138(9):962–966. doi: 10.1001/archsurg.138.9.962. [DOI] [PubMed] [Google Scholar]
- Dömötör O., Enyedy É.A. Binding mechanisms of half-sandwich Rh (III) and Ru (II) arene complexes on human serum albumin: a comparative study. JBIC, J. Biol. Inorg. Chem. 2019;24:703–719. doi: 10.1007/s00775-019-01683-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff M.R., Jr., Kumar C.V. The metallomics approach: use of Fe (II) and Cu (II) footprinting to examine metal binding sites on serum albumins. Metallomics. 2009;1(6):518–523. doi: 10.1039/b910253a. [DOI] [PubMed] [Google Scholar]
- Elzoghby A.O., Samy W.M., Elgindy N.A. Albumin-based nanoparticles as potential controlled release drug delivery systems. J. Contr. Release. 2012;157(2):168–182. doi: 10.1016/j.jconrel.2011.07.031. [DOI] [PubMed] [Google Scholar]
- Erstad B.L., Gales B.J., Rappaport W.D. The use of albumin in clinical practice. Arch. Intern. Med. 1991;151(5):901–911. [PubMed] [Google Scholar]
- Evans T. Review article: albumin as a drug—biological effects of albumin unrelated to oncotic pressure. Aliment Pharmacol. Therapeut. 2002;16(s5):6–11. doi: 10.1046/j.1365-2036.16.s5.2.x. [DOI] [PubMed] [Google Scholar]
- Fanali G., Fesce R., Agrati C., Ascenzi P., Fasano M. Allosteric modulation of myristate and Mn (III) heme binding to human serum albumin. FEBS J. 2005;272(18):4672–4683. doi: 10.1111/j.1742-4658.2005.04883.x. [DOI] [PubMed] [Google Scholar]
- Fanali G., Bocedi A., Ascenzi P., Fasano M. Modulation of heme and myristate binding to human serum albumin by anti‐HIV drugs. FEBS J. 2007;274(17):4491–4502. doi: 10.1111/j.1742-4658.2007.05978.x. [DOI] [PubMed] [Google Scholar]
- Fanali G., Di Masi A., Trezza V., Marino M., Fasano M., Ascenzi P. Human serum albumin: from bench to bedside. Mol. Aspect. Med. 2012;33(3):209–290. doi: 10.1016/j.mam.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Fasano M., Baroni S., Vannini A., Ascenzi P., Aime S. Relaxometric characterization of human hemalbumin. JBIC, J. Biol. Inorg. Chem. 2001;6(5–6):650–658. doi: 10.1007/s007750100242. [DOI] [PubMed] [Google Scholar]
- Fasano M., Bocedi A., Mattu M., Coletta M., Ascenzi P. Nitrosylation of rabbit ferrous heme-hemopexin. JBIC, J. Biol. Inorg. Chem. 2004;9(7):800–806. doi: 10.1007/s00775-004-0598-0. [DOI] [PubMed] [Google Scholar]
- Fasano M., Curry S., Terreno E., Galliano M., Fanali G., Narciso P., et al. The extraordinary ligand binding properties of human serum albumin. IUBMB Life. 2005;57(12):787–796. doi: 10.1080/15216540500404093. [DOI] [PubMed] [Google Scholar]
- Fehske K.J., Jähnchen E., Müller W.E., Stillbauer A. Azapropazone binding to human serum albumin. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1980;313(2):159–163. doi: 10.1007/BF00498574. [DOI] [PubMed] [Google Scholar]
- Fehske K.J., Müller W.E., Wollert U. The location of drug binding sites in human serum albumin. Biochem. Pharmacol. 1981;30(7):687–692. doi: 10.1016/0006-2952(81)90151-9. [DOI] [PubMed] [Google Scholar]
- Fehske K., Schläfer U., Wollert U., Müller W. Characterization of an important drug binding area on human serum albumin including the high-affinity binding sites of warfarin and azapropazone. Mol. Pharmacol. 1982;21(2):387–393. [PubMed] [Google Scholar]
- Fitos I., Visy J., Simonyi M., Hermansson J. Stereoselective allosteric binding interaction on human serum albumin between ibuprofen and lorazepam acetate. Chirality. 1999;11(2):115–120. doi: 10.1002/(SICI)1520-636X(1999)11:2<115::AID-CHIR6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Fiume L., Stefano G.D., Busi C., Mattioli A., Bonino F., Torrani‐Cerenzia M., et al. Liver targeting of antiviral nucleoside analogues through the asialoglycoprotein receptor. J. Viral Hepat. 1997;4(6):363–370. doi: 10.1046/j.1365-2893.1997.00067.x. [DOI] [PubMed] [Google Scholar]
- Fiume L., Bolondi L., Busi C., Chieco P., Kratz F., Lanza M., et al. Doxorubicin coupled to lactosaminated albumin inhibits the growth of hepatocellular carcinomas induced in rats by diethylnitrosamine. J. Hepatol. 2005;43(4):645–652. doi: 10.1016/j.jhep.2005.02.045. [DOI] [PubMed] [Google Scholar]
- Gautam A., Gailer J. More effective mobilization of Hg2+ from human serum albumin compared to Cd2+ by L-cysteine at near-physiological conditions. Toxics. 2023;11(7):599. doi: 10.3390/toxics11070599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelderblom H., Verweij J., Nooter K., Sparreboom A. Cremophor EL: the drawbacks and advantages of vehicle selection for drug formulation. Eur. J. Cancer. 2001;37(13):1590–1598. doi: 10.1016/s0959-8049(01)00171-x. [DOI] [PubMed] [Google Scholar]
- Ghuman J., Zunszain P.A., Petitpas I., Bhattacharya A.A., Otagiri M., Curry S. Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 2005;353(1):38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- Giannoukakis N. vol. 4. 2003. pp. 1245–1249. (CJC-1131. ConjuChem. Current Opinion in Investigational Drugs (London, England: 2000)). 10. [PubMed] [Google Scholar]
- Goble S., Bear H.D. Emerging role of taxanes in adjuvant and neoadjuvant therapy for breast cancer: the potential and the questions. Surg. Clin. 2003;83(4):943–971. doi: 10.1016/S0039-6109(03)00071-9. [DOI] [PubMed] [Google Scholar]
- Goldin A., Beckman J.A., Schmidt A.M., Creager M.A. Advanced glycation end products sparking the development of diabetic vascular injury. Circulation. 2006;114(6):597–605. doi: 10.1161/CIRCULATIONAHA.106.621854. [DOI] [PubMed] [Google Scholar]
- GRAM Hansen P., Eriksen J., Mourits‐Andersen T., Olesen L. Glycosylated haemoglobin (HbA1c) in iron‐and vitamin B12 deficiency. J. Intern. Med. 1990;227(2):133–136. doi: 10.1111/j.1365-2796.1990.tb00131.x. [DOI] [PubMed] [Google Scholar]
- Gundry R.L., Fu Q., Jelinek C.A., Van Eyk J.E., Cotter R.J. Investigation of an albumin-enriched fraction of human serum and its albuminome. Proteonomics Clin. Appl. 2007;1(1):73. doi: 10.1002/prca.200600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunturi S.B., Narayanan R., Khandelwal A. In silico ADME modelling 2: computational models to predict human serum albumin binding affinity using ant colony systems. Bioorg. Med. Chem. 2006;14(12):4118–4129. doi: 10.1016/j.bmc.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Gupta D., Lis C.G. Pretreatment serum albumin as a predictor of cancer survival: a systematic review of the epidemiological literature. Nutr. J. 2010;9(69):10–1186. doi: 10.1186/1475-2891-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag R., Kratz F. Polymer therapeutics: concepts and applications. Angew. Chem. Int. Ed. 2006;45(8):1198–1215. doi: 10.1002/anie.200502113. [DOI] [PubMed] [Google Scholar]
- Hall L.M., Hall L.H., Kier L.B. Modeling drug albumin binding affinity with e-state topological structure representation. J. Chem. Inf. Comput. Sci. 2003;43(6):2120–2128. doi: 10.1021/ci030019w. [DOI] [PubMed] [Google Scholar]
- Hall M.L., Jorgensen W.L., Whitehead L. Automated ligand-and structure-based protocol for in silico prediction of human serum albumin binding. J. Chem. Inf. Model. 2013;53(4):907–922. doi: 10.1021/ci3006098. [DOI] [PubMed] [Google Scholar]
- Harper M.E., Dugaiczyk A. Linkage of the evolutionarily-related serum albumin and alpha-fetoprotein genes within q11-22 of human chromosome 4. Am. J. Hum. Genet. 1983;35(4):565. [PMC free article] [PubMed] [Google Scholar]
- Hennemann G., Krenning E., Otten M., Docter R., Bos G., Visser T. Raised total thyroxine and free thyroxine index but normal free thyroxine: a serum abnormality due to inherited increased affinity of iodothyronines for serum binding protein. Lancet. 1979;313(8117):639–642. doi: 10.1016/s0140-6736(79)91080-8. [DOI] [PubMed] [Google Scholar]
- Hervé F., Urien S., Albengres E., Duché J.-C., Tillement J.-P. Drug binding in plasma. Clin. Pharmacokinet. 1994;26(1):44–58. doi: 10.2165/00003088-199426010-00004. [DOI] [PubMed] [Google Scholar]
- Heys S., Walker L., Deehan D., Eremin E. Serum albumin: a prognostic indicator in patients with colorectal cancer. Age. 1998;38(3) 0.00005. [PubMed] [Google Scholar]
- Himmelfarb J., McMonagle E. Albumin is the major plasma protein target of oxidant stress in uremia. Kidney Int. 2001;60(1):358–363. doi: 10.1046/j.1523-1755.2001.00807.x. [DOI] [PubMed] [Google Scholar]
- Huang J., Cheng A., Kumar R., Fang Y., Chen G., Zhu Y., et al. Hypoalbuminemia predicts the outcome of COVID‐19 independent of age and co‐morbidity. J. Med. Virol. 2020;92(10):2152–2158. doi: 10.1002/jmv.26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Saura Y., Tsuda Y., Yamada H. Stereoselectivity and enantiomer‐enantiomer interactions in the binding of ibuprofen to human serum albumin. Chirality. 1997;9(7):643–649. doi: 10.1002/(SICI)1520-636X(1997)9:7<643::AID-CHIR1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Karimi M., Bahrami S., Ravari S.B., Zangabad P.S., Mirshekari H., Bozorgomid M., et al. Albumin nanostructures as advanced drug delivery systems. Expet Opin. Drug Deliv. 2016;13(11):1609–1623. doi: 10.1080/17425247.2016.1193149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami A., Kubota K., Yamada N., Tagami U., Takehana K., Sonaka I., et al. Identification and characterization of oxidized human serum albumin. FEBS J. 2006;273(14):3346–3357. doi: 10.1111/j.1742-4658.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- Kendall F.E. Studies on human serum proteins II. Crystallization of human serum albumin. J. Biol. Chem. 1941;138(1):97–109. [Google Scholar]
- Kianfar E. Protein nanoparticles in drug delivery: animal protein, plant proteins and protein cages, albumin nanoparticles. J. Nanobiotechnol. 2021;19(1):159. doi: 10.1186/s12951-021-00896-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Bullard K.M., Herman W.H., Beckles G.L. Association between iron deficiency and A1C levels among adults without diabetes in the national health and nutrition examination survey, 1999–2006. Diabetes Care. 2010;33(4):780–785. doi: 10.2337/dc09-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston M., Ali M.A., Atiyeh M., Donnelly R. Diabetes mellitus in chronic active hepatitis and cirrhosis. Gastroenterology. 1984;87(3):688–694. [PubMed] [Google Scholar]
- Koenig R.J., Peterson C.M., Jones R.L., Saudek C., Lehrman M., Cerami A. Correlation of glucose regulation and hemoglobin AIc in diabetes mellitus. N. Engl. J. Med. 1976;295(8):417–420. doi: 10.1056/NEJM197608192950804. [DOI] [PubMed] [Google Scholar]
- Koga M., Kasayama S. Clinical impact of glycated albumin as another glycemic control marker. Endocr. J. 2010;57(9):751–762. doi: 10.1507/endocrj.k10e-138. [DOI] [PubMed] [Google Scholar]
- Koga M., Murai J., Saito H., Matsumoto S., Kasayama S. Usefullness of glycated albumin as a glycemic control marker after iron treatment for diabetic patients with iron deficiency anemia. J. Jpn. Diabet. Soc. 2009;52:341–345. [Google Scholar]
- Koga M., Saito H., Mukai M., Matsumoto S., Kasayama S. Influence of iron metabolism indices on glycated haemoglobin but not glycated albumin levels in premenopausal women. Acta Diabetol. 2010;47(1):65–69. doi: 10.1007/s00592-009-0123-6. [DOI] [PubMed] [Google Scholar]
- Kosecki S.M., Rodgers P.T., Adams M.B. Glycemic monitoring in diabetics with sickle cell plus β-thalassemia hemoglobinopathy. Ann. Pharmacother. 2005;39(9):1557–1560. doi: 10.1345/aph.1G010. [DOI] [PubMed] [Google Scholar]
- Kouzuma T., Uemastu Y., Usami T., Imamura S. Study of glycated amino acid elimination reaction for an improved enzymatic glycated albumin measurement method. Clin. Chim. Acta. 2004;346(2):135–143. doi: 10.1016/j.cccn.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Kragh-Hansen U. Molecular aspects of ligand binding to serum albumin. Pharmacol. Rev. 1981;33(1):17–53. [PubMed] [Google Scholar]
- Kragh-Hansen U. Relations between high-affinity binding sites for L-tryptophan, diazepam, salicylate and Phenol Red on human serum albumin. Biochem. J. 1983;209:135–142. doi: 10.1042/bj2090135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragh-Hansen U. Evidence for a large and flexible region of human serum albumin possessing high affinity binding sites for salicylate, warfarin, and other ligands. Mol. Pharmacol. 1988;34(2):160–171. [PubMed] [Google Scholar]
- Kragh-Hansen U., Chuang V.T.G., Otagiri M. Practical aspects of the ligand-binding and enzymatic properties of human serum albumin. Biol. Pharm. Bull. 2002;25(6):695–704. doi: 10.1248/bpb.25.695. [DOI] [PubMed] [Google Scholar]
- Kratz F. Drug conjugates with albumin and transferrin. Expert Opin. Ther. Pat. 2002;12(3):433–439. [Google Scholar]
- Kratz F. Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. J. Contr. Release. 2008;132(3):171–183. doi: 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Kratz F., Beyer U. Serum proteins as drug carriers of anticancer agents: a review. Drug Deliv. 1998;5(4):281–299. doi: 10.3109/10717549809065759. [DOI] [PubMed] [Google Scholar]
- Kratz F., Elsadek B. Clinical impact of serum proteins on drug delivery. J. Contr. Release. 2012;161(2):429–445. doi: 10.1016/j.jconrel.2011.11.028. [DOI] [PubMed] [Google Scholar]
- Krebs N.F., Hambidge K., Jacobs M., Rasbach J.O. The effects of a dietary zinc supplement during lactation on longitudinal changes in maternal zinc status and milk zinc concentrations. Am. J. Clin. Nutr. 1985;41(3):560–570. doi: 10.1093/ajcn/41.3.560. [DOI] [PubMed] [Google Scholar]
- Kurtzhals P. Pharmacology of insulin detemir. Endocrinol Metab. Clin. N. Am. 2007;36:14–20. doi: 10.1016/s0889-8529(07)80004-1. [DOI] [PubMed] [Google Scholar]
- Lalloz M., Byfield P., Himsworth R. Hyperthyroxinaemia: abnormal binding of T4 by an inherited albumin variant. Clin. Endocrinol. 1983;18(1):11–24. doi: 10.1111/j.1365-2265.1983.tb03181.x. [DOI] [PubMed] [Google Scholar]
- Lee P., Wu X. Modifications of human serum albumin and their binding effect. Curr. Pharmaceut. Des. 2015;21(14):1862–1865. doi: 10.2174/1381612821666150302115025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.P., Golden M.P., VAN Herle A.J., Lippe B.M., Kaplan S.A. Inherited abnormal thyroid hormone-binding protein causing selective increase of total serum thyroxine. J. Clin. Endocrinol. Metabol. 1979;49(2):292–299. doi: 10.1210/jcem-49-2-292. [DOI] [PubMed] [Google Scholar]
- Lejon S., Frick I.-M., Björck L., Wikström M., Svensson S. Crystal structure and biological implications of a bacterial albumin binding module in complex with human serum albumin. J. Biol. Chem. 2004;279(41):42924–42928. doi: 10.1074/jbc.M406957200. [DOI] [PubMed] [Google Scholar]
- Lien Y.-C., Hsieh C.-C., Wu Y.-C., Hsu H.-S., Hsu W.-H., Wang L.-S., et al. Preoperative serum albumin level is a prognostic indicator for adenocarcinoma of the gastric cardia. J. Gastrointest. Surg. 2004;8(8):1041–1048. doi: 10.1016/j.gassur.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Lipscomb W.N., Sträter N. Recent advances in zinc enzymology. Chem. Rev. 1996;96(7):2375–2434. doi: 10.1021/cr950042j. [DOI] [PubMed] [Google Scholar]
- Lis C.G., Grutsch J.F., Vashi P.G., Lammersfeld C.A. Is serum albumin an independent predictor of survival in patients with breast cancer? J. Parenter. Enteral Nutr. 2003;27(1):10–15. doi: 10.1177/014860710302700110. [DOI] [PubMed] [Google Scholar]
- Loureiro A., G Azoia N., C Gomes A., Cavaco-Paulo A. Albumin-based nanodevices as drug carriers. Curr. Pharmaceut. Des. 2016;22(10):1371–1390. doi: 10.2174/1381612822666160125114900. [DOI] [PubMed] [Google Scholar]
- Melder R.J., Osborn B.L., Riccobene T., Kanakaraj P., Wei P., Chen G., et al. Pharmacokinetics and in vitro and in vivo anti-tumor response of an interleukin-2-human serum albumin fusion protein in mice. Cancer Immunol. Immunother. 2005;54(6):535–547. doi: 10.1007/s00262-004-0624-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez C.M., McClain C.J., Marsano L.S. Albumin therapy in clinical practice. Nutr. Clin. Pract. 2005;20(3):314–320. doi: 10.1177/0115426505020003314. [DOI] [PubMed] [Google Scholar]
- Mera K., Anraku M., Kitamura K., Nakajou K., Maruyama T., Otagiri M. The structure and function of oxidized albumin in hemodialysis patients: its role in elevated oxidative stress via neutrophil burst. Biochem. Biophys. Res. Commun. 2005;334(4):1322–1328. doi: 10.1016/j.bbrc.2005.07.035. [DOI] [PubMed] [Google Scholar]
- Mikkelsen M., Jacobsen P., Henningsen K. Possible localization of Gc-system on chromosome 4. Loss of long arm 4 material associated with father-child incompatibility within the Gc-system. Hum. Hered. 1977;27(2):105–107. doi: 10.1159/000152857. [DOI] [PubMed] [Google Scholar]
- Minghetti P.P., Ruffner D., Kuang W., Dennison O., Hawkins J.W., Beattie W.G., et al. Molecular structure of the human albumin gene is revealed by nucleotide sequence within q11-22 of chromosome 4. J. Biol. Chem. 1986;261(15):6747–6757. [PubMed] [Google Scholar]
- Musante L., Bruschi M., Candiano G., Petretto A., Dimasi N., Del Boccio P., et al. Characterization of oxidation end product of plasma albumin ‘in vivo’. Biochem. Biophys. Res. Commun. 2006;349(2):668–673. doi: 10.1016/j.bbrc.2006.08.079. [DOI] [PubMed] [Google Scholar]
- Nakajou K., Watanabe H., Kragh-Hansen U., Maruyama T., Otagiri M. The effect of glycation on the structure, function and biological fate of human serum albumin as revealed by recombinant mutants. Biochim. Biophys. Acta Gen. Subj. 2003;1623(2–3):88–97. doi: 10.1016/j.bbagen.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Narazaki R., Otagiri M. Covalent binding of a bucillamine derivative with albumin in sera from healthy subjects and patients with various diseases. Pharmaceut. Res. 1997;14(3):351–353. doi: 10.1023/a:1012006306915. [DOI] [PubMed] [Google Scholar]
- Neelam S., Gokara M., Sudhamalla B., Amooru D.G., Subramanyam R. Interaction studies of coumaroyltyramine with human serum albumin and its biological importance. J. Phys. Chem. B. 2010;114(8):3005–3012. doi: 10.1021/jp910156k. [DOI] [PubMed] [Google Scholar]
- Neužil J., Stocker R. Bilirubin attenuates radical-mediated damage to serum albumin. FEBS Lett. 1993;331(3):281–284. doi: 10.1016/0014-5793(93)80353-v. [DOI] [PubMed] [Google Scholar]
- Nicoletti F.P., Howes B.D., Fittipaldi M., Fanali G., Fasano M., Ascenzi P., et al. Ibuprofen induces an allosteric conformational transition in the heme complex of human serum albumin with significant effects on heme ligation. J. Am. Chem. Soc. 2008;130(35):11677–11688. doi: 10.1021/ja800966t. [DOI] [PubMed] [Google Scholar]
- Nishio H., Dugaiczyk A. Complete structure of the human alpha-albumin gene, a new member of the serum albumin multigene family. Proc. Natl. Acad. Sci. USA. 1996;93(15):7557–7561. doi: 10.1073/pnas.93.15.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y., Yamamoto Y., Mori Y., Satoh K., Takashima H., Ohkawa M., et al. Usefulness of Technetium-99m human serum albumin lymphoscintigraphy in chyluria. Clin. Nucl. Med. 1998;23(7):429–431. doi: 10.1097/00003072-199807000-00006. [DOI] [PubMed] [Google Scholar]
- Niwa Y., Iio A., Niwa G., Sakane T., Tsunematsu T., Kanoh T. Serum albumin metabolism in rheumatic diseases: relationship to corticosteroids and peptic ulcer. J. Clin. Lab. Immunol. 1990;31(1):11–16. [PubMed] [Google Scholar]
- Oelshlegel F., Brewer G., Knutsen C., Prasad A., Schoomaker E. Studies on the interaction of zinc with human hemoglobin. Arch. Biochem. Biophys. 1974;163(2):742–748. doi: 10.1016/0003-9861(74)90536-0. [DOI] [PubMed] [Google Scholar]
- Oettl K., Stauber R. Physiological and pathological changes in the redox state of human serum albumin critically influence its binding properties. Br. J. Pharmacol. 2007;151(5):580–590. doi: 10.1038/sj.bjp.0707251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate-Ocana L.F., Aiello-Crocifoglio V., Gallardo-Rincon D., Herrera-Goepfert R., Brom-Valladares R., Carrillo J.F., et al. Serum albumin as a significant prognostic factor for patients with gastric carcinoma. Ann. Surg Oncol. 2007;14(2):381–389. doi: 10.1245/s10434-006-9093-x. [DOI] [PubMed] [Google Scholar]
- Otagiri M. A molecular functional study on the interactions of drugs with plasma proteins. Drug Metabol. Pharmacokinet. 2005;20(5):309–323. doi: 10.2133/dmpk.20.309. [DOI] [PubMed] [Google Scholar]
- Otagiri M., Chuang V.T.G. Pharmaceutically important pre-and posttranslational modifications on human serum albumin. Biol. Pharm. Bull. 2009;32(4):527–534. doi: 10.1248/bpb.32.527. [DOI] [PubMed] [Google Scholar]
- Panzer S., Kronik G., Lechner K., Bettelheim P., Neumann E., Dudczak R. Glycosylated hemoglobins (GHb): an index of red cell survival. Blood. 1982;59(6):1348–1350. [PubMed] [Google Scholar]
- Papatheodorou L., Weiss N. Vascular oxidant stress and inflammation in hyperhomocysteinemia. Antioxidants Redox Signal. 2007;9(11):1941–1958. doi: 10.1089/ars.2007.1750. [DOI] [PubMed] [Google Scholar]
- Peters T., Jr. Academic press; 1995. All about Albumin: Biochemistry, Genetics, and Medical Applications. [Google Scholar]
- Peters T., Jr. 3 serum albumin. Plasma Proteins 2E V1: Struct. Funct. Genet. Control. 2012;1:133. [Google Scholar]
- Petersen C.E., Ha C.-E., Jameson D.M., Bhagavan N.V. Mutations in a specific human serum albumin thyroxine binding site define the structural basis of familial dysalbuminemic hyperthyroxinemia. J. Biol. Chem. 1996;271(32):19110–19117. doi: 10.1074/jbc.271.32.19110. [DOI] [PubMed] [Google Scholar]
- Petitpas I., Bhattacharya A.A., Twine S., East M., Curry S. Crystal structure analysis of warfarin binding to human serum albumin anatomy of drug site I. J. Biol. Chem. 2001;276(25):22804–22809. doi: 10.1074/jbc.M100575200. [DOI] [PubMed] [Google Scholar]
- Pomier K.M., Ahmed R., Melacini G. Interactions of intrinsically disordered proteins with the unconventional chaperone human serum albumin: from mechanisms of amyloid inhibition to therapeutic opportunities. Biophys. Chem. 2022;282 doi: 10.1016/j.bpc.2021.106743. [DOI] [PubMed] [Google Scholar]
- Quinlan G.J., Mumby S., Martin G.S., Bernard G.R., Gutteridge J.M., Evans T.W. Albumin influences total plasma antioxidant capacity favorably in patients with acute lung injury. Crit. Care Med. 2004;32(3):755–759. doi: 10.1097/01.ccm.0000114574.18641.5d. [DOI] [PubMed] [Google Scholar]
- Rabenstein D.L., Isab A.A. The complexation of zinc in intact human erythrocytes studied by 1 H spin-echo NMR. FEBS Lett. 1980;121(1):61–64. doi: 10.1016/0014-5793(80)81267-1. [DOI] [PubMed] [Google Scholar]
- Rahimizadeh P., Yang S., Lim S.I. Albumin: an emerging opportunity in drug delivery. Biotechnol. Bioproc. Eng. 2020;25:985–995. [Google Scholar]
- Rang H., Dale M., Ritter J., Moore P. In: How Drugs Act: Molecular Aspects. Pharmacology. fourth ed. rang H.P., Dale M.M., Ritter J.M., editors. Churchill Livingstone Edinburg; London, NY, Philadelphia, Sydney, Toronto: 1999. p. 25. [Google Scholar]
- Reichlin T., Hochholzer W., Bassetti S., Steuer S., Stelzig C., Hartwiger S., et al. Early diagnosis of myocardial infarction with sensitive cardiac troponin assays. N. Engl. J. Med. 2009;361(9):858–867. doi: 10.1056/NEJMoa0900428. [DOI] [PubMed] [Google Scholar]
- Rink T., Heuser T., Fitz H., Schroth H.-J., Weller E., Zippel H.H. Lymphoscintigraphic sentinel node imaging and gamma probe detection in breast cancer with Tc-99m nanocolloidal albumin: results of an optimized protocol. Clin. Nucl. Med. 2001;26(4):293–298. doi: 10.1097/00003072-200104000-00002. [DOI] [PubMed] [Google Scholar]
- Roche M., Rondeau P., Singh N.R., Tarnus E., Bourdon E. The antioxidant properties of serum albumin. FEBS Lett. 2008;582(13):1783–1787. doi: 10.1016/j.febslet.2008.04.057. [DOI] [PubMed] [Google Scholar]
- Roy D., Quiles J., Gaze D., Collinson P., Kaski J., Baxter G. Role of reactive oxygen species on the formation of the novel diagnostic marker ischaemia modified albumin. Heart. 2006;92(1):113–114. doi: 10.1136/hrt.2004.049643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler P.J., Tucker A., Viles J.H. Involvement of a lysine residue in the N‐terminal Ni2+ and Cu2+ binding site of serum albumins. Eur. J. Biochem. 1994;220(1):193–200. doi: 10.1111/j.1432-1033.1994.tb18614.x. [DOI] [PubMed] [Google Scholar]
- Sakata N., Moh A., Takebayashi S. Contribution of superoxide to reduced antioxidant activity of glycoxidative serum albumin. Heart Ves. 2002;17(1):22–29. doi: 10.1007/s003800200038. [DOI] [PubMed] [Google Scholar]
- Sampath V., Zhao X.-J., Caughey W.S. Anesthetic-like Interactions of nitric oxide with albumin and hemeproteins a mechanism for control of protein function. J. Biol. Chem. 2001;276(17):13635–13643. doi: 10.1074/jbc.M006588200. [DOI] [PubMed] [Google Scholar]
- Sbarouni E., Georgiadou P., Voudris V. Ischemia modified albumin changes–review and clinical implications. Clin. Chem. Lab. Med. 2011;49(2):177–184. doi: 10.1515/CCLM.2011.037. [DOI] [PubMed] [Google Scholar]
- Schleicher E.D., Olgemöller B., Wiedenmann E., Gerbitz K. Specific glycation of albumin depends on its half-life. Clin. Chem. 1993;39(4):625–628. [PubMed] [Google Scholar]
- Sekula B., Ciesielska A., Rytczak P., Koziołkiewicz M., Bujacz A. Structural evidence of the species-dependent albumin binding of the modified cyclic phosphatidic acid with cytotoxic properties. Biosci. Rep. 2016;36(3) doi: 10.1042/BSR20160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seve P., Ray‐Coquard I., Trillet‐Lenoir V., Sawyer M., Hanson J., Broussolle C., et al. Low serum albumin levels and liver metastasis are powerful prognostic markers for survival in patients with carcinomas of unknown primary site. Cancer. 2006;107(11):2698–2705. doi: 10.1002/cncr.22300. [DOI] [PubMed] [Google Scholar]
- Shaw C.F., III The protein chemistry of antiarthritic gold (I) thiolates and related complexes. Comments Mod. Chem. 1989;8(6):233–267. [Google Scholar]
- Shojai S., Haeri Rohani S.-A., Moosavi-Movahedi A.A., Habibi-Rezaei M. Human serum albumin in neurodegeneration. Rev. Neurosci. 2022;33(7):803–817. doi: 10.1515/revneuro-2021-0165. [DOI] [PubMed] [Google Scholar]
- Siddiqui A., Heinzerling J., Livingston E.H., Huerta S. Predictors of early mortality in veteran patients with pancreatic cancer. Am. J. Surg. 2007;194(3):362–366. doi: 10.1016/j.amjsurg.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Silverberg J.D., Premachandra B. Familial hyperthyroxinemia due to abnormal thyroid hormone binding. Ann. Intern. Med. 1982;96(2):183–186. doi: 10.7326/0003-4819-96-2-183. [DOI] [PubMed] [Google Scholar]
- Simard J.R., Zunszain P.A., Hamilton J.A., Curry S. Location of high and low affinity fatty acid binding sites on human serum albumin revealed by NMR drug-competition analysis. J. Mol. Biol. 2006;361(2):336–351. doi: 10.1016/j.jmb.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Simons T. Intracellular free zinc and zinc buffering in human red blood cells. J. Membr. Biol. 1991;123(1):63–71. doi: 10.1007/BF01993964. [DOI] [PubMed] [Google Scholar]
- Soeters P.B., Wolfe R.R., Shenkin A. Hypoalbuminemia: pathogenesis and clinical significance. J. Parenter. Enteral Nutr. 2019;43(2):181–193. doi: 10.1002/jpen.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokołowska M., Wszelaka-Rylik M., Poznański J., Bal W. Spectroscopic and thermodynamic determination of three distinct binding sites for Co (II) ions in human serum albumin. J. Inorg. Biochem. 2009;103(7):1005–1013. doi: 10.1016/j.jinorgbio.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Sokołowska M., Pawlas K., Bal W. Bioinorganic Chemistry and Applications. 2010. Effect of common buffers and heterocyclic ligands on the binding of Cu (II) at the multimetal binding site in human serum albumin; p. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler J.S. S-nitrosothiols in the blood roles, amounts, and methods of analysis. Circ. Res. 2004;94(4):414–417. doi: 10.1161/01.RES.0000122071.55721.BC. [DOI] [PubMed] [Google Scholar]
- Stehle G., Sinn H., Wunder A., Schrenk H.H., Stewart J.C.M., Hartung G., et al. Plasma protein (albumin) catabolism by the tumor itself—implications for tumor metabolism and the genesis of cachexia. Crit. Rev. Oncol.-Hematol. 1997;26(2):77–100. doi: 10.1016/s1040-8428(97)00015-2. [DOI] [PubMed] [Google Scholar]
- Stockigt J.R., Topliss D.J., Barlow J.W., White E.L., Hurley D.M., Taft P. Familial euthyroid thyroxine excess: an appropriate response to abnormal thyroxine binding associated with albumin. J. Clin. Endocrinol. Metabol. 1981;53(2):353–359. doi: 10.1210/jcem-53-2-353. [DOI] [PubMed] [Google Scholar]
- Subramanian G.M., Fiscella M., Lamousé-Smith A., Zeuzem S., McHutchison J.G. Albinterferon α-2b: a genetic fusion protein for the treatment of chronic hepatitis C. Nat. Biotechnol. 2007;25(12):1411–1419. doi: 10.1038/nbt1364. [DOI] [PubMed] [Google Scholar]
- Sudlow G., Birkett D., Wade D. The characterization of two specific drug binding sites on human serum albumin. Mol. Pharmacol. 1975;11(6):824–832. [PubMed] [Google Scholar]
- Sudlow G., Birkett D., Wade D. Further characterization of specific drug binding sites on human serum albumin. Mol. Pharmacol. 1976;12(6):1052–1061. [PubMed] [Google Scholar]
- Suga K., Kume N., Matsunaga N., Motoyama K., Hara A., Ogasawara N. Assessment of leg oedema by dynamic lymphoscintigraphy with intradermal injection of technetium-99m human serum albumin and load produced by standing. Eur. J. Nucl. Med. 2001;28(3):294–303. doi: 10.1007/s002590000418. [DOI] [PubMed] [Google Scholar]
- Sugio S., Kashima A., Mochizuki S., Noda M., Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12(6):439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- Sun L.-C., Chu K.-S., Cheng S.-C., Lu C.-Y., Kuo C.-H., Hsieh J.-S., et al. Preoperative serum carcinoembryonic antigen, albumin and age are supplementary to UICC staging systems in predicting survival for colorectal cancer patients undergoing surgical treatment. BMC Cancer. 2009;9(1):288. doi: 10.1186/1471-2407-9-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunthornthepvarakul T., Angkeow P., Weiss R.E., Hayashi Y., Refetoff S. An identical missense mutation in the albumin gene results in familial dysalbuminemic hyperthyroxinemia in eight unrelated families. Biochem. Biophys. Res. Commun. 1994;202(2):781–787. doi: 10.1006/bbrc.1994.1998. [DOI] [PubMed] [Google Scholar]
- Tao H-y, Wang R-q, Sheng W-j, Zhen Y-s. The development of human serum albumin-based drugs and relevant fusion proteins for cancer therapy. Int. J. Biol. Macromol. 2021;187:24–34. doi: 10.1016/j.ijbiomac.2021.07.080. [DOI] [PubMed] [Google Scholar]
- Terawaki H., Yoshimura K., Hasegawa T., Matsuyama Y., Negawa T., Yamada K., et al. Oxidative stress is enhanced in correlation with renal dysfunction: examination with the redox state of albumin. Kidney Int. 2004;66(5):1988–1993. doi: 10.1111/j.1523-1755.2004.00969.x. [DOI] [PubMed] [Google Scholar]
- Tibaduiza E.C., Bobilya D.J. Zinc transport across an endothelium includes vesicular cotransport with albumin. J. Cell. Physiol. 1996;167(3):539–547. doi: 10.1002/(SICI)1097-4652(199606)167:3<539::AID-JCP17>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Tullis J.L. Albumin: 1. Background and use. JAMA. 1977;237(4):355–360. doi: 10.1001/jama.237.4.355. [DOI] [PubMed] [Google Scholar]
- Van Campenhout A., Van Campenhout C., Lagrou A.R., Moorkens G., De Block C., Manuel-y-Keenoy B. Iron-binding antioxidant capacity is impaired in diabetes mellitus. Free Radic. Biol. Med. 2006;40(10):1749–1755. doi: 10.1016/j.freeradbiomed.2006.01.010. [DOI] [PubMed] [Google Scholar]
- van Reyk D.M., Brown A.J., Hult'en L.M., Dean R.T., Jessup W. Oxysterols in biological systems: sources, metabolism and pathophysiological relevance. Redox Rep. 2006;11(6):255–262. doi: 10.1179/135100006X155003. [DOI] [PubMed] [Google Scholar]
- Vorum H. Reversible ligand binding to human serum albumin. Theoretical and clinical aspects. Dan. Med. Bull. 1999;46(5):379–399. [PubMed] [Google Scholar]
- Vorum H., Honoré B. Influence of fatty acids on the binding of warfarin and phenprocoumon to human serum albumin with relation to anticoagulant therapy. J. Pharm. Pharmacol. 1996;48(8):870–875. doi: 10.1111/j.2042-7158.1996.tb03990.x. [DOI] [PubMed] [Google Scholar]
- Walter P.B., Fung E.B., Killilea D.W., Jiang Q., Hudes M., Madden J., et al. Oxidative stress and inflammation in iron‐overloaded patients with β‐thalassaemia or sickle cell disease. Br. J. Haematol. 2006;135(2):254–263. doi: 10.1111/j.1365-2141.2006.06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Tanase S., Nakajou K., Maruyama T., Kragh-Hansen U., Otagiri M. Role of Arg-410 and Tyr-411 in human serum albumin for ligand binding and esterase-like activity. Biochem. J. 2000;349(3):813–819. doi: 10.1042/bj3490813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R., Sunthornthepvarakul T., Angkeow P., Marcus-Bagley D., Cox N., Alper C.A., et al. Linkage of familial dysalbuminemic hyperthyroxinemia to the albumin gene in a large Amish kindred. J. Clin. Endocrinol. Metabol. 1995;80(1):116–121. doi: 10.1210/jcem.80.1.7829599. [DOI] [PubMed] [Google Scholar]
- Wiedermann C.J. Hypoalbuminemia as surrogate and culprit of infections. Int. J. Mol. Sci. 2021;22(9):4496. doi: 10.3390/ijms22094496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding G., Feldhoff R.C., Vesell E.S. Concentration-dependent effects of fatty acids on warfarin binding to albumin. Biochem. Pharmacol. 1977;26(12):1143–1146. doi: 10.1016/0006-2952(77)90058-2. [DOI] [PubMed] [Google Scholar]
- Wilkinson P., Jeremy R., Brooks F.P., Hollander J.L. The mechanism of hypoalbuminemia in rheumatoid arthritis. Ann. Intern. Med. 1965;63(1):109–114. doi: 10.7326/0003-4819-63-1-109. [DOI] [PubMed] [Google Scholar]
- Wu T.-W., Wu J., Li R.-K., Mickle D., Carey D. Albumin-bound bilirubins protect human ventricular myocytes against oxyradical damage. Biochem. Cell. Biol. 1991;69(10–11):683–688. doi: 10.1139/o91-102. [DOI] [PubMed] [Google Scholar]
- Wu D., Gucwa M., Czub M.P., Cooper D.R., Shabalin I.G., Fritzen R., et al. Structural and biochemical characterisation of Co 2+-binding sites on serum albumins and their interplay with fatty acids. Chem. Sci. 2023;14(23):6244–6258. doi: 10.1039/d3sc01723k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder A., Muller-Ladner U., Stelzer E., Neumann E., Sinn H., Gay S., et al. Albumin-based drug delivery as novel therapeutic approach for rheumatoid arthritis. Arthritis Res. Ther. 2003;5(Suppl. 3):1. doi: 10.4049/jimmunol.170.9.4793. [DOI] [PubMed] [Google Scholar]
- Xue C., Zhang R., Liu H., Yao X., Liu M., Hu Z., et al. QSAR models for the prediction of binding affinities to human serum albumin using the heuristic method and a support vector machine. J. Chem. Inf. Comput. Sci. 2004;44(5):1693–1700. doi: 10.1021/ci049820b. [DOI] [PubMed] [Google Scholar]
- Yabu Y., Miyai K., Kobayashi A., Miki K., Doi K., Takamatsu J., et al. A new type of albumin with predominantly increased binding affinity for 3, 3′, 5-triiodothyronine in a patient with Graves' disease. J. Endocrinol. Invest. 1987;10(2):163–169. doi: 10.1007/BF03347183. [DOI] [PubMed] [Google Scholar]
- Yamasaki K., Maruyama T., Kragh-Hansen U., Otagiri M. Characterization of site I on human serum albumin: concept about the structure of a drug binding site. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1996;1295(2):147–157. doi: 10.1016/0167-4838(96)00013-1. [DOI] [PubMed] [Google Scholar]
- Yeun J.Y., Kaysen G.A. Factors influencing serum albumin in dialysis patients. Am. J. Kidney Dis. 1998;32(6):S118–S125. doi: 10.1016/s0272-6386(98)70174-x. [DOI] [PubMed] [Google Scholar]
- Zeeshan F., Madheswaran T., Panneerselvam J., Taliyan R., Kesharwani P. Human serum albumin as multifunctional nanocarrier for cancer therapy. J. Pharmaceut. Sci. 2021;110(9):3111–3117. doi: 10.1016/j.xphs.2021.05.001. [DOI] [PubMed] [Google Scholar]
- Zhu L., Yang F., Chen L., Meehan E.J., Huang M. A new drug binding subsite on human serum albumin and drug–drug interaction studied by X-ray crystallography. J. Struct. Biol. 2008;162(1):40–49. doi: 10.1016/j.jsb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Zsila F. Subdomain IB is the third major drug binding region of human serum albumin: toward the three-sites model. Mol. Pharm. 2013;10(5):1668–1682. doi: 10.1021/mp400027q. [DOI] [PubMed] [Google Scholar]
- Zunszain P.A., Ghuman J., McDonagh A.F., Curry S. Crystallographic analysis of human serum albumin complexed with 4Z, 15E-bilirubin-IXα. J. Mol. Biol. 2008;381(2):394–406. doi: 10.1016/j.jmb.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.











