Abstract
Liquid–liquid phase separation (LLPS) is a novel principle for interpreting precise spatiotemporal coordination in living cells through biomolecular condensate (BMC) formation via dynamic aggregation. LLPS changes individual molecules into membrane-free, droplet-like BMCs with specific functions, which coordinate various cellular activities. The formation and regulation of LLPS are closely associated with oncogenesis, tumor progressions and metastasis, the specific roles and mechanisms of LLPS in tumors still need to be further investigated at present. In this review, we comprehensively summarize the conditions of LLPS and identify mechanisms involved in abnormal LLPS in cancer processes, including tumor growth, metastasis, and angiogenesis from the perspective of cancer hallmarks. We have also reviewed the clinical applications of LLPS in oncologic areas. This systematic summary of dysregulated LLPS from the different dimensions of cancer hallmarks will build a bridge for determining its specific functions to further guide basic research, finding strategies to intervene in LLPS, and developing relevant therapeutic approaches.
Keywords: Biomolecular condensate, Membrane-less organelle, Cancer, Novel therapeutics, Liquid–liquid phase separation
Background
The spatial and temporal coordination of biochemical reactions is crucial for cellular physiology [1]. While membrane-bound organelles are essential for spatially organized cellular processes, the discovery of membrane-less organelles (MLOs) has shed light on new mechanisms for tightly controlling processes within cells [2]. MLOs, as known as biomolecular condensates (BMCs), include the nucleolus [2], Cajal bodies [3], nucleoli [4], stress granules (SGs) [5–7], and super-enhancers (SEs)[8–10] etc. These structures typically range from 0.1 to 3 µm [11]and play key roles in facilitating or modulating specific cellular processes. BMCs and MLOs are both formed by the process of phase separation, and in most scenarios, these two concepts are equivalent.
Until the emergence of the concept of liquid–liquid phase separation (LLPS), the formation and organization of MLOs remained unclear [12]. Thus, LLPS provides a reasonable framework to explain the formation mechanism of MLOs and BMCs. This dynamic process involves the transition of biomolecules from a homogeneous environment to sparse and dense phases [11, 13, 14], aiming to reach the lowest-entropy state. Notably, LLPS occurs when multivalent biopolymers instantaneously interact with each other [15–17], forming liquid-like entities such as bodies, puncta, granules, droplets, and condensates [18].
Normal BMCs ensure basic cellular functions, whereas their aberrant forms result in cellular dysfunction and possible tumorigenesis. Studies have demonstrated that LLPS are crucial in the regulation of tumor onset, progression [19], including promoting cancer cells proliferations and metastasis. Further, the hallmarks and enabling characteristics of cancer in the 2022 version provide a framework for further oncological studies[20]. However, understanding of the regarding phase separation processes involved in each hallmark is still limited. Therefore, unveiling a novel dimension of its biological functions is in need.
In this review, we include all cutting-edge and typical articles related to liquid–liquid separation in oncology. Firstly, we describe the methods used to investigate LLPS, followed by their role in promoting the formation of BMCs/MLOs. Subsequently, we examine the current understanding of how LLPS influences tumorigenesis, progression and their emerging role in cancer treatment. Finally, we comprehensively summarize the latest insights into methods to interfere with aberrant forms of BMCs.
Mechanisms and methods associated with the phenomenon of LLPS
Concepts and mechanisms
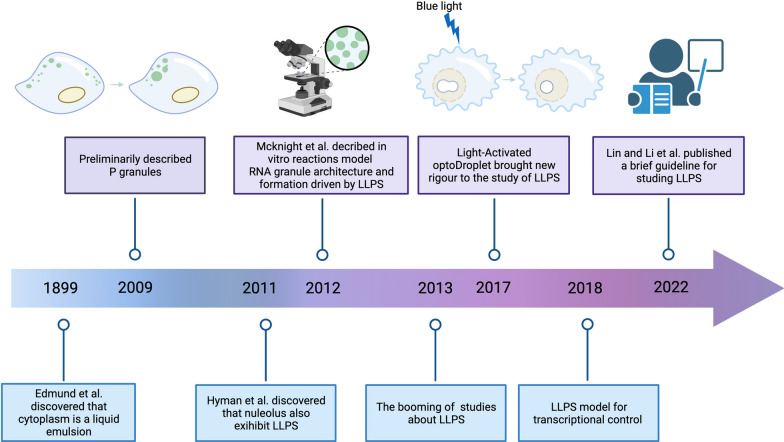
Phase separation is defined as the spontaneous aggregation of molecules when their concentration exceeds a certain threshold, thus forming a membrane-less compartment [21]. Typically, the interactions between macromolecules in LLPS are typically non-covalent and of low affinity [22, 23]. This process is often driven by the modification of intrinsically disordered regions (IDRs) within proteins [24, 25]. The concept of LLPS was first introduced in the biochemical field of biochemistry in 2009 by Hyman and colleagues with various milestone events followed subsequently (Fig. 1), offering a novel perspective on various MLOs distributed in cells (Fig. 2) [26]. Although several in silico tools help forecast the potential of phase-separated molecules (Table 1), comprehensive summaries of the characteristics and conditions that induce LLPS are limited.
Fig. 1.
History of LLPS research developments. Milestone discoveries are outlined
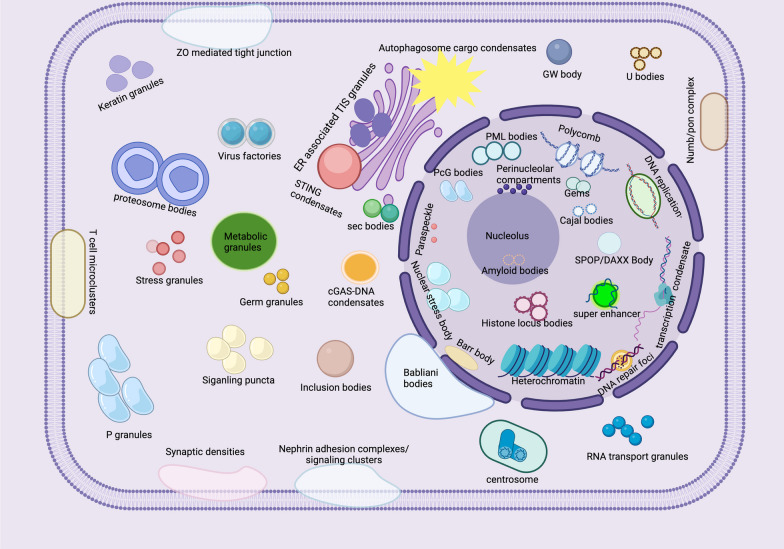
Fig. 2.
Intra-cellular MLOs within a eukaryotic cell. MLOs are distributed in the nucleus, nuclear membrane, cytoplasm, and plasma membranes of cells. Nucleolus, perinucleolar compartments, paraspeckles, Cajal bodies, transcription condensates, Gems, DNA repair foci, nuclear stress bodies, PcG bodies, histone locusbody, PML bodies, DNA replication bodies, polycombs, SPOP/DAXX bodies, super enhancers, heterochromatin, and amyloid bodies (located in nucleolus) are located in the nuclear by the LLPS. Whereas some MLOs are distributed in the nuclear membrane (Babliani bodies), cytoplasm (such as sec bodies, cGAS-DNA condensates, ER associated TIS granules, autophagosome cargo condensates, stress granule, P granules, U bodies, Virus factories, Numb/pon complex, RNA transport granules, centrosome, inclusion bodies, siganling puncta, GW bodies, germ granules, transport RNP, and proteosome bodies, metabolic granules, keratin granules), and cell plasma membrane (such as immune synapse densities, Numb/pon complex, Nephrin adhesion complexes/ signaling clusters, T cell microclusters, and ZO mediated tight junction)
Table 1.
Overview of databases related to liquid–liquid phase separation (LLPS)
| Category | Database | Availability | Details of databases | References |
|---|---|---|---|---|
| Prediction of LLPS related proteins | SGnn | http://sgnn.ppmclab.com | Proteins bearing prion-like domains (PrLDs) | [27] |
| PhaSepDB | http://db.phasep.pro/ | Phase-separation related proteins | [28] | |
| D2P2 | https://d2p2.pro/search | Phase-separation related proteins | [29] | |
| PLAAC | http://plaac.wi.mit.edu/ | Prion-Like Amino Acid Composition | [30] | |
| DrLLPS | http://llps.biocuckoo.cn/ |
Proteins in this database are classified as drivers, regulators and potential Clients |
[31] | |
| PhaSePro | https://phasepro.elte.hu | A manually curated database of LLPS driver proteins in various organisms, with emphasis on the biophysical properties that govern phase separation. | [32] | |
| BioGRID | https://thebiogrid.org/ | Database of Protein, Genetic and Chemical Interactions | [33] | |
| LLPSDB | http://biocomp.ucas.ac.cn/llpsdb/home.aspx | A database of proteins undergoing LLPS in vitro | [34] | |
| HUMAN CELL MAP | https://cell-map.org/ or https://humancellmap.org/ |
Summarizes for each compartment the enrichment of expected domains and motifs as well as GO-terms Provides channels to analyze spatiotemporal correlations between proteins in different organelles |
[35] | |
| MLOsMetaDB | http://mlos.leloir.org.ar/ | Unified resource of MLOs and LLPS associated proteins | [36] | |
| catGRANULE | http://s.tartaglialab.com/ | A website good at predicting LLPS propensity of dosage-sensitive proteins | [37] | |
| PScore | https://github.com/haocai1992/PScore-online#pscore-online | A machine learning algorithm that predicts the likelihood of phase separated proteins | [38] | |
| Prediction of LLPS related RNAs | RPS | http://rps.renlab.org/#/Home | A comprehensive database of RNAs involved in liquid–liquid phase separation | [39] |
| RNAPhaSep | http://www.rnaphasep.cn/#/Home | A resource of RNAs undergoing phase separation | [40] | |
| RNA granule database | http://rnagranuledb.lunenfeld.ca/ | A database containing RNA granules | [41] | |
| Integreation of LLPS related diseases | DisPhaseDB | http://disphasedb.leloir.org.ar | An integrative database of diseases related variations in liquid–liquid phase separation proteins | [42] |
| Prediction of specific structures or features of LLPS | IUPred2A | https://iupred2a.elte.hu/ | Combination of the iupred database and the ANCHOR database, which can predict the disordered and disordered binding regions of proteins | [43] |
| PONDR | http://www.pondr.com | Predictor of natural disordered regions | [44] | |
| MobiDB | https://mobidb.org | Provides information about intrinsically disordered regions and related features | [45] | |
| CIDER | http://pappulab.wustl.edu/CIDER/ | Calculation of many different parameters associated with disordered protein sequences | [46] | |
| ZipperDB | https://services.mbi.ucla.edu/zipperdb/ | Predictions of fibril-forming segments within protein | [47] | |
| Metadisorder | http://iimcb.genesilico.pl/metadisorder/ | Prediction of protein disorder | [48] | |
| DisMeta | https://montelionelab.chem.rpi.edu/dismeta/ | Prediction of protein disorder | [49] | |
| Expasy | https://web.expasy.org/compute_pi/ | Computation of the theoretical pI (isoelectric point) and Mw (molecular weight) | [50] | |
| AMYCO | http://bioinf.uab.es/amycov04/ | Evaluation of mutation impact on prion-like proteins aggregation propensity | [51] | |
| MFDp2 | http://biomine.ece.ualberta.ca/MFDp2/ | Accurate sequence-based prediction of protein disorder which also outputs well-described sequence-derived information that allows profiling the predicted disorder | [52] |
Structural characteristics and critical components that triggers LLPS
The concept of a driver (or scaffold)/client is widely accepted. Proteins, DNA, and RNA can also be used as scaffolds. With multiple binding sites, these macromolecules facilitate weak interactions and trigger LLPS. The detailed structures are summarized below.
Multi-foldable domains
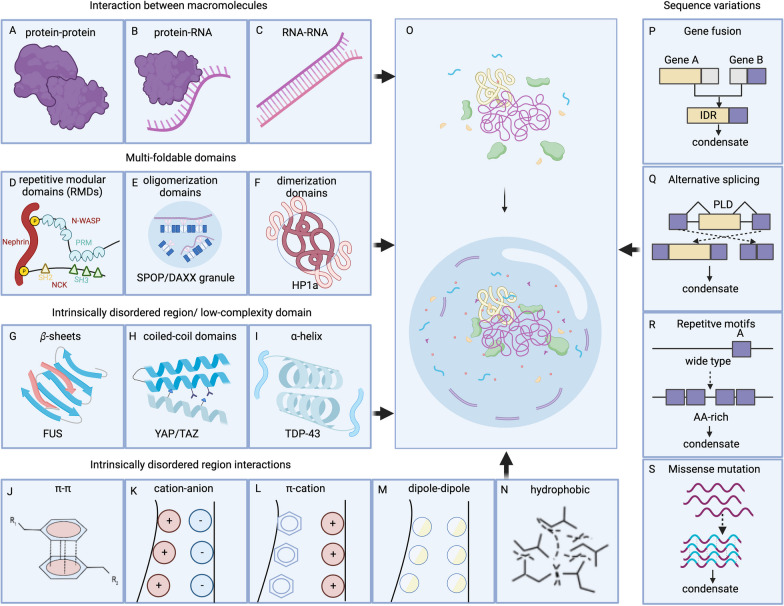
One of the most common structural features that facilitates LLPS is multivalency, which involves the interaction of various macromolecules (Figs. 3A–C). By using multiple, similar domains to mediate the interactions, these macromolecules effectively trigger LLPS and attract client molecules to form condensates. For example, the proline-rich motif (PRM) domain characteristic of the neural Wiskott-Aldrich syndrome (N-WASP) interacts with the SH3 domain of NCK, thereby inducing LLPS [53]. A similar principle applies to the nephrin/Nck/N-WASP system, wherein the phosphotyrosines of nephrin interact with the SH2 and SH3 domains of NCK to bind to the PRMs (Fig. 3D) [54]. Similarly, higher-order polymerized structures are formed via the tandem dimerization domains of the speckled POZ protein (SPOP) and its interaction with cullin-3-RING ubiquitin ligase and other substrates, promoting its localization in nuclear speckles[55] (Fig. 3E). Dimerization or oligomerization of proteins can also contribute to LLPS. For example, when the dimerization of HP1a is disrupted, the mobility of the droplets increases, hindering the maturation of heterochromatin formations (Fig. 3F)[56].
Fig. 3.
Basic condensates promoting features. A-C Interactions between macromolecules that facilitate phase separation. D The SH2 domain of NCK binds to Nephrin, and NCK possesses three SH3 domains that can bind the proline-rich motifs (PRMs) of N-WASP, showing a typical repetitive molecular domain (RMD) that contributes to LLPS. E Oligomerization of SPOP and its interactions with substrates can induce phase separation. F Dimerization of HP1a promotes LLPS. G–I Several classic IDRs, which consist of LCDs. J–N Fundamental interacting force between IDRs. O Formation of BMCs, from dissociation to assembly. P–S Four types of sequence variations that drive phase separation
IDR/low-complexity domains contribute to LLPS
IDRs are distinctive features of certain proteins of the condensates, accounting for 33–55% of eukaryotic proteomes [57, 58]. Like IDRs, low-complexity domains (LCDs) are also distinctive features of proteins comprised by highly biased amino acid compositions [59]. IDRs and LCDs lack stable tertiary structures and have flexible conformations, making them prone to undergo LLPS [11, 60–62]. β sheets in TDP34/FUS (Fig. 3G), coiled-coil domains in YAP/TAZ (Fig. 3H) and alpha-helix in TDP43 (Fig. 3I), exemplify the role of LCDs in LLPS [63–69]. While IDR interactions involve pi-pi interactions, salt bridges between opposing charge residues, pi-cation interactions, dipole-dipole interactions (Van der Waals forces), and hydrophobic forces (Figs. 3J–N) represent different forms of LLPS [70].
Nucleic acids regulate LLPS
Nucleic acids, especially the single-stranded nucleic acids, tend to aggregate to form droplets, whereas double-stranded nucleic acids tend to form gel-like aggregates [71]. Via electrostatic interactions and the pairing of repeating molecules, certain RNAs achieve polyvalency, effectively inducing LLPS in combination with proteins, as observed in the RG/RGG-rich domains of the SERPINE1 mRNA-binding protein 1 (SERBP1) system [72]. In contrast, the RNA concentration does not show a strong positive correlation with the phase transition ability [73]. RNA modifications and non-coding RNAs can also induce LLPS spontaneously [74] or by attaching to proteins, facilitating clients recruitment for the condensate assembly [75–77].
Head-to-tail polymerization
Occasionally, stable structural domains in proteins, such as SAM and DIX, retain their ability to trigger local condensation [78, 79]. Among the dishevelled and axin components of the Wnt signaling, the DIX domain can assemble in a head-to-tail manner and promote Wnt signaling [80, 81]. The SAM domain of the tankyrase protein forms similar puncta in a head-to-tail manner to bind and ribosylate poly ADP AXIN, thus promoting Wnt signaling [82]. These structural conditions facilitate the formation of condensates (Fig. 3O).
Sequence variations at the gene levels
Disease-related genomic changes regulate LLPS. The NUP98 fusion protein in leukemia, carrying IDRs, serves as a good model for gene fusion [83] (Fig. 3P). Similar results have been obtained with anaplastic lymphoma kinase (ALK) and BCR-ABL1 fusions [84, 85]. Linear motifs that modulate ligand recognition within IDRs control the function of alternatively spliced (AS) proteins [86, 87] and modulate their assemblies (Fig. 3Q). On the contrary, repetitive motifs can induce pathogenic repeat expansions (Fig. 3R). Missense mutations in IDRs and polymerization/modular domains may influence the phase transition status bilaterally (Fig. 3S). For instance, F291S and Y283S mutations in the heterogeneous nuclear ribonucleoprotein A2 scarcely affect the aggregation, whereas D290V and P298L mutations improve the condensation [88].
External conditions and physicochemical properties affect LLPS
In this section, we focus on the conditions and the post-translational modifications (PTMs) which play a crucial role in regulating the dynamic transitions of molecules within the cell.
The interplay of various intracellular conditions, such as the concentration of proteins, pH level, and changes of the cellular milieu, alter the strength of polyvalent interactions. These conditions are key regulators of transitions within the cell. Furthermore, the concentrations of macromolecules are critical. When the concentration exceeds a critical threshold, the interaction between these macromolecules outweighs the forces that maintain homogeneity of the system, making the solution susceptible to phase separation. Conversely, when the concentrations are below this threshold, the components remain evenly distributed [89, 90]. The alterations of pH value can significantly impact LLPS by changing the surface charges of amino acids, the α-carbonyl groups, and the α-amino terminal protonation status. pH alterations affect the stability of specific proteins and change the secondary structure from ordered to disordered. Altering the protonation of amino acids directly influences the chemical properties of macromolecules, further altering their intermolecular interactions and triggering LLPS. For example, the decreased cytoplasmic pH, induced by external stimuli, can promote LLPS of naturally disordered proteins, as observed with Sup35 in yeast cells [91]. The increase in salt concentration and the addition of substances such as PEG3000 and glycerol can also modulate LLPS [73, 92]. Additionally, weak electrostatic interactions, driven by IDRs, are highly sensitive to changes in pH and ionic strength, potentially explaining LLPS induction due to environmental changes [17, 93]. In addition, temperature and stress levels can also trigger or disrupt LLPS by affecting the solubility of macromolecules [11]. Moreover, prion-like domains in proteins can sense pressure, influencing the solubility and phase behavior [94, 95].
The PTMs are crucial in the regulation of phase transitions by altering molecular interactions or directly modifying the potency of BMCs [96–98]. PTMs can induce changes of biomolecules in the spatial structures and state of proteins [96, 99]. PTMs of RNA-binding proteins (RBPs) can directly weaken or enhance the interactions between components, contributing to the formation of RNP granules, serving as an example of an MLO that is composed of RBP and RNA [96]. PTMs can promote or inhibit polyvalent interactions by influencing the condition of proteins, thus affecting the occurrence of LLPS [100]. Notably, the Lys residues within the IDRs are particularly prone to get SUMOylation, a modification that significantly contributes to the formation of the promyelocytic leukemia nuclear bodies (NBs). De-SUMOylation can lead to the release of a constituent protein and the separation of NBs during mitosis [101, 102].
Given the complexity of physicochemical conditions, the manipulation of PTMs is an intriguing approach to influence LLPS. Thus, it is pivotal to understand the possible mechanisms in cancer-related PTMs associated with LLPS (Table 2).
Table 2.
Summary of cancer-related PTMs involved in LLPS
| PTM | Disease association | Participants | Biological role | Regulation of LLPS | References |
|---|---|---|---|---|---|
| Ubiquitination | Non-small-cell lung cancer | USP42 | Drives nuclear speckle mrna splicing and promote tumorigenesis | Promotion | [8] |
| Multiple cancer types | p62 | Promotes tumor cell survival by upregulating p62 liquid droplet formation and degradation | Promotion | [103] | |
| Multiple cancer types | SPOP/DAXX | Co-localizes with DAXX in Liquid Nuclear Organelles and facilitates DAXX Ubiquitination | Promotion | [104] | |
| Phosphorylation | Multiple cancer types | TAZ | Formation of transcription compartment to promote gene expression | Promotion | [68] |
| Methylation | Leukaemia | YTHDC1-m6A condensates | Facilitates a phase-separated nuclear body and suppresses myeloid leukemica differentiation | Promotion | [105] |
| Multiple cancer types | UTX (namely KDM6A) | Involved in chromatin-regulatory activity in tumour suppression | Promotion | [106] | |
| Sumoylation | Colon cancer | RNF168 | Genomic instability and DNA damage repair | Promotion | [107] |
| Acetylation | Multiple cancer types | KAT8-IRF1 | KAT8-IRF1 condensate formation boosts PD-L1 transcription | Promotion | [108] |
| Neddylation | Acute promyelocytic leukemia (APL) | PML/RARa | Induce abberent LLPS and disrupt function of PML nuclear bodies to drive APL | Inhibition | [109] |
Deregulated phase separation in cancer
Emerging evidence has robustly revealed that aberrant BMCs are involved in various biochemical processes in human diseases and various oncogenic signaling pathways [19] (Table 3). Next, we review the role of LLPS in tumors based on several hallmarks (Fig. 4).
Table 3.
Oncogenic signaling assosciated condensates that were involved in LLPS
| Signaling Pathway | Cancer type | Biomolecule/ condensate | Biological role | Ref |
|---|---|---|---|---|
| EGFR/RAS signaling | Lung cancer | EGFR condensates | Regulating pro-tumor activation of Ras | [110, 111] |
| KRAS signaling | Lung cancer | EML4-ALK condensates | Modulating the KRAS signaling pathway, amplifying the oncogenic potential of this cascade, ultimately leading to dysregu- lated cellular proliferation and survival | [112, 113] |
| JAK-STAT3 signaling | Lung cancer | EZH2/STAT3 | Myristoylation modification of EZH2 enables its phase separation, compartmentalize STAT3 within the condensates and leads to the sustained activation and enhanced transcriptional activity of STAT3 | [113] |
| PI3K-AKT-mTOR signaling pathway | Lung cancer | stress granule | dynamically interacting with a key component of lung oncogenic pathway, mTOR and its regulators, influencing its localization, activity, and downstream signaling | [114] |
| Hippo signaling pathway | Pan-cancer | YAP, TAZ, TEAD | Undergoing LLPS, accumulating in the nucleus coregulator with increased activity in various cancers | [68, 115] |
| Hepatocellular carcinoma | G6PC (glycogen compartments) | YAP signaling activation | [116] | |
| Hepatocellular carcinoma | YAP/TEAD transcriptional condensates | Acting as signaling hubs for the tumor microenvironment | [117] | |
| Hepatocellular carcinoma | Laforin-Mst1/2 condensates | Increasing hepatocarcinogenesis | [116] | |
| p53 signaling | Pan-cancer | p53, 53BP1 | 53BP1 can form phase separation droplets, which enrich tumor suppressor protein p53. Cancer-associated mutation of p53 can accelerate the protein aggregation and amyloid formation by destroying the folding of p53 core domain | [118, 119] |
| Wnt/β-catenin signaling | Breast and prostate cancer | DACT1 | WNT signaling inhibition | [120] |
| TGF-β signaling | Colorectal cancer | SMAD3 | forming nuclear foci when the signaling pathway is activated | [121] |
| cAMP/PKA signaling | Atypical liver cancer fibrolamellar carcinoma | DnaJB1-PKAcat fusion | Tumorigenic cAMP signaling | [122] |
| Hepatocellular carcinoma | RIα condensates | Promoting cell proliferation and transformation | [122] | |
| RAS signaling | Pan-cancer | EML4-ALK fusion | RAS signaling overactivation | [123, 124] |
| Pan-cancer | CCDC6-RET fusion | RAS signaling overactivation | [123, 124] | |
| Pan-cancer | LAT, GRB2, SOS | Activating Ras in tumour development | [125] | |
| MAPK signaling | RTK-driven human cancer | SHP2 | Stimulation of downstream MAPK signaling pathways and ERK1/2 activation | [126] |
| Wnt/β-Catenin signaling | Colorectal cancer | Destruction complex | Regulating development and stemness | [127] |
| NRF2/NF-κB signaling | Lung cancer | p62 bodies | Accelerating cancer development | [128] |
| NF-κB pathway signaling | Virus-associated cancer | p65/inclusion body | The trapped p65 (subunit of NF-κB) by phase separation of viral replication machinery cannot translocate into the nucleus to activate the downstream transcription of proinflammatory cytokine genes and other antiviral genes | [129] |
| cGAS-STING signaling | Pan-cancer | NF2m-IRF3 condensates | Regulating tumor immunity | [130, 131] |
| IL-6/STAT3 signaling | Hepatocellular carcinoma | Paraspeckles | IL-6/STAT3 signaling promotes paraspeckles formation, which favors overactivation of STAT3 | [132] |
Fig. 4.
Summary of deregulated phase separations in cancer. A RTK granule formations activate RTK/MAPK signaling pathways to promote tumor proliferation. B DDX21phase separation activates MCM5, facilitating EMT signaling and modulating metastasis of colon cancer. C LLPS of 53BP1 diminish downstream targets of p53 to evade growth suppressions. D The accumulation of 53BP1 in the nuclear foci is enhanced after DNA damage, activating p53 and regulating cellular senescence. E SUMO ALT-associated PML bodies on the telomeres facilitate the replicative immortality of cancer cells. F Nuclear condensates (nYACs) generated through the LLPS of YTHDC1 (binding with m6A-mRNA) are significantly increased in AML cells. G Mutations in the FERM domain of NF2 (NF2m) robustly inhibited STING-initiated antitumor immunity by forming NF2m-IRF3 condensates. H PML nuclear bodies (NBs) serve as comprehensive ROS sensors associated with antioxidative pathways. I EBNA2 becomes part of BMCs and regulates EBV gene transcriptions. J BRD4 forms condensates with SEs to regulate angiogenesis. K NUP98-HOXA9 fusion proteins attenuate aberrant chromatin organizations. L m6A-modified androgen receptor (AR) mRNA phase separated with YTHDF3 responds to AR pathway inhibition (ARPI) stress in prostate cancers. M LLPS of GIRGL-CAPRIN1-GLS1 mRNA suppresses GLS1 translation and adapts to an adverse glutamine-deficient environment. N icFSP1 induces FSP1 condensates to trigger ferroptosis in the dedifferentiation of cancer cells
LLPS promotes the proliferation of cancer cells
Cancer cells can undergo unrestricted division [20, 133–136], which can occur through gene mutations that activate oncogenic receptor tyrosine kinases (RTKs) and the downstream MAPK signaling involving RAS proteins.
Adaptor proteins involved in RTK and RAS signaling, such as LAT, GRB2 and SOS, undergo phase separation during RTK activation [137]. This phenomenon increases the interaction time between SOS and RTK/RAS, providing a mechanism for kinetic proofreading during RTK activation [125, 138] and preventing the spontaneous membrane localization of SOS, and the downstream activation of RAS. Interestingly, carcinogenic RTK mutations resulting from chromosomal rearrangements cause the loss of membrane localization but not its ability to stimulate downstream pathways. Mechanically, these condensates can assemble the RAS-activating complex GRB2/SOS1, which activates the RAS-MAPK signaling in a membrane-independent manner [123]. Moreover, RTK fusion oncoprotein granules enable the activation of RTK signaling [123, 139]. The close binding to RTK oncoprotein condensates allows GRB2 to concentrate key downstream molecules, achieving the constitutive activation of RAS-MAPK signaling in cancer cells (Fig. 4A). Therefore, BMCs provide a new method for modulating cancer-promoting signaling in a spatially restricted manner.
LLPS promotes the metastasis of tumors
The ability to invade and metastasize allows the tumors to develop distantly, and the epithelial–mesenchymal transition (EMT) programs are commonly involved [140]. Activated by EMT, the transcription coactivators YAP and TAZ facilitate metastasis [141, 142]. Hu et al. found that YAP fusion proteins undergo LLPS in the nucleus and that the IDR provided by the partner of YAP is required for assembly. This aggregation promotes the YAP/TAZ-specific transcriptions and attenuates metastasis [68]. Similarly, another study revealed that the phase separation of DDX21 activates MCM5, thus triggering EMT signaling and modulating the colon cancer metastasis (Fig. 4B)[143]. Besides, SGs are also involved in malignant invasion and metastasis. EMT markers Cadherin, Vimentin, Snail and Slug are suppressed under SG core component G3BP1 depletion, implying the role of G3BP1 in tumor metastasis [144]. Moreover, G3BP modulates mRNA stability under stress conditions and facilitates the invasion of cancer cells [145]. These carcinogenic mechanisms provide new explanations for tumor metastasis, as well as the inspiring ideas for models of cancer progression regulation by the BMCs.
LLPS helps evade tumor growth suppression, regulate the aging process, and achieve replicative immortality of tumor cells
Cancer cells not only promote their growth but also modify tumor-suppression pathways [20]. By inhibiting tumor suppressors such as SPOP, p53, and RB1 [146–148], cancer cells escape intrinsic growth limitations. P53, one of the most well-known tumor suppressors, inhibits tumorigenesis via transcriptional activation, which leads to the disorders of apoptosis, cell cycle, and cell senescence. Tumor-associated stress significantly triggers p53 aggregation [149–154]. These findings demonstrate that the disruption of particular BMCs may cause cancer (Fig. 4C). Further studies are needed to validate this approach with other tumor suppressors and to test its potential applications.
Cellular senescence is considered an anticancer mechanism that maintain homeostasis and is associated with cell cycle arrest. The initiation and maintenance of cellular senescence rely on the frequent damage to the P53/Rb signaling pathway. Increased accumulation of 53BP1 in the nuclear foci after DNA damage can activate p53 and has recently been shown to regulate the cellular senescence via LLPS (Fig. 4D) [155].
Cancer cells can overcome the cell senescence and death via telomerase or alternative methods for lengthening telomeres (ALT) [156–158]. Multivalent interactions between SUMO and SUMO-interacting motifs were observed in the formation of ALT-associated PML bodies on telomeres in cancer stem cells (Fig. 4E) [159]. The fusion of PML bodies enables the clustering of telomere elements and the recruitment of DNA helicases, and other molecular machinery to extend the length of telomeres [160]. This finding suggests that cancer stem cells achieve replicative immortality through the unchecked cell division, and that this process is associated with LLPS.
LLPS modulates epigenetic reprogramming of various BMCs
Common epigenetic modifications include histone modifications, DNA methylation, and RNA interference [161, 162]. Interactions between epigenetic modifications and their corresponding reader proteins also exhibit polyvalent interactions. M6A, known as the most common mRNA modification [163], alters the mRNA structure and interacts with multiple other mRNA modifications and proteins. This modification facilitates YTHDF protein phase separation, further contributing to the forming of various RNA–protein granules, including P bodies and SGs [74, 164]. In addition, YTHDC1 can undergo LLPS in the nucleus by interacting with m6A-modified mRNAs. This interaction results in the formation of nuclear YTHDC1-m6A condensates (nYACs), which are significantly enhanced in acute myeloid leukemia (AML) cells (Fig. 4F) [105].
LLPS helps cancer cells escape immune destruction and participate in tumor-associated inflammation
The immune system employs the RLR-MAVS and cGAS-STING signaling pathways for protection against microbial invasion and support tumor immune surveillance [165–167]. However, tumors often escape immune clearance surveillance. Recent findings by Meng et al. revealed that neurofibromin 2 (NF2) facilitates innate immunity by eliminating tank-binding kinase 1 (TBK1) activation. It is the missense mutations in the FERM domain of NF2 (NF2m) that robustly inhibit the STING-initiated antitumor immunity via the NF2m-IRF3 condensates formations (Fig. 4G), suppressing the TBK1 activation [130]. This offers novel insights into NF2-related cancer treatments.
Notably, inflammation often plays a dual role in cancer. Overproduced in various inflammatory tissues, the reactive oxygen species (ROS) may accelerate the genetic mutations of cells, making them more aggressive and malignant [168]. However, recent research indicates that the PML NB may function as a sensor for ROS in two ways: protecting cancer cells from excessive ROS or promoting ROS-induced apoptosis (Fig. 4H). Given the lack of in-depth research in this field, further tumor microenvironment exploration is required to understand these processes fully.
Tumor-associated viruses, such as human papillomavirus, Kaposi sarcoma herpesvirus, and Epstein–Barr virus (EBV), influence tumor progression through LLPS [169–171]. In EBV proteins such as EBNA2 and EBNALP, LLPS regulates host gene expression, forming biomolecule condensates at Runx3 and MYC SE sites to regulate viral and cellular gene transcription (Fig. 4I). Further, the LLPS of EBNA2 can influence the alternative splicing of the pre-MPPE1 gene in cancer [170].
LLPS induce vasculature of the tumors
Vascularization, also known as angiogenesis, is crucial for supplying tumors with nutrients and oxygen for growth. Vascular endothelial growth factor (VEGF) is the leading factor responsible for rapid nutrient supply. Mounting evidence has indicated a correlation between BMC formation and angiogenesis. For example, the constitutive expression of the transcription factor (TF) MYC in metastasizing cells can lead to VEGF transcription by potentially forming phase-separated transcription condensates, promoting promotes angiogenesis [172]. Similarly, the use of 1,6-hexanediol, an inhibitor of LLPS, has recently been shown to regulate angiogenesis by inhibiting cyclin A1-related endothelial functions as well as condensates with BRD4, indicating that targeting condensates can block critical reactions (Fig. 4J) [173, 174].
Genomic arrangements initiate LLPS
Genomic instability contributes to tumor progression. Genomic translocations and rearrangements can lead to the fusion between the IDR of one protein and the DNA- or chromatin-binding domain of another [175]. This fusion acts as a TF, eliciting LLPS and attracting additional partners to initiate transcriptional programs that ultimately contribute to tumorigenesis. A typical example is the NUP98 fusion oncoprotein (FO), which occurs in 50% of patients with chemotherapy-resistant AML [176–179]. FOs demonstrate that malignancies establish cancerous TF condensates [83, 180, 181] and attenuate aberrant chromatin organization (Fig. 4K).
LLPS of SGs assist in avoiding cell death of cancer cells under the stress
Cancer cells can escape apoptosis by forming SGs (a form of MLOs) when exposed to extreme conditions, such as high temperatures, toxins, mechanical damage, or other stresses. For example, the Y-box binding protein 1 (YB-1) interacts with the 5'-untranslated region (UTR) of G3BP1[182], leading to the increased expression of G3BP1 and SGs, which is elevated in human sarcomas [183–185]. Consequently, these cancer cells survive hyperproliferation, chemotherapy and other various stressful conditions. Additional studies on prostate cancer have demonstrated that the m6A-modified androgen receptor (AR) mRNA phase separated with YTHDF3, while the unmodified AR mRNA phase separated with G3BP1 to survive AR pathway inhibition stress (Fig. 4L)[186]. Collectively, SGs may serve as novel targets for cancer biology investigations.
LLPS regulates cellular metabolisms of cancer cells
Malignant cells undergo metabolic reprogramming [187], thereby attracting considerable interest in tumor-related research in the past decades [188]. For example, the reduction of glutaminase-1 (GLS1) enables cancer cells to survive under prolonged glutamine deprivation stress [189, 190]. Wang et al. reported that the lncRNA GIRGL promotes the LLPS of GIRGL-CAPRIN1-GLS1 mRNA to suppress GLS1 translation, thus adapting to an adverse glutamine-deficient environment (Fig. 4M)[191]. CAPRIN1, an RNA-binding protein involved in the SG formation via LLPS, plays a role in this metabolic adaptation. Therefore, alteration of cell adaptation to an adverse metabolic environment is possible by targeting condensates.
Potential role of LLPS in the phenotypic plasticity of tumorigenesis
Tumor cells often exhibit phenotypic plasticity to evade terminal differentiation. This plasticity includes the dedifferentiation, the differentiation inhibition, and the transdifferentiation [20]. During dedifferentiation, specific malignant cells become sensitized to ferroptosis [192–194], a form of cell death. Nakamura et al. [195] first demonstrated that the novel FSP1 inhibitor, icFSP1 impairs cell proliferation and induces FSP1 condensation to trigger ferroptosis in cancer cells (Fig. 4N). This highlights the role of iron in tumor progression and the dependence of cancer cells on iron in drug-resistant states.
Clinical applications of LLPS in oncologic fields
Potential of LLPS in cancer treatments
Considering that various regulatory mechanisms of LLPS are closely associated with tumorigenesis, it is imperative to explore therapeutic approaches against abnormal LLPS. These strategies can be categorized into three main approaches (Table 4).
Table 4.
Summary of strategies and drugs that use phase separation to intervene in tumorigenesis and progression
| Targeting strategy | Drug/molecules | Tumor types | Associated protein/condensate | Mechanism of action | References |
|---|---|---|---|---|---|
| Disruptions of the BMCs’ formations | IIA4B20, IIA6B17, mycmycin-1/2 | Pan-cancer | Myc | Preventing the Myc/Max dimerization inhibit Myc-induced malignant transformation | [196] |
| YK-4–279 | EWS | EWS-FLI1 fusion | Binding to the IDR of the oncogenic transcription factor EWS-FLI1 and prevents the interaction between EWS-FLI1 and RNA helicase A, thereby slowing down EWS cell growth | [197] | |
| elvitegravir | Lung cancer | SRC1 | Directly binding to the highly disordered SRC1 and effectively inhibit YAP oncogene transfer by disrupting liquid–liquid separation in SRC1/YAP/TEAD condensates | [117] | |
| C108 | Breast cancer | G3BP2 (SG core component) | Diminishing the role of SG core component G3BP2 in breast cancer initiation and improve the efficacy of chemotherapy drugs | [198] | |
| 2142–R8 peptide | Pan-cancer | KAT8–IRF1 condensates | disrupt the formation of KAT8–IRF1 condensates, subsequently suppressing PD-L1 expression and enhancing antitumor immunity in vitro and in vivo | [198] | |
| BAY 249716 | Pan-cancer | p53 | Inducing condensate formation of DNA-binding defective mutants; dissolve nuclear condensates of structural mutants; covalent binders | [199] | |
| BAY 1892005 | |||||
| Avrainvillamide | Aml | NPM1 | Restoring nucleolar localization of cytoplasmic NPM1 mutant; covalent binder | [200] | |
| SHP099 | RTK-driven human cancer | SHP2 | Stabilizing SHP2 in an auto-inhibited conformation and suppressing RAS–ERK signalling to inhibit the proliferation of receptor-tyrosine-kinase-driven human cancer cells | [201] | |
| ET070 | RTK-driven human cancer | SHP2 | Inhibiting the phase separation ability of SHP2 mutants by locking SHP2 in the “off” conformation | [126] | |
| JQ1 | Breast cancer and colon cancer | BET family of bromodomain proteins | Partitions into transcriptional condensates; dissolving MED1 nuclear condensates | [202] | |
| EPI-001 | Prostate cancer | Androgen receptor | Dissolving androgen receptor-rich transcriptional condensates | [203] | |
| Leptomycin B | Leukemia | CRM1 | Inhibiting formation of aberrant NUP98–HOXA9 transcriptional condensates | [204] | |
| Ribavirin | Prostate cancer | OCT4/AR/FOXA1, OCT4/NRF1 | Inhibiting the formation of OCT4-AR axis by modulating OCT4 condensates in the nucleus | [205] | |
| Tin (IV) oxochloride-derived cluster | Pan-cancer | IDR of TAF2 in TFIID | Disrupting transcription initiation by selectively impairing the function of TFIID | [206] | |
| PRIMA-1; ReACp53 | Ovarian carcinoma | p53 mutants | Induction of cell cycle arrest in cancer cells with mutant p53 by restoring the native conformation of aggregated mutant p53 proteins | [207] | |
| PCG | Breast cancer | IDR of BRD4 | Suppression of BRD4-dependent gene transcription | [208] | |
| bis-ANS | Colon cancer | LCD of TDP-43 | high concentrations of bis-ANS inhibit TDP-43 condensate assembly, whereas low concentrations facilitate the formation of liquid droplets | [209] | |
| Modifications of PTMs and physicochemical conditions | SI-2 | Multiple myeloma | SRC3/NSD2 condensate | Phase separation of SRC3 mediated by histone methyltransferase NSD2 leads to resistance to bleitinib in multiple myeloma, whereas the inhibitor SI-2 Inhibits formation of drug-resistant SRC3/NSD2 condensates and improves the therapeutic efficacy of bleitinib | [210] |
| Olaparib | Pan-cancer | PARP1/2 DNA repair condensate | Inhibiting PARP1/2 and thus interferes with the formation of PARylation related DNA repair condensates | [211] | |
| GSK-J4 | Osteosarcoma | HOXB8/FOSL1 CRC | The H3K27 demethylase inhibitor GSK-J4, inhibits the CRC phase separation and results in metastasis suppression and re-sensitivity to chemotherapy drugs | [212] | |
| icFSP1 | Melanoma and lung cancer | hFSP1 | Inducing phase separation of myristoylated hFSP1, thus promoting ferroptosis and inhibit tumor proliferations | [195] | |
| GSK-626616 | Pan-cancer | DYRK3 |
Inhibit PRAS40 phosphorylation and restrain mTORC1 signaling in SGs |
[213] | |
| JQ1 | AML | BRD4 | Release the Mediator complex from SEs | [214] | |
| SGC0946 | MLL leukemia | DOT1L | Inhibit histone H3K79 methylation and histone H4 acetylation | [215] | |
| THZ1 | Pan-cancer | CDK7 | Inhibit RNAPII phosphorylation | [216] | |
| THZ531 | Pan-cancer | CDK12 and CDK13 | Inhibit RNAPII phosphorylation | [217] | |
| Drug interventions and distributions of the dynamics of condensates | Cisplatin | Breast cancer and colon cancer | MED1 transcriptional condensates | Partitions into transcriptional condensates; dissolves MED1 and BRD4 nuclear condensates | [202] |
| Tamoxifen | breast cancer and colon cancer | Estrogen receptor | Seletively partitions into transcriptional condensates | [202] | |
| mitoxantrone | breast cancer and colon cancer | Estrogen receptor | Seletively partitions into transcriptional condensates | [202] | |
| PML-retinoic acid receptor α | APL | PML bodies | Hindering the assembly of PML bodies and result in the suppression of differentiation genes. Successful APL treatment involves the restoration of PML nuclear bodies using empirically discovered drugs | [109] |
BMC, biomolecular condensate; EWS, Ewing sarcoma; IDR, intrinsically disordered region; SG, stress granule, RTK, receptor-tyrosine-kinase; CRC, core regulatory circuitry; MLL, mixed lineage leukemia; PML, Promyelocytic leukemia protein; APL, acute promyelocytic leukemia
Disruptions of the formation of BMCs
The direct disruption of the driving force behind LLPS offers a straightforward approach (Fig. 5A). For example, certain drugs can intervene in the LLPS process by targeting IDRs of proteins. Notably, the anti-HIV drug elvitegravir directly binds to the highly disordered steroid receptor coactivator 1, effectively inhibiting oncogene YAP transcription by disrupting SRC1/YAP/TEAD condensates (Table 4) [117]. Similarly, Yu et al. reported that the nuclear translocation of YAP and LLPS are affected by IFN treatment in cancer cells. Therefore, interrupting the LLPS of YAP can inhibit cancer cell proliferation and enhance the immune response, indicating its potential as a predictive biomarker in immune checkpoint blockade [67]. Further, altering interactions between LCDs indirectly modulates the transcriptional subunits, thus offering a promising approach for targeting disease-causing TFs.
Fig. 5.
Potential approaches to developing new cancer treatments by regulating BMCs. A Targeting driving forces to disrupt condensate formation. B Changing the modifications of components or physicochemical interaction. C Drug concentrations influenced by dynamic condensates
Modifications of PTMs and physicochemical conditions
As previously mentioned, certain post-transitional modifications and physiochemical conditions contribute to LLP dynamics (Fig. 5B). For example, nYACs protect mRNAs from degradation and strengthen the role of YTHDC1 in leukemogenesis, which inspires us to disrupt m6A to violate deleterious condensates[105]. Further, studies have reported that modulating PTMs in LLPS proteins is also significant [25, 96, 102, 218–221]. In the case of colon cancer, SENP1 has been reported to decrease RNF168 SUMOylation, inhibit nuclear condensate formation, and promote DNA damage repair (DDR) and drug resistance. Given these observations, strategies to curb the harmful effects of protein aggregation by influencing protein modifications warrant further investigation.
Drug interventions of the dynamics of condensates
Drugs can significantly influence the dynamics of the condensates, affecting their anticancer effects and potentially leading to drug resistance (Fig. 5C). For example, in luminal breast cancer, tamoxifen accumulates in MED1 condensates, preventing the incorporation of ERα into these condensates, partially inhibiting cancer progression. However, when MED1 is overexpressed, larger condensates dilute the drug concentration, ultimately leading to the development of resistance [202]. Several drugs, such as cisplatin, mitoxantrone, and THZ1, selectively partition into BMCs formed by MED1 (Table 4). Drug resistance can occur via selective partitioning into BMCs or changes in properties. Notably, cisplatin exerts its anticancer activity by dissolving SEs, indicating that changes in the condensate properties may improve therapeutic outcomes[202]. This finding highlights the potential of altering the properties of condensates to improve therapeutic outcomes. In some cases, promoting the formation of BMCs may have therapeutic effects. For example, in APL, fusion proteins of PML-retinoic acid receptor α (RARA) hinder the assembly of PML bodies and result in the suppression of differentiation genes. Successful APL treatment involves the restoration of PML nuclear bodies using empirically discovered drugs (Table 4) [222].
Roles of LLPS in vesicular trafficking and drugs’ delivery
Although LLPS and traditional vesicles are two different concepts with distinctive definitions, the vesicular trafficking role of LLPS is still rarely described and attractive. Conventional approaches typically utilize nanoscale carriers that are confined within the compartments of the intranuclear body. Nevertheless, recent findings have demonstrated that micron-scale polypeptide clusters, formed through phase separation, possess the ability to traverse the cell membrane via a non-canonical endocytic pathway. These clusters undergo glutathione-induced release of their cargo and exhibit the capacity to rapidly incorporate various macromolecules into microdroplets, such as RNA, small peptides and enzymes [223]. Loaded with polysomes, they can provide new approaches for vaccine carriers based on mRNAs and intracellular transportations for cancer treatments.
Likewise, as previously mentioned, droplets of drugs formed by LLPS can unexpectedly raise the inner drug concentration up to 600 times higher than that outside the condensate [202]. Furthermore, MED1 predominantly acts on oncogene promoters, thereby enabling cisplatin to ultimately target the corresponding DNA through its toxic platinum atoms, effectively assaulting the vital components of the cancer cells. Besides, the phosphopeptide KYp has been observed to induce LLPS level at the cell membrane, thus enhancing the permeation and internalization of the peptide drug [224]. KYp has the ability to interact with alkaline phosphatase, resulting in the dephosphorylation and in situ self-assembly at the cell membrane [224]. The process induces the aggregation of alkaline phosphatase and the separation of proteolipid phases at the membrane, ultimately enhancing membrane leakage and facilitating the entry of the peptide drug. These great discoveries provide inspirations for designing drug delivery systems and more similar ideas are worth exploring.
Conclusions and future perspectives
In the past decades, crucial advances have been made in figuring out the role of LLPS in a variety of cellular processes and biological functions. Since the update of the new version of “Hallmarks of cancer 2022”, cancer hallmarks and their enabling characteristics help distill the oncogenic complexity into an evidently logical science, which have been gradually proven to be closely associated with LLPS. In this review, we summarize the mechanism of LLPS formations, recent discoveries and the individual role of LLPS in oncology. These findings collectively reveal its vital role in solving undruggable targets and multiple clinical problems.
The role of specific proteins and post-translational mechanisms in the formation and regulation of LLPS are being investigated. These efforts aim to identify abnormal conditions and gain insights into the mechanisms regulating the formation of the condensates. These studies have already begun to help find new strategies for targeting disease-related condensates. Notably, while previous drugs were designed to inhibit each protein directly, LLPS offers a novel and unexpected possibility of interfering with the pathological process and does not necessitate targeting each protein individually. This approach achieves disruption of the condensates formed by IDRs of TFs.
Despite the steady progress in targeting BMCs using LLPS, several fundamental questions need to be answered. For example, what are the functional differences between LLPS-formed assemblies and typical protein complexes? What factors contribute to dynamic condensation and decondensation, and how do different BMCs communicate in vitro and in vivo? Moreover, the role of PTMs in tumorigenesis requires further exploration (Table 5). Clarifying these aspects will improve our understanding of the conversion of physiological into pathological condensates in cancer. Future research will require collaborative efforts, innovative approaches, and a holistic approach to studying cancer-associated LLPS, which may lead to novel anti-tumor therapies directly targeting BMCs.
Table 5.
Outlook and reflection on the future of the field
| Critical issues in the current development of oncogenic LLPS | Outlook and reflection on the future/ possible solutions to the questions |
|---|---|
|
What are the functional differences between LLPS-formed assemblies and typical protein complexes? What factors contribute to dynamic condensation and decondensation, and how do different BMCs communicate in vitro and in vivo? |
The target protein molecules and signaling pathways discovered through LLPS method are a class of molecules that can form condensates spontaneously due to their own unique properties or under different environmental conditions. LLPS is essentially an energy saving process in the organisms. Further functional differences between LLPS‐formed assemblies and canonical protein complexes deserve investigations |
| Is there other function of PTMs in tumorigenesis and tumor progressions? | Further studies on phase separation on the basis of proteomics and PTMs are needed |
| Detections of BMCs/ MLOs in tumor samples and clinicopathologic associations with cancer patients are deficient | Clinicopathologic tests should be involved in further studies |
| How do environment conditions inducing condensate assemblies being applied to clinical practice? | Perhaps changing the environment conditions can dynamically alter the condensation and decondensation of the BMC, which will make sense in drug deliveries. A greater understanding of the opportunities for targeting LLPS condensates in the pharmaceutical intervention should be obtained |
| Is there any new convenient method to probe and control (induce, dissolve, or tune) the endogenous condensates? | The partitioning of anticancer drugs in subcellular condensates is also dominant for drug efficacy. According to this characteristic, we can detect the distribution of drugs in cells or by linking drugs to molecules that can specifically aggregate in liquid droplets |
| How to make use of LLPS to enhance the efficiency of drugs in clinical practice? |
Acknowledgements
The figures are created with Biorender.com.
Abbreviations
- ALT
Alternative lengthening of telomeres
- ALK
Anaplastic lymphoma kinase
- AML
Acute myeloid leukemia
- AR
Androgen receptor
- AS
Alternatively spliced
- APL
Acute promyelocytic leukemia
- BMCs
Biomolecular condensates
- DDR
DNA damage repair
- EBV
Epstein–Barr virus
- EMT
Epithelial–mesenchymal transition
- FO
Fusion oncoproteins
- GLS1
Glutaminase-1
- IDR
Intrinsically disordered regions
- LCDs
Low-complexity domains
- LLPS
Liquid-liquid phase separation
- MLOs
Membrane-less organelles
- NB
Nuclear body
- NF2
Neurofibromin 2
- N-WASP
Neural Wiskott—Aldrich Syndrome
- PRM
Proline-rich motif
- PTM
Post-translational modification
- RARA
Retinoic acid receptor α
- RBPs
RNA-binding proteins
- ROS
Reactive oxygen species
- RTKs
Receptor tyrosine kinases
- SEs
Super-enhancers
- SERBP1
SERPINE1 mRNA-binding protein 1
- SG
Stress granule
- SPOP
Speckled POZ protein
- TF
Transcription factor
- UTR
Untranslated region
- VEGF
Vascular endothelial growth factor
Author contributions
L-WZ conceived and drafted the manuscript and drew the figures. K-DY and C–CL discussed the concepts and provided valuable suggestions and revised the manuscript.
Funding
The authors acknowledge funding from the National Natural Science Foundation of China (grant number: 82325042, 82203860, 82072916), and National Key R&D Program of China (grant number: 2023YFC3404100, 2023YFC2506400), and Shanghai Municipal Education Commission Scientific Research Innovation Project (grant number: 2023-05-50), Wu Jieping Medical Foundation Research Project (grant number: 32067502023-18-29).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing Interests
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA controls PolyQ protein phase transitions. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pederson T The nucleolus, Cold Spring Harb Perspect Biol, 3 (2011) [DOI] [PMC free article] [PubMed]
- 3.Shimobayashi SF, Ronceray P, Sanders DW, Haataja MP, Brangwynne CP. Nucleation landscape of biomolecular condensates. Nature. 2021;599:503–506. doi: 10.1038/s41586-021-03905-5. [DOI] [PubMed] [Google Scholar]
- 4.Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021;22:165–182. doi: 10.1038/s41580-020-0272-6. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. Elife. 2016;5:e18413. doi: 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M, Tao X, Jacob MD, Bennett CA, Ho JJD, Gonzalgo ML, Audas TE, Lee S. Stress-induced low complexity RNA activates physiological amyloidogenesis. Cell Rep. 2018;24:1713–1721.e1714. doi: 10.1016/j.celrep.2018.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gui X, Luo F, Li Y, Zhou H, Qin Z, Liu Z, Gu J, Xie M, Zhao K, Dai B, Shin WS, He J, He L, Jiang L, Zhao M, Sun B, Li X, Liu C, Li D. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun. 2019;10:2006. doi: 10.1038/s41467-019-09902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Wang T, Shi Y, Bai L, Wang S, Guo D, Zhang Y, Qi Y, Chen C, Zhang J, Zhang Y, Liu Q, Yang Q, Wang Y, Liu H. USP42 drives nuclear speckle mRNA splicing via directing dynamic phase separation to promote tumorigenesis. Cell Death Differ. 2021;28:2482–2498. doi: 10.1038/s41418-021-00763-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao SE, Regev O. Splicing at the phase-separated nuclear speckle interface: a model. Nucleic Acids Res. 2021;49:636–645. doi: 10.1093/nar/gkaa1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall'Agnese A, Hannett NM, Spille JH, Afeyan LK, Zamudio AV, Shrinivas K, Abraham BJ, Boija A, Decker TM, Rimel JK, Fant CB, Lee TI, Cisse II, Sharp PA, Taatjes DJ, Young RA. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543–548. doi: 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 13.Xiao Q, McAtee CK, Su X. Phase separation in immune signalling. Nat Rev Immunol. 2022;22:188–199. doi: 10.1038/s41577-021-00572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 15.Roden C, Gladfelter AS. RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol. 2021;22:183–195. doi: 10.1038/s41580-020-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langdon EM, Gladfelter AS. A new lens for RNA localization: liquid-liquid phase separation. Annu Rev Microbiol. 2018;72:255–271. doi: 10.1146/annurev-micro-090817-062814. [DOI] [PubMed] [Google Scholar]
- 17.Alberti S. Phase separation in biology. Curr Biol. 2017;27:R1097–r1102. doi: 10.1016/j.cub.2017.08.069. [DOI] [PubMed] [Google Scholar]
- 18.Wang B, Zhang L, Dai T, Qin Z, Lu H, Zhang L, Zhou F. Liquid-liquid phase separation in human health and diseases. Signal Transduct Target Ther. 2021;6:290. doi: 10.1038/s41392-021-00678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong X, Tang R, Xu J, Wang W, Zhao Y, Yu X, Shi S. Liquid-liquid phase separation in tumor biology. Signal Transduct Target Ther. 2022;7:221. doi: 10.1038/s41392-022-01076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 21.Mehta S, Zhang J. Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat Rev Cancer. 2022;22:239–252. doi: 10.1038/s41568-022-00444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nozawa RS, Yamamoto T, Takahashi M, Tachiwana H, Maruyama R, Hirota T, Saitoh N. Nuclear microenvironment in cancer: control through liquid-liquid phase separation. Cancer Sci. 2020;111:3155–3163. doi: 10.1111/cas.14551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uversky VN. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 24.Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, Fuxreiter M. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boija A, Klein IA, Young RA. Biomolecular condensates and cancer. Cancer Cell. 2021;39:174–192. doi: 10.1016/j.ccell.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 27.Iglesias V, Santos J, Santos-Suárez J, Pintado-Grima C, Ventura S. SGnn: a web server for the prediction of prion-like domains recruitment to stress granules upon heat stress. Front Mol Biosci. 2021;8:718301. doi: 10.3389/fmolb.2021.718301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou C, Wang X, Xie H, Chen T, Zhu P, Xu X, You K, Li T. PhaSepDB in 2022: annotating phase separation-related proteins with droplet states, co-phase separation partners and other experimental information. Nucleic Acids Res. 2022;51(2023):D460–d465. doi: 10.1093/nar/gkac783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztányi Z, Uversky VN, Obradovic Z, Kurgan L, Dunker AK, Gough J. D2P2: database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lancaster AK, Nutter-Upham A, Lindquist S, King OD. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014;30:2501–2502. doi: 10.1093/bioinformatics/btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning W, Guo Y, Lin S, Mei B, Wu Y, Jiang P, Tan X, Zhang W, Chen G, Peng D, Chu L, Xue Y. DrLLPS: a data resource of liquid-liquid phase separation in eukaryotes. Nucleic Acids Res. 2020;48:D288–d295. doi: 10.1093/nar/gkz1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mészáros B, Erdős G, Szabó B, Schád É, Tantos Á, Abukhairan R, Horváth T, Murvai N, Kovács OP, Kovács M, Tosatto SCE, Tompa P, Dosztányi Z, Pancsa R. PhaSePro: the database of proteins driving liquid-liquid phase separation. Nucleic Acids Res. 2020;48:D360–d367. doi: 10.1093/nar/gkz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oughtred R, Rust J, Chang C, Breitkreutz BJ, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, Dolma S, Coulombe-Huntington J, Chatr-Aryamontri A, Dolinski K, Tyers M. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Peng X, Li Y, Tang W, Zhu J, Huang J, Qi Y, Zhang Z. LLPSDB: a database of proteins undergoing liquid-liquid phase separation in vitro. Nucleic Acids Res. 2020;48:D320–d327. doi: 10.1093/nar/gkz778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Go CD, Knight JDR, Rajasekharan A, Rathod B, Hesketh GG, Abe KT, Youn JY, Samavarchi-Tehrani P, Zhang H, Zhu LY, Popiel E, Lambert JP, Coyaud É, Cheung SWT, Rajendran D, Wong CJ, Antonicka H, Pelletier L, Palazzo AF, Shoubridge EA, Raught B, Gingras AC. A proximity-dependent biotinylation map of a human cell. Nature. 2021;595:120–124. doi: 10.1038/s41586-021-03592-2. [DOI] [PubMed] [Google Scholar]
- 36.Orti F, Fernández ML, Marino-Buslje C MLOsMetaDB, a meta-database to centralize the information on Liquid-liquid phase separation proteins and Membraneless organelles, bioRxiv, (2023) [DOI] [PMC free article] [PubMed]
- 37.Bolognesi B, Lorenzo Gotor N, Dhar R, Cirillo D, Baldrighi M, Tartaglia GG, Lehner B. A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 2016;16:222–231. doi: 10.1016/j.celrep.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, Lin H, Forman-Kay JD. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife. 2018;7:e31486. doi: 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu M, Li H, Luo X, Cai J, Chen T, Xie Y, Ren J, Zuo Z. RPS: a comprehensive database of RNAs involved in liquid-liquid phase separation. Nucleic Acids Res. 2022;50:D347–d355. doi: 10.1093/nar/gkab986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu H, Fu H, Cui T, Ning L, Shao H, Guo Y, Ke Y, Zheng J, Lin H, Wu X, Liu G, He J, Han X, Li W, Zhao X, Lu H, Wang D, Hu K, Shen X. RNAPhaSep: a resource of RNAs undergoing phase separation. Nucleic Acids Res. 2022;50:D340–d346. doi: 10.1093/nar/gkab985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Youn JY, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman-Kay JD, Gingras AC. Properties of stress granule and P-body proteomes. Mol Cell. 2019;76:286–294. doi: 10.1016/j.molcel.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Navarro AM, Orti F, Martínez-Pérez E, Alonso M, Simonetti FL, Iserte JA, Marino-Buslje C. DisPhaseDB: an integrative database of diseases related variations in liquid-liquid phase separation proteins. Comput Struct Biotechnol J. 2022;20:2551–2557. doi: 10.1016/j.csbj.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mészáros B, Erdos G, Dosztányi Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46:W329–w337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 1804;2010:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piovesan D, Del Conte A, Clementel D, Monzon AM, Bevilacqua M, Aspromonte MC, Iserte JA, Orti FE, Marino-Buslje C, Tosatto SCE. MobiDB: 10 years of intrinsically disordered proteins. Nucleic Acids Res. 2023;51:D438–d444. doi: 10.1093/nar/gkac1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holehouse AS, Das RK, Ahad JN, Richardson MO, Pappu RV. CIDER: resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys J. 2017;112:16–21. doi: 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson MJ, Sievers SA, Karanicolas J, Ivanova MI, Baker D, Eisenberg D. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci U S A. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozlowski LP, Bujnicki JM. MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics. 2012;13:111. doi: 10.1186/1471-2105-13-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang YJ, Acton TB, Montelione GT. DisMeta: a meta server for construct design and optimization. Methods Mol Biol. 2014;1091:3–16. doi: 10.1007/978-1-62703-691-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duvaud S, Gabella C, Lisacek F, Stockinger H, Ioannidis V, Durinx C. Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 2021;49:W216–w227. doi: 10.1093/nar/gkab225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nastou KC, Nasi GI, Tsiolaki PL, Litou ZI, Iconomidou VA. AmyCo: the amyloidoses collection. Amyloid. 2019;26:112–117. doi: 10.1080/13506129.2019.1603143. [DOI] [PubMed] [Google Scholar]
- 52.Mizianty MJ, Uversky V, Kurgan L. Prediction of intrinsic disorder in proteins using MFDp2. Methods Mol Biol. 2014;1137:147–162. doi: 10.1007/978-1-4939-0366-5_11. [DOI] [PubMed] [Google Scholar]
- 53.Case LB, Zhang X, Ditlev JA, Rosen MK. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science. 2019;363:1093–1097. doi: 10.1126/science.aau6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, Russo PS, Jiang QX, Nixon BT, Rosen MK. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, Tjian R. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361:2555. doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD, Narlikar GJ. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature. 2019;575:390–394. doi: 10.1038/s41586-019-1669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30:137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- 58.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 59.Pessina F, Gioia U, Brandi O, Farina S, Ceccon M, Francia S, d'Adda di Fagagna F. DNA damage triggers a new phase in neurodegeneration. Trends Genet. 2021;37:337–354. doi: 10.1016/j.tig.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 60.Borcherds W, Bremer A, Borgia MB, Mittag T. How do intrinsically disordered protein regions encode a driving force for liquid-liquid phase separation? Curr Opin Struct Biol. 2021;67:41–50. doi: 10.1016/j.sbi.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin Y, Currie SL, Rosen MK. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J Biol Chem. 2017;292:19110–19120. doi: 10.1074/jbc.M117.800466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, Pappu RV, Alberti S, Hyman AA. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.e616. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hughes MP, Goldschmidt L, Eisenberg DS. Prevalence and species distribution of the low-complexity, amyloid-like, reversible, kinked segment structural motif in amyloid-like fibrils. J Biol Chem. 2021;297:101194. doi: 10.1016/j.jbc.2021.101194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J, Zhang Y, Chen X, Ma L, Li P, Yu H. Protein phase separation and its role in chromatin organization and diseases. Biomed Pharmacother. 2021;138:111520. doi: 10.1016/j.biopha.2021.111520. [DOI] [PubMed] [Google Scholar]
- 65.Hughes MP, Sawaya MR, Boyer DR, Goldschmidt L, Rodriguez JA, Cascio D, Chong L, Gonen T, Eisenberg DS. Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science. 2018;359:698–701. doi: 10.1126/science.aan6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barrio M, Huguet J, Robert B, Rietveld IB, Céolin R, Tamarit JL. Pressure-temperature phase diagram of the dimorphism of the anti-inflammatory drug nimesulide. Int J Pharm. 2017;525:54–59. doi: 10.1016/j.ijpharm.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 67.Yu M, Peng Z, Qin M, Liu Y, Wang J, Zhang C, Lin J, Dong T, Wang L, Li S, Yang Y, Xu S, Guo W, Zhang X, Shi M, Peng H, Luo X, Zhang H, Zhang L, Li Y, Yang XP, Sun S. Interferon-γ induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol Cell. 2021;81:1216–1230.e1219. doi: 10.1016/j.molcel.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 68.Lu Y, Wu T, Gutman O, Lu H, Zhou Q, Henis YI, Luo K. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat Cell Biol. 2020;22:453–464. doi: 10.1038/s41556-020-0485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conicella AE, Zerze GH, Mittal J, Fawzi NL. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin Y, Fichou Y, Longhini AP, Llanes LC, Yin P, Bazan GC, Kosik KS, Han S. Liquid-liquid phase separation of tau driven by hydrophobic interaction facilitates fibrillization of tau. J Mol Biol. 2021;433:166731. doi: 10.1016/j.jmb.2020.166731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vieregg JR, Lueckheide M, Marciel AB, Leon L, Bologna AJ, Rivera JR, Tirrell MV. Oligonucleotide-peptide complexes: phase control by hybridization. J Am Chem Soc. 2018;140:1632–1638. doi: 10.1021/jacs.7b03567. [DOI] [PubMed] [Google Scholar]
- 72.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baudin A, Moreno-Romero AK, Xu X, Selig EE, Penalva LOF, Libich DS. Structural characterization of the RNA-binding protein SERBP1 reveals intrinsic disorder and atypical RNA binding modes. Front Mol Biosci. 2021;8:744707. doi: 10.3389/fmolb.2021.744707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, Jaffrey SR. m(6)A enhances the phase separation potential of mRNA. Nature. 2019;571:424–428. doi: 10.1038/s41586-019-1374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fox AH, Nakagawa S, Hirose T, Bond CS. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem Sci. 2018;43:124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 77.Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 78.Bienz M. Head-to-tail polymerization in the assembly of biomolecular condensates. Cell. 2020;182:799–811. doi: 10.1016/j.cell.2020.07.037. [DOI] [PubMed] [Google Scholar]
- 79.Bienz M. Signalosome assembly by domains undergoing dynamic head-to-tail polymerization. Trends Biochem Sci. 2014;39:487–495. doi: 10.1016/j.tibs.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 80.Schwarz-Romond T, Merrifield C, Nichols BJ, Bienz M. The Wnt signalling effector Dishevelled forms dynamic protein assemblies rather than stable associations with cytoplasmic vesicles. J Cell Sci. 2005;118:5269–5277. doi: 10.1242/jcs.02646. [DOI] [PubMed] [Google Scholar]
- 81.Gammons M, Bienz M. Multiprotein complexes governing Wnt signal transduction. Curr Opin Cell Biol. 2018;51:42–49. doi: 10.1016/j.ceb.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 82.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 83.Chandra B, Michmerhuizen NL, Shirnekhi HK, Tripathi S, Pioso BJ, Baggett DW, Mitrea DM, Iacobucci I, White MR, Chen J, Park CG, Wu H, Pounds S, Medyukhina A, Khairy K, Gao Q, Qu C, Abdelhamed S, Gorman SD, Bawa S, Maslanka C, Kinger S, Dogra P, Ferrolino MC, Di Giacomo D, Mecucci C, Klco JM, Mullighan CG, Kriwacki RW. Phase separation mediates NUP98 fusion oncoprotein leukemic transformation. Cancer Discov. 2022;12:1152–1169. doi: 10.1158/2159-8290.CD-21-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qin Z, Sun H, Yue M, Pan X, Chen L, Feng X, Yan X, Zhu X, Ji H. Phase separation of EML4-ALK in firing downstream signaling and promoting lung tumorigenesis. Cell Discov. 2021;7:33. doi: 10.1038/s41421-021-00270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fawal M, Espinos E, Jean-Jean O, Morello D. Looking for the functions of RNA granules in ALK-transformed cells. BioArchitecture. 2011;1:91–95. doi: 10.4161/bioa.1.2.16269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weatheritt RJ, Luck K, Petsalaki E, Davey NE, Gibson TJ. The identification of short linear motif-mediated interfaces within the human interactome. Bioinformatics. 2012;28:976–982. doi: 10.1093/bioinformatics/bts072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tompa P, Fuxreiter M, Oldfield CJ, Simon I, Dunker AK, Uversky VN. Close encounters of the third kind: disordered domains and the interactions of proteins. BioEssays. 2009;31:328–335. doi: 10.1002/bies.200800151. [DOI] [PubMed] [Google Scholar]
- 88.Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hancock R. A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. J Struct Biol. 2004;146:281–290. doi: 10.1016/j.jsb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 90.Weber SC, Brangwynne CP. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr Biol. 2015;25:641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nüske E, Richter D, Baumeister W, Grill SW, Pappu RV, Hyman AA, Alberti S. Phase separation of a yeast prion protein promotes cellular fitness. Science. 2018;359:5654. doi: 10.1126/science.aao5654. [DOI] [PubMed] [Google Scholar]
- 92.Wang Y, Annunziata O. Comparison between protein-polyethylene glycol (PEG) interactions and the effect of PEG on protein-protein interactions using the liquid-liquid phase transition. J Phys Chem B. 2007;111:1222–1230. doi: 10.1021/jp065608u. [DOI] [PubMed] [Google Scholar]
- 93.Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamaguchi A, Kitajo K. The effect of PRMT1-mediated arginine methylation on the subcellular localization, stress granules, and detergent-insoluble aggregates of FUS/TLS. PLoS ONE. 2012;7:e49267. doi: 10.1371/journal.pone.0049267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gal J, Zhang J, Kwinter DM, Zhai J, Jia H, Jia J, Zhu H. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging. 2011;32(2323):e2327–2340. doi: 10.1016/j.neurobiolaging.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hofweber M, Dormann D. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem. 2019;294:7137–7150. doi: 10.1074/jbc.TM118.001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tanikawa C, Ueda K, Suzuki A, Iida A, Nakamura R, Atsuta N, Tohnai G, Sobue G, Saichi N, Momozawa Y, Kamatani Y, Kubo M, Yamamoto K, Nakamura Y, Matsuda K. Citrullination of RGG motifs in FET proteins by PAD4 regulates protein aggregation and ALS susceptibility. Cell Rep. 2018;22:1473–1483. doi: 10.1016/j.celrep.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 98.Bah A, Forman-Kay JD. Modulation of intrinsically disordered protein function by post-translational modifications. J Biol Chem. 2016;291:6696–6705. doi: 10.1074/jbc.R115.695056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal AC, Nielsen ML. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol. 2017;24:325–336. doi: 10.1038/nsmb.3366. [DOI] [PubMed] [Google Scholar]
- 101.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, 3rd, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xia Q, Li Y, Xu W, Wu C, Zheng H, Liu L, Dong L. Enhanced liquidity of p62 droplets mediated by Smurf1 links Nrf2 activation and autophagy. Cell Biosci. 2023;13:37. doi: 10.1186/s13578-023-00978-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N, Granata D, Marzahn MR, Lindorff-Larsen K, Salvatella X, Schulman BA, Mittag T. Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol Cell. 2018;72:19–36.e18. doi: 10.1016/j.molcel.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheng Y, Xie W, Pickering BF, Chu KL, Savino AM, Yang X, Luo H, Nguyen DT, Mo S, Barin E, Velleca A, Rohwetter TM, Patel DJ, Jaffrey SR, Kharas MG. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell. 2021;39:958–972.e958. doi: 10.1016/j.ccell.2021.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shi B, Li W, Song Y, Wang Z, Ju R, Ulman A, Hu J, Palomba F, Zhao Y, Le JP, Jarrard W, Dimoff D, Digman MA, Gratton E, Zang C, Jiang H. UTX condensation underlies its tumour-suppressive activity. Nature. 2021;597:726–731. doi: 10.1038/s41586-021-03903-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wei M, Huang X, Liao L, Tian Y, Zheng X. SENP1 decreases RNF168 phase separation to promote DNA damage repair and drug resistance in colon cancer. Cancer Res. 2023;83(17):2908–2923. doi: 10.1158/0008-5472.CAN-22-4017. [DOI] [PubMed] [Google Scholar]
- 108.Wu Y, Zhou L, Zou Y, Zhang Y, Zhang M, Xu L, Zheng L, He W, Yu K, Li T, Zhang X, Chen Z, Zhang R, Zhou P, Zhang N, Zheng L, Kang T. Disrupting the phase separation of KAT8-IRF1 diminishes PD-L1 expression and promotes antitumor immunity. Nat Cancer. 2023;4:382–400. doi: 10.1038/s43018-023-00522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shao X, Chen Y, Xu A, Xiang D, Wang W, Du W, Huang Y, Zhang X, Cai M, Xia Z, Wang Y, Cao J, Zhang Y, Yang B, He Q, Ying M. Deneddylation of PML/RARα reconstructs functional PML nuclear bodies via orchestrating phase separation to eradicate APL. Cell Death Differ. 2022;29:1654–1668. doi: 10.1038/s41418-022-00955-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Martinelli E, Morgillo F, Troiani T, Ciardiello F. Cancer resistance to therapies against the EGFR-RAS-RAF pathway: the role of MEK. Cancer Treat Rev. 2017;53:61–69. doi: 10.1016/j.ctrv.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 111.Liang SI, van Lengerich B, Eichel K, Cha M, Patterson DM, Yoon TY, von Zastrow M, Jura N, Gartner ZJ. Phosphorylated EGFR dimers are not sufficient to activate ras. Cell Rep. 2018;22:2593–2600. doi: 10.1016/j.celrep.2018.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grabarz A, Barascu A, Guirouilh-Barbat J, Lopez BS. Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am J Cancer Res. 2012;2:249–268. [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang J, Zeng Y, Xing Y, Li X, Zhou L, Hu L, Chin YE, Wu M. Myristoylation-mediated phase separation of EZH2 compartmentalizes STAT3 to promote lung cancer growth. Cancer Lett. 2021;516:84–98. doi: 10.1016/j.canlet.2021.05.035. [DOI] [PubMed] [Google Scholar]
- 114.Lanz MC, Dibitetto D, Smolka MB. DNA damage kinase signaling: checkpoint and repair at 30 years. Embo j. 2019;38:e101801. doi: 10.15252/embj.2019101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li W, Shu X, Zhang X, Zhang Z, Sun S, Li N, Long M. Potential Roles of YAP/TAZ mechanotransduction in spaceflight-induced liver dysfunction. Int J Mol Sci. 2023;24(3):2197. doi: 10.3390/ijms24032197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu Q, Li J, Zhang W, Xiao C, Zhang S, Nian C, Li J, Su D, Chen L, Zhao Q, Shao H, Zhao H, Chen Q, Li Y, Geng J, Hong L, Lin S, Wu Q, Deng X, Ke R, Ding J, Johnson RL, Liu X, Chen L, Zhou D. Glycogen accumulation and phase separation drives liver tumor initiation. Cell. 2021;184:5559–5576.e5519. doi: 10.1016/j.cell.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 117.Zhu G, Xie J, Fu Z, Wang M, Zhang Q, He H, Chen Z, Guo X, Zhu J. Pharmacological inhibition of SRC-1 phase separation suppresses YAP oncogenic transcription activity. Cell Res. 2021;31:1028–1031. doi: 10.1038/s41422-021-00504-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kamagata K, Kanbayashi S, Honda M, Itoh Y, Takahashi H, Kameda T, Nagatsugi F, Takahashi S. Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains. Sci Rep. 2020;10:580. doi: 10.1038/s41598-020-57521-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. Embo j. 2019;38:e101379. doi: 10.15252/embj.2018101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Esposito M, Fang C, Cook KC, Park N, Wei Y, Spadazzi C, Bracha D, Gunaratna RT, Laevsky G, DeCoste CJ, Slabodkin H, Brangwynne CP, Cristea IM, Kang Y. TGF-β-induced DACT1 biomolecular condensates repress Wnt signalling to promote bone metastasis. Nat Cell Biol. 2021;23:257–267. doi: 10.1038/s41556-021-00641-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zamudio AV, Dall'Agnese A, Henninger JE, Manteiga JC, Afeyan LK, Hannett NM, Coffey EL, Li CH, Oksuz O, Sabari BR, Boija A, Klein IA, Hawken SW, Spille JH, Decker TM, Cisse BJ, II, Abraham TI, Lee DJ, Taatjes J, Schuijers RA. Young, mediator condensates localize signaling factors to key cell identity genes. Mol Cell. 2019;76:753–766. doi: 10.1016/j.molcel.2019.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang JZ, Lu TW, Stolerman LM, Tenner B, Yang JR, Zhang JF, Falcke M, Rangamani P, Taylor SS, Mehta S, Zhang J. Phase Separation of a PKA regulatory subunit controls camp compartmentation and oncogenic signaling. Cell. 2020;182:1531–1544.e1515. doi: 10.1016/j.cell.2020.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tulpule A, Guan J, Neel DS, Allegakoen HR, Lin YP, Brown D, Chou YT, Heslin A, Chatterjee N, Perati S, Menon S, Nguyen TA, Debnath J, Ramirez AD, Shi X, Yang B, Feng S, Makhija S, Huang B, Bivona TG. Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. Cell. 2021;184:2649–2664.e2618. doi: 10.1016/j.cell.2021.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sampson J, Richards MW, Choi J, Fry AM, Bayliss R. Phase-separated foci of EML4-ALK facilitate signalling and depend upon an active kinase conformation. EMBO Rep. 2021;22:e53693. doi: 10.15252/embr.202153693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang WYC, Alvarez S, Kondo Y, Lee YK, Chung JK, Lam HYM, Biswas KH, Kuriyan J, Groves JT. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science. 2019;363:1098–1103. doi: 10.1126/science.aau5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhu G, Xie J, Kong W, Xie J, Li Y, Du L, Zheng Q, Sun L, Guan M, Li H, Zhu T, He H, Liu Z, Xia X, Kan C, Tao Y, Shen HC, Li D, Wang S, Yu Y, Yu ZH, Zhang ZY, Liu C, Zhu J. Phase separation of disease-associated SHP2 mutants underlies MAPK hyperactivation. Cell. 2020;183:490–502.e418. doi: 10.1016/j.cell.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schaefer KN, Peifer M. Wnt/beta-catenin signaling regulation and a role for biomolecular condensates. Dev Cell. 2019;48:429–444. doi: 10.1016/j.devcel.2019.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cloer EW, Siesser PF, Cousins EM, Goldfarb D, Mowrey DD, Harrison JS, Weir SJ, Dokholyan NV, Major MB. p62-Dependent Phase Separation of Patient-Derived KEAP1 Mutations and NRF2. Mol Cell Biol. 2018;38(22):00644–717. doi: 10.1128/MCB.00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jobe F, Simpson J, Hawes P, Guzman E, Bailey D. Respiratory syncytial virus sequesters NF-κB subunit p65 to cytoplasmic inclusion bodies to inhibit innate immune signaling. J Virol. 2020;94(22):10–1128. doi: 10.1128/JVI.01380-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Meng F, Yu Z, Zhang D, Chen S, Guan H, Zhou R, Wu Q, Zhang Q, Liu S, Venkat Ramani MK, Yang B, Ba XQ, Zhang J, Huang J, Bai X, Qin J, Feng XH, Ouyang S, Zhang YJ, Liang T, Xu P. Induced phase separation of mutant NF2 imprisons the cGAS-STING machinery to abrogate antitumor immunity. Mol Cell. 2021;81:4147–4164. doi: 10.1016/j.molcel.2021.07.040. [DOI] [PubMed] [Google Scholar]
- 131.Todoric J, Antonucci L, Di Caro G, Li N, Wu X, Lytle NK, Dhar D, Banerjee S, Fagman JB, Browne CD, Umemura A, Valasek MA, Kessler H, Tarin D, Goggins M, Reya T, Diaz-Meco M, Moscat J, Karin M. Stress-activated NRF2-MDM2 cascade controls neoplastic progression in pancreas. Cancer Cell. 2017;32:824–839.e828. doi: 10.1016/j.ccell.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang S, Zhang Q, Wang Q, Shen Q, Chen X, Li Z, Zhou Y, Hou J, Xu B, Li N, Cao X. NEAT1 paraspeckle promotes human hepatocellular carcinoma progression by strengthening IL-6/STAT3 signaling. Oncoimmunology. 2018;7:e1503913. doi: 10.1080/2162402X.2018.1503913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dalla-Favera R, Martinotti S, Gallo RC, Erikson J, Croce CM. Translocation and rearrangements of the c-myc oncogene locus in human undifferentiated B-cell lymphomas. Science. 1983;219:963–967. doi: 10.1126/science.6401867. [DOI] [PubMed] [Google Scholar]
- 135.Tabin CJ, Bradley SM, Bargmann CI, Weinberg RA, Papageorge AG, Scolnick EM, Dhar R, Lowy DR, Chang EH. Mechanism of activation of a human oncogene. Nature. 1982;300:143–149. doi: 10.1038/300143a0. [DOI] [PubMed] [Google Scholar]
- 136.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 137.Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang WY, Yan Q, Lin WC, Chung JK, Hansen SD, Christensen SM, Tu HL, Kuriyan J, Groves JT. Phosphotyrosine-mediated LAT assembly on membranes drives kinetic bifurcation in recruitment dynamics of the Ras activator SOS. Proc Natl Acad Sci U S A. 2016;113:8218–8223. doi: 10.1073/pnas.1602602113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yamazaki T, Zaal K, Hailey D, Presley J, Lippincott-Schwartz J, Samelson LE. Role of Grb2 in EGF-stimulated EGFR internalization. J Cell Sci. 2002;115:1791–1802. doi: 10.1242/jcs.115.9.1791. [DOI] [PubMed] [Google Scholar]
- 140.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, Daidone MG, Dupont S, Basso G, Bicciato S, Piccolo S. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 142.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gao H, Wei H, Yang Y, Li H, Liang J, Ye J, Zhang F, Wang L, Shi H, Wang J, Han A. Phase separation of DDX21 promotes colorectal cancer metastasis via MCM5-dependent EMT pathway. Oncogene. 2023;42:1704–1715. doi: 10.1038/s41388-023-02687-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang Y, Fu D, Chen Y, Su J, Wang Y, Li X, Zhai W, Niu Y, Yue D, Geng H. G3BP1 promotes tumor progression and metastasis through IL-6/G3BP1/STAT3 signaling axis in renal cell carcinomas. Cell Death Dis. 2018;9:501. doi: 10.1038/s41419-018-0504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Taniuchi K, Nishimori I, Hollingsworth MA. The N-terminal domain of G3BP enhances cell motility and invasion by posttranscriptional regulation of BART. Mol Cancer Res. 2011;9:856–866. doi: 10.1158/1541-7786.MCR-10-0574. [DOI] [PubMed] [Google Scholar]
- 146.Clark A, Burleson M. SPOP and cancer: a systematic review. Am J Cancer Res. 2020;10:704–726. [PMC free article] [PubMed] [Google Scholar]
- 147.Ghodke I, Remisova M, Furst A, Kilic S, Reina-San-Martin B, Poetsch AR, Altmeyer M, Soutoglou E. AHNAK controls 53BP1-mediated p53 response by restraining 53BP1 oligomerization and phase separation. Mol Cell. 2021;81:2596–2610.e2597. doi: 10.1016/j.molcel.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Massagué J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rangel LP, Costa DC, Vieira TC, Silva JL. The aggregation of mutant p53 produces prion-like properties in cancer. Prion. 2014;8:75–84. doi: 10.4161/pri.27776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ano Bom AP, Rangel LP, Costa DC, de Oliveira GA, Sanches D, Braga CA, Gava LM, Ramos CH, Cepeda AO, Stumbo AC, De Moura Gallo CV, Cordeiro Y, Silva JL. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: implications for cancer. J Biol Chem. 2012;287:28152–28162. doi: 10.1074/jbc.M112.340638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, Cornelis A, Rozenski J, Zwolinska A, Marine JC, Lambrechts D, Suh YA, Rousseau F, Schymkowitz J. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- 152.Higashimoto Y, Asanomi Y, Takakusagi S, Lewis MS, Uosaki K, Durell SR, Anderson CW, Appella E, Sakaguchi K. Unfolding, aggregation, and amyloid formation by the tetramerization domain from mutant p53 associated with lung cancer. Biochemistry. 2006;45:1608–1619. doi: 10.1021/bi051192j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ishimaru D, Andrade LR, Teixeira LS, Quesado PA, Maiolino LM, Lopez PM, Cordeiro Y, Costa LT, Heckl WM, Weissmüller G, Foguel D, Silva JL. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry. 2003;42:9022–9027. doi: 10.1021/bi034218k. [DOI] [PubMed] [Google Scholar]
- 154.De Smet F, Saiz Rubio M, Hompes D, Naus E, De Baets G, Langenberg T, Hipp MS, Houben B, Claes F, Charbonneau S, Delgado Blanco J, Plaisance S, Ramkissoon S, Ramkissoon L, Simons C, van den Brandt P, Weijenberg M, Van England M, Lambrechts S, Amant F, D'Hoore A, Ligon KL, Sagaert X, Schymkowitz J, Rousseau F. Nuclear inclusion bodies of mutant and wild-type p53 in cancer: a hallmark of p53 inactivation and proteostasis remodelling by p53 aggregation. J Pathol. 2017;242:24–38. doi: 10.1002/path.4872. [DOI] [PubMed] [Google Scholar]
- 155.Oda T, Gotoh N, Kasamatsu T, Handa H, Saitoh T, Sasaki N. DNA damage-induced cellular senescence is regulated by 53BP1 accumulation in the nuclear foci and phase separation. Cell Prolif. 2023;56:e13398. doi: 10.1111/cpr.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 157.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 158.Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 159.Zhang H, Zhao R, Tones J, Liu M, Dilley RL, Chenoweth DM, Greenberg RA, Lampson MA. Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol Biol Cell. 2020;31:2048–2056. doi: 10.1091/mbc.E19-10-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Min J, Wright WE, Shay JW. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 2019;33:814–827. doi: 10.1101/gad.324905.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang L, Gao Y, Zheng X, Liu C, Dong S, Li R, Zhang G, Wei Y, Qu H, Li Y, Allis CD, Li G, Li H, Li P. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol Cell. 2019;76:646–659.e646. doi: 10.1016/j.molcel.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 163.Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, Yuan W, Kan Q, Sun Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12:121. doi: 10.1186/s13045-019-0805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC, Li P. Multivalent m(6)A motifs promote phase separation of YTHDF proteins. Cell Res. 2019;29:767–769. doi: 10.1038/s41422-019-0210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, Duggan R, Wang Y, Barber GN, Fitzgerald KA, Alegre ML, Gajewski TF. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li XD, Mauceri H, Beckett M, Darga T, Huang X, Gajewski TF, Chen ZJ, Fu YX, Weichselbaum RR. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Marcus A, Mao AJ, Lensink-Vasan M, Wang L, Vance RE, Raulet DH. Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the nk cell response. Immunity. 2018;49:754–763.e754. doi: 10.1016/j.immuni.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vladimirova O, De Leo A, Deng Z, Wiedmer A, Hayden J, Lieberman PM. Phase separation and DAXX redistribution contribute to LANA nuclear body and KSHV genome dynamics during latency and reactivation. PLoS Pathog. 2021;17:e1009231. doi: 10.1371/journal.ppat.1009231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Peng Q, Wang L, Qin Z, Wang J, Zheng X, Wei L, Zhang X, Zhang X, Liu C, Li Z, Wu Y, Li G, Yan Q, Ma J. Phase separation of epstein-barr virus EBNA2 and its coactivator EBNALP controls gene expression. J Virol. 2020;94(7):10–1128. doi: 10.1128/JVI.01771-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Peng Q, Wang L, Wang J, Liu C, Zheng X, Zhang X, Wei L, Li Z, Wu Y, Wen Y, Cao P, Liao Q, Yan Q, Ma J. Epstein-Barr virus EBNA2 phase separation regulates cancer-associated alternative RNA splicing patterns. Clin Transl Med. 2021;11:e504. doi: 10.1002/ctm2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Baudino TA, McKay C, Pendeville-Samain H, Nilsson JA, Maclean KH, White EL, Davis AC, Ihle JN, Cleveland JL. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002;16:2530–2543. doi: 10.1101/gad.1024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jiang Y, Lei G, Lin T, Zhou N, Wu J, Wang Z, Fan Y, Sheng H, Mao R. 1,6-Hexanediol regulates angiogenesis via suppression of cyclin A1-mediated endothelial function. BMC Biol. 2023;21:75. doi: 10.1186/s12915-023-01580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shi Y, Liao Y, Liu Q, Ni Z, Zhang Z, Shi M, Li P, Li H, Rao Y. BRD4-targeting PROTAC as a unique tool to study biomolecular condensates. Cell Discov. 2023;9:47. doi: 10.1038/s41421-023-00544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cai D, Liu Z, Lippincott-Schwartz J. Biomolecular condensates and their links to cancer progression. Trends Biochem Sci. 2021;46:535–549. doi: 10.1016/j.tibs.2021.01.002. [DOI] [PubMed] [Google Scholar]
- 176.Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, Zhou X, Li Y, Rusch MC, Easton J, Huether R, Gonzalez-Pena V, Wilkinson MR, Hermida LC, Davis S, Sioson E, Pounds S, Cao X, Ries RE, Wang Z, Chen X, Dong L, Diskin SJ, Smith MA, Guidry Auvil JM, Meltzer PS, Lau CC, Perlman EJ, Maris JM, Meshinchi S, Hunger SP, Gerhard DS, Zhang J. Pan-cancer genome and transcriptome analyses of 1699 paediatric leukaemias and solid tumours. Nature. 2018;555:371–376. doi: 10.1038/nature25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bolouri H, Farrar JE, Triche T, Jr, Ries RE, Lim EL, Alonzo TA, Ma Y, Moore R, Mungall AJ, Marra MA, Zhang J, Ma X, Liu Y, Liu Y, Auvil JMG, Davidsen TM, Gesuwan P, Hermida LC, Salhia B, Capone S, Ramsingh G, Zwaan CM, Noort S, Piccolo SR, Kolb EA, Gamis AS, Smith MA, Gerhard DS, Meshinchi S. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24:103–112. doi: 10.1038/nm.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Struski S, Lagarde S, Bories P, Puiseux C, Prade N, Cuccuini W, Pages MP, Bidet A, Gervais C, Lafage-Pochitaloff M, Roche-Lestienne C, Barin C, Penther D, Nadal N, Radford-Weiss I, Collonge-Rame MA, Gaillard B, Mugneret F, Lefebvre C, Bart-Delabesse E, Petit A, Leverger G, Broccardo C, Luquet I, Pasquet M, Delabesse E. NUP98 is rearranged in 3.8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia. 2017;31:565–572. doi: 10.1038/leu.2016.267. [DOI] [PubMed] [Google Scholar]
- 179.McNeer NA, Philip J, Geiger H, Ries RE, Lavallée VP, Walsh M, Shah M, Arora K, Emde AK, Robine N, Alonzo TA, Kolb EA, Gamis AS, Smith M, Gerhard DS, Guidry-Auvil J, Meshinchi S, Kentsis A. Genetic mechanisms of primary chemotherapy resistance in pediatric acute myeloid leukemia. Leukemia. 2019;33:1934–1943. doi: 10.1038/s41375-019-0402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/MCB.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 182.Somasekharan SP, El-Naggar A, Leprivier G, Cheng H, Hajee S, Grunewald TG, Zhang F, Ng T, Delattre O, Evdokimova V, Wang Y, Gleave M, Sorensen PH. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol. 2015;208:913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Oda Y, Sakamoto A, Shinohara N, Ohga T, Uchiumi T, Kohno K, Tsuneyoshi M, Kuwano M, Iwamoto Y. Nuclear expression of YB-1 protein correlates with P-glycoprotein expression in human osteosarcoma. Clin Cancer Res. 1998;4:2273–2277. [PubMed] [Google Scholar]
- 184.Oda Y, Kohashi K, Yamamoto H, Tamiya S, Kohno K, Kuwano M, Iwamoto Y, Tajiri T, Taguchi T, Tsuneyoshi M. Different expression profiles of Y-box-binding protein-1 and multidrug resistance-associated proteins between alveolar and embryonal rhabdomyosarcoma. Cancer Sci. 2008;99:726–732. doi: 10.1111/j.1349-7006.2008.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fujiwara-Okada Y, Matsumoto Y, Fukushi J, Setsu N, Matsuura S, Kamura S, Fujiwara T, Iida K, Hatano M, Nabeshima A, Yamada H, Ono M, Oda Y, Iwamoto Y. Y-box binding protein-1 regulates cell proliferation and is associated with clinical outcomes of osteosarcoma. Br J Cancer. 2013;108:836–847. doi: 10.1038/bjc.2012.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Somasekharan SP, Saxena N, Zhang F, Beraldi E, Huang JN, Gentle C, Fazli L, Thi M, Sorensen PH, Gleave M. Regulation of AR mRNA translation in response to acute AR pathway inhibition. Nucleic Acids Res. 2022;50:1069–1091. doi: 10.1093/nar/gkab1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Carmona-Fontaine C, Deforet M, Akkari L, Thompson CB, Joyce JA, Xavier JB. Metabolic origins of spatial organization in the tumor microenvironment. Proc Natl Acad Sci U S A. 2017;114:2934–2939. doi: 10.1073/pnas.1700600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tajan M, Hock AK, Blagih J, Robertson NA, Labuschagne CF, Kruiswijk F, Humpton TJ, Adams PD, Vousden KH. A role for p53 in the adaptation to glutamine starvation through the expression of SLC1A3. Cell Metab. 2018;28:721–736.e726. doi: 10.1016/j.cmet.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rubin H. Deprivation of glutamine in cell culture reveals its potential for treating cancer. Proc Natl Acad Sci U S A. 2019;116:6964–6968. doi: 10.1073/pnas.1815968116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang R, Cao L, Thorne RF, Zhang XD, Li J, Shao F, Zhang L, Wu M. LncRNA GIRGL drives CAPRIN1-mediated phase separation to suppress glutaminase-1 translation under glutamine deprivation. Sci Adv. 2021;7:eabe5708. doi: 10.1126/sciadv.abe5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B, 3rd, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, Kaffenberger SD, Eaton JK, Shimada K, Aguirre AJ, Viswanathan SR, Chattopadhyay S, Tamayo P, Yang WS, Rees MG, Chen S, Boskovic ZV, Javaid S, Huang C, Wu X, Tseng YY, Roider EM, Gao D, Cleary JM, Wolpin BM, Mesirov JP, Haber DA, Engelman JA, Boehm JS, Kotz JD, Hon CS, Chen Y, Hahn WC, Levesque MP, Doench JG, Berens ME, Shamji AF, Clemons PA, Stockwell BR, Schreiber SL. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, Galeas J, Dhruv HD, Berens ME, Schreiber SL, McCormick F, McManus MT. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–250. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nakamura T, Hipp C, Mourão ASD, Borggräfe J, Aldrovandi M, Henkelmann B, Wanninger J, Mishima E, Lytton E, Emler D, Proneth B, Sattler M, Conrad M. Phase separation of FSP1 promotes ferroptosis. Nature. 2023;619:371–377. doi: 10.1038/s41586-023-06255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci U S A. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Erkizan HV, Kong Y, Merchant M, Schlottmann S, Barber-Rotenberg JS, Yuan L, Abaan OD, Chou TH, Dakshanamurthy S, Brown ML, Uren A, Toretsky JA. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing's sarcoma. Nat Med. 2009;15:750–756. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Gupta N, Badeaux M, Liu Y, Naxerova K, Sgroi D, Munn LL, Jain RK, Garkavtsev I. Stress granule-associated protein G3BP2 regulates breast tumor initiation. Proc Natl Acad Sci U S A. 2017;114:1033–1038. doi: 10.1073/pnas.1525387114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lemos C, Schulze L, Weiske J, Meyer H, Braeuer N, Barak N, Eberspächer U, Werbeck N, Stresemann C, Lange M, Lesche R, Zablowsky N, Juenemann K, Kamburov A, Luh LM, Leissing TM, Mortier J, Steckel M, Steuber H, Eis K, Eheim A, Steigemann P. Identification of small molecules that modulate mutant p53 condensation. Science. 2020;23:101517. doi: 10.1016/j.isci.2020.101517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mukherjee H, Chan KP, Andresen V, Hanley ML, Gjertsen BT, Myers AG. Interactions of the natural product (+)-avrainvillamide with nucleophosmin and exportin-1 Mediate the cellular localization of nucleophosmin and its AML-associated mutants. ACS Chem Biol. 2015;10:855–863. doi: 10.1021/cb500872g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chen YN, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, Antonakos B, Chen CH, Chen Z, Cooke VG, Dobson JR, Deng Z, Fei F, Firestone B, Fodor M, Fridrich C, Gao H, Grunenfelder D, Hao HX, Jacob J, Ho S, Hsiao K, Kang ZB, Karki R, Kato M, Larrow J, La Bonte LR, Lenoir F, Liu G, Liu S, Majumdar D, Meyer MJ, Palermo M, Perez L, Pu M, Price E, Quinn C, Shakya S, Shultz MD, Slisz J, Venkatesan K, Wang P, Warmuth M, Williams S, Yang G, Yuan J, Zhang JH, Zhu P, Ramsey T, Keen NJ, Sellers WR, Stams T, Fortin PD. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535:148–152. doi: 10.1038/nature18621. [DOI] [PubMed] [Google Scholar]
- 202.Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall'Agnese A, Oksuz O, Henninger JE, Shrinivas K, Sabari BR, Sagi I, Clark VE, Platt JM, Kar M, McCall PM, Zamudio AV, Manteiga JC, Coffey EL, Li CH, Hannett NM, Guo YE, Decker TM, Lee TI, Zhang T, Weng JK, Taatjes DJ, Chakraborty A, Sharp PA, Chang YT, Hyman AA, Gray NS, Young RA. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang F, Biswas M, Massah S, Lee J, Lingadahalli S, Wong S, Wells C, Foo J, Khan N, Morin H, Saxena N, Kung SHY, Sun B, Parra Nuñez AK, Sanchez C, Chan N, Ung L, Altıntaş UB, Bui JM, Wang Y, Fazli L, Oo HZ, Rennie PS, Lack NA, Cherkasov A, Gleave ME, Gsponer J, Lallous N. Dynamic phase separation of the androgen receptor and its coactivators key to regulate gene expression. Nucleic Acids Res. 2023;51:99–116. doi: 10.1093/nar/gkac1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Oka M, Mura S, Yamada K, Sangel P, Hirata S, Maehara K, Kawakami K, Tachibana T, Ohkawa Y, Kimura H, Yoneda Y. Chromatin-prebound Crm1 recruits Nup98-HoxA9 fusion to induce aberrant expression of Hox cluster genes. Elife. 2016;5:e09540. doi: 10.7554/eLife.09540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Takayama KI, Kosaka T, Suzuki T, Hongo H, Oya M, Fujimura T, Suzuki Y, Inoue S. Subtype-specific collaborative transcription factor networks are promoted by OCT4 in the progression of prostate cancer. Nat Commun. 2021;12:3766. doi: 10.1038/s41467-021-23974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhang Z, Boskovic Z, Hussain MM, Hu W, Inouye C, Kim HJ, Abole AK, Doud MK, Lewis TA, Koehler AN, Schreiber SL, Tjian R. Chemical perturbation of an intrinsically disordered region of TFIID distinguishes two modes of transcription initiation. Elife. 2015;4:e07777. doi: 10.7554/eLife.07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rangel LP, Ferretti GDS, Costa CL, Andrade S, Carvalho RS, Costa DCF, Silva JL. p53 reactivation with induction of massive apoptosis-1 (PRIMA-1) inhibits amyloid aggregation of mutant p53 in cancer cells. J Biol Chem. 2019;294:3670–3682. doi: 10.1074/jbc.RA118.004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang C, Lu H, Liu X, Gao X, Tian W, Chen H, Xue Y, Zhou Q. A natural product targets BRD4 to inhibit phase separation and gene transcription. iScience. 2022;25:103719. doi: 10.1016/j.isci.2021.103719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Babinchak WM, Dumm BK, Venus S, Boyko S, Putnam AA, Jankowsky E, Surewicz WK. Small molecules as potent biphasic modulators of protein liquid-liquid phase separation. Nat Commun. 2020;11:5574. doi: 10.1038/s41467-020-19211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Liu J, Xie Y, Guo J, Li X, Wang J, Jiang H, Peng Z, Wang J, Wang S, Li Q, Ye L, Zhong Y, Zhang Q, Liu X, Lonard DM, Wang J, O'Malley BW, Liu Z. Targeting NSD2-mediated SRC-3 liquid-liquid phase separation sensitizes bortezomib treatment in multiple myeloma. Nat Commun. 2021;12:1022. doi: 10.1038/s41467-021-21386-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Singatulina AS, Hamon L, Sukhanova MV, Desforges B, Joshi V, Bouhss A, Lavrik OI, Pastré D. PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 2019;27:1809–1821.e1805. doi: 10.1016/j.celrep.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 212.Lu B, Zou C, Yang M, He Y, He J, Zhang C, Chen S, Yu J, Liu KY, Cao Q, Zhao W. Pharmacological inhibition of core regulatory circuitry liquid-liquid phase separation suppresses metastasis and chemoresistance in osteosarcoma. Adv Sci (Weinh) 2021;8:e2101895. doi: 10.1002/advs.202101895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 214.Bhagwat AS, Roe JS, Mok BYL, Hohmann AF, Shi J, Vakoc CR. BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep. 2016;15:519–530. doi: 10.1016/j.celrep.2016.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gilan O, Lam EY, Becher I, Lugo D, Cannizzaro E, Joberty G, Ward A, Wiese M, Fong CY, Ftouni S, Tyler D, Stanley K, MacPherson L, Weng CF, Chan YC, Ghisi M, Smil D, Carpenter C, Brown P, Garton N, Blewitt ME, Bannister AJ, Kouzarides T, Huntly BJ, Johnstone RW, Drewes G, Dawson SJ, Arrowsmith CH, Grandi P, Prinjha RK, Dawson MA. Functional interdependence of BRD4 and DOT1L in MLL leukemia. Nat Struct Mol Biol. 2016;23:673–681. doi: 10.1038/nsmb.3249. [DOI] [PubMed] [Google Scholar]
- 216.Nilson KA, Guo J, Turek ME, Brogie JE, Delaney E, Luse DS, Price DH. THZ1 Reveals roles for Cdk7 in Co-transcriptional capping and pausing. Mol Cell. 2015;59:576–587. doi: 10.1016/j.molcel.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang T, Kwiatkowski N, Olson CM, Dixon-Clarke SE, Abraham BJ, Greifenberg AK, Ficarro SB, Elkins JM, Liang Y, Hannett NM, Manz T, Hao M, Bartkowiak B, Greenleaf AL, Marto JA, Geyer M, Bullock AN, Young RA, Gray NS. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat Chem Biol. 2016;12:876–884. doi: 10.1038/nchembio.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wheeler RJ. Therapeutics-how to treat phase separation-associated diseases, Emerg Top. Life Sci. 2020;4:307–318. doi: 10.1042/ETLS20190176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Luo YY, Wu JJ, Li YM. Regulation of liquid-liquid phase separation with focus on post-translational modifications. Chem Commun (Camb) 2021;57:13275–13287. doi: 10.1039/D1CC05266G. [DOI] [PubMed] [Google Scholar]
- 220.Chong PA, Vernon RM, Forman-Kay JD. RGG/RG Motif regions in RNA binding and phase separation. J Mol Biol. 2018;430:4650–4665. doi: 10.1016/j.jmb.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 221.Dao TP, Kolaitis RM, Kim HJ, O'Donovan K, Martyniak B, Colicino E, Hehnly H, Taylor JP, Castañeda CA. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol Cell. 2018;69:965–978.e966. doi: 10.1016/j.molcel.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.de Thé H, Pandolfi PP, Chen Z. Acute promyelocytic leukemia: a paradigm for oncoprotein-targeted cure. Cancer Cell. 2017;32:552–560. doi: 10.1016/j.ccell.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 223.Sun Y, Lau SY, Lim ZW, Chang SC, Ghadessy F, Partridge A, Miserez A. Phase-separating peptides for direct cytosolic delivery and redox-activated release of macromolecular therapeutics. Nat Chem. 2022;14:274–283. doi: 10.1038/s41557-021-00854-4. [DOI] [PubMed] [Google Scholar]
- 224.Guo RC, Zhang XH, Fan PS, Song BL, Li ZX, Duan ZY, Qiao ZY, Wang H. In vivo self-assembly induced cell membrane phase separation for improved peptide drug internalization. Angew Chem Int Ed Engl. 2021;60:25128–25134. doi: 10.1002/anie.202111839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.