Abstract
The spore coat of Bacillus subtilis has a unique morphology and consists of polypeptides of different sizes, whose synthesis and assembly are precisely regulated by a cascade of transcription factors and regulatory proteins. We examined the factors that regulate cotS gene expression and CotS assembly into the coat layer of B. subtilis by Northern blot and Western blot analysis. Transcription of cotS mRNA was not detected in sporulating cells of ςK and gerE mutants by Northern blot analysis. By Western blot analysis using anti-CotS antibody, CotS was first detected in protein samples solubilized from wild-type cells at 5 h after the start of sporulation. CotS was not detected in the vegetative cells and spores of a gerE mutant or in the spores of mutants deficient in ςE, ςF, ςG, or ςK. CotS was detected in the sporangium but not in the spores of a cotE mutant. The sequence of the promoter region of cotS was similar to the consensus sequences for binding of ςK and GerE. These results demonstrate that ςK and GerE are required for cotS expression and that CotE is essential for the assembly of CotS in the coat. Immunoelectron microscopic observation using anti-CotS antibody revealed that CotS is located within the spore coat, in particular in the inner coats of dormant spores.
Endospore formation by Bacillus subtilis is a good model system with which to study fundamental issues of cell biology concerning how the genes involved in cell differentiation are temporally regulated and how structural protein components are assembled at particular sites within a cell. After a final round of chromosomal replication in B. subtilis, cells asymmetrically divide into two compartments, the mother cell and the forespore. The forespore is engulfed by the mother cell double-membrane septum as a discrete cell within the mother cell, and the forespore is then surrounded by a cortex layer and coat layers to make a mature spore. The programs of gene expression are distinct in each compartment. Sporulation genes (spo) and other related genes that are active in the forespore compartment are governed by RNA polymerase sigma factors ςF and ςG. Gene expression in the mother cell is governed by ςE and ςK and by the regulatory proteins SpoIIID and GerE (39).
Bacterial spores are morphologically complex structures in which the spore core (cytoplasm), a primordial germ cell wall, a cortical layer (cortex), and a proteinous spore coat are observed. The spore coat, surrounding the cortex, consists of an electron-dense thick outer layer and a thinner, lamella-like inner layer (4), with dozens of proteins ranging in size from 8 to 65 kDa (32). The cortex and coat layers are essential for the remarkable resistance properties of spores. The spore coat, particularly, provides a protective barrier against lysozyme, solvents, and other harsh chemicals and is in part responsible for the prompt response of spores to components capable of triggering germination (4–7, 12, 18, 22, 36, 38).
A recent systematic search of the B. subtilis genome listed at least 22 genes that are necessary for the formation of the spore coat (21). Correct formation of the coat is under dual control. A cascade of transcription factors regulates the temporal appearance of the coat components (39), and the action of morphogenetic proteins controls proper assembly of those components to organize the two layers of the coat (38). Temporal control of spore coat genes (cot) involves a cascade of four regulatory factors in the sequence, ςE-SpoIIID-ςK-GerE (38). ςE and ςK are RNA polymerase sigma factors, whereas SpoIIID and GerE are DNA-binding proteins. The cot genes and their transcription regulators can be divided into four classes based on their appearance during sporulation (24). The class 1 genes, spoIIID and cotE, are expressed after the onset of sporulation under the control of ςE. In class 2, sigK and cotJABC are expressed by the action of ςE and SpoIIID. The class 3 genes, gerE, cotA, cotD, cotF, and cotH, are controlled by ςK. The last class of genes, cotB, cotC, cotG, cotYZ, and cotVXW, are expressed under the control of ςK and GerE. CotE is a morphogenic protein required for the assembly of proteins of the electron-dense outer layer of the spore coat and serves as a basement protein on which the proteins of the outer coat assemble (38). GerE is required for assembly of most of the lamella-like inner coat layer (22).
The cotS operon of B. subtilis consists of cotS, which encodes a spore coat protein (CotS) of 41 kDa, and open reading frame orfX (named ytxN in the B. subtilis genome project [21]) (2). The cotS operon is transcribed at about the fifth hour of sporulation (T5) and has a putative promoter sequence similar to the consensus sequence for ςK-dependent promoters. Disruption of the cotS gene results in no alteration of growth, sporulation, spore germination, or spore resistance to organic solvents (2). A similar observation has been made for other cot genes (32). In this study, we examined what regulatory factors direct CotS protein synthesis and which factors direct its assembly into the spore coat. We first purified recombinant CotS having a His6 tag from Escherichia coli and prepared antibody against the protein. Using this antibody, we demonstrated that expression of cotS depended on ςK and gerE and that assembly of CotS into the spore coat depended on CotE. Furthermore, immunoelectron microscopy revealed that CotS localized to the inner coat and/or on the outside of the cortex of the mature spore.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and general techniques.
The B. subtilis strains used in this study are listed in Table 1 and were all grown in DS medium (30). E. coli was grown in LB medium. The conditions for sporulation of B. subtilis and method for purification of mature spores have been described previously (2). Recombinant DNA methods were as described by Sambrook et al. (28). Methods for preparing competent cells, for transformation, and for the preparation of chromosomal DNA from B. subtilis were as described by Cutting and Vander Horn (11).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype or description | Source (reference) |
|---|---|---|
| B. subtilis | ||
| 168trpC2 | trpC2 | BGSCa |
| 1S101 | cotA::cat trpC2 | BGSC |
| 1S102 | cotB::cat trpC2 | BGSC |
| 1S103 | cotC::cat trpC2 | BGSC |
| 1S104 | cotD::cat trpC2 | BGSC |
| 1S105 | cotEΔ::cat trpC2 | BGSC |
| 1S106 | cotF::cat trpC2 | BGSC |
| 1S108 | cotT::pΔ E194 pheA1 trpC2 | BGSC |
| CB701 | cotS::cat trpC2 | This work |
| 1G12 | gerE36 leu-2 trpC2 | BGSC |
| 1S38 | spoIIIC94 trpC2 (ςK mutant) | BGSC |
| 1S60 | leuB8 spoIIG41 tal-1 (ςE mutant) | BGSC |
| SC1159 | spoIIAC1 (ςF mutant) | S. Cutting (10) |
| spoIIIGΔ1 | spoIIIGΔ1 trpC2 (ςG mutant) | J. Sekiguchi (31) |
| E. coli JM109 | relA supE44 endA1 hsdR17 gyrA96 mcrA mcrB+ thi Δ (lac-proAB)/F′ (traD36 proAB+ lacIqlacZΔM15) | J. Sambrook (28) |
| Plasmids | ||
| pCX18S | CmrcotS5′ lacZ′ | A. Abe (2) |
| pTUE1122 | AmprlacI tac promoter His6 | A. Nakane (23) |
| pBCS1 | AmprlacI tac promoter cotS His6 | This work |
BGSC, Bacillus Genetic Stock Center.
Preparation of the cotS mutant.
Plasmid pCX18S, which had been prepared by ligation of a central portion of the cotS gene (302 bp) between the PstI and HindIII sites of plasmid pCX18 (2.4 kb) (2), was transformed into B. subtilis 168trpC2 to obtain the cotS gene disruption mutant CB701. The correct integration of pCX18 in CB701 was verified by restriction analysis of DNA amplified from cotS by PCR.
RNA preparation and Northern analysis.
B. subtilis cells were grown in DS medium, and 5-ml samples were harvested every hour throughout sporulation. The RNA was then prepared as described by Igo and Losick (17). Each 10 μg of the RNA preparation was analyzed by size fractionation through a 1% (wt/vol) agarose gel containing 2.2 M formaldehyde and transferred to a positively charged Hybond-N+ membrane (Amersham). The membrane was stained with 0.04% methylene blue solution containing 0.5 M sodium acetate (pH 5.2) to measure the concentrations of 16S and 23S RNAs in the preparations as described by Herrin and Schmidt (16). The RNA on the membrane was hybridized to a DNA probe corresponding to nucleotides −64 to +467 of the translation initiation codon of cotS. This DNA fragment was amplified by PCR using two primers, 5′-GCTTCTAGAGGGTGGCTGAAAA-3′ and 5′-TAATACGACTCACTATAGGGCGATCCTGCAGCTTCCAACGG-3′. Hybridization was performed and hybrids were detected according to the procedure provided by Boehringer Mannheim.
Preparation of whole proteins from sporulating cells.
Cultures (5 ml) were harvested every hour throughout sporulation and washed with 10 mM sodium phosphate buffer (pH 7.2). The pellets were suspended in 100 μl of lysozyme buffer (25 mM Tris-HCl [pH 8.0], 10 mM EDTA, 50 mM glucose, 1% lysozyme), kept on ice for 5 min, and then boiled for 5 min in 2% (vol/vol) sodium dodecyl sulfate (SDS)–5% (vol/vol) 2-mercaptoethanol–10% (vol/vol) glycerol–62.5 mM Tris-HCl (pH 6.8)–0.05% (wt/vol) bromophenol blue.
Preparation of spores and solubilization of coat proteins.
Cultures (5 ml) were harvested at T18 and washed with 10 mM sodium phosphate buffer (pH 7.2). The pellets were suspended in 100 μl of lysozyme solution (10 mM sodium acetate [pH 7.2], 1% lysozyme) and incubated for 15 min at 37°C. After addition of 1.0 ml of 10 mM sodium phosphate buffer (pH 7.2), the suspensions were centrifuged to remove soluble proteins from mother cells. The spores in the pellet fraction were boiled in 2% (vol/vol) SDS–5% (vol/vol) 2-mercaptoethanol–10% (vol/vol) glycerol–62.5 mM Tris-HCl (pH 6.8)–0.05% (wt/vol) bromophenol blue for 5 min.
SDS-PAGE and immunoblotting.
Protein samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (12.0% acrylamide) as described previously (1). For immunoblotting, proteins were transferred onto a polyvinyl difluoridine membrane (Immobilon; 0.45-μm pore size; Millipore), and detected by using rabbit immunoglobulin G (IgG) against CotS as the first antibody and donkey anti-rabbit IgG–horseradish peroxidase conjugate as the second antibody (Amersham). Anti-ςA antibody and anti-ςK antibody were provided by M. Fujita and Y. Sadaie (National Institute of Genetics, Mishima, Japan).
Purification of recombinant CotS protein.
The DNA sequence from 67 bp upstream of the initiation codon of cotS to the stop codon was amplified by PCR using two primers, 5′-GCTTCTAGAGGGTGGCTGAAAA-3′ and 5′-TCAGATCTATTCGCCTCCCGAT-3′. An XbaI-to-BglII fragment carrying the amplified cotS gene was then inserted between the XbaI and BglII sites of plasmid pTUE1122, yielding the recombinant plasmid pBCS1. pTUE1122 is a multicopy E. coli plasmid containing the tac promoter, the lacI gene, and a multicloning site followed by a His6 tag coding sequence (23). Consequently, the cotS gene in pBCS1 encodes a product with an additional amino acid sequence, RSHHHHHH, at its C-terminal end. Transformants carrying pBCS1 were grown in 200 ml of L broth supplemented with 50 μg of ampicillin per ml at 37°C for 3 h, the culture was made 1 mM in isopropyl-β-d-thiogalactopyranoside, and the cells were incubated for a further 3 h at 37°C. The His-tagged recombinant CotS protein was purified by affinity chromatography on Ni-nitrilotriacetic acid agarose beads (Qiagen Inc., Chatsworth, Calif.) as described previously (35) and was further purified by electroelution from an SDS-gel after SDS-PAGE as described previously (1).
Preparation of antibody against CotS.
One milliliter of purified CotS (0.2 μg/μl) and 16 mg of killed Mycobacterium tuberculosis cells (Difco) were well mixed with 2 ml of complete Freund’s adjuvant (Difco), and 3 ml of the emulsion was injected into a healthy rabbit. After 2 weeks, CotS solutions which were prepared with incomplete Freund’s adjuvant (Difco) were injected; 2 weeks after the second immunization, antiserum for CotS was obtained.
Electron and immunoelectron microscopy.
Electron and immunoelectron microscopic observations were carried out essentially as described by Sakae et al. (27), with minor modifications as follows. The spores prepared as described previously (2) were fixed with 4% freshly depolymerized paraformaldehyde–0.5% glutaraldehyde buffered at pH 7.0 with 50 mM cacodylate buffer at 4°C for 48 h. Fixed spores were then suspended in 2% agar, and small cubes were cut and dehydrated with a graded series of 30, 50, 70, 80, 90, and 100% ethanol for 1 h at each ethanol concentration at −20°C. The samples were suspended in Lowicryl K4M (Chemische Werke Lowi GmbH)–100% ethanol at 2:1, 1:1, and 1:2 ratios for 2 h at each ratio at 4°C and finally in Lowicryl K4M overnight at 4°C. Samples were then placed in gelatin capsules filled with Lowicryl K4M and polymerized by UV irradiation (model TUV-100 polymerizer; Dosake EM) for 24 h at 4°C. Ultrathin sections were cut with a glass knife on a Reichert Ultracut-E ultramicrotome and mounted on Formvar-coated nickel grids (Veco Ni 200). The grids were placed on droplets of 0.05% Tween 20–0.05 M Tris-HCl buffered saline (pH 7.2) for 5 min and on Block Ace (Dainippon Pharmaceutical Co., Ltd.) for 1 h at room temperature. The grids were washed, treated with the anti-CotS antibody at a dilution of 1:120 in 1/10 Block Ace for 2 h at room temperature, and then treated with 10-nm gold-conjugated goat anti-rabbit IgG antibody (Zymed Laboratories) at a dilution of 1:40 in 1/10 Block Ace for 1 h at room temperature. The sections were stained with 4% uranyl acetate for 15 min at room temperature. Control sections were treated in a similar way by using preimmune serum from the same rabbit instead of anti-CotS antibody. The sections were observed with a JEM-1200EX electron microscope operating at 80 kV.
RESULTS
Expression of CotS during development.
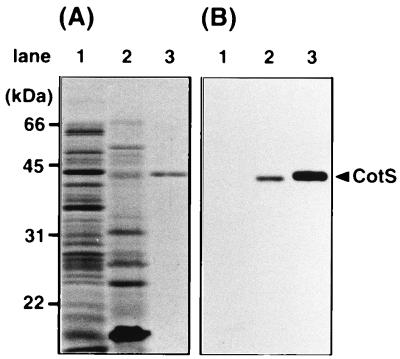
Since cotS was identified as the gene encoding the spore coat protein CotS (2), we tried to determine immunologically where during the developmental cycle of B. subtilis that CotS is present. The anti-CotS antibody reacted with purified CotS (Fig. 1B, lane 3) and also with CotS protein solubilized from mature spores by treatment with 2% SDS and 5% 2-mercaptoethanol (Fig. 1B, lane 2). However, CotS was not detected in protein samples solubilized from vegetative cells (Fig. 1B, lane 1). These results indicated that CotS protein is present in spores but not in the vegetative cells.
FIG. 1.
Detection of CotS protein by immunoblotting using anti-CotS antibody. The protein samples were solubilized from vegetative cells of B. subtilis 168trpC2 (lane 1), the SDS-mercaptoethanol-soluble fraction prepared from dormant spores of B. subtilis 168trpC2 (lane 2), and purified CotS protein with a His6 tag (lane 3). The samples were analyzed by SDS-PAGE (12% gel). (A) Coomassie brilliant blue stain; (B) immunoblotting using anti-CotS antibody. The arrowhead shows the migration position of CotS.
Immunocytochemical localization of CotS.
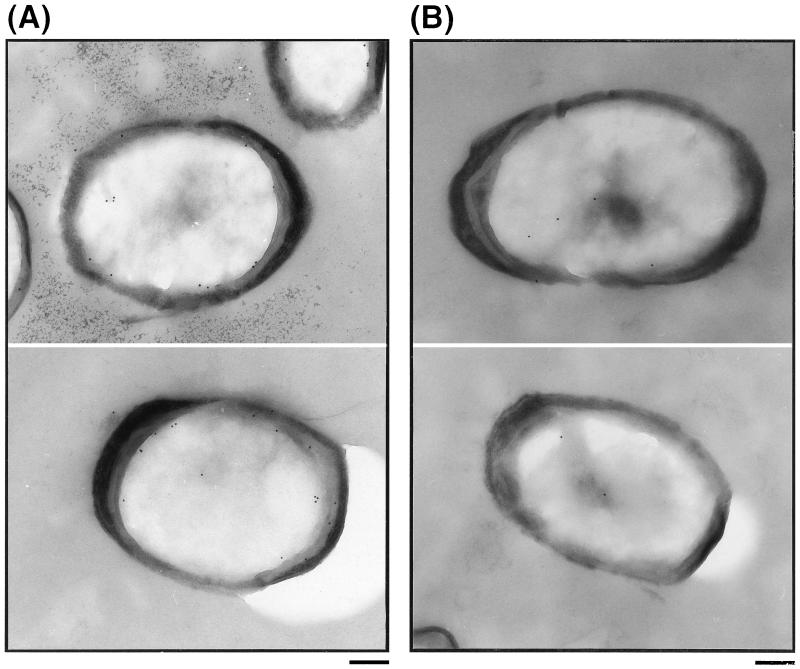
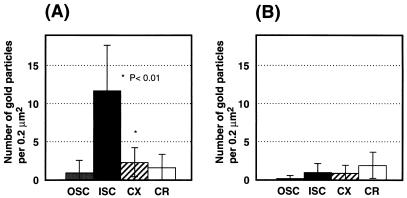
Most spore coat proteins which have been analyzed are presumably located in the outer coat layer, while CotD, CotH, and CotT are thought to be present in the inner coat layer (5, 6, 38). The morphologies of spores of the wild-type and a cotS mutant were examined by electron microscopy and found to exhibit similar structures in outer and inner coat layers, cortex, and core regions (data not shown). Therefore, the location of CotS in spores was determined by immunochemical staining using anti-CotS antibody and colloidal gold-labeled second antibody (Fig. 2). In wild-type spores, many gold particles were observed within the spore integument, especially on the inner coat and on the outside region of the cortex, whereas only a few gold particles were seen in the core region (Fig. 2A). In contrast, negligible numbers of gold particles were observed in cotS mutant spores (Fig. 2B). Similarly, only a few gold particles were seen with wild-type and cotS mutant spores in assays using preimmune serum (data not shown). To confirm the localization of CotS in wild-type spores, a statistical analysis of 20 photographs of wild-type and cotS mutant spores was carried out as described previously (15). The numbers of gold particles per 0.2 μm2 were clearly highest in the inner spore coat of the wild-type strain and were much higher than in the inner coats of cotS mutant spores (Fig. 3). The analysis suggests that CotS is localized in the inner coat and/or subcoat regions of the spores.
FIG. 2.
Immunoelectron microscopic localization of CotS protein in wild-type spores (A) and cotS mutant spores. Thin sections of purified wild-type spores (A) and cotS mutant spores (CB701) (B) were stained with anti-CotS antibody and a colloidal gold (10 nm)-IgG complex. Bars = 200 nm.
FIG. 3.
Number of gold particles in each region of wild-type and cotS mutant spores. Each value represents the mean from the analysis of 20 spores (± standard deviation) sectioned and stained as described in the legend to Fig. 2. (A) Wild-type spores; (B) cotS mutant (CB701) spores. OSC, outer spore coat; ISC, inner spore coat; CX; cortex; CR, core.
Northern blot analysis of cotS mRNA.
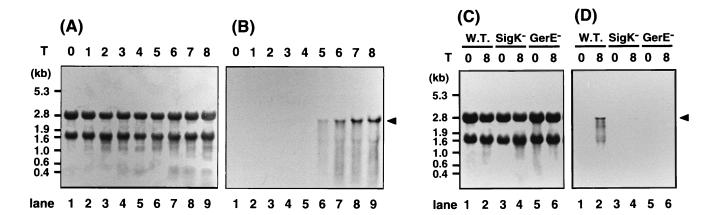
Previous work demonstrated that a cotS transcript appears at about T5 and that the cotS operon has a ςK-controlled promoter (2). To address questions concerning the dependence of cotS expression on ςK and other factors, if any, we first examined expression of a cotS-lacZ fusion. Expression of the fusion in wild-type cells was detected first in cells at T5 and increased during subsequent development; however, no cotS-lacZ expression was detected in a ςK mutant (data not shown). To confirm directly that cotS gene expression is regulated by ςK, we then analyzed cotS mRNA in the cells of the wild type, a ςK mutant, and a gerE mutant. Cultures of the wild-type cells were harvested every hour throughout sporulation, and the total RNA was extracted for Northern blot analysis (Fig. 4). All samples contained essentially the same amount of 16S RNA and 23S RNA (Fig. 4A and C). The cotS mRNA first appeared as a 2.7-kb band at T5 (Fig. 4B), and the amount of cotS mRNA increased until at least T8. In contrast to the results with wild-type cells, the cotS transcript was not detected in the extracts of ςK and gerE mutant cells prepared at T8 (Fig. 4D, lanes 4 and 6). These results indicate that both ςK and GerE are essential for transcription of cotS.
FIG. 4.
Northern blot analysis of cotS mRNA. B. subtilis 168trpC2 (wild type) (A and B) and spoIIIC94 (ςK mutant; SigK−) and gerE36 (gerE mutant; GerE−) (C and D) were grown in DS medium; the cultures were harvested at the indicated times; RNA was prepared, electrophoresed, and transferred to a membrane. (A and C) 16S and 23S RNAs were visualized by staining with methylene blue. (B and D) The gene product of cotS was detected by using a digoxigenin-labeled antisense RNA of cotS as a probe. Tn shown at the top indicates the harvesting time of cells, where n is the number of hours after the end of exponential phase of growth. The arrowhead indicates the band for cotS mRNA. W.T., wild-type cell.
Factors affecting synthesis and assembly of CotS.
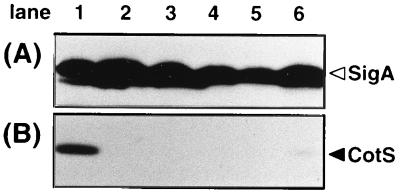
The dependence of cotS expression on ςK and GerE was confirmed by Western blot analysis. Mutants deficient in sporulation-specific sigma factors and a gerE mutant were grown in DS medium and harvested at T8. The SDS-soluble protein samples prepared from those cells were analyzed by Western blotting using anti-CotS antibody. ςA, a major sigma factor expressed during vegetative growth and sporulation (34), was analyzed as a control by using anti-ςA antibody and was detectable in all samples (Fig. 5A). However, CotS protein was not detectable in the samples from any of the mutants (Fig. 5B). Since ςK is the last in a cascade of sigma factors (34), these results strongly suggested that the synthesis of CotS depended on both ςK and GerE.
FIG. 5.
CotS protein in mutants lacking sporulation-specific transcription factors. The sigma factor-deficient cells and gerE mutant cells were harvested from DS medium at T8. Whole-protein samples were solubilized from the sporulating cells and were analyzed by SDS-PAGE (12% gel) and immunoblotting using anti-ςA (A) and anti-CotS (B) antibodies. B. subtilis 168trpC2 (lane 1), spoIIG41 (ςE mutant) (lane 2), spoIIAC1 (ςF mutant) (lane 3), spoIIIGΔ1 (ςG mutant) (lane 4), spoIIIC94 (ςK mutant) (lane 5), and gerE36 (gerE mutant) (lane 6) were analyzed. The arrowheads indicate the bands for ςA and CotS.
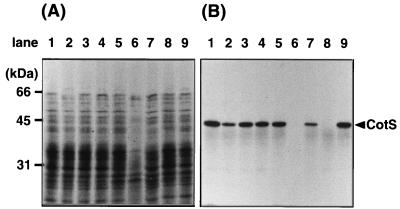
We also examined the effects of mutations in other cot genes on accumulation of CotS into the spore coat (Fig. 6). The 40-kDa CotS was detected in spores that had mutations in cotA, -B, -C, -D, -F, or -T but not in cotS or -E mutants (Fig. 6B). CotE is a morphogenic protein and is involved in the proper assembly of the spore coat (13, 24). It is particularly noteworthy in our results that the cotE gene product appeared to be involved in incorporation of CotS protein into the coat layer.
FIG. 6.
CotS protein in the spores of cot mutants. The cot mutant cells were harvested from DS medium at T18. They were then incubated in the presence of lysozyme and washed with buffer to obtain spore preparations. The protein samples solubilized from the spores were analyzed by SDS-PAGE (12% gel). (A) Coomassie brilliant blue stain; (B) immunoblotting using anti-CotS antibody. B. subtilis 168trpC2 (wild type) (lane 1), 1S101 (cotA mutant) (lane 2), 1S102 (cotB mutant) (lane 3), 1S103 (cotC mutant) (lane 4), 1S104 (cotD mutant) (lane 5), 1S105 (cotE mutant) (lane 6), 1S106 (cotF mutant) (lane 7), CB701 (cotS mutant) (lane 8), and 1S108(cotT mutant) (lane 9) were analyzed. The arrowhead shows the migration position of CotS.
Immunoblot analysis of CotS expression in gerE and cotE mutants.
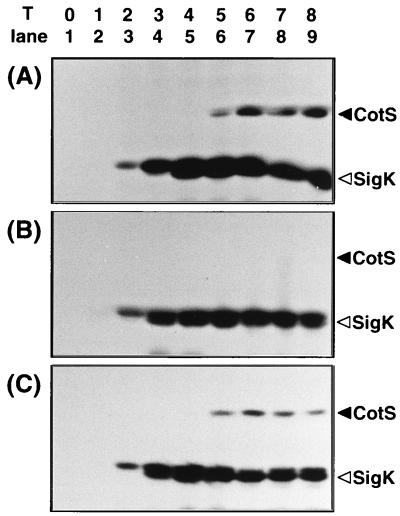
The failure to detect CotS in gerE and cotE mutants suggested two possibilities: (i) GerE and CotE positively regulate CotS synthesis and (ii) they are responsible for assembly of CotS protein into the coat layer. If the latter is the case, CotS protein synthesized in these mutants may have been degraded by protease(s) in the mother cell compartment. To test these possibilities, the level of CotS protein during sporulation was monitored by Western blotting (Fig. 7). ςK, a sporulation-specific sigma factor which is expressed at T2 and later, was also analyzed as a control (19, 33). CotS was detected in the protein samples solubilized from the wild-type cells at T5 to T8 (Fig. 7A), consistent with the results of Northern blot analysis as described above. CotS protein was not detected in the vegetative and sporulating cells of the gerE mutant (Fig. 7B). In contrast, CotS was detected among the mother cell proteins of the cotE mutant from T5 onward (Fig. 7C), although CotS was not detected in the spores of cotE mutant cells at T18 (Fig. 6B, lane 6). These results indicate that while GerE is required for cotS expression, CotE is essential for the assembly of CotS in the coat layer.
FIG. 7.
Immunoblot analysis of CotS expression. B. subtilis 168trpC2 (A), 1G12 (gerE mutant) (B), and 1S105 (cotE mutant) (C) were grown in DS medium, and the cultures were harvested every hour throughout sporulation (T0 to T8). Whole-protein samples were solubilized from the sporulating cells and were analyzed by immunoblotting using anti-CotS and anti-ςK antibodies. The arrowheads indicate the migration positions of ςK and CotS.
DISCUSSION
Previously we speculated that expression of the cotS operon is dependent on ςK and the regulatory protein GerE (2). To test this idea, we examined expression of cotS by Northern blot and Western blot analyses. CotS protein was first detectable at T5 in wild-type cells, whereas almost no CotS protein was synthesized in ςE, ςF, ςG, ςK, and gerE mutants. Northern blot analysis revealed a 2.8-kb mRNA as the product of cotS, but the cotS transcript was barely detectable in both the ςK and GerE mutants at T8 (Fig. 4). CotS was detected in the sporangium but not in the spores of cotE mutants by Western blot analysis. These observations suggest that expression of cotS was dependent on both ςK and GerE and that assembly of CotS into the coat layer required CotE.
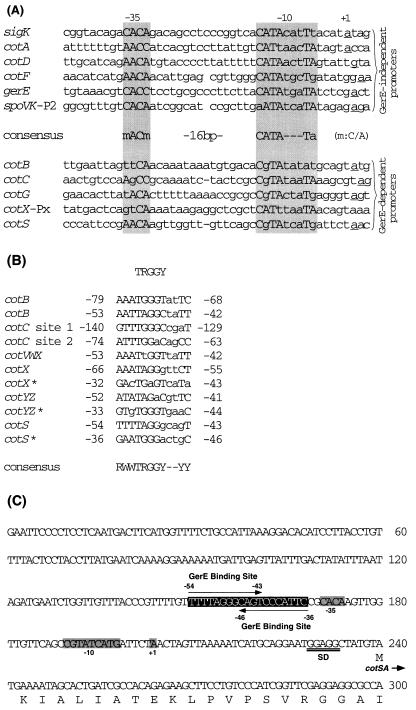
Alignment of promoter regions of genes transcribed by RNA polymerase containing ςK is summarized in Fig. 8A. Six of these genes have GerE-independent promoters, whereas the others have GerE-dependent promoters (14, 25, 37, 39, 40). GerE-independent promoters have a good consensus sequence in their −35 and −10 regions (CATA---Ta). In contrast, GerE-dependent promoters do not have this consensus sequence. Comparison of the cotS promoter with the consensus sequence indicates that the cotS promoter belongs to the GerE-dependent promoter family.
FIG. 8.
Alignment of promoter regions of genes transcribed by ςK RNA polymerase. (A) Sequences near the transcription start sites of genes transcribed by RNA polymerase containing ςK. Promoters for six genes transcribed in the absence of GerE and five genes whose transcription required GerE in addition to ςK are shown separately. Nucleotides in each promoter that match the consensus sequence are shown between the groups (m = C or A). The underlined nucleotides correspond to the transcription start point. (B) Alignment of nucleotide sequences of GerE-binding sites. The consensus sequence proposed by Zheng et al. (39) is shown at the top. Numbers refer to positions relative to the transcriptional start site. ∗, sequence from the opposite DNA strand. The bottom line shows an enlarged consensus sequence for GerE binding based on the sequence shown. R, purine; W, A or T; Y, pyrimidine. (C) Nucleotide sequence of the cotS promoter, showing putative −35 and −10 regions, the transcription start point (+1), and a ribosome-binding site (SD) (2). The boxed sequence is a putative GerE− binding site in the cotS operon. References for the sequences of these promoters are as follows: sigK (spoIVCB), 20; cotA, 29; cotB and cotD, 39; cotC, 39 and 40; cotF, 9; gerE, 8; spoVK, 14; cotVXW, cotX, and cotYZ, 36; cotG, 26; and cotS, 2.
Runoff transcription and DNase I footprinting studies of cotX, cotY, and cotZ of B. subtilis, whose transcription is dependent on ςK and GerE, indicate that the consensus sequence of GerE-binding sites in these genes is RWWTRGGY--YY (R, purine; W, A or T; Y, pyrimidine) (37). Comparison of the cotS promoter region with the GerE consensus sequence indicated that the two have similar sequences upstream of the −35 region (Fig. 8B). Two putative consensus regions are possible. One is a 12-bp stretch extending from positions −54 to −43 on the transcribed strand, and the other extends from positions −36 to −46 on the nontranscribed strand. Consequently, we note the presence of a possible GerE-binding site in the cotS regulatory region (Fig. 8C).
One of the most striking results in this report concerns the interaction of CotE and CotS. Proper formation of the spore coat depends on the CotE protein (3), and spores from a cotE mutant lack the outer coat (37). Immunoelectron microscopic analysis indicated that CotS is localized in the inner coat and/or on the outside region of the cortex of dormant spores. We speculate that CotE protein also functions in assembly of a certain inner coat protein in addition to outer coat morphogenesis. Spore coat components synthesized in the mother cell compartment at an intermediate stage of sporulation could interact with CotE and then be assembled into the inner-laminated layer or outer layer to organize a rigid mature spore coat.
ACKNOWLEDGMENTS
We thank Adam Driks for helpful discussion and Michael G. Bramucci for critical reading of the manuscript. We thank Yoshito Sadaie and Masaya Fujita for providing antisera against ςA and ςK. We also thank Sayoko Nakao for technical assistance.
REFERENCES
- 1.Abe A, Ogawa S, Kohno T, Watabe K. Purification of Bacillus subtilis spore coat protein by electrophoretic elution procedure and determination of NH2-terminal amino acid sequences. Microbiol Immunol. 1993;37:809–812. doi: 10.1111/j.1348-0421.1993.tb01709.x. [DOI] [PubMed] [Google Scholar]
- 2.Abe A, Koide H, Kohno T, Watabe K. A Bacillus subtilis spore coat polypeptide gene, cotS. Microbiology. 1995;141:1433–1442. doi: 10.1099/13500872-141-6-1433. [DOI] [PubMed] [Google Scholar]
- 3.Adam D, Roels S, Beall B, Moran C P, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 4.Aronson A I, Fitz-James P C. Structure and morphogenesis of the bacterial spore coat. Bacteriol Rev. 1976;40:360–402. doi: 10.1128/br.40.2.360-402.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronson A I, Song H-Y, Bourne N. Gene structure and precursor processing of a novel Bacillus subtilis spore coat protein. Mol Microbiol. 1989;3:437–444. doi: 10.1111/j.1365-2958.1989.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 6.Beall B, Driks A, Losick R, Moran C P., Jr Cloning and characterization of a gene required for the assembly of the Bacillus subtilis spore coat. J Bacteriol. 1993;175:1705–1716. doi: 10.1128/jb.175.6.1705-1716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourne N, Fitz-James P C, Aronson A I. Structural and germination defects of Bacillus subtilis spores with altered contents of a spore coat protein. J Bacteriol. 1991;173:6618–6625. doi: 10.1128/jb.173.20.6618-6625.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutting S, Panzer S, Losick R. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J Mol Biol. 1989;207:393–404. doi: 10.1016/0022-2836(89)90262-3. [DOI] [PubMed] [Google Scholar]
- 9.Cutting S, Zheng L, Losick R. Gene encoding two alkali-soluble components of the spore coat from Bacillus subtilis. J Bacteriol. 1991;173:2915–2919. doi: 10.1128/jb.173.9.2915-2919.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of gene in the signal transduction pathway governs pro-ςK processing in Bacillus subtilis. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 11.Cutting S M, Vander Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1990. pp. 27–74. [Google Scholar]
- 12.Donovan W, Zheng L, Sandman K, Losick R. Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol. 1987;196:1–10. doi: 10.1016/0022-2836(87)90506-7. [DOI] [PubMed] [Google Scholar]
- 13.Driks A, Roels S, Beall B, Moran C P, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 14.Foulger D, Errington E. Sequential activation of dual promoters by different sigma factors maintains spoVJ expression during successive developmental stages of Bacillus subtilis. Mol Microbiol. 1991;5:1363–1373. doi: 10.1111/j.1365-2958.1991.tb00783.x. [DOI] [PubMed] [Google Scholar]
- 15.Fujita Y, Yasuda Y, Tochikubo K. Permeability of gentamicin and polymyxin B into the inside of Bacillus subtilis spores. Microbiol Immunol. 1990;34:1013–1023. doi: 10.1111/j.1348-0421.1990.tb01523.x. [DOI] [PubMed] [Google Scholar]
- 16.Herrin D L, Schmidt G W. Rapid, reversible staining of Northern blots prior to hybridization. BioTechniques. 1988;6:196–197. [PubMed] [Google Scholar]
- 17.Igo M M, Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986;191:615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- 18.Jenkinson H F, Sawyer W D, Mandelstam J. Synthesis and order of assembly of spore coat proteins in Bacillus subtilis. J Gen Microbiol. 1981;123:1–16. [Google Scholar]
- 19.Krooss L, Kunkel B, Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989;243:526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel B, Sandman K, Panzer S, Youngman P, Losick R. The promoter for a sporulation gene in the spoIVC locus of Bacillus subtilis and its use in studies of temporal and spatial control of gene expression. J Bacteriol. 1988;170:3513–3522. doi: 10.1128/jb.170.8.3513-3522.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunst F, Ogasawara N, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–258. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 22.Moir A. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J Bacteriol. 1981;146:1106–1116. doi: 10.1128/jb.146.3.1106-1116.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakane A, Takamatsu H, Oguro A, Sadaie Y, Nakamura K, Yamane K. Acquisition of azide-resistance by elevated SecA ATPase activity confers azide-resistance upon cell growth and protein translocation in Bacillus subtilis. Microbiology. 1995;141:113–121. doi: 10.1099/00221287-141-1-113. [DOI] [PubMed] [Google Scholar]
- 24.Ricca E, Baccigalupi L, Naclerio G, Cutting S. Spore coat differentiation in Bacillus subtilis. Res Microbiol. 1997;148:5–9. doi: 10.1016/S0923-2508(97)81894-3. [DOI] [PubMed] [Google Scholar]
- 25.Roels S, Losick R. Adjacent and divergently oriented operons under the control of the sporulation regulatory protein GerE in Bacillus subtilis. J Bacteriol. 1995;177:6263–6275. doi: 10.1128/jb.177.21.6263-6275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sacco M, Ricca E, Losick R, Cutting S. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J Bacteriol. 1995;177:372–377. doi: 10.1128/jb.177.2.372-377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakae Y, Yasuda Y, Tochikubo K. Immunoelectron microscopic localization of one of the spore germination proteins, GerAB, in Bacillus subtilis spores. J Bacteriol. 1995;177:6294–6296. doi: 10.1128/jb.177.21.6294-6296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Sandman K, Kroos L, Cutting S, Youngman P, Losick R. Identification of the promoter for a spore coat protein genes in Bacillus subtilis and studies on the regulation of its induction at a late stage of sporulation. J Mol Biol. 1988;200:461–473. doi: 10.1016/0022-2836(88)90536-0. [DOI] [PubMed] [Google Scholar]
- 30.Schaefer P, Millet J, Aubert J. Catabolite repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:707–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekiguchi J, Akeo K, Yamamoto H, Khasanov F K, Alonso J C, Kuroda A. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which affects germination in Bacillus subtilis. J Bacteriol. 1995;177:5582–5589. doi: 10.1128/jb.177.19.5582-5589.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setlow P. Spore structure proteins. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 801–809. [Google Scholar]
- 33.Stragier P, Kunkel B, Kroos L, Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989;234:507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- 34.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 35.Takamatsu H, Bunai K, Horinaka T, Oguro A, Nakamura K, Watabe K, Yamane K. Identification of a region required for binding to presecretory protein in Bacillus subtilis Ffh, a homologue of the 54-kDa subunit of mammalian signal recognition particle. Eur J Biochem. 1997;248:575–582. doi: 10.1111/j.1432-1033.1997.00575.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Fitz-James P C, Aronson A I. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J Bacteriol. 1993;175:3757–3766. doi: 10.1128/jb.175.12.3757-3766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Ichikawa H, Halberg R, Kroos L, Aronson A I. Regulation of the transcription of a cluster of Bacillus subtilis spore coat genes. J Mol Biol. 1994;240:405–415. doi: 10.1006/jmbi.1994.1456. [DOI] [PubMed] [Google Scholar]
- 38.Zheng L, Donovan W P, Fitz-James P C, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988;2:1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]
- 39.Zheng L, Losick R. Cascade regulation of spore coat gene expression in Bacillus subtilis. J Mol Biol. 1990;212:645–660. doi: 10.1016/0022-2836(90)90227-d. [DOI] [PubMed] [Google Scholar]
- 40.Zheng L, Halberg R, Roels S, Ichikawa H, Kroos L, Losick R. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J Mol Biol. 1992;226:1037–1050. doi: 10.1016/0022-2836(92)91051-p. [DOI] [PubMed] [Google Scholar]