Abstract
O antigen is part of the lipopolysaccharide present in the outer membrane of gram-negative bacteria. The surface-exposed O antigen is subject to selection by the host immune system, which may account for the maintenance of many different O-antigen forms. Characteristically, all genes specific to O-antigen synthesis are clustered in a region close to the his and gnd genes on the chromosome of Escherichia coli and related species. Shigella sonnei, essentially a clone of E. coli (E. coli clone Sonnei), is an important human pathogen and is unusual in that its O-antigen gene cluster is located on a plasmid. Our results suggest that it once had a normal chromosomal O-antigen gene cluster which has been largely deleted. We suggest that the O antigen encoded by the plasmid-borne genes offered a selective advantage in adapting to a new environment and that the chromosomal O-antigen genes were eventually inactivated. We also identified, by PCR and sequencing, a potential ancestor of E. coli Sonnei among the 166 known E. coli serotype strains.
Shigella spp. cause diseases ranging from diarrhea to bacillary dysentery. Four species—Shigella dysenteriae, Shigella flexneri, Shigella boydii, and Shigella sonnei—are recognized, but all are sufficiently similar to Escherichia coli to be placed in the same species (4, 6, 11, 22, 25). Thus, Shigella strains should be treated as E. coli clones, with host specificity and a particular mode of pathogenesis. We refer to Shigella sonnei as E. coli clone Sonnei.
The O antigen is an extremely variable surface polysaccharide. There are 166 known O antigens recognized in the E. coli typing scheme (18) and about 37 among Shigella strains (5, 7, 8, 17). The genes for O-antigen synthesis are normally grouped together on the chromosome in a gene cluster which maps close to gnd in both E. coli and Salmonella enterica. We, among others, have undertaken an extensive study of the genetic basis of O-antigen variation by sequencing and identifying the O-antigen genes, mostly in S. enterica and E. coli (see reference 26 for a review). It has been found that, in general, O-antigen genes are of low G+C content (usually less than 40%). We have suggested that this deviation in G+C content from those of typical S. enterica or E. coli genes (51%) indicates that the O-antigen DNA originated in species other than S. enterica or E. coli and was captured by lateral transfer (15).
Most E. coli strains have a colanic acid (CA) gene cluster upstream of the O-antigen gene cluster, separated by two genes including galF (Fig. 1) (28). CA is an extracellular polysaccharide which is widely found within E. coli and other species of the family Enterobacteriaceae, including S. enterica (12). CA contains fucose, and the gene cluster contains the manC and manB genes (Fig. 1) (28), which encode phosphomannomutase and mannose-1-phosphate guanyltransferase, both involved in the synthesis of GDP-mannose, GDP-fucose, and GDP-colitose (2, 10, 15, 28). When an O antigen has one or more of these sugars, there are manB and manC genes in the O-antigen gene cluster. For example in S. enterica LT2, there are copies of these two genes in both the CA and O-antigen gene clusters (29). The two copies of the man genes can exhibit major sequence divergence without any functional divergence (1, 16). The O-antigen gene manB is of particular interest, as in S. enterica C1 (16) and E. coli O7 (20) it differs in G+C content from the manC gene of the same O-antigen gene cluster and closely resembles in G+C content and sequence the manB gene of the CA gene cluster of the same species (61% G+C content in S. enterica and 55% in E. coli). It appears that the manB genes of the S. enterica C1 and E. coli O7 clusters have been obtained from a CA gene cluster (16, 20).
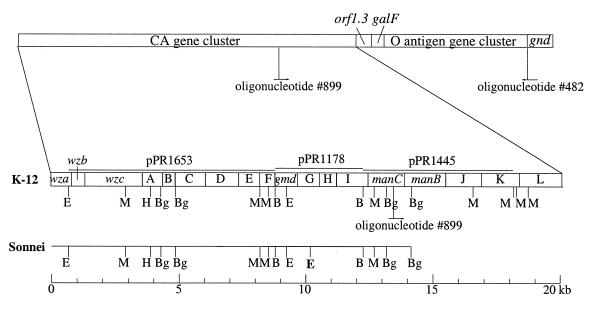
FIG. 1.
Organization of the E. coli K-12 CA and O-antigen gene clusters and comparison of the CA gene clusters of K-12 and clone Sonnei (strain M564 [ATCC 11060]). Restriction sites (E, EcoRI; M, MluI; H, HindIII; Bg, BglII) in the CA region are indicated. The EcoRI site at about position 10.1, which is not present in the K-12 CA DNA, is indicated by boldface type. Regions covered by plasmids used in Southern blotting are shown above the K-12 CA map. The priming sites of oligonucleotides 899 and 482 are indicated.
Clone Sonnei has an O antigen not otherwise found in E. coli but which is identical to that of serotype 17 of Plesiomonas shigelloides. The O-antigen genes of Sonnei are unusual in that they occur on a plasmid; it has recently been shown that they will hybridize to the chromosomal O-antigen genes of P. shigelloides serotype O17 (30) and that for at least a few hundred base pairs the Sonnei O-antigen gene cluster is identical in sequence to that of P. shigelloides O17 (14). Sonnei, and indeed all Shigella and many other human pathogenic clones of E. coli, are thought to have relatively recently adapted to the diarrhea-causing mode of pathogenesis in humans (9, 21). It appears that the Sonnei O antigen has been transferred from P. shigelloides, and one of us has suggested that the acquisition of an O antigen from another species may be related to this adoption of a new niche in the last 10,000 years (27).
Nothing was known of the chromosomal O-antigen genes presumably present in Sonnei prior to acquisition of the plasmid-encoded gene cluster. Sonnei strains which have lost the plasmid lack O antigen, suggesting that O-antigen genes at the chromosomal site are nonfunctional. We report the cloning and sequencing of remnant chromosomal O-antigen genes from Sonnei and show that the upstream part of the CA gene cluster and the downstream part of the original O-antigen gene cluster have been fused by a deletion involving recombination between the manB genes in each cluster.
Location of Sonnei CA genes by Southern blotting.
We first attempted long PCR amplification of the clone Sonnei chromosomal O-antigen gene cluster by using primers for the JUMPstart sequence present at the 5′ end of the O-antigen gene clusters (13) and the gnd gene (Fig. 1), but this failed to give a product. We then carried out PCR to amplify the galF and gnd genes (Fig. 1) but succeeded only with the gnd gene and concluded that there could be a deletion from the CA gene cluster extending into the O-antigen cluster. We next looked at the CA gene cluster in Sonnei by Southern blotting with clones of E. coli K-12 CA DNA (28) as probes.
The inserts of plasmids pPR1653, pPR1178, and pPR1445, which together cover most of the E. coli K-12 CA gene cluster (Fig. 1) (28), were labelled and used as probes for hybridization to blotted restriction fragments of Sonnei and K-12 chromosomal DNA. The inserts of pPR1653 and pPR1178, carrying the K-12 CA DNA from positions 728 to 8780 and 8780 to 12310 (K-12 CA map positions; GenBank accession no. U38473), hybridized to Sonnei DNA (data not shown); comparison of the Southern blots for BamHI, BglII, MluI, MluI-HindIII, EcoRI-BglII, EcoRI-BamHI, and BamHI-HindIII shows that the K-12 sites in this region are conserved in Sonnei but that there is one extra EcoRI site at about position 10100 in Sonnei (Fig. 1). Hybridization of the insert of pPR1445 shows that restriction sites from position 12310 to the BglII site at position 14204 are also present in Sonnei (Fig. 1). Thus, CA DNA from the 5′ end of the gene cluster to at least position 14204 is present in Sonnei, but most of the remainder appears to have been lost.
Sonnei DNA between the CA and gnd genes.
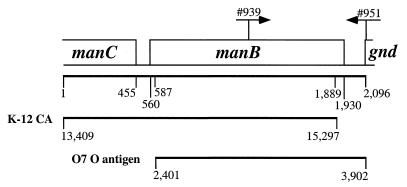
To study the clone Sonnei DNA between the CA gene cluster and the gnd gene, primers binding to K-12 CA DNA positions 13387 to 13408 (primer 899) and gnd DNA positions 39 to 4 (primer 482) were used to PCR amplify Sonnei chromosomal DNA; a 2.1-kb DNA fragment was obtained and was then cloned into pGEM-T to make plasmid pPR1790. The insert of plasmid pPR1790 was sequenced, and the sequence was compared with those in databases. The sequence from positions 1 to 586 has 98.5% identity at the DNA level with K-12 CA DNA from positions 13409 to 13994, which comprises the 3′ half of manC and the first nine codons of manB (Fig. 2) (28). The sequence from positions 587 to 1889, which comprises all but the first 9 and last 12 codons of manB, is almost identical to those of both K-12 CA DNA (positions 13995 to 15297) (28) and E. coli O7 DNA (positions 2401 to 3702), with 97 and 96% identity, respectively (Fig. 2) (19). The DNA from positions 1890 to 2096, which includes the last 12 codons of manB, the intergenic region, and the first codon of gnd (Fig. 2), has 93.7% identity with E. coli O7 DNA (19) (positions 3703 to 3907).
FIG. 2.
The clone Sonnei chromosomal O-antigen region. Gene names and relevant base positions are given. Priming sites for the oligonucleotides used in PCR against all known E. coli serotypes are indicated by arrows. Regions with high-level DNA identity to K-12 CA or O7 O-antigen DNA are indicated by lines below the diagram.
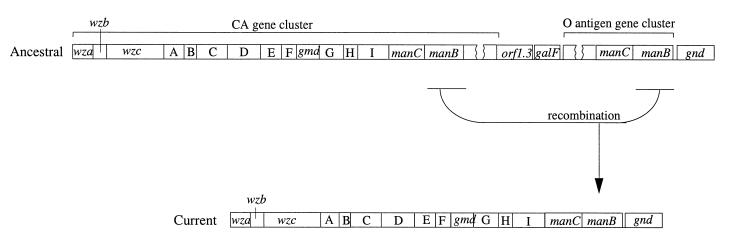
The Sonnei DNA that we have sequenced has the CA form from positions 1 to 1889 and the O7 O-antigen form from positions 587 to 2096, with overlap between positions 587 and 1889, where the CA and O7 forms are very similar. The simplest hypothesis is that the parent of the Sonnei clone had an O-antigen gene cluster resembling that of E. coli O7, with a manB gene nearly identical (except for the last 12 codons) to that of the CA gene cluster, and that recombination between the manB genes in this segment of nearly identical sequence led to deletion of three genes of the CA cluster, two intervening genes, and all but the manB gene of the O-antigen cluster (Fig. 3).
FIG. 3.
Potential mechanism for the inactivation of the clone Sonnei chromosomal O-antigen gene cluster.
O antigen of the ancestral Sonnei strain.
In E. coli O7, the gene order at the end of the O-antigen gene cluster is manC, manB, and gnd, and the clone Sonnei sequence is consistent with the parent being an O7 E. coli strain. To determine if O7 strains are the only potential parents for Sonnei, we carried out PCR on representative strains for each of the 166 known E. coli O antigens (7, 23, 24) by using oligonucleotides 939 (5′ TTTGATGGCGATTTTGACCGC) and 951 (5′ CATTGTTTACTCCTGTCAGGG). Oligonucleotide 939 binds to positions 1289 to 1339 in the middle of the Sonnei manB gene, and 951 binds to positions 2096 to 2076, part of the intergenic region between manB and gnd including the first codon of gnd (Fig. 2). Chromosomal DNA was isolated with the Promega genomic isolation kit and checked by gel electrophoresis. PCR amplification of the mdh gene with the same primers as used by Boyd et al. (3) was done as a positive control for each of the 166 strains. In addition to O7, strains from 14 other serotypes produced PCR bands of the same size as that of Sonnei (with the differences in sequence from that of Sonnei in parentheses): O6 (3.51%), O7 (5.56%), O11 (2.92%), O14 (4.24%), O20 (2.92%), O34 (3.65%), O39 (3.36%), O41 (4.09%), O43 (4.09%), O58 (4.53%), O62 (1.61%), O66 (12.28%), O88 (3.51%), O125 (4.09%), and O131 (3.8%). These strains must each have a manB gene as the final gene in the O-antigen gene cluster. We conclude that Sonnei may have arisen from an O62 strain, as its manB sequence shows only 1.61% difference from that of Sonnei in the sequenced region.
Conclusions.
We have shown that clone Sonnei has a remnant O-antigen gene cluster on the chromosome, most of the original cluster having apparently been deleted by homologous recombination between manB genes in the O antigen and adjacent CA gene clusters. Sonnei has its functional O-antigen genes on a plasmid, and they are thought to have been acquired from P. shigelloides by lateral transfer on this plasmid. The finding that a presumably typical chromosomal O-antigen gene cluster has been lost by deletion indicates that the current situation in Sonnei arose by a gain of O-antigen genes on a plasmid followed by inactivation of the original chromosomal O-antigen gene cluster. Diseases of humans caused by enteric bacterial infections are thought to have emerged after agricultural settlement, about 8,000 B.C., because the natures of their infection and transmission make them unlikely to be successful in the previous hunter-gatherer society (9, 21). Thus, Sonnei is thought to have emerged as a human pathogenic clone of E. coli in the last 10,000 years. We suggest that capture of the O-antigen gene cluster from P. shigelloides and loss of function of the original O-antigen gene cluster occurred in that period as part of the adaptation to a new niche.
Nucleotide sequence accession numbers.
The Sonnei sequence has been deposited in GenBank under accession no. AF031957; those of E. coli strains are as follows: AF053603 (O6), AF053592 (O11), AF053595 (O14), AF053596 (O20), AF053597 (O34), AF053598 (O39), AF053599 (O41), AF053600 (O43), AF053601 (O58), AF053602 (O62), AF053605 (O66), AF053604 (O88), AF053593 (O125), and AF053594 (O131).
Acknowledgments
We thank Heather Curd for excellent technical assistance.
This investigation was supported by a grant from the Australian Research Council.
REFERENCES
- 1.Aoyama K M, Haase A M, Reeves P R. Evidence for effect of random genetic drift on G+C content after lateral transfer of fucose pathway genes to Escherichia coli K-12. Mol Biol Evol. 1994;11:829–838. doi: 10.1093/oxfordjournals.molbev.a040166. [DOI] [PubMed] [Google Scholar]
- 2.Bastin D A, Reeves P R. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene. 1995;164:17–23. doi: 10.1016/0378-1119(95)00459-j. [DOI] [PubMed] [Google Scholar]
- 3.Boyd E F, Nelson K, Wang F-S, Whittam T S, Selander R K. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 1994;91:1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner D J, Fanning G R, Miklos G V, Steigerwalt A G. Polynucleotide sequence relatedness among Shigella species. Int J Syst Bacteriol. 1973;23:1–7. [Google Scholar]
- 5.Echeverria P, Hoge C W, Bodhidatta L, Serichantalergs O, Dalsgaard A, Eampokalap B, Perrault J, Pazzaglia G, O’Hanley P, English C. Molecular characterization of Vibrio cholerae O139 isolates from Asia. Am J Trop Med Hyg. 1995;52:124–127. doi: 10.4269/ajtmh.1995.52.124. [DOI] [PubMed] [Google Scholar]
- 6.Ewing W H. Serological relationships between Shigella and coliform cultures. J Bacteriol. 1953;66:333–340. doi: 10.1128/jb.66.3.333-340.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewing W H. Edwards and Ewing’s identification of the Enterobacteriaceae. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. [Google Scholar]
- 8.Farmer J J, III, Kelly M T. Enterobacteriaceae. In: Balows A, Hausler W J J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C: American Society for Microbiology; 1991. pp. 360–383. [Google Scholar]
- 9.Fenner F. The effects of changing social organisation on the infectious diseases of man. In: Boyden S W, editor. The impact of civilization on the biology of man. Toronto, Canada: University of Toronto Press; 1970. pp. 48–68. [Google Scholar]
- 10.Gabriel O. Biosynthesis of sugar residues for glycogen, peptidoglycan, lipopolysaccharide, and related systems. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 504–511. [Google Scholar]
- 11.Goullet P. Esterase electrophoretic pattern between Shigella species and Escherichia coli. J Gen Microbiol. 1980;117:493–500. doi: 10.1099/00221287-117-2-493. [DOI] [PubMed] [Google Scholar]
- 12.Grant W D, Sutherland I W, Wilkinson J F. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J Bacteriol. 1969;100:1187–1193. doi: 10.1128/jb.100.3.1187-1193.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobbs M, Reeves P R. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol Microbiol. 1994;12:855–856. doi: 10.1111/j.1365-2958.1994.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 14.Houng H H, Zapor M J, Hartman A B, Hale T L, Venkatesan M M. The roles of IS630 sequence in the expression of the form I antigen of Shigella sonnei: molecular and evolutionary aspects. In: van der Zeijst B A M, Hoekstra W P M, van Embden J D A, van Alphen A J W, editors. Ecology of pathogenic bacteria: molecular and evolutionary aspects. Amsterdam, The Netherlands: Elsevier; 1997. pp. 282–283. [Google Scholar]
- 15.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee S J, Romana L K, Reeves P R. Sequence and structural analysis of the rfb (O antigen) gene cluster from a group C1 Salmonella enterica strain. J Gen Microbiol. 1992;138:1843–1855. doi: 10.1099/00221287-138-9-1843. [DOI] [PubMed] [Google Scholar]
- 17.Le Minor L, Richard C, editors. Shigella: Méthod de laboratoire pour l’identification des entérobactéries. Paris, France: Institut Pasteur; 1993. [Google Scholar]
- 18.Lior H. Classification of Escherichia coli. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Tucson, Ariz: CAB International; 1994. pp. 31–72. [Google Scholar]
- 19.Marolda C L, Valvano M A. Identification, expression, and DNA sequence of the GDP-mannose biosynthesis genes encoded by the O7 rfb gene cluster of strain VW187 (Escherichia coli O7:K1) J Bacteriol. 1993;175:148–158. doi: 10.1128/jb.175.1.148-158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marolda C L, Welsh J, Dafoe L, Valvano M A. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J Bacteriol. 1990;172:3590–3599. doi: 10.1128/jb.172.7.3590-3599.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKeown T. The origins of human disease. Oxford, England: Basil Blackwell; 1988. [Google Scholar]
- 22.Ochman H, Whittam T S, Caugant D A, Selander R K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- 23.Ørskov I, Ørskov F, Rowe B. Six new E. coli O groups: O165, O166, O167, O168, O169 and O170. Acta Pathol Microbiol Immunol Scand. 1984;92:189–193. doi: 10.1111/j.1699-0463.1984.tb02819.x. [DOI] [PubMed] [Google Scholar]
- 24.Ørskov I, Wachsmuth K, Taylor D N, Echeverria P, Rowe B, Sakazaki R, Ørskov F. Two new Escherichia coli O groups: O172 from “Shiga-like” toxin II-producing strains (EHEC) and O173 from enteroinvasive E. coli (EIEC) APMIS. 1991;99:30–32. doi: 10.1111/j.1699-0463.1991.tb05114.x. [DOI] [PubMed] [Google Scholar]
- 25.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reeves P R. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 27.Reeves P R. Specialized clones and lateral transfer in pathogens. In: van der Zeijst B A M, Hoekstra W P M, van Embden J D A, van Alphen A J W, editors. Ecology of pathogenic bacteria: molecular and evolutionary aspects. Amsterdam, The Netherlands: Elsevier; 1997. pp. 237–254. [Google Scholar]
- 28.Stevenson G, Andrianopoulos K, Hobbs H, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevenson G, Lee S J, Romana L K, Reeves P R. The cps gene cluster of Salmonella strain LT2 includes a second mannose pathway: sequence of two genes and relationship to genes in the rfb gene cluster. Mol Gen Genet. 1991;227:173–180. doi: 10.1007/BF00259668. [DOI] [PubMed] [Google Scholar]
- 30.Viret J-F, Cryz S J, Jr, Lang A B, Favre D. Molecular cloning and characterisation of the genetic determinants that express the complete Shigella serotype D (Shigella sonnei) lipopolysaccharide in heterologous live attenuated vaccine strains. Mol Microbiol. 1993;7:239–252. doi: 10.1111/j.1365-2958.1993.tb01115.x. [DOI] [PubMed] [Google Scholar]