Abstract
Secretogranin 2 (Scg2) is a member of the secretogranin/chromogranin family of proteins that is involved in neuropeptide and hormone packaging to secretory granules and serves as a precursor for several secreted pleiotropic peptides. A recent study in zebrafish showed that the teleost Scg2 orthologs, scg2a and scg2b, play an important role in mating behavior, but its modes of action and regulatory mechanisms remain unclear. In this study, we identify scg2a in another teleost species, medaka, by transcriptomic analysis as a gene that is expressed in an ovarian secretion-dependent manner in a group of neurons relevant to female sexual receptivity, termed FeSP neurons. Investigation of scg2a expression in the FeSP neurons of estrogen receptor (Esr)-deficient medaka revealed that it is dependent on estrogen signaling through Esr2b, the major determinant of female-typical mating behavior. Generation and characterization of scg2a-deficient medaka showed no overt changes in secretory granule packaging in FeSP neurons. This, along with the observation that Scg2a and neuropeptide B, a major neuropeptide produced by FeSP neurons, colocalize in a majority of secretory granules, suggests that Scg2a mainly serves as a precursor for secreted peptides that act in conjunction with neuropeptide B. Further, scg2a showed sexually biased expression in several brain nuclei implicated in mating behavior. However, we found no significant impact of scg2a deficiency on the performance of mating behavior in either sex. Collectively, our results indicate that, although perhaps not essential for mating behavior, scg2a acts in an estrogen/Esr2b signaling-dependent manner in neurons that are relevant to female sexual receptivity.
Keywords: estrogen, mating behavior, neuropeptide B, secretogranin 2, secretory granule
Significance Statement.
Secretogranin 2 (Scg2) is an emerging regulator of mating behavior in teleosts that functions in neuropeptide/hormone packaging to secretory granules and acts as a precursor for the neuropeptide secretoneurin (SN). By investigating the expression of scg2a (a teleost ortholog of Scg2) in a group of behaviorally relevant female-specific peptidergic neurons in medaka, we revealed an estrogen-dependent expression mechanism for this gene and determined that the likely function of Scg2a in these neurons is solely as a precursor to SN. Although we were unable to confirm a behavioral role for scg2a in medaka, this work significantly advances our understanding of the regulation and function of scg2a in the nervous system, which may be conserved across species.
Introduction
Secretogranin 2 (Scg2) is a member of the secretogranin/chromogranin family of proteins, expressed broadly throughout the nervous and endocrine systems. Scg2 is targeted to the trans-Golgi network, where it functions to facilitate the sorting and packaging of neuropeptides and hormones to secretory granules, also known as dense-core secretory granules or large dense-core vesicles (1, 2). Scg2 can also be processed in the lumen of secretory granules and serves as a precursor to several distinct pleiotropic peptides, of which secretoneurin (SN) is the best studied (3). Numerous physiological functions have been ascribed to Scg2/SN in mammals and teleosts, including the regulation of the hypothalamic–pituitary–gonadal (HPG) axis (4–7), feeding (8), neuroinflammation (3, 9), and angiogenesis (3, 10).
Whereas mammals and amphibians have a single Scg2 gene, teleosts have two, scg2a and scg2b (which produce SNa and SNb, respectively), as the result of a whole-genome duplication event early in teleost evolution (7). A recent study in zebrafish (Danio rerio) showed that genetic deletion of both scg2a and scg2b resulted in fewer courtship behaviors and decreased spawning success (11), suggesting an important role for these genes in regulating teleost mating behavior. However, their modes of action and regulatory mechanisms remain unclear.
In another teleost species, medaka (Oryzias latipes), we recently identified a group of neurons that occur exclusively in females in the magnocellular/gigantocellular portion of the magnocellular preoptic nucleus (PMm/PMg), which is considered homologous to the mammalian paraventricular nucleus (PVN) (12). These neurons were originally identified as expressing a neuropeptide B gene (npba), but subsequent transcriptomic work showed that they express multiple neuropeptide genes dependent upon ovarian signals and were thus termed female-specific, sex steroid-responsive peptidergic (FeSP) neurons (12–14). These neurons are presumed to be a critical action site of ovarian estrogen on the neural circuitry governing female mating behavior, and so far, behavioral studies on female medaka deficient for several key estrogen-responsive genes expressed in these neurons—npba, esr2b (encoding an estrogen receptor [Esr] subtype), and ptger4b (encoding a prostaglandin E2 receptor)—all showed abnormal receptivity to male courtship (14–16). In particular, esr2b-deficient females were not receptive to males at all, and instead courted other females, suggesting that estrogen/Esr2b signaling in FeSP neurons is the major determinant of the female pattern of mating behavior (16). It is therefore vital to understand what genes act downstream of estrogen/Esr2b signaling in FeSP neurons and how these genes contribute to the establishment of female-typical mating behavior.
In the present study, we identify scg2a as a gene expressed in FeSP neurons dependent on ovarian estrogen. Further characterization of scg2a expression in FeSP neurons reveals that it lies downstream of Esr2b. To assess its significance, we generate scg2a knockout medaka and examine the effects of scg2a deficiency on neuropeptide packaging to secretory granules in FeSP neurons and mating behavior.
Results
FeSP neurons express scg2a dependent on ovarian estrogen
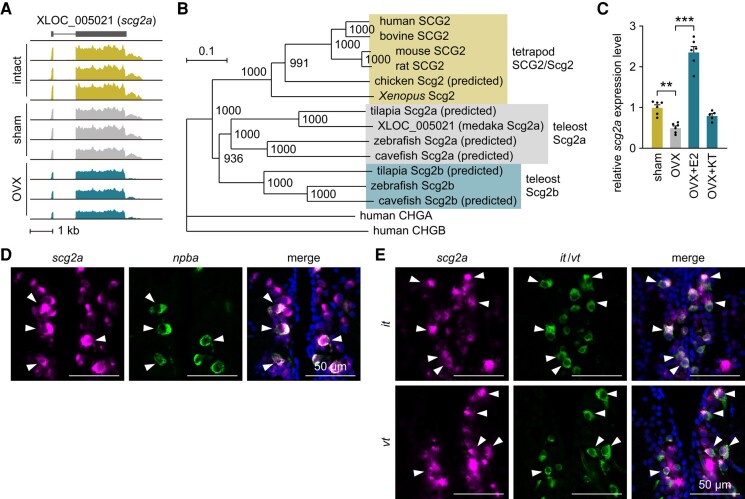
We previously defined the ovarian-dependent transcriptome of FeSP neurons by comparing RNA sequencing (RNA-seq) profiles of FeSP neurons isolated and purified from intact, sham-operated, and ovariectomized female medaka (14). We identified a putative neuropeptide gene (gene ID: XLOC_005021) in the list of genes significantly down-regulated by ovariectomy (Fig. 1A). Further annotation revealed that XLOC_005021 was identical to a medaka Scg2 cDNA found in the National BioResource Project (NBRP) Medaka expressed sequence tag (EST) database (clone ID: olbrno23_o04) and a predicted medaka Scg2 cDNA in the GenBank database (GenBank accession number: XM_004075713). Phylogenetic tree analysis showed that it encodes the medaka ortholog of Scg2a in other teleost species (Fig. 1B).
Fig. 1.
FeSP neurons express scg2a dependent on ovarian estrogen. A) Integrative genomics viewer image of RNA-seq reads from FeSP neurons in the PMm/PMg of intact, sham-operated (sham), and ovariectomized (OVX) females (n = 3 for each treatment) for XLOC_005021 (scg2a). B) Phylogenetic tree showing the relationship of medaka Scg2a to other known granins. The number at each node indicates bootstrap values for 1,000 replicates. Scale bar represents 0.1 substitutions per site. For species names and GenBank accession numbers, see Table S2. CHGA, chromogranin A; CHGB, chromogranin B. C) Levels of scg2a expression in FeSP neurons of females that were sham-operated (sham) or ovariectomized and treated with vehicle only (OVX), estradiol-17β (OVX + E2), or 11-ketotestosterone (OVX + KT) as determined by real-time PCR (n = 6 for all groups except OVX + KT, where n = 5). Mean value of the sham group was arbitrarily set to 1. D and E) Representative micrographs showing the expression of scg2a in FeSP neurons (expressing npba) (D) and it- and vt-expressing neurons (E) in the PMm/PMg. Left and middle panels show images of scg2a (magenta) and npba/it/vt (green) expression, respectively, in the same section; right panels show the merged images with nuclear counterstaining (blue). Arrowheads indicate representative neurons coexpressing scg2a and npba/it/vt. Scale bars represent 50 μm. Statistical differences were assessed by Bonferroni's post hoc test (C). **P < 0.01; ***P < 0.001.
In order to validate the ovarian-dependent expression of scg2a in FeSP neurons and determine the ovarian hormone responsible, we analyzed FeSP neurons independently isolated and purified from females that were sham-operated, ovariectomized, or ovariectomized and treated with estradiol-17β (E2; the major estrogen in vertebrates, including teleosts) or 11-ketotestosterone (KT; the primary, nonaromatizable androgen in teleosts) for scg2a expression by real-time PCR. The results showed that ovariectomy caused a significant decrease in scg2a expression (P = 0.0022), which was recovered by E2 treatment (P < 0.0001), but not KT (Fig. 1C), indicating an induction of scg2a expression in FeSP neurons by ovarian estrogen.
Next, in order to visualize the expression of scg2a in FeSP neurons, we performed double in situ hybridization for scg2a and npba, a typical neuropeptide produced by FeSP neurons (12, 15). All npba-expressing neurons in the PMm/PMg were found to coexpress scg2a (Fig. 1D). In addition to FeSP neurons, scg2a expression was widely observed in neurons throughout the PMm/PMg. In medaka, this brain nucleus is also known to contain populations of isotocin (it) and vasotocin (vt)—the teleost orthologs of mammalian oxytocin and vasopressin, respectively—neurons, which are distinct from FeSP neurons (12, 17). Double in situ hybridization for scg2a and it/vt showed that all it- and vt-expressing neurons in the PMm/PMg also coexpress scg2a (Fig. 1E). These results demonstrate that scg2a is expressed widely throughout the PMm/PMg in all FeSP, isotocin, and vasotocin neurons.
The expression of scg2a in FeSP neurons is mediated by Esr2b
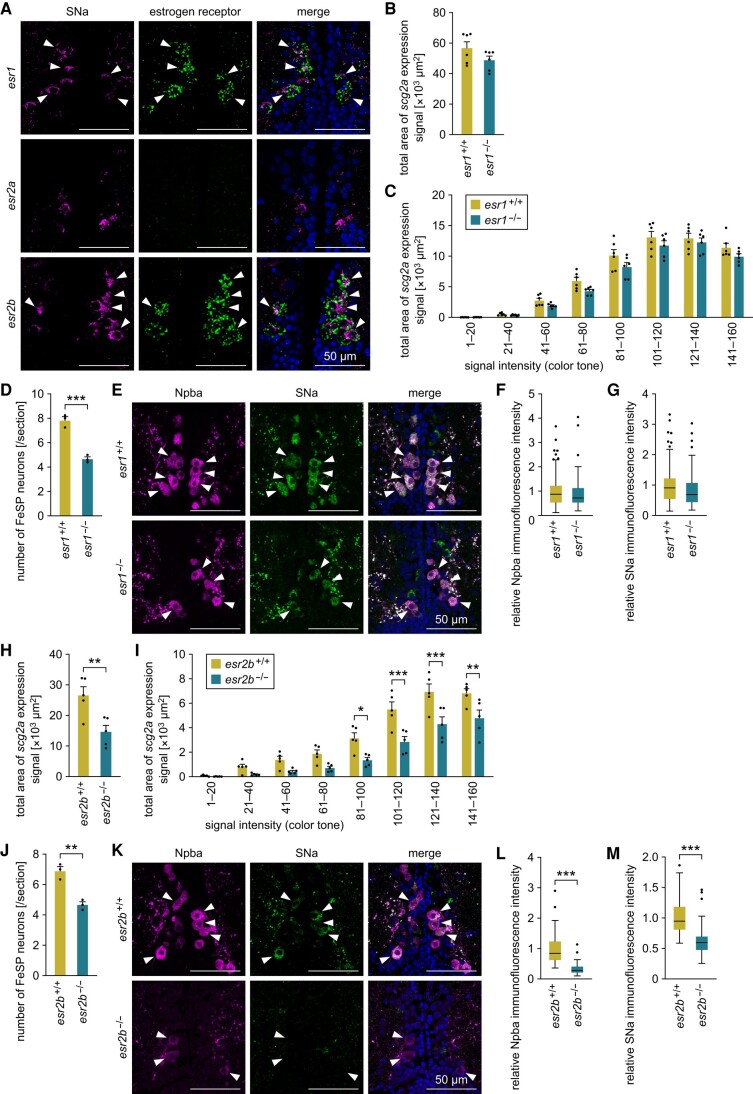
To further explore the estrogenic induction of scg2a expression in FeSP neurons, we examined the colocalization of the SNa polypeptide and Esr transcripts in the PMm/PMg by double labeling with immunohistochemistry and in situ hybridization. Nearly all SNa-positive neurons were found to express esr1 and esr2b, but not esr2a (Fig. 2A), which is in accordance with a previous study detailing the expression of sex steroid receptors in FeSP neurons (12).
Fig. 2.
The expression of scg2a in FeSP neurons is mediated by Esr2b. A) Representative micrographs showing the expression of estrogen receptors in Scg2a-expressing neurons in the PMm/PMg. Left and middle panels show images of SNa immunofluorescence (magenta) and estrogen receptor (esr1/esr2a/esr2b) transcript (green), respectively, in the same section; right panels show the merged images with nuclear counterstaining (blue). Arrowheads indicate representative Scg2a-expressing neurons that coexpress the indicated estrogen receptor. B) Total area of scg2a expression signal in the PMm/PMg of esr1+/+ and esr1−/− females (n = 6 for each genotype). C) Area of scg2a expression signal at different intensities in the PMm/PMg of esr1+/+ and esr1−/− females (n = 6 for each genotype). The x-axis indicates the signal intensity, with smaller numbers representing stronger intensity. D) Number of FeSP neurons per coronal section of the PMm/PMg of esr1+/+ and esr1−/− females (n = 3 for each genotype). E) Representative micrographs showing Npba (left panels; magenta) and SNa (middle panels; green) immunofluorescence in FeSP neurons (arrowheads) of esr1+/+ and esr1−/− females. Right panels show the merged images with nuclear counterstaining (blue). F, G) Relative intensity of Npba (F) and SNa (G) immunofluorescence in FeSP neurons of esr1+/+ (n = 94 neurons) and esr1−/− (n = 70 neurons) females. Mean intensity in esr1+/+ FeSP neurons was arbitrarily set to 1. H) Total area of scg2a expression signal in the PMm/PMg of esr2b+/+ and esr2b−/− females (n = 5 for each genotype). I) Area of scg2a expression signal at different intensities in the PMm/PMg of esr2b+/+ and esr2b−/− females (n = 5 for each genotype). The x-axis indicates the signal intensity, with smaller numbers representing stronger intensity. J) Number of FeSP neurons per coronal section of the PMm/PMg of esr2b+/+ and esr2b−/− females (n = 3 for each genotype). K) Representative micrographs showing Npba (left panels; magenta) and SNa (middle panels; green) immunofluorescence in FeSP neurons (arrowheads) of esr2b+/+ and esr2b−/− females. Right panels show the merged images with nuclear counterstaining (blue). L, M) Relative intensity of Npba (L) and SNa (M) immunofluorescence in FeSP neurons of esr2b+/+ (n = 70 neurons) and esr2b−/− (n = 72 neurons) females. Mean intensity in esr2b+/+ FeSP neurons was arbitrarily set to 1. Scale bars in all panels represent 50 μm. Statistical differences were assessed by unpaired t-test (B, D, F–H, J, M), unpaired t-test with Bonferroni–Dunn correction (C, I), and unpaired t-test with Welch's correction (L). *P < 0.05; **P < 0.01; ***P < 0.001.
Considering these results, we utilized knockout medaka for either esr1 (generated in this study) or esr2b (16) to determine the Esr subtype responsible for scg2a expression in FeSP neurons. esr1 knockout medaka were generated using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated genome editing. An esr1 knockout line was developed containing a 482-bp deletion (Δ482) that resulted in early truncation of the DNA-binding domain and complete loss of the ligand-binding domain (Fig. S1A and B). esr1−/− females had normal body weight and length but exhibited excessively enlarged ovaries, with significantly higher gonad weight (P = 0.0011) and gonad/body weight ratio (P < 0.0001) than their esr1+/+ female siblings (Fig. S1C–F).
We first analyzed the expression of scg2a in the PMm/PMg of esr1+/+ and esr1−/− females by in situ hybridization and found no statistical difference in the total area of scg2a expression signal between genotypes (Fig. 2B). Because the signal intensity of scg2a expression varied greatly among cells, we further quantified scg2a expression by factoring in the signal intensity. Although no significant differences were detected, esr1−/− females showed a lower signal area than esr1+/+ females in all intensity groups (Fig. 2C), suggesting slightly reduced scg2a expression in the PMm/PMg of esr1−/− females. We then performed double-label immunohistochemistry for SNa and Npba to specifically determine the effects of esr1 deficiency on FeSP neurons. The FeSP neurons of esr1−/− females labeled for these polypeptides were significantly reduced in number compared to esr1+/+ females (P = 0.0008), but retained a normal morphology, with a characteristically large cell body and nucleus (Fig. 2D and E). No difference was observed in the immunofluorescence intensity of Npba and SNa between FeSP neurons of esr1+/+ and esr1−/− females (Fig. 2F and G). Taken together, these results suggest that Esr1 does not mediate scg2a expression in FeSP neurons, and instead, the slight decrease in scg2a expression in the PMm/PMg was likely due to a reduction in the total number of FeSP neurons.
We next analyzed the expression of scg2a in the PMm/PMg of esr2b+/+ and esr2b−/− females by in situ hybridization and found a significant reduction in the total area of scg2a expression signal (P = 0.0088; Fig. 2H). Analysis by signal intensity showed that esr2b−/− females had significantly reduced signal area in several intensity groups (81–100, P = 0.0103; 101–120, P < 0.0001; 121–140, P < 0.0001; 141–160, P = 0.0028; Fig. 2I). Immunohistochemical analysis for Npba and SNa demonstrated that, as we have previously reported (16), the FeSP neurons of esr2b−/− females were significantly reduced in number (P = 0.0032) and smaller in size compared to esr2b+/+ females (Fig. 2J and K). Furthermore, the immunofluorescence intensity of both Npba and SNa was drastically reduced in the FeSP neurons of esr2b−/− females (P < 0.0001 for both polypeptides; Fig. 2L and M). Together, these results show that Esr2b is the main Esr subtype mediating scg2a expression in FeSP neurons.
Although FeSP neurons do not project to the pituitary (15), we additionally examined the effects of esr1 and esr2b deficiency on the localization of the SNa polypeptide in the pituitary. Immunohistochemical analysis for SNa on the pituitary revealed a dense plexus of SNa-immunoreactive axons in the central portion of the pituitary (immunoreactivity was also observed in cells in the periphery of the pituitary, but it was not abolished in scg2a knockout fish [see below] and thus appears to be nonspecific; Fig. S2A). Neither esr1 nor esr2b deficiency influenced the SNa-immunoreactive axons in the pituitary (Fig. S2B and C), suggesting that the production and transport of SNa in neurons that project to the pituitary are not dependent on estrogen signaling through these receptors.
Scg2a is not involved in neuropeptide packaging to secretory granules in FeSP neurons
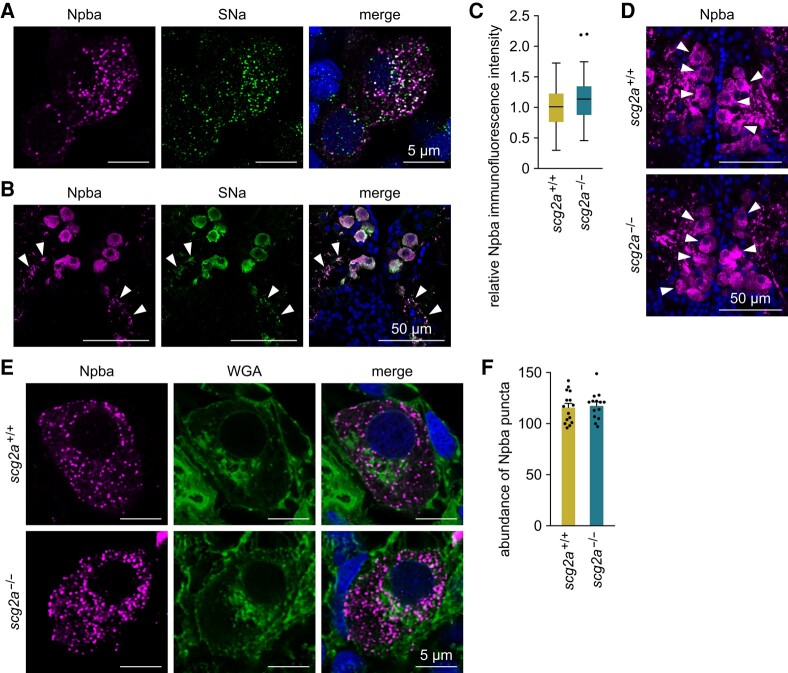
Scg2 has dual functions, being involved in neuropeptide packaging to secretory granules and serving as a precursor for SN (1, 2). To determine its function in FeSP neurons, we first examined the colocalization of Npba and SNa polypeptides at the subcellular level by high-resolution immunohistochemistry. Immunofluorescence for both Npba and SNa was observed in discrete puncta throughout the cell bodies of FeSP neurons, and the majority of Npba puncta were found to be closely associated with SNa immunoreactivity in secretory granules (percentage of Npba puncta that colocalized with SNa = 90.5 ± 1.8%; Pearson's correlation coefficient Rp = 0.40 ± 0.05; Fig. 3A). Immunofluorescence for SNa as well as Npba was also observed in axons emanating from FeSP neurons (Fig. 3B), indicating their axonal transport. These data suggest that Scg2a functionally interacts with Npba, either as cargo for secretory granule packaging or as neuropeptides.
Fig. 3.
Scg2a is not involved in neuropeptide packaging to secretory granules in FeSP neurons. A) Representative micrographs showing the distribution and colocalization of Npba and SNa polypeptides in FeSP neurons of wild-type females. Left and middle panels show images of Npba (magenta) and SNa (green) immunofluorescence, respectively, in the same section; right panel shows the merged image with nuclear counterstaining (blue). Scale bars represent 5 μm. B) Representative micrographs showing Npba (left panel; magenta) and SNa (middle panel; green) immunofluorescence in the axons of FeSP neurons (arrowheads). Right panel shows the merged image with nuclear counterstaining (blue). Scale bars represent 50 μm. C) Relative intensity of Npba immunofluorescence in FeSP neurons of scg2a+/+ (n = 87 neurons) and scg2a−/− (n = 80 neurons) females. Mean intensity in scg2a+/+ FeSP neurons was arbitrarily set to 1. D) Representative micrographs showing Npba immunofluorescence (magenta) in FeSP neurons (arrowheads) of scg2a+/+ and scg2a−/− females. Cell nuclei are shown in blue. Scale bars represent 50 μm. E) Representative micrographs showing the distribution of Npba puncta (left panels; magenta) in scg2a+/+ and scg2a−/− FeSP neurons, counterstained with WGA (middle panels; green). Right panels show merged images with nuclear counterstaining (blue). Scale bars represent 5 μm. F) Abundance of Npba puncta in FeSP neurons of scg2a+/+ (n = 15 neurons) and scg2a−/− (n = 14 neurons) females. Data in panels C–F were obtained from the Δ14 knockout line. Statistical differences were assessed by unpaired t-test (C, F).
To further test these possibilities, we generated scg2a knockout medaka using CRISPR/Cas9 and examined the phenotype of their FeSP neurons. Two independent scg2a knockout lines, one containing a 14-bp deletion (Δ14) and the other a 3-bp deletion and 23-bp insertion (Δ20), were generated to ensure the validity of our results and control for possible off-target effects. Each line contained a frameshift mutation that caused early truncation of the Scg2a protein, eliminating the mature SNa polypeptide (Fig. S3A and B). In both scg2a knockout lines, we found no statistical difference in body weight, body length, gonad weight, or gonad/body weight ratio between genotypes in either males or females (Fig. S3C–J).
We first compared the abundance of Npba in the FeSP neurons of scg2a+/+ and scg2a−/− females by immunohistochemistry. No significant difference was found in the immunofluorescence intensity of Npba between genotypes in either Δ14 (Fig. 3C and D) or Δ20 lines (Fig. S4A and B). To more closely examine the possibility that Scg2a affects Npba-containing secretory granules within FeSP neurons, we next performed high-resolution immunohistochemistry for Npba with wheat germ agglutinin (WGA) counterstain to visualize the outer cellular membrane, endoplasmic reticulum, and Golgi apparatus. Again, no observable difference was found in the intracellular distribution and abundance of Npba puncta between genotypes in either Δ14 (Fig. 3E and F) or Δ20 lines (Fig. S4C and D). Taken together, these results indicate that Scg2a does not affect neuropeptide packaging to secretory granules in FeSP neurons and instead serves mainly as a precursor to SNa, which likely acts in conjunction with Npba as a cotransmitter.
scg2a shows region-specific sex differences in expression in the medaka brain
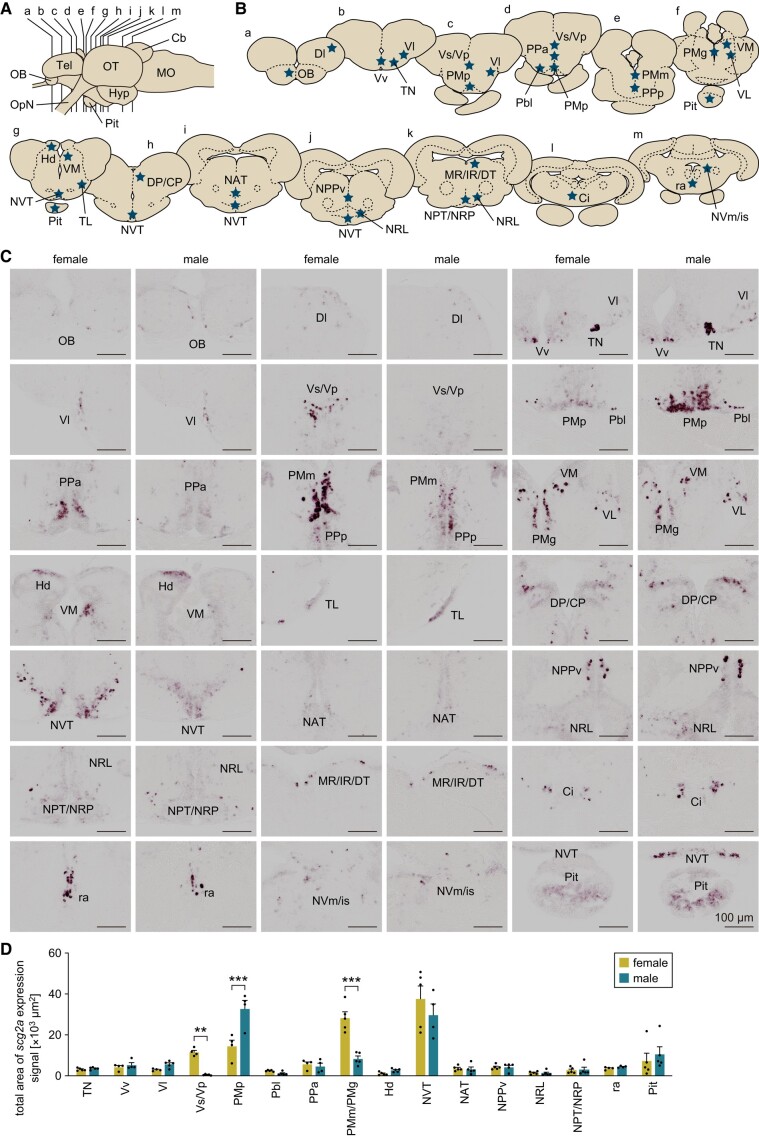
As a prerequisite for studying the behavioral impact of scg2a, we next determined the spatial pattern of scg2a expression in the female and male brain by semiquantitative in situ hybridization. Expression of scg2a was detected in the following regions: the olfactory bulb; Dl in the dorsal telencephalon; TN, Vv, Vl, and Vs/Vp in the ventral telencephalon; PMp, Pbl, PPa, PPp, and PMm/PMg in the preoptic area (POA); VM, VL, and DP/CP in the thalamus; Hd in the habenula; TL, NVT, NAT, NPPv, NRL, and NPT/NRP in the hypothalamus; MR/IR/DT in the midbrain tegmentum; Ci, ra, and NVm/is in the brain stem; and the pituitary (Fig. 4A–C). For abbreviations of brain nuclei, see Table S1. Among these regions, Vs/Vp and PMm/PMg showed a marked female bias in the total area of scg2a expression signal (P < 0.0001 and P = 0.0061, respectively), with the signal in Vs/Vp occurring almost exclusively in females (Fig. 4C and D). Both Vs/Vp and PMm/PMg harbor female-specific populations of npba-expressing neurons (FeSP neurons) (12), which are likely responsible for this sex bias. In contrast, a male bias in scg2a expression was observed in the PMp (P < 0.0001; Fig. 4C and D). These results indicate that scg2a is widely expressed in the brain of both females and males, with marked sex differences in several brain nuclei.
Fig. 4.
scg2a shows region-specific sex differences in expression in the medaka brain. A) Lateral view (anterior to the left) of the medaka brain showing the approximate levels of coronal sections presented in panel B. Cb, cerebellum; Hyp, hypothalamus; MO, medulla oblongata; OB, olfactory bulb; OpN, optic nerve; OT, optic tectum; Pit, pituitary; Tel, telencephalon. B) Coronal sections showing the location of brain nuclei in which scg2a is expressed (stars). C) Representative micrographs depicting the expression of scg2a in the respective brain nuclei and pituitary of females and males. Scale bars represent 100 µm. For abbreviations of brain nuclei, see Table S1. D) Total area of scg2a expression signal in the respective brain nuclei and pituitary of females and males (n = 5 per sex). In brain nuclei other than these, signal was scarce and/or weak and could not be reliably quantitated. Statistical differences were assessed by unpaired t-test with Bonferroni–Dunn correction (D). **P < 0.01; ***P < 0.001.
scg2a-deficient female medaka show no alterations in mating behavior
Finally, we assessed the effect of scg2a deficiency on female and male mating behavior. Medaka mating behavior consists of a stereotyped and quantifiable series of actions (18, 19), beginning with the male approaching and closely following the female, and then performing courtship displays by swimming in a semicircle in front of the female. If the female is receptive, the male will grasp her with his dorsal and anal fins (termed “wrapping”), and they quiver together (“quivering”) until sperm and eggs are released (“spawning”). If the female is not receptive, she will either assume a rejection posture or swim rapidly away.
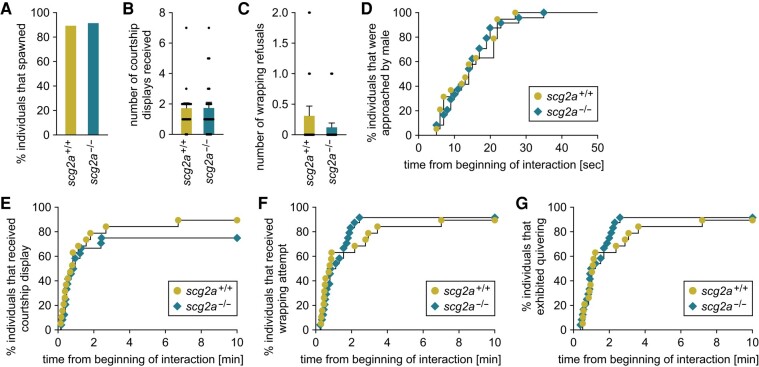
Most scg2a+/+ and scg2a−/− females of both Δ14 and Δ20 knockout lines spawned successfully, and there were no differences between genotypes in the number of courtship displays received from males or wrapping refusals (Figs. 5A–C and S5A–C). scg2a+/+ and scg2a−/− females from both lines showed no significant differences in the latencies from the beginning of the interaction to the first approach by the male, the first courtship display by the male, the first wrapping by the male, and quivering (Figs. 5D–G and S5D–G). Similar to females, most scg2a+/+ and scg2a−/− males of both Δ14 and Δ20 lines spawned, and no differences between genotypes were found in the number of courtship displays or wrapping refusals (Figs. S6A–C and S7A–C). Again, we found no significant differences between genotypes from both lines in the latencies from the beginning of the interaction to the first approach, the first courtship display, the first wrapping, and quivering (Figs. S6D–G and S7D–G). These data indicate that scg2a deficiency does not affect the performance of mating behavior in medaka.
Fig. 5.
scg2a-deficient female medaka show no alterations in mating behavior. scg2a+/+ (n = 19) and scg2a−/− (n = 24) females of the Δ14 line were tested for mating behavior. A) Percentage of individuals that spawned within the test period (10 min). B, C) Number of courtship displays received (B) and wrapping refusals (C). D–G) Latency from the beginning of interaction to the first approach (D), first courtship display (E), first wrapping attempt (F), and quivering (G). Statistical differences were assessed by Fisher's exact test (A), unpaired t-test (B), unpaired t-test with Welch's correction (C), and Gehan–Breslow–Wilcoxon test (D–G).
Discussion
In the present study, we identified scg2a by transcriptomic analysis as a gene expressed in multipeptidergic neurons in the medaka PMm/PMg, termed FeSP neurons. Although neurons equivalent to FeSP neurons have not been identified in other species, the PMm/PMg is considered homologous to the PVN in mammals (12), where the expression of both Npb and Scg2 has been reported (1, 20, 21). Neurons coexpressing Npb and Scg2 may thus be conserved in this brain region across species. In addition to FeSP neurons, we also observed the expression of scg2a in isotocin and vasotocin neurons in the PMm/PMg. This is consistent with data from other species, where SN immunoreactivity has been shown to colocalize with isotocin and vasotocin in the POA of goldfish (Carassius auratus) and electric fish (Brachyhypopomus gauderio) (22, 23) and with oxytocin in magnocellular neurons of the PVN in rats (20). Moreover, the expression of Scg2 in the rat PVN is concomitantly up-regulated with oxytocin and vasopressin during lactation and after salt loading, respectively, inferring that Scg2 may have important cofunctions with these neuropeptides (20, 24). These lines of evidence suggest that Scg2/SN may share roles with isotocin/oxytocin, vasotocin/vasopressin, and perhaps also Npb in the PMm/PMg/PVN across vertebrate species.
Our results also revealed that scg2a expression in FeSP neurons is highly dependent on ovarian estrogen. Since FeSP neurons coexpress Scg2a/SNa and two Esr subtypes, esr1 and esr2b, and esr2b-deficient females, but not esr1-deficient females, have reduced SNa levels in their FeSP neurons, ovarian estrogen most likely acts directly on FeSP neurons via Esr2b to induce scg2a expression. Esr2b is the primary factor responsible for the feminization/demasculinization of medaka mating behavior (16), raising the possibility that scg2a represents a component of the Esr2b-dependent mechanism governing female mating behavior. However, we found no effect of scg2a deficiency on mating behavior, suggesting that scg2a is dispensable, is compensated for by other factors, or has other functions unrelated to mating behavior, as discussed below. It should be noted that evidence from other studies suggests that estrogenic regulation of scg2a is variable and highly dependent on cell type and species. For example, in the goldfish pituitary, no effect of estrogen was found on scg2a expression at any stage of their reproductive cycle (25), whereas in rats, estrogen down-regulates Scg2 expression in the pituitary but up-regulates its expression in the hypothalamus (26, 27). Our survey of the medaka scg2a locus failed to identify any typical estrogen-responsive element-like sequences. This suggests that estrogenic regulation of scg2a in FeSP neurons (and other cell types) is likely indirect, consistent with the species- and cell type-specific effects reported previously.
In contrast to esr2b, deficiency of esr1 had only a small effect on the expression of scg2a in the PMm/PMg, stemming from a reduced total number of FeSP neurons. We also found that esr1 deficiency causes enlargement of the ovary, again in contrast to esr2b, which does not affect ovarian morphology or function (16). Given that there was no change in Npba expression, which is highly estrogen dependent (12, 15), in the FeSP neurons of esr1-deficient females, their reduced number of FeSP neurons may be specifically attributable to the loss of neural Esr1 signaling, rather than secondary to abnormal ovarian function, such as reduced estrogen levels. This presumption is consistent with two previous reports on esr1-deficient medaka that found no resulting changes in fecundity or pituitary gonadotropin expression levels (28, 29), although one of these reports, unlike our results, found no significant changes in ovarian weight.
High-resolution microscopic analysis of FeSP neurons in scg2a-deficient medaka revealed no noticeable changes in the subcellular localization and total number of Npba-containing secretory granules in FeSP neurons, suggesting that Scg2a is not involved in the packaging of Npba to secretory granules in these neurons. This is in stark contrast to the consistently reported function of Scg2 and other granins in the sorting and packaging of peptides to secretory granules (2, 30–32). However, the effects of granins on peptide packaging to secretory granules have mostly been studied in endocrine cells and peripheral neurons, and no studies have confirmed that these effects hold true for central neurons (33). Our results suggest that granins may be dispensable for peptide packaging to secretory granules in central neurons, unlike in endocrine cells or peripheral neurons. In support of this, it has been reported that Scg2 processing to SN is highest in the brain and limited in the adrenal medulla and pituitary (3). Additionally, teleost and mammalian Scg2 have quite low sequence similarity (only 10% identity and 40% similarity) except for the region encoding the mature SN polypeptide. It is therefore possible that the functions of Scg2 that are assumed to involve regions other than SN, including peptide packaging to secretory granules, are not conserved among species. Another possibility, although less likely, is that the mutant Scg2a proteins of our knockout medaka partially retain some function in neuropeptide packaging. Human SCG2 has N- and C-terminal α-helical domains, each of which helps to target SCG2 to secretory granules (31). The amino acid sequences of these domains are somewhat conserved between humans and medaka, and the sequence corresponding to the N-terminal α-helical domain is still present in the mutant Scg2a proteins of our knockout medaka, raising the possibility that they could still be targeted to secretory granules. However, considering that regions outside the N-terminus are required for the full functionality of human SCG2 (31), the mutant Scg2a proteins should lack the putative function of neuropeptide packaging.
Instead, our findings that SNa and Npba colocalize in a majority of secretory granules and that SNa is axonally transported suggest that Scg2a/SNa functions as a secretory neuropeptide with Npba in FeSP neurons, likely sharing many action sites. The best-studied role for SNa in teleosts is as a component of the HPG axis, where it regulates the synthesis and release of luteinizing hormone from the pituitary via paracrine and neuroendocrine modes of action (34). However, this role does not apply to SNa in FeSP neurons because they do not project to the pituitary, but instead to a wide range of other brain regions and the spinal cord (15). A possible function of SNa in FeSP neurons is paracrine regulation of radial glial cells, which lie throughout the POA, including the PMm/PMg, and express cyp19a1b, a gene encoding the enzyme responsible for the conversion of testosterone to E2 (35, 36). In goldfish, SNa has been shown to down-regulate the expression of cyp19a1b in preoptic radial glial cells (37). Given that FeSP neurons and their expression of scg2a are highly sensitive to estrogen, SNa may function as a feedback molecule in these neurons to regulate the sex steroidal microenvironment of the PMm/PMg.
We found that both female and male scg2a-deficient medaka exhibited normal mating behaviors. This finding was rather unexpected given the following observations: (i) scg2a is expressed in an estrogen-dependent manner in FeSP neurons, which have been implicated in female sexual receptivity (14, 15), (ii) expression of scg2a in FeSP neurons lies downstream of Esr2b, the primary factor responsible for feminization/demasculinization of mating behavior in medaka (16), and (iii) in addition to the PMm/PMg, scg2a also shows sexually dimorphic expression in the Vs/Vp and PMp, brain regions implicated in mating behavior in teleosts by classic legion and stimulation studies (38–41). While it is possible that Scg2a/SNa is dispensable for or has no role in mating behavior in medaka, these observations suggest that there may be an unidentified mechanism compensating for the genetic loss of scg2a. One possibility is compensation by scg2b, the other scg2 paralog in teleosts. Indeed, in zebrafish, double knockout females of both scg2a and scg2b exhibited impaired mating behavior, whereas single knockout females of either gene did not (although they had lower spawning success when paired with males of the same genotype) (11). Seemingly inconsistent with this possibility, however, are the observations that the reduced spawning success of scg2a/scg2b double knockout females could only be restored by administration with SNa, but not with SNb, and that the expression level of scg2b in the telencephalon, hypothalamus, and pituitary of scg2a knockout females was down-regulated (11). Another possibility is a functional redundancy in the neuropeptides expressed in FeSP neurons that may compensate for the loss of SNa signaling. We have recently provided evidence that in addition to npba and scg2a, FeSP neurons express several other neuropeptide genes, including tac1, tac4a, and capt4pt (14). Among the neuropeptides encoded by these genes, substance P (encoded by tac1) has been shown to cofunction with SNa as a neuroprotective factor in the retina (42) and colocalize with SN in neurons that project to preganglionic sympathetic neurons (43). However, further studies are needed to understand the multipeptidergic nature of FeSP neurons.
Additionally, in teleosts, the knockout of many neuropeptide genes that have been shown in mammals to play an indispensable role in regulating reproduction has little to no effect, resulting in normal gonadal development and spawning (44). Two examples of this include gnrh2/gnrh3 double knockout and gnrh3/kiss1/kiss2 triple knockout zebrafish, which were both found to maintain normal reproductive performance. Notably, attempts to identify mechanisms compensating for the genetic loss of these reproductively relevant neuropeptides commonly identified an up-regulation of scg2 expression (45, 46). Scg2/SN may thus be a component of a complex neuropeptide assembly that governs teleost reproduction, which can compensate for one another after genetic ablation. In line with this assumption, we observed that scg2a is abundantly expressed in many brain nuclei where peptidergic neurons are located, including TN, Vs/Vp, PMp, PPa, and NVT. It may be worthwhile in future research to explore the possibility that SNa functions in concert with a variety of neuropeptides.
In summary, here we found that Scg2a acts downstream of estrogen/Esr2b signaling in behaviorally relevant FeSP neurons, where it most likely serves as a precursor for secreted neuropeptides. Our results also revealed that scg2a is expressed in a sexually dimorphic fashion in other brain regions relevant to mating behavior, which would be worth further study. Lastly, based on these findings and the report showing the involvement of Scg2 in mating behavior in zebrafish (11), we analyzed the mating behavior of medaka deficient for scg2a but were unable to obtain evidence for its behavioral role. Further studies are needed to determine whether scg2a has no behavioral function or if there is an unidentified compensatory mechanism for its loss of function.
Materials and methods
Animals
Wild-type medaka of the d-rR strain, npba-GFP transgenic medaka, which express GFP under the control of regulatory regions of npba (15), esr2b knockout medaka (16), and esr1 and scg2a knockout medaka generated in this study were raised at 28°C with a 14-h light/10-h dark photoperiod. Fish were fed 3–4 times per day with live brine shrimp and a commercial food pellet (Otohime; Marubeni Nisshin Feed, Tokyo, Japan). Sexually mature, spawning adults (aged 3–5 months) were used for experimental analysis. All samplings were performed 1–3 h after the beginning of the light period. All animal procedures were performed in accordance with guidelines of the Institutional Animal Care and Use Committee of the University of Tokyo.
In silico cloning and phylogenetic tree analysis
A BLAST search using the sequence obtained by RNA-seq (gene ID: XLOC_005021) as a query was performed to identify the corresponding cDNA in the GenBank nucleotide database, and this sequence was further verified by identification of the corresponding EST clone in the medaka EST database at NBRP Medaka (http://www.shigen.nig.ac.jp/medaka/). The deduced amino acid sequence of medaka scg2a was aligned with those of its putative orthologs/paralogs from different species by using ClustalW. The resulting alignment was used to construct a bootstrapped (1,000 replicates) neighbor-joining tree using Genetyx software (Genetyx Corporation, Tokyo, Japan). Chromogranins A and B in humans were used as outgroups to root the tree. The species names and GenBank accession numbers of the sequences used are listed in Table S2.
Ovariectomy and hormone treatments
The procedure for ovariectomy and hormone treatments has been previously described (14). Briefly, the ovary was removed from each npba-GFP transgenic female (15) under tricaine methane sulfonate anesthesia (0.02%) through a small incision in the ventrolateral abdominal wall. After removal of the ovary, the incision was sutured with nylon thread. Sham-operated females underwent the same operation except for removal of the ovary. Ovariectomized females were immersed in water containing 100 ng/mL of E2, KT, or vehicle (ethanol) for 6 days following a 3-day recovering period. Sham-operated females were treated with only vehicle and used as controls.
Isolation and purification of FeSP neurons
The procedure for the isolation and purification of FeSP neurons has been described elsewhere (14). Briefly, the PMm/PMg region containing GFP-labeled FeSP neurons was dissected from the brain of sham-operated, ovariectomized, ovariectomized plus E2-treated, and ovariectomized plus KT-treated npba-GFP females under a fluorescence stereomicroscope (M165FC; Leica Microsystems, Wetzlar, Germany). Dissected brain tissue was dissociated by papain treatment, triturated through Pasteur pipettes, and suspended in Leibovitz's L-15 medium containing 5% fetal bovine serum. Individual GFP-labeled FeSP neurons were handpicked with a glass pipette mounted on a micromanipulator (Narishige, Tokyo, Japan) under a fluorescence stereomicroscope (M 165FC; Leica Microsystems).
Real-time PCR
The cDNA template for real-time PCR was prepared as previously reported (14) from FeSP neurons isolated and purified as described above. In brief, total RNA was purified from 50 neurons per sample using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA) and reverse transcribed using SuperScript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) and the oligo(dT) primer (5′-TATAGAATTCGCGGCCGCTCGCGATAATACGACTCACTATAGGGCG(T)24-3′). After the addition of a poly(A) tail with terminal deoxynucleotidyl transferase (Roche Diagnostics, Basel, Switzerland), second-strand cDNA was synthesized using MightyAmp DNA polymerase (Takara Bio, Shiga, Japan) and the tagging primer (5′-TATAGAATTCGCGGCCGCTCGCGA(T)24-3′). The cDNA was then amplified by 18 cycles of suppression PCR using MightyAmp DNA polymerase (Takara Bio) and the 5′-aminated primer (5′-NH2-GTATAGAATTCGCGGCCGCTCGCGAT-3′).
Real-time PCR was performed on the LightCycler 480 System II using the LightCycler 480 SYBR Green I Master (Roche Diagnostics). Melt curve analysis was performed to verify that a single amplicon was obtained in each sample. Normalization was performed using the geometric mean of actb (GenBank accession number: NM_001104808) and eef1a (GenBank accession number: NM_001104662), according to Vandesompele et al. (47). The primers used for real-time PCR are listed in Table S3.
Double-label in situ hybridization
A partial DNA fragment corresponding to nucleotides 756–1,910 (1,155 bp) of the medaka scg2a cDNA (GenBank accession number: XM_004075713) was PCR-amplified and transcribed in vitro to generate a digoxigenin (DIG)-labeled cRNA probe using DIG RNA Labeling Mix and T7 RNA polymerase (Roche Diagnostics). Similarly, partial DNA fragments corresponding to nucleotides 1–645 (645 bp) of the medaka npba cDNA (NM_001308979), 1–738 (738 bp) of the medaka it cDNA (NM_001278830), and 1–845 (845 bp) of the medaka vt cDNA (NM_001278891) were PCR-amplified and transcribed to generate fluorescein-labeled cRNA probes using Fluorescein RNA Labeling Mix and T7 RNA polymerase (Roche Diagnostics).
Double-label in situ hybridization was performed according to Kawabata-Sakata et al. (48), with some modifications. Briefly, whole brains dissected from females were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin. Serial coronal sections of 10-μm thickness were cut and hybridized with the above-mentioned DIG-labeled scg2a probe in conjunction with a fluorescein-labeled probe for npba, it, or vt. The DIG-labeled probe was visualized using a mouse anti-DIG antibody (RRID: AB_304362; Abcam, Cambridge, UK) and the Alexa Fluor 555 Tyramide SuperBoost Kit, goat anti-mouse IgG (Thermo Fisher Scientific) according to the manufacturer's instructions. The fluorescein-labeled probe was visualized using a rabbit anti-fluorescein antibody (RRID: AB_444949; Abcam) and an Alexa Fluor 488-conjugated goat anti-rabbit IgG antibody (RRID: AB_10563748; Thermo Fisher Scientific). Cell nuclei were counterstained with DAPI.
Production and validation of antibody
A polyclonal anti-SNa antibody was raised in rabbit against a synthetic 34-amino acid polypeptide representing full-length medaka SNa (Medical and Biological Laboratories, Aichi, Japan). The specificity of the anti-SNa antibody was confirmed by the following observations: (i) a double-labeling experiment with immunohistochemistry and in situ hybridization showed that the antibody labeled only neurons expressing the scg2a transcript (Fig. S8A), (ii) preabsorption of the antibody with SNa polypeptide (432 ng/μL, for 16 h) prior to the immunohistochemical procedure resulted in the absence of labeled neurons (Fig. S8B), and (iii) scg2a knockout fish from both Δ14 and Δ20 lines showed a complete loss of neurons labeled with this antibody (Fig. S8C). The procedures for immunohistochemistry and double labeling with immunohistochemistry and in situ hybridization are described below.
Double labeling with immunohistochemistry and in situ hybridization
Double labeling with immunohistochemistry and in situ hybridization was performed to examine the colocalization of the SNa polypeptide and Esr transcripts in the PMm/PMg and confirm the specificity of the anti-SNa antibody, the procedure for which has been described elsewhere (15). Briefly, whole brains from females were fixed in 4% PFA and embedded in 5% agarose (type IX-A; Sigma-Aldrich, St Louis, MO, USA) supplemented with 20% sucrose. Frozen coronal sections of 20-μm thickness were cut and hybridized with the aforementioned scg2a probe or Esr (esr1, XM_020714493; esr2a, NM_001104702; esr2b, NM_001128512) probe described previously (49), which was labeled with fluorescein using Fluorescein RNA Labeling Mix and T7/SP6 RNA polymerase (Roche Diagnostics). After blocking with phosphate-buffered saline (PBS) containing 2% normal goat serum (NGS), sections were incubated overnight at 4°C with the anti-SNa antibody (diluted 1:1,000 and 1:200 in antibody diluent [PBS containing 2% NGS, 0.1% bovine serum albumin, and 0.02% keyhole limpet hemocyanin] for double labeling with scg2a and Esr, respectively). The sections were then reacted overnight at 4°C with a horseradish peroxidase-conjugated anti-fluorescein antibody (diluted 1:1,000 and 1:200 for the detection of scg2a and Esr, respectively; RRID: AB_2737388; PerkinElmer, Waltham, MA, USA) and Alexa Fluor 555-conjugated goat antirabbit IgG (diluted 1:1,000; RRID: AB_2535849; Thermo Fisher Scientific) in Tris-buffered saline containing 1.5% blocking reagent (Roche Diagnostics) and DAPI. The anti-fluorescein antibody was visualized using the TSA Plus Fluorescein System (PerkinElmer) according to the manufacturer's instructions.
Generation of knockout medaka
esr1 and scg2a knockout medaka were generated by CRISPR/Cas9-mediated genome editing. For esr1 knockout, two CRISPR RNAs (crRNAs) were designed to target the region encoding the DNA-binding domain (Fig. S1A); for scg2a knockout, a single crRNA was designed to target the region encoding the mature SNa polypeptide (Fig. S3A). These protein domains were predicted using InterPro (https://www.ebi.ac.uk/interpro/). crRNA and transactivating CRISPR RNA (tracrRNA) were synthesized by Fasmac (Kanagawa, Japan). The crRNA, tracrRNA, and Cas9 protein were comicroinjected into medaka embryos at the one-cell stage.
Potential founder fish were screened by outcrossing with wild-type fish and testing progeny for mutations at the target site using T7 endonuclease I assay (50) followed by direct sequencing. For the esr1 knockout line, a founder was identified that yielded a high proportion of progeny carrying a 482-bp deletion that caused early truncation of the DNA-binding domain and complete loss of the ligand-binding domain (Fig. S1B). For the scg2a knockout lines, two founders were identified that produced progeny carrying deletions that caused frameshifts leading to premature truncation of the Scg2a protein and loss of the mature SNa polypeptide: the progeny of one founder carried a 14-bp deletion (Δ14) and progeny of the other carried a 3-bp deletion and 23-bp insertion (Δ20; Fig. S3B). These progenies were intercrossed to establish knockout lines. Each line was maintained by breeding heterozygous individuals to obtain wild-type and knockout siblings for experimental use. The genotype of each fish was first determined by direct sequencing and thereafter by PCR/electrophoresis (for esr1 knockouts) or high-resolution melt analysis (for scg2a and esr2b knockouts) using the primers listed in Table S3. In each subsequent experiment, siblings raised under the same conditions were used to control for genetic and environmental factors.
Single-label in situ hybridization
Single-label in situ hybridization was performed as described previously (48). Briefly, whole brains, with pituitaries attached, of both sexes were fixed in 4% PFA and embedded in paraffin. Serial 10-μm coronal sections were cut and hybridized with the aforementioned DIG-labeled scg2a probe. Hybridization signals were visualized using an alkaline phosphatase-conjugated anti-DIG antibody (RRID: AB_514497; Roche Diagnostics) and 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) substrate (Roche Diagnostics). Color development was allowed to proceed for 90 min. Brain nuclei were identified using the medaka brain atlases (51, 52). Images were acquired with a virtual slide microscope (VS120; Olympus, Tokyo, Japan) and the total area of scg2a expression signal in each brain region was calculated using Olyvia software (Olympus).
For the analysis factoring in signal intensity, the different signal intensities of BCIP/NBT were represented by the 256 colors in the 8-bit color spectrum, where 1 corresponded to the highest scg2a signal intensity and 256 to no signal. Colors 161–256 were considered background or no signal and were excluded from the analysis. The area for the remaining 160 colors was quantified in 8 groups of 20 colors each (1–20, 21–40, 41–60, 61–80, 81–100, 101–120, 121–140, and 141–160).
Immunohistochemistry
For single-label immunohistochemistry for Npba, whole brains from females were fixed in 4% PFA, embedded in paraffin, and cut into 10-μm coronal sections. After blocking with PBS containing 2% NGS, the sections were reacted overnight at 4°C with the anti-Npba antibody (RRID: AB_2810229) (15) diluted 1:1,000 in antibody diluent. The sections were then incubated overnight at 4°C with Alexa Fluor 555-conjugated goat anti-rabbit IgG (RRID: AB_2535849; Thermo Fisher Scientific) diluted 1:1,000 in PBS. For double-label immunohistochemistry for Npba and SNa, sections were then reacted overnight at 4°C with the abovementioned anti-SNa antibody directly labeled with Alexa Fluor 488 Conjugation Kit (Fast) - Lightning-Link (Abcam), diluted 1:100 in antibody diluent. For immunohistochemistry combined with WGA staining, sections were further permeabilized for 10 min with PBS containing 0.2% Triton X-100 and incubated for 1 h at room temperature with Alexa Flour 488-conjugated WGA (Thermo Fisher Scientific) diluted 1:100 in Hank's balanced salt solution. Cell nuclei were counterstained with DAPI.
For single-label immunohistochemistry for SNa, frozen coronal sections of 20-μm thickness were made from the female brains with pituitaries attached as described above. After blocking as described above, the sections were incubated overnight at 4°C with the anti-SNa antibody diluted 1:1,000 in antibody diluent and then incubated overnight at 4°C with Alexa Fluor 488-conjugated goat anti-rabbit IgG (Thermo Fisher Scientific) diluted 1:1,000 in PBS. Cell nuclei were counterstained with DAPI.
Confocal microscopy and image analysis
All fluorescence images were acquired using a confocal laser-scanning microscope (TCS SP8; Leica Microsystems) with either a 40× or a 63× oil immersion objective. The following excitation and emission wavelengths were used: 405 and 410–480 nm for DAPI; 488 and 495–545 nm for Alexa Flour 488 and fluorescein; and 552 and 562–700 nm for Alexa Flour 555. For all confocal microscopy observations except those detailed below, sections were mounted with ProLong Diamond Antifade Mountant (Thermo Fisher Scientific). The intensity of the immunofluorescence signals was quantified using LAS X software (Leica Microsystems).
For obtaining the high-resolution images shown in Figs. 3A, E and S4C, sections were mounted with ProLong Glass Antifade Mountant (Thermo Fisher Scientific) and photographed using a 63× oil immersion objective with a zoom factor of 4.19, pinhole diameter of 0.63 Airy unit, and spatial resolution of 1,024 × 1,024 pixels (resulting pixel size: 43.05 nm). Image stacks were deconvolved using Huygens Essential software (Scientific Volume Imaging, Hilversum, The Netherlands). The colocalization between Npba and SNa fluorophores was quantified by manually counting the coincidence of Npba and SNa puncta in FeSP neurons (n = 9) using LAS X software (Leica Microsystems) and calculating Pearson's correlation coefficients (Rp) (53) using the ImageJ plugin Coloc 2 (https://imagej.nih.gov/ij/) on the cell body area of each individual FeSP neuron (n = 8), excluding the nucleus. Npba puncta were manually counted using the ImageJ plugin Cell Counter (https://imagej.nih.gov/ij/).
Mating behavior analysis
The mating behavior analysis was performed essentially as described elsewhere (15). In brief, on the day before behavioral analysis, each focal male/female (from Δ14 or Δ20 scg2a knockout line) was paired with a stimulus fish of the opposite sex (wild-type d-rR strain) in a 2-L rectangular tank and separated by a perforated transparent partition. The partition was removed 1 h after the onset of the light period of the following day, and fish were allowed to interact for 10 min. All interactions were recorded with a digital video camera (iVIS HF S11, Canon, Tokyo, Japan, or HC-W870M, Panasonic Corporation, Osaka, Japan). The recorded videos were analyzed for the following parameters: percentage of fish that spawned within the test period; number of courtship displays by the male prior to spawning; number of wrapping refusals by the female; and latency from the beginning of interaction to the first approach from the male, first courtship display by the male, first wrapping attempt by the male, and quivering. Videos were played back at normal speed using VLC media player (https://www.videolan.org/vlc/).
Statistical analysis
For continuous data, results are presented as mean ± standard error of the mean, with individual data points shown as dots except for immunofluorescence quantification in FeSP neurons, where data are plotted as box-and-whisker plots by Tukey’s method for visual clarity. Categorical data are presented as percentages. Behavioral time-series data were analyzed using Kaplan–Meier plots with the inclusion of fish that did not exhibit the given behavior within the test period, following Jahn-Eimermacher et al. (54).
Statistical analyses were performed using GraphPad Prism (GraphPad software, San Diego, CA, USA). Continuous data between two groups were compared by the unpaired two-tailed Student's t-test. Welch's correction was applied if the F-test indicated a significant difference in variance between groups. The Bonferroni–Dunn correction was applied for multiple comparisons between two groups. Differences in continuous data between more than two groups were evaluated by one-way ANOVA followed by Bonferroni's post hoc test. Homogeneity of variance was verified for all datasets by the Brown–Forsythe test. Differences between Kaplan–Meier curves were tested for statistical significance using the Gehan–Breslow–Wilcoxon test. Fisher's exact test was used to compare categorical data.
Supplementary Material
Acknowledgments
The authors thank NBRP Medaka for providing the EST clone used in this study. The authors also thank Dr Junpei Yamashita for performing ovariectomy and Akira Hirata, Tomiko Iba, Kaoru Furukawa, and Ayu Kuwakubo for assistance with medaka husbandry.
Contributor Information
Thomas Fleming, Department of Aquatic Bioscience, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Masaya Tachizawa, Department of Aquatic Bioscience, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Yuji Nishiike, Department of Aquatic Bioscience, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Ai Koiwa, Department of Aquatic Bioscience, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Yuki Homan, Department of Aquatic Bioscience, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Kataaki Okubo, Department of Aquatic Bioscience, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) in Japan and the Japan Society for the Promotion of Science (JSPS; MEXT/JSPS grant numbers 20J13802 [to T.F.], 16H04979, 17H06429, and 19H03044 [to K.O.]).
Author Contributions
T.F., M.T., and K.O. designed research. T.F., M.T., Y.N., A.K., and Y.H. performed research. T.F., M.T., Y.N., A.K., Y.H., and K.O. analyzed data. T.F. and K.O. wrote the paper.
Data Availability
All data supporting the findings of this study are included within the article and the Supplementary material.
References
- 1. Hotta K, Hosaka M, Tanabe A, Takeuchi T. 2009. Secretogranin II binds to secretogranin III and forms secretory granules with orexin, neuropeptide Y, and POMC. J Endocrinol. 202:111–121. [DOI] [PubMed] [Google Scholar]
- 2. Bartolomucci A, et al. 2011. The extended granin family: structure, function, and biomedical implications. Endocr Rev. 32:755–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Troger J, et al. 2017. Granin-derived peptides. Prog Neurobiol. 154:37–61. [DOI] [PubMed] [Google Scholar]
- 4. Zhao E, Basak A, Trudeau VL. 2006. Secretoneurin stimulates goldfish pituitary luteinizing hormone production. Neuropeptides. 40:275–282. [DOI] [PubMed] [Google Scholar]
- 5. Zhao E, et al. 2009. The secretogranin II-derived peptide secretoneurin stimulates luteinizing hormone secretion from gonadotrophs. Endocrinology. 150:2273–2282. [DOI] [PubMed] [Google Scholar]
- 6. Zhao E, et al. 2010. Secretoneurin is a potential paracrine factor from lactotrophs stimulating gonadotropin release in the goldfish pituitary. Am J Physiol Regul Integr Comp Physiol. 299:R1290–R1297. [DOI] [PubMed] [Google Scholar]
- 7. Zhao E, Hu H, Trudeau VL. 2010. Secretoneurin as a hormone regulator in the pituitary. Regul Pept. 165:117–122. [DOI] [PubMed] [Google Scholar]
- 8. Mikwar M, et al. 2016. Stimulatory effect of the secretogranin-II derived peptide secretoneurin on food intake and locomotion in female goldfish (Carassius auratus). Peptides. 78:42–50. [DOI] [PubMed] [Google Scholar]
- 9. Kähler CM, Kaufmann G, Kähler ST, Wiedermann CJ. 2002. The neuropeptide secretoneurin stimulates adhesion of human monocytes to arterial and venous endothelial cells in vitro. Regul Pept. 110:65–73. [DOI] [PubMed] [Google Scholar]
- 10. Tao B, et al. 2018. Secretogranin-II plays a critical role in zebrafish neurovascular modeling. J Mol Cell Biol. 10:388–401. [DOI] [PubMed] [Google Scholar]
- 11. Mitchell K, et al. 2020. Targeted mutation of secretogranin-2 disrupts sexual behavior and reproduction in zebrafish. Proc Natl Acad Sci U S A. 117:12772–12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hiraki T, et al. 2014. Neuropeptide B is female-specifically expressed in the telencephalic and preoptic nuclei of the medaka brain. Endocrinology. 155:1021–1032. [DOI] [PubMed] [Google Scholar]
- 13. Kikuchi Y, et al. 2019. Sexually dimorphic neuropeptide B neurons in medaka exhibit activated cellular phenotypes dependent on estrogen. Endocrinology. 160:827–839. [DOI] [PubMed] [Google Scholar]
- 14. Fleming T, et al. 2022. Prostaglandin E2 receptor Ptger4b regulates female-specific peptidergic neurons and female sexual receptivity in medaka. Commun Biol. 5:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hiraki-Kajiyama T, et al. 2019. Neuropeptide B mediates female sexual receptivity in medaka fish, acting in a female-specific but reversible manner. eLife. 8:e39495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nishiike Y, et al. 2021. Estrogen receptor 2b is the major determinant of sex-typical mating behavior and sexual preference in medaka. Curr Biol. 31:1699–1710.e6. [DOI] [PubMed] [Google Scholar]
- 17. Kawabata Y, Hiraki T, Takeuchi A, Okubo K. 2012. Sex differences in the expression of vasotocin/isotocin, gonadotropin-releasing hormone, and tyrosine and tryptophan hydroxylase family genes in the medaka brain. Neuroscience. 218:65–77. [DOI] [PubMed] [Google Scholar]
- 18. Ono Y, Uematsu T. 1957. Mating ethogram in Oryzias latipes. J Fac Sci Hokkaido Univ. 13:197–202. [Google Scholar]
- 19. Walter RO, Hamilton JB. 1970. Head-up movements as an indicator of sexual unreceptivity in female medaka, Oryzias latipes. Anim Behav. 18:125–127. [Google Scholar]
- 20. Mahata SK, et al. 1993. Concomitant changes of messenger ribonucleic acid levels of secretogranin II, VGF, vasopressin and oxytocin in the paraventricular nucleus of rats after adrenalectomy and during lactation. J Neuroendocrinol. 5:323–330. [DOI] [PubMed] [Google Scholar]
- 21. Jackson VR, Lin SH, Wang Z, Nothacker HP, Civelli O. 2006. A study of the rat neuropeptide B/neuropeptide W system using in situ techniques. J Comp Neurol. 497:367–383. [DOI] [PubMed] [Google Scholar]
- 22. Canosa LF, et al. 2011. Forebrain mapping of secretoneurin-like immunoreactivity and its colocalization with isotocin in the preoptic nucleus and pituitary gland of goldfish. J Comp Neurol. 519:3748–3765. [DOI] [PubMed] [Google Scholar]
- 23. Pouso P, et al. 2015. The secretogranin-II derived peptide secretoneurin modulates electric behavior in the weakly pulse type electric fish, Brachyhypopomus gauderio. Gen Comp Endocrinol. 222:158–166. [DOI] [PubMed] [Google Scholar]
- 24. Mahata SK, Mahata M, Steiner HJ, Fischer-Colbrie R, Winkler H. 1992. In situ hybridization: mRNA levels of secretogranin II, neuropeptides and carboxypeptidase H in brains of salt-loaded and brattleboro rats. Neuroscience. 48:669–680. [DOI] [PubMed] [Google Scholar]
- 25. Samia M, et al. 2004. Seasonal cyclicity of secretogranin-II expression and its modulation by sex steroids and GnRH in the female goldfish pituitary. Gen Comp Endocrinol. 139:198–205. [DOI] [PubMed] [Google Scholar]
- 26. Anouar Y, Duval J. 1992. Direct estradiol down-regulation of secretogranin II and chromogranin A mRNA levels in rat pituitary cells. Mol Cell Endocrinol. 88:97–104. [DOI] [PubMed] [Google Scholar]
- 27. Naimin SSK, Mulchahey WJJ, Leboeuf RD, Neill JD. 1993. Regulation of expression of secretogranin II mRNA in female rat pituitary and hypothalamus. Neuroendocrinology. 57:422–431. [DOI] [PubMed] [Google Scholar]
- 28. Tohyama S, et al. 2017. Establishment of estrogen receptor 1 (ESR1)-knockout medaka: ESR1 is dispensable for sexual development and reproduction in medaka, Oryzias latipes. Dev Growth Differ. 59:552–561. [DOI] [PubMed] [Google Scholar]
- 29. Kayo D, Zempo B, Tomihara S, Oka Y, Kanda S. 2019. Gene knockout analysis reveals essentiality of estrogen receptor β1 (Esr2a) for female reproduction in medaka. Sci Rep. 9:8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beuret N, Stettler H, Renold A, Rutishauser J, Spiess M. 2004. Expression of regulated secretory proteins is sufficient to generate granule-like structures in constitutively secreting cells. J Biol Chem. 279:20242–20249. [DOI] [PubMed] [Google Scholar]
- 31. Courel M, Vasquez MS, Hook VY, Mahata SK, Taupenot L. 2008. Sorting of the neuroendocrine secretory protein Secretogranin II into the regulated secretory pathway: role of N- and C-terminal α-helical domains. J Biol Chem. 283:11807–11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Courel M, et al. 2010. Pro-hormone secretogranin II regulates dense core secretory granule biogenesis in catecholaminergic cells. J Biol Chem. 285:10030–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dominguez N, van Weering JRT, Borges R, Toonen RFG, Verhage M. 2018. Dense-core vesicle biogenesis and exocytosis in neurons lacking chromogranins A and B. J Neurochem. 144:241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trudeau VL, et al. 2012. Is secretoneurin a new hormone? Gen Comp Endocrinol. 175:10–18. [DOI] [PubMed] [Google Scholar]
- 35. Okubo K, et al. 2011. Sex differences in aromatase gene expression in the medaka brain. J Neuroendocrinol. 23:412–423. [DOI] [PubMed] [Google Scholar]
- 36. Takeuchi A, Okubo K. 2013. Post-proliferative immature radial glial cells female-specifically express aromatase in the medaka optic tectum. PLoS One. 8:e73663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. da Fonte DF, Xing L, Mikwar M, Trudeau VL. 2018. Secretoneurin-A inhibits aromatase B (cyp19a1b) expression in female goldfish (Carassius auratus) radial glial cells. Gen Comp Endocrinol. 257:106–112. [DOI] [PubMed] [Google Scholar]
- 38. Demski LS, Bauer DH, Gerald JW. 1975. Sperm release evoked by electrical stimulation of the fish brain: a functional-anatomical study. J Exp Zool. 191:215–231. [DOI] [PubMed] [Google Scholar]
- 39. Kyle AL, Peter RE. 1982. Effects of forebrain lesions on spawning behaviour in the male goldfish. Physiol Behav. 28:1103–1109. [DOI] [PubMed] [Google Scholar]
- 40. Koyama Y, Satou M, Oka Y, Ueda K. 1984. Involvement of the telencephalic hemispheres and the preoptic area in sexual behavior of the male goldfish, Carassius auratus: a brain-lesion study. Behav Neural Biol. 40:70–86. [DOI] [PubMed] [Google Scholar]
- 41. Satou M, et al. 1984. Telencephalic and preoptic areas integrate sexual behavior in hime salmon (landlocked red salmon, Oncorhynchus nerka): results of electrical brain stimulation experiments. Physiol Behav. 33:441–447. [DOI] [PubMed] [Google Scholar]
- 42. Schmid E, et al. 2012. Secretoneurin, substance P and neuropeptide Y in the oxygen-induced retinopathy in C57Bl/6N mice. Peptides. 37:252–257. [DOI] [PubMed] [Google Scholar]
- 43. Klimaschewski L, Benndorf K, Kirchmair R, Fischer-Colbrie R, Heym C. 1995. Secretoneurin-immunoreactivity in nerve terminals apposing identified preganglionic sympathetic neurons in the rat: colocalization with substance P and enkephalin. J Chem Neuroanat. 9:55–63. [DOI] [PubMed] [Google Scholar]
- 44. Trudeau VL. 2018. Facing the challenges of neuropeptide gene knockouts: why do they not inhibit reproduction in adult teleost fish? Front Neurosci. 12:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Y, et al. 2017. Genetic evidence for multifactorial control of the reproductive axis in zebrafish. Endocrinology. 158:604–611. [DOI] [PubMed] [Google Scholar]
- 46. Marvel M, Spicer OS, Wong TT, Zmora N, Zohar Y. 2018. Knockout of the Gnrh genes in zebrafish: effects on reproduction and potential compensation by reproductive and feeding-related neuropeptides. Biol Reprod. 99:565–577. [DOI] [PubMed] [Google Scholar]
- 47. Vandesompele J, et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kawabata-Sakata Y, Nishiike Y, Fleming T, Kikuchi Y, Okubo K. 2020. Androgen-dependent sexual dimorphism in pituitary tryptophan hydroxylase expression: relevance to sex differences in pituitary hormones. Proc Biol Sci. 287:20200713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hiraki T, et al. 2012. Female-specific target sites for both oestrogen and androgen in the teleost brain. Proc Biol Sci. 279:5014–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. 2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 19:1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Anken R, Bourrat F. 1998. Brain atlas of the medakafish. Paris: INRA Editions. [Google Scholar]
- 52. Ishikawa Y, Yoshimoto M, Ito H. 1999. A brain atlas of a wild-type inbred strain of the medaka, Oryzias latipes. Fish Biol J Medaka. 10:1–26. [Google Scholar]
- 53. Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. 1992. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 103:857–862. [DOI] [PubMed] [Google Scholar]
- 54. Jahn-Eimermacher A, Lasarzik I, Raber J. 2011. Statistical analysis of latency outcomes in behavioral experiments. Behav Brain Res. 221:271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are included within the article and the Supplementary material.