Abstract
Undifferentiated melanoma should be considered in the differential diagnosis of sarcomatoid cutaneous malignancies to ensure that patients receive the correct treatment. Dermatopathologists should recognize the pitfalls of relying too heavily on immunohistochemistry to establish this diagnosis and consider ancillary tests, including single nucleotide polymorphism (SNP) copy number arrays and targeted next generation sequencing (NGS), when a definitive diagnosis cannot be rendered on a primary or metastatic tumor. This technology can also help to exclude a collision of melanoma and sarcoma when both differentiated and undifferentiated components are juxtaposed. We describe an exceedingly rare, illustrative example of undifferentiated sarcomatoid melanoma presenting as a pedunculated nodule. The clinical context and presence of a small differentiated component helped to establish the diagnosis; however, the transition from differentiated to undifferentiated melanoma was accompanied by an abrupt loss of S100, Sox10, MITF, MelanA and HMB45 with gain of CD10 and p63 staining. SNP copy number array and NGS revealed shared chromosomal copy number changes and overlapping mutations with additional aberrances detected exclusively in the sarcomatoid component; thus, excluding a collision tumor and confirming our putative impression of melanoma with progression to an undifferentiated sarcomatoid phenotype.
Keywords: melanoma, atypical fibroxanthoma, pleomorphic dermal sarcoma, high risk skin cancer, molecular pathology, immunohistochemistry
Introduction
Undifferentiated malignant melanomas are exceedingly unusual and potentially underreported given the difficulties inherent in establishing the diagnosis. A primary melanoma with sarcomatoid features and a loss of melanoma-related antigen expression must be distinguished from atypical fibroxanthoma (AFX), pleomorphic dermal sarcoma (PDS), sarcomatoid squamous cell carcinoma (SSCC), and undifferentiated pleomorphic sarcoma (UPS) since treatments are different. For instance, a sentinel lymph node biopsy will likely be offered to a patient with a deep sarcomatoid melanoma. Likewise, metastatic melanoma with loss of differentiation must also be distinguished from a soft tissue sarcoma. We present an unusual and illustrative example of undifferentiated sarcomatoid melanoma and discuss the most useful clinical, histologic and molecular findings that help dermatopathologists establish this diagnosis.
Case Report
A 73-year-old man presented with a rapidly-growing, pedunculated 6 × 5 × 3 cm nodule on his lower left lateral thigh. An excisional biopsy was performed, and gross examination of the specimen revealed a focally necrotic intradermal mass that was partially contiguous with the epidermis. On microscopic examination, a small portion of superficial spreading melanoma was identified in contiguity with a considerably larger, subjacent nodule of spindled and epithelioid cells with bizarre nuclei, atypical mitoses and admixed giant cells. Some portions of the tumor showed a gradual transition from nests of invasive melanoma to fasciculated spindled cells with intermediate grade nuclei to enlarged cells with anaplastic nuclei indistinguishable from those of undifferentiated pleomorphic sarcoma. The tumor in its entirety had a Breslow depth of 7.9 mm and was ulcerated (AJCC stage pT4b). MelanA immunohistochemistry stained the in situ and superficial intradermal component of the differentiated melanoma, but was not expressed by the spindled and sarcomatoid components (Figure 1).
Figure 1. Malignant melanoma with an undifferentiated sarcomatoid component.
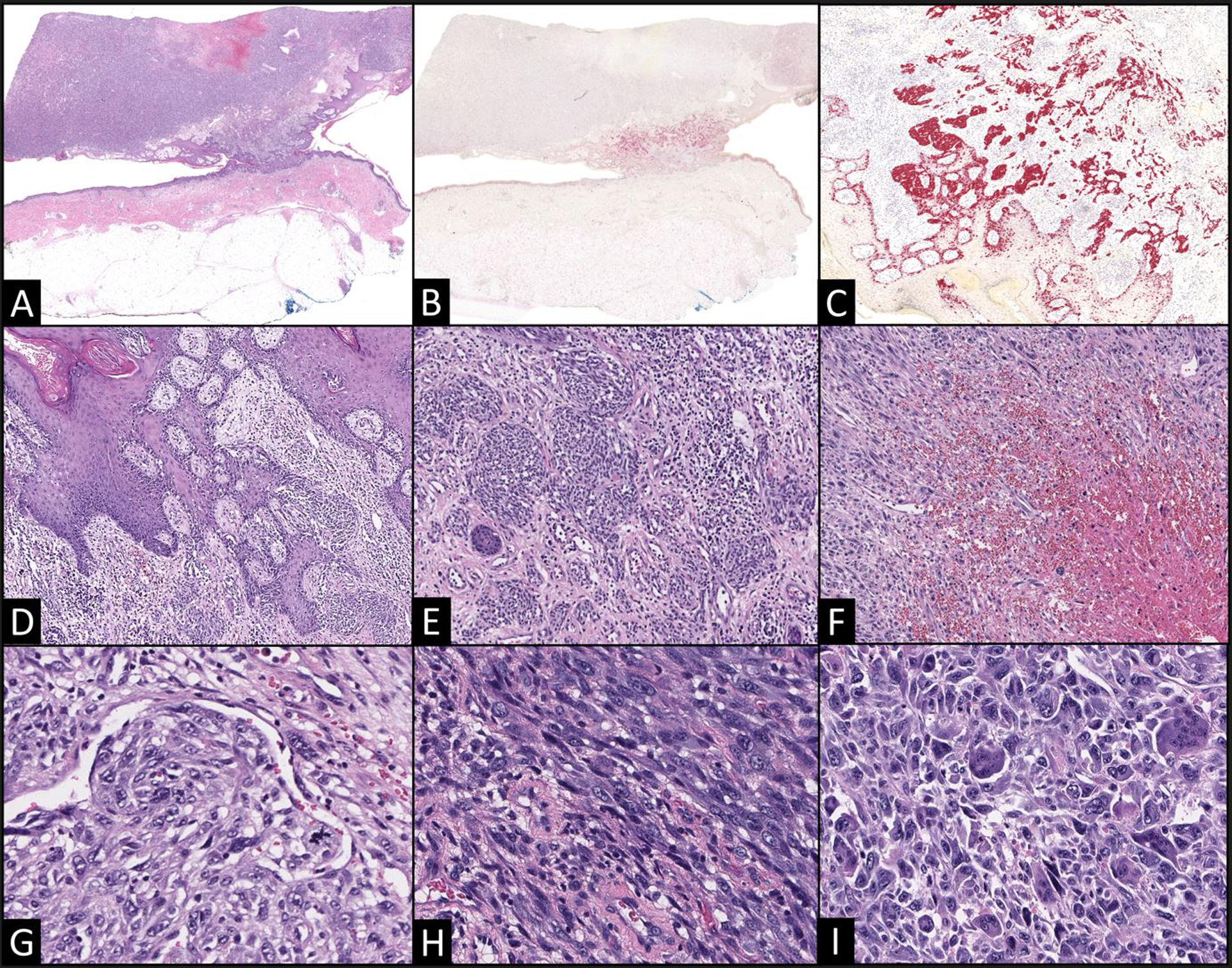
(A) A large pedunculated, centrally necrotic mass with a narrow stalk is focally contiguous with the epidermis (5x). (B) MelanA immunohistochemistry highlights the intraepidermal and superficial intradermal components; however, staining is not identified in the deep nodular component (5x). Higher power views (40x) of the superficial nested component reveal superficial spreading melanoma (C MelanA; D–E H&E). (F) However, portions of the tumor nearer to its necrotic center are comprised of disorganized sheets of cells with enlarged, hyperchromatic nuclei. Despite these distinct appearances, a gradual transition in cytomorphology was apparent from the superficial nested melanoma (G, 200x) to fascicles of spindled cells with increased nucleomegaly and pleomorphism (H, 200x) to sheets of sarcomatoid cells with bizarre nuclear features and accompanying multinucleated giant cells (I, 200x).
The anatomic site, generous excision sample that permitted for the evaluation of the entire lesion, dermal localization with limited involvement of the subcutis, and continuity of the differentiated and undifferentiated cell populations strongly supported a presumptive diagnosis of melanoma with an undifferentiated sarcomatoid component. PET/CT revealed an FDG-avid, presumptive left inguinal node metastasis measuring 1.9 × 1.5 cm but no evidence of distant metastases. In anticipation of the planned core needle biopsy of the left inguinal lymph node, a battery of immunohistochemical stains was performed in order to determine the phenotype of the tumor. The differentiated melanoma component expressed S100, Sox10, HMB45, and MITF. Although S100 highlighted background dendritic cells throughout the undifferentiated sarcomatoid component, the sarcomatoid cells were negative for S100 as well as the other aforementioned markers. Additional immunohistochemical stains revealed dense CD10 expression in only the sarcomatoid component with a majority of cells co-expressing p63 (Figure 2). Neither of these markers was expressed by the differentiated component and neither component expressed p40, CK5, AE1/AE3, BRAF, desmin, smooth muscle actin or CD34.
Figure 2. Expression of CD10 and p63 in undifferentiated sarcomatoid melanoma is a potential diagnostic pitfall.
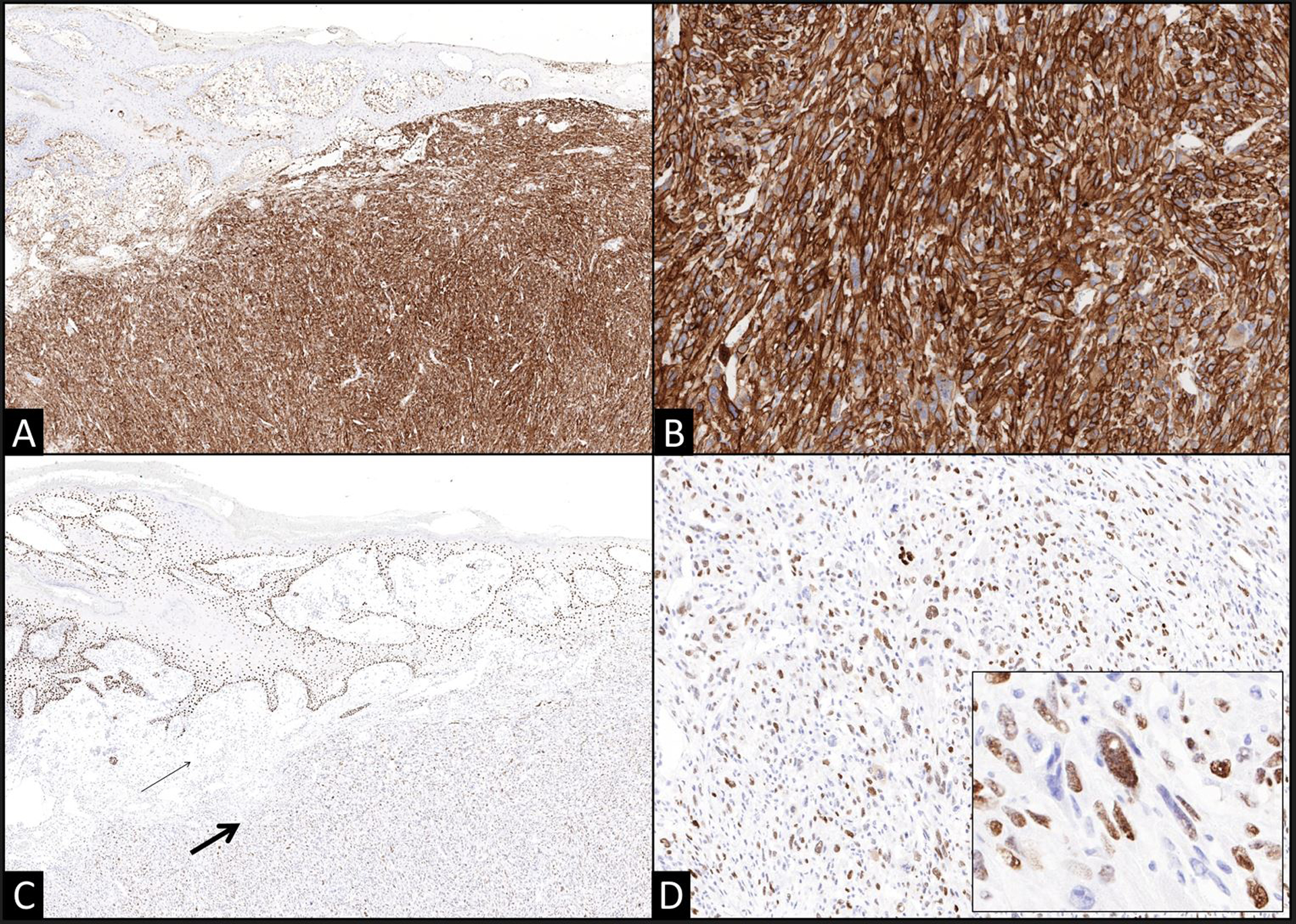
(A) The undifferentiated sarcomatoid component exhibits strong and diffuse CD10 expression (A 40x; B 100x) and unequivocal p63 expression in a majority of the lesional cells (thick arrow) in contrast with the superficial intradermal component of the melanoma (thin arrow) (C 40x; D 100x, inset 200x). The nested in situ and superficial invasive components did not express either of these markers.
Rapid sample-to-answer PCR testing using the Idylla™ NRAS-BRAF Mutation Assay cartridge (Biocartis, Mechelen, Belgium) revealed an NRAS p.Q61L somatic variant in the sarcomatoid component but not the differentiated melanoma component. Subsequent follow-up testing by next-generation sequencing (NGS) using the 50 gene Cancer Hotspot Panel v2 (ThermoFisher Scientific, Waltham, Massachusetts USA) revealed the NRAS p.Q61L in the sarcomatoid component, with a variant allele frequency of 43.9%, and in the differentiated melanoma, with a variant allele frequency of 11.8%.. Re-evaluation of the Idylla PCR amplification curves suggested the presence of a low-level p.Q61L variant in the differentiated melanoma component below the established cut-off. No additional BRAF or NRAS mutations were identified in either component by the Idylla NRAS-BRAF assay or the NGS panel. A TP53 p.P278S variant (NM_002524.4 c.182A>T) was detected in both components with a VAF of 13.6% in the differentiated melanoma and 52.4% in the sarcomatoid component, which reflected the NRAS variant VAF in the two components (Table 1).
Table 1.
Single nucleotide variants (SNVs) detected by Idylla/NGS.
| Sarcomatoid | Melanoma | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Reference Sequence | Variant | Consequence | VAF | Idylla | VAF | Idylla |
| NRAS | NM_002524.4 | c.182A>T | p.Q61L | 43.9% | + | 11.8% | +/− |
| TP53 | NM_000546.5 | c.832C>T | p.P278S | 52.4% | N/A | 13.6% | N/A |
Single nucleotide polymorphism (SNP) chromosomal microarray (OncoScan FFPE Assay kit, Affymetrix, Santa Clara, California, USA) was performed on both components of the tumor and revealed multiple copy number changes and copy neutral losses of heterozygosity (CN-LOH) found in both components (8p loss, 8q gain, 9p CN-LOH, 11q loss, 12q CN-LOH, 20p gain and chromothripsis/shattering of chromosome 22) and several additional copy number changes unique to the sarcomatoid component (1p gain, 6p gain, 6q loss, gain of chromosome 7, loss of chromosomes 10 and 13 and loss of 17p). These findings suggested a clonal evolution of the differentiated melanoma tumor into a more advanced sarcomatoid component of the tumor that harbored additional chromosomal abnormalities. No copy number changes were detected involving TP63 (3q28) or TERT (5p15), genes not included in the NGS panel used. The 9p CN-LOH detected in both components of the tumor included CDKN2A (p16).
The patient underwent core needle biopsy of his left inguinal lymphadenopathy, which revealed an undifferentiated metastatic sarcomatoid malignancy harboring an NRAS p.Q61L mutation that was detected by NRAS-BRAF Mutation Assay cartridge and identical to the previously identified mutation. The patient completed 5 cycles of pembrolizumab and at 3 months follow-up has developed no new PET-avid lesions.
Discussion
Undifferentiated sarcomatoid melanomas are exceedingly rare and can pose several diagnostic conundrums. The diagnosis in our case was possible given the presence of a residual differentiated component and supportive clinical context. Ancillary tests confirmed the putative relationship between the differentiated and undifferentiated components, excluding the unlikely prospect of a collision scenario between melanoma and AFX or PDS. The distinction between undifferentiated sarcomatoid melanoma and AFX or PDS would have been of much greater difficulty in the case of a rapidly growing tumor on sun-exposed skin, particularly on the head and neck, of a patient of advanced age, since the vast majority of AFX and PDS arise in this clinical setting.1 UPS would enter the differential diagnosis when evaluating a metastatic or primary tumor situated in the subcutis. In these instances, and particularly when a partial sample is provided for diagnosis, a component of differentiated melanoma may not be present. Serial step-leveled sections and immunohistochemistry should be performed in an effort to identify any evidence of a differentiated component before raising consideration for a mesenchymal malignancy.2 In some instances, it may be prudent to defer a definitive diagnosis until the tumor can be evaluated in its entirety.
Overreliance on immunohistochemistry in the workup of an undifferentiated sarcomatoid malignancy is fraught with diagnostic pitfalls. As our example illustrates, sarcomatoid melanomas can express CD10 and p63. CD10 has gained recognition as a marker that is helpful for distinguishing AFX and PDS from other cutaneous sarcomatoid malignancies including SSCC and melanoma3–7; however, CD10 expression has been reported in a subset of melanomas, and some studies have associated CD10 expression in melanomas with disease progression.2,3,8,9 The two most recent examples of CD10-positive melanomas reported in the literature were also undifferentiated sarcomatoid melanomas with polypoid, pedunculated clinical appearances.2 p63 immunohistochemistry is generally helpful for distinguishing SSCC from melanoma and AFX/PDS given its sensitivity for SSCC and crisp nuclear labeling; however, it should be interpreted cautiously in concert with markers that offer a greater specificity for SSCC, including the p40 isoform10,11 and cytokeratins. p63 expression has previously been reported in mesenchymal malignancies10,11 and cannot be used in isolation to exclude AFX, PDS, and UPS. p63 immunohistochemical staining is infrequently assessed in melanomas, and is rarely reported as positive.12 However, its expression is well-documented in a subpopulation of melanomas and evidence suggests that its expression may be associated with antiapoptotic activity and a poorer prognosis.13,14 Additional pitfalls in the assessment of undifferentiated sarcomatoid malignancies of the skin include aberrant MITF expression noted in a subset of AFX, PDS, and UPS.15,16 S100 labeling of an impressive quantity of dendritic cells within AFX and PDS has been described in a pattern similar to that which was observed in our melanoma and should not be construed as positive staining.15,16 Close inspection of the stain may be necessary to distinguish staining of background dendritic cells from S100 expression by the malignant cells.
Judicious use of molecular studies can help to exclude the possibility of a collision scenario of melanoma and sarcoma. The distinct transition in both the morphology and immunophenotype between the components of melanoma in our case example prompted some consideration for this possibility. Collisions of AFX and melanoma, AFX and melanocytic nevi, and AFX and squamous cell carcinoma are rare but nonetheless well-documented.17–22 In our case, the overlap in SNP copy number changes and oncogenic mutations suggested a common origin for both the differentiated and undifferentiated sarcomatoid components of this melanoma. Furthermore, additional copy number variations were identified in the sarcomatoid component, which was in keeping with our understanding that it had evolved from the preexisting differentiated melanoma. This concept was further supported histomorphologically in portions of the tumor that showed a gradual transition from a nested epithelioid morphology to spindled and bizarre pleomorphic cells. Similar molecular methods have been used previously to distinguish a desmoplastic melanoma with sarcomatoid dedifferentiation from a collision of desmoplastic melanoma and PDS.23 In this previous case, the identification of neurofibromin 1 (NF1) mutations in both components and high proportion of shared chromosomal copy number changes and mutations similarly excluded the possibility of a collision tumor.
Molecular studies can also be helpful in the evaluation of presumptive metastases or partial samples of a cutaneous undifferentiated sarcomatoid malignancy when no differentiated component is identified.24 Genomic analyses of AFX and PDS have revealed similarities and differences with melanoma. As in melanoma, AFX and PDS show a high ultraviolet mutational burden, sometimes involving the TERT promoter and TP53 coding regions.25–27 They also show a wide range of DNA copy number alterations, with greater copy number variations in PDS than in AFX, which has fostered discussion as to whether PDS represents progression of AFX and not a distinct neoplasm.28 Furthermore, these entities show occasional chromosome 9p deletions27–29 and rarer MYC amplifications that are seen in cutaneous melanomas.30 To our knowledge, however, HRAS and KRAS oncogenic mutations have been discovered in only a small number of PDS and not in AFX.27,31 Furthermore, NRAS mutations, as identified in the sarcomatoid component of our case, have not been reported in these neoplasms and should raise strong consideration for melanoma since approximately two thirds of melanomas, including undifferentiated melanomas, have RAS-RAF-MAPK pathway activating mutations.15,24 Furthermore, NRAS mutations may be slightly overrepresented among undifferentiated melanomas.24 The finding of a BRAF mutation in an undifferentiated malignancy should raise even stronger consideration for melanoma since BRAF mutations have not been identified in UPS and are in general rarely identified in sarcomas.32 Some melanoma variants are strongly associated with additional mutations, including KIT mutations present in mucosal and acral lentiginous melanomas, and NF1 mutations which have been associated with a subset of desmoplastic melanomas23. In contrast, AFX has a higher frequency of mutations involving NOTCH1/2, FAT1, COL11A1, CSMD3 and ERBB4.27,29
In summary, dermatopathologists must consider the possibility of melanoma when evaluating an undifferentiated sarcomatoid malignancy and not rely too heavily on immunohistochemistry when there are clinical or histologic features that would otherwise raise strong consideration for melanoma. The distinction between undifferentiated melanoma and AFX, PDS, and UPS is necessary for management since treatment differs tremendously. AFX and PDS are clinically indolent in comparison to melanoma. Excision of an AFX is essentially curative. Conversely, patients with melanomas may be offered sentinel lymph node biopsies at the time of excision, receive BRAF testing in the setting of metastatic disease, and undergo surgical and medical therapies commensurate with a melanoma diagnosis and in lieu of those reserved for UPS. This critical distinction can be nonetheless problematic depending on the clinical context, especially when a differentiated component of melanoma cannot be identified in the provided specimen. Therefore, it may be prudent to defer definitive diagnosis or pursue ancillary molecular tests when evaluating partial samples of significantly larger lesions or biopsies of presumptive metastases. Rare cases including our own illustrate how SNP and NGS can be leveraged to establish a firm diagnosis by identifying common copy number variations and or mutations characteristic of melanoma or sarcoma across both components. NGS and rapid BRAF/NRAS testing offer an added potential benefit of unveiling therapeutic targets. Dermatopathologists must appreciate the limitations and benefits of clinical, histologic, immunohistochemical, and molecular findings when evaluating an undifferentiated cutaneous sarcomatoid malignancies in order for patients to receive the correct treatment.
Figure 3. SNP Chromosomal Microarray findings support the clinicopathologic impression of a clonal evolution from differentiated to undifferentiated sarcomatoid melanoma.
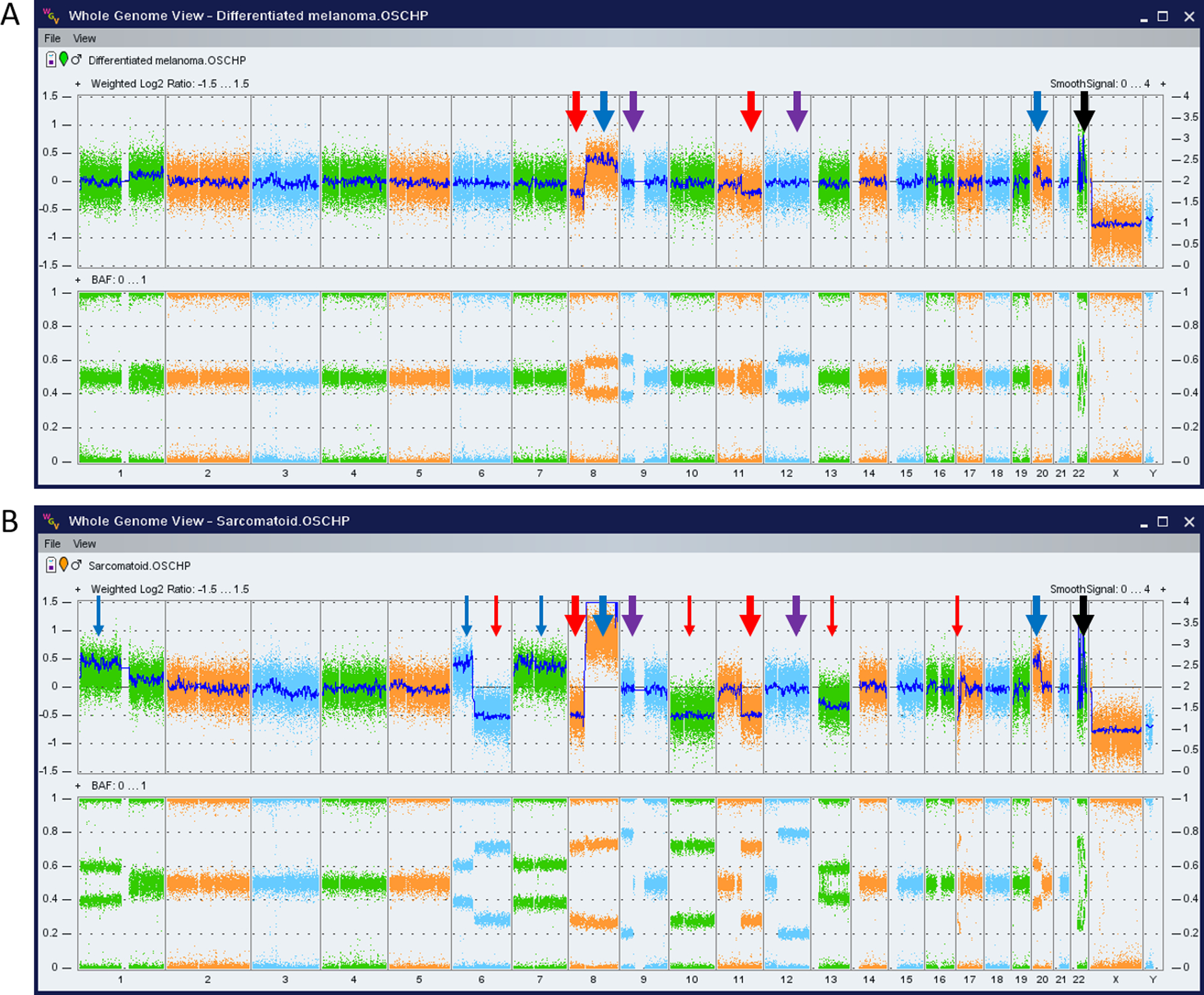
Whole genome views of the microarray data for the differentiated melanoma (A) and the undifferentiated sarcomatoid component (B) with thick arrows indicating findings present in both components (losses of 8p and 11q, gains of 8q and 20p, CN-LOH of 9p and 12q, and chromothripsis of chromosome 22) suggesting common clonal origins. Thin arrows indicate additional alterations present exclusively in the sarcomatoid component that suggest a later stage in clonal evolution (gains of 1p, 6p, chromosome 7 and losses of 6q, chromosomes 10 and 13 and part of 17p). The color of the arrows indicate copy number losses/deletions (red), copy number gains (blue), copy neutral loss of heterozygosity (purple) and chromothripsis/chromosome shattering (black). The top portion of each whole genome view shows copy number state expressed as log2 ratio (y-axis on the left) and smoothed signal/estimated copy number (y-axis to the right). The bottom portion of each whole genome view contains the B-allele frequency (BAF), with allelic imbalances due to copy number alterations or copy neutral losses of heterozygosity indicated by the lack of heterozygous calls in the middle of plot (BAF of 0.5). Chromosomes numbers are shown along the bottom x-axis of each plot.
Acknowledgements:
The authors wish to thank the staff of the Pathology Shared Resource Laboratory, a section of the laboratory for Clinical Genomics and Advanced Technology (CGAT). The data presented in this manuscript was in part generated through CGAT in the Department of Pathology and Laboratory Medicine of the Geisel School of Medicine at Dartmouth, the Dartmouth-Hitchcock Medical Center and the Norris Cotton Cancer Center. (NCI Cancer Support Grant #5P30CA023108-37).
References
- 1.Koch M, Freundl AJ, Agaimy A, et al. Atypical Fibroxanthoma - Histological Diagnosis, Immunohistochemical Markers and Concepts of Therapy. Anticancer Res. 2015;35(11):5717–5735. [PubMed] [Google Scholar]
- 2.Erstine EM, Tetzlaff MT, Ko JS, Prieto VG, Cheah AL, Billings SD. Living on the Edge: Diagnosing Sarcomatoid Melanoma Using Histopathologic Cues at the Edge of a Dedifferentiated Tumor: A Report of 2 Cases and Review of the Literature. Am J Dermatopathol. 2017;39(8):593–598. [DOI] [PubMed] [Google Scholar]
- 3.de Feraudy S, Mar N, McCalmont TH. Evaluation of CD10 and procollagen 1 expression in atypical fibroxanthoma and dermatofibroma. The American journal of surgical pathology. 2008;32(8):1111–1122. [DOI] [PubMed] [Google Scholar]
- 4.Hanlon A, Stasko T, Christiansen D, Cyrus N, Galan A. LN2, CD10, and Ezrin Do Not Distinguish Between Atypical Fibroxanthoma and Undifferentiated Pleomorphic Sarcoma or Predict Clinical Outcome. Dermatol Surg. 2017;43(3):431–436. [DOI] [PubMed] [Google Scholar]
- 5.Aiad HA, Hanout HM. Immunohistochemical Expression of CD10 in Cutaneous Basal and Squamous Cell Carcinomas. J Egypt Natl Canc Inst. 2007;19(3):195–201. [PubMed] [Google Scholar]
- 6.Sari Aslani F, Akbarzadeh-Jahromi M, Jowkar F. Value of CD10 Expression in Differentiating Cutaneous Basal from Squamous Cell Carcinomas and Basal Cell Carcinoma from Trichoepithelioma. Iran J Med Sci. 2013;38(2):100–106. [PMC free article] [PubMed] [Google Scholar]
- 7.Shafaei S, Sharifian M, Hajian-Tilaki K. Immunohistochemical expression of CD10 in cutaneous basal and squamous cell carcinomas. Caspian J Intern Med. 2015;6(2):103–107. [PMC free article] [PubMed] [Google Scholar]
- 8.Weinreb I, Cunningham KS, Perez-Ordonez B, Hwang DM. CD10 is expressed in most epithelioid hemangioendotheliomas: a potential diagnostic pitfall. Arch Pathol Lab Med. 2009;133(12):1965–1968. [DOI] [PubMed] [Google Scholar]
- 9.Oba J, Nakahara T, Hashimoto-Hachiya A, et al. CD10-Equipped Melanoma Cells Acquire Highly Potent Tumorigenic Activity: A Plausible Explanation of Their Significance for a Poor Prognosis. PLoS One. 2016;11(2):e0149285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha Lan TT, Chen SJ, Arps DP, et al. Expression of the p40 isoform of p63 has high specificity for cutaneous sarcomatoid squamous cell carcinoma. J Cutan Pathol. 2014;41(11):831–838. [DOI] [PubMed] [Google Scholar]
- 11.Henderson SA, Torres-Cabala CA, Curry JL, et al. p40 is more specific than p63 for the distinction of atypical fibroxanthoma from other cutaneous spindle cell malignancies. The American journal of surgical pathology. 2014;38(8):1102–1110. [DOI] [PubMed] [Google Scholar]
- 12.Kanner WA, Brill LB 2nd, Patterson JW, Wick MR. CD10, p63 and CD99 expression in the differential diagnosis of atypical fibroxanthoma, spindle cell squamous cell carcinoma and desmoplastic melanoma. J Cutan Pathol. 2010;37(7):744–750. [DOI] [PubMed] [Google Scholar]
- 13.Matin RN, Chikh A, Chong SL, et al. p63 is an alternative p53 repressor in melanoma that confers chemoresistance and a poor prognosis. J Exp Med. 2013;210(3):581–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monti P, Ghiorzo P, Menichini P, et al. TP63 mutations are frequent in cutaneous melanoma, support UV etiology, but their role in melanomagenesis is unclear. Oncol Rep. 2017;38(4):1985–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choy B, Hyjek E, Montag AG, et al. High prevalence of MiTF staining in undifferentiated pleomorphic sarcoma: caution in the use of melanocytic markers in sarcoma. Histopathology. 2017;70(5):734–745. [DOI] [PubMed] [Google Scholar]
- 16.Helbig D, Mauch C, Buettner R, Quaas A. Immunohistochemical expression of melanocytic and myofibroblastic markers and their molecular correlation in atypical fibroxanthomas and pleomorphic dermal sarcomas. J Cutan Pathol. 2018;45(12):880–885. [DOI] [PubMed] [Google Scholar]
- 17.Specchio F, Argenziano G, Zalaudek I, et al. Photoletter to the editor: Collision tumor of melanoma and atypical fibroxanthoma of the scalp. J Dermatol Case Rep. 2014;8(3):84–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGregor DH, Cherian R, Romanas MM, Ulusarac O, Mathur SC, Feldman MM. Amelanotic malignant melanoma: two collision tumors presenting as basal cell carcinoma and atypical fibroxanthoma. Ann Clin Lab Sci. 2008;38(2):157–162. [PubMed] [Google Scholar]
- 19.Wilsher MJ. Collision tumour: atypical fibroxanthoma and invasive melanoma. Pathology. 2009;41(7):699–701. [DOI] [PubMed] [Google Scholar]
- 20.Hyatt AM, Mutasim DF, Spicknall KE. Collision of atypical fibroxanthoma and acantholytic squamous cell carcinoma in situ. Am J Dermatopathol. 2012;34(5):563–564. [DOI] [PubMed] [Google Scholar]
- 21.Arsenovic N, Sen S, Naik V, Reed M, Moreira R. Trichilemmal cyst with carcinoma in situ within an atypical fibroxanthoma. Am J Dermatopathol. 2009;31(6):587–590. [DOI] [PubMed] [Google Scholar]
- 22.Steel A, Debbaneh M, Cassarino D. An Atypical Fibroxanthoma and Intradermal Nevus Collision Tumor-Potential for Misdiagnosis. Am J Dermatopathol. 2019;41(8):e87–e89. [DOI] [PubMed] [Google Scholar]
- 23.Kiuru M, McDermott G, Berger M, Halpern AC, Busam KJ. Desmoplastic melanoma with sarcomatoid dedifferentiation. The American journal of surgical pathology. 2014;38(6):864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agaimy A, Specht K, Stoehr R, et al. Metastatic Malignant Melanoma With Complete Loss of Differentiation Markers (Undifferentiated/Dedifferentiated Melanoma): Analysis of 14 Patients Emphasizing Phenotypic Plasticity and the Value of Molecular Testing as Surrogate Diagnostic Marker. The American journal of surgical pathology. 2016;40(2):181–191. [DOI] [PubMed] [Google Scholar]
- 25.Griewank KG, Schilling B, Murali R, et al. TERT promoter mutations are frequent in atypical fibroxanthomas and pleomorphic dermal sarcomas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2014;27(4):502–508. [DOI] [PubMed] [Google Scholar]
- 26.Dei Tos AP, Maestro R, Doglioni C, et al. Ultraviolet-induced p53 mutations in atypical fibroxanthoma. Am J Pathol. 1994;145(1):11–17. [PMC free article] [PubMed] [Google Scholar]
- 27.Griewank KG, Wiesner T, Murali R, et al. Atypical fibroxanthoma and pleomorphic dermal sarcoma harbor frequent NOTCH1/2 and FAT1 mutations and similar DNA copy number alteration profiles. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2018;31(3):418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helbig D, Quaas A, Mauch C, et al. Copy number variations in atypical fibroxanthomas and pleomorphic dermal sarcomas. Oncotarget. 2017;8(65):109457–109467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai K, Harwood CA, Purdie KJ, et al. Genomic analysis of atypical fibroxanthoma. PLoS One. 2017;12(11):e0188272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaiser T, Hirsch D, Orouji A, et al. MYC gene amplification is a rare event in atypical fibroxanthoma and pleomorphic dermal sarcoma. Oncotarget. 2018;9(30):21182–21189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakamoto A, Oda Y, Itakura E, et al. H-, K-, and N-ras gene mutation in atypical fibroxanthoma and malignant fibrous histiocytoma. Hum Pathol. 2001;32(11):1225–1231. [DOI] [PubMed] [Google Scholar]
- 32.Cipriani NA, Letovanec I, Hornicek FJ, et al. BRAF mutation in ‘sarcomas’: a possible method to detect de-differentiated melanomas. Histopathology. 2014;64(5):639–646. [DOI] [PubMed] [Google Scholar]


