Genome editing methods generally involve two critical steps: targeting a DNA sequence of interest and introducing the desired genetic modification. Capabilities for programmable DNA targeting have advanced rapidly over the past decade, owing primarily to the retooling of bacterial RNA-guided CRISPR-Cas systems. However, efficient and precise modification of the target site remains challenging, particularly for large-scale perturbations such as the insertion of gene-sized DNA payloads. To address this limitation, an emerging body of research is now drawing inspiration from mobile genetic elements (MGEs), DNA segments that have exploited diverse targeting and integration strategies to propagate in the genomes of their hosts for billions of years. Of particular interest are retroelements, which mobilize using an RNA intermediate. Recent investigations into the structure and function of retroelement-encoded protein-RNA complexes have resolved key molecular details of their targeting and integration mechanisms (1, 2), unveiling exciting opportunities to reengineer them for programmable DNA insertion.
MGEs are present in all organisms and encode enzymes that facilitate the movement of their own DNA within and between genomes. Two major categories of MGEs—DNA transposons and retroelements—can be distinguished by their spreading mechanisms. DNA transposons typically encode transposase enzymes that mobilize DNA by excising sequences from their original location and integrating them into a target locus (“cut-and-paste”). By contrast, retroelements rely on the host cell’s RNA polymerase to generate an RNA copy of the element, which in turn serves as a template for complementary DNA (cDNA) synthesis by a retroelement-encoded reverse transcriptase (RT). This cDNA product is then incorporated into the genome through a range of mechanisms, creating a new copy of the original DNA segment (“copy-and-paste”).
MGEs are natural candidates for genome editing applications. Their ubiquitous presence reflects a fine-tuned capacity for safe and efficient DNA integration into host genomes. However, MGEs generally exhibit either random or fixed specificity for integration sites, which limits their utility for programmable DNA insertion. This contrasts with CRISPR-based genome editing, in which the Cas9 nuclease can be directed by a user-defined guide RNA (gRNA) to target almost any DNA sequence of interest. But, in conventional genome editing with Cas9, DNA targeting and editing constitute separate processes: Cas9 recognizes and cleaves the target sequence to generate a DNA double-strand break (DSB), activating cellular DNA repair pathways that incorporate permanent sequence edits. This process introduces substantial heterogeneity and can lead to unwanted by-products, such as large chromosomal deletions and translocations, resulting in safety concerns around DSB-generating genome editing methods (3). CRISPR-Cas systems and MGEs thus exhibit complementary strengths and weaknesses with regard to target site specificity and genotoxicity. The ideal genome engineering tool would merge their capabilities, directly coupling programmable DNA targeting with DSB-free modification of the target locus.
Indeed, approaches that combine CRISPR-mediated DNA targeting with transposase-mediated DNA insertion have emerged in the past 5 years (4–6). These methods use various strategies to recruit transposase enzymes to gRNA-specified target sites, where they catalyze the integration of a donor DNA molecule supplied on a plasmid. Targeted DNA insertion with CRISPR-associated transposases (CASTs) represents a promising area of investigation, with demonstrated success for DNA cargoes up to 15 kilobases (6). Analogous retroelement-mediated approaches, which could encode the donor molecule on RNA rather than DNA, might offer additional advantages for clinical applications. Whereas DNA cargoes for genome editing are often delivered to target tissues by adeno-associated viruses (AAVs) or other vectors, RNA cargoes can be delivered by encapsulation in lipid nanoparticles. Lipid nanoparticles are less immunogenic than AAVs, and they present less risk for off-target editing owing to the transient nature of their RNA cargoes (7), which are degraded by the recipient cell. Thus, retroelement-based genome editing tools, which do not require any DNA components and can be packaged in lipid nanoparticles, offer a distinct safety advantage over tools that require an exogenously provided DNA donor molecule.
Retroelements have been a part of the genome engineering toolkit for over two decades, though until recently they have demonstrated limited utility for targeted DNA insertion. For example, targetrons are modified bacterial group II introns that home to user-specified DNA sequences through base-pairing interactions with the intron RNA, which catalyzes reverse splicing into the target site (8). Although targetrons have been used for targeted gene disruption, poor tolerance of synthetic cargoes has hindered efforts to engineer them for transgene insertion. Retrons, a more recent addition to the toolkit, are bacterial retroelements that have been engineered to produce donor DNAs from RNA templates for homology-directed repair of CRISPR-Cas9 cleavage sites (9). However, retron-mediated approaches have only been applied to small-scale edits, and their reliance on the generation of a DSB poses safety risks for clinical gene editing. Targetrons and retrons have thus demonstrated the promise of harnessing retroelements for biotechnology, but they fall short of the ultimate capability for a retroelement-mediated genome editing tool: to directly write any new DNA sequence into any genomic target site.
The development of prime editing represents the most important breakthrough to date toward achieving this capability. In prime editing, a nickase variant of Cas9 (nCas9) is directed by a prime editing guide RNA (pegRNA) to cleave a single DNA strand at the genomic target site, exposing a free DNA end from which an RT enzyme polymerizes a new DNA sequence using the pegRNA as a template. Cellular repair factors incorporate the new DNA strand into the genomic target site, resulting in permanent integration of the pegRNA-specified sequence (10). Prime editing avoids the generation of DSBs and can install all types of point mutations, small insertions, and small deletions. Nonetheless, editing efficiencies remain low for many applications, and the template size that can be accommodated is limited to ~40 base pairs or less. Recent advances have addressed this size limitation by employing multiple pegRNAs or combining prime editing with site-specific recombinases (11)—another class of enzymes used by MGEs to perform DNA integration—but at the cost of adding complexity and reaction steps to the overall editing pathway.
Notably, the mechanism of DNA writing utilized by prime editors, known as target-primed reverse transcription (TPRT), is the same mechanism used to mobilize entire genes by a family of retroelements that is widespread in nature (12). Harnessing these elements, known as non–long terminal repeat (LTR) retrotransposons, has the potential to substantially advance gene insertion technology. But doing so will require a detailed understanding of their native mobility mechanisms, including how they select their respective RNA substrates and genomic targets and how they write their cargoes into target sites. Fortunately, the mobilization of non-LTR retrotransposons is relatively well studied owing to their pervasiveness—for example, the long interspersed nuclear element 1 (LINE-1) retrotransposon makes up nearly 20% of the human genome (13). Intriguingly, whereas LINE-1 undergoes largely untargeted mobilization, the R2 non-LTR retrotransposon, which is abundant in arthropods, exhibits stringent target site preference for the 28S ribosomal RNA locus (14). But how the R2 protein assembles with R2 RNA to initiate TPRT at this site, and whether R2 can perform TPRT with a synthetic RNA template, was unclear until earlier this year.
Two recent studies used cryo–electron microscopy to determine the three-dimensional structure of the R2 RT from Bombyx mori (domestic silk moth) bound to R2 RNA and the 28S DNA target (1, 2). This revealed specific molecular interactions in the RT-RNA-DNA complex, indicating that the retrotransposon selects its RNA template and DNA target through sequence-dependent recognition by the R2 protein. These structural details were used to engineer a synthetic R2 RNA that included the minimal recognition sequence and a “cargo” RNA appendage. The R2 RT recognized this engineered substrate and could be redirected by Cas9 to incorporate the cargo at non-28S target sequences in biochemical reactions (1).
A strong foundation has been laid for leveraging retroelements as targeted gene insertion tools. However, important gaps in knowledge remain. For example, genomic integration of cDNAs generated by the R2 RT requires the synthesis of a second cDNA strand (14), but the mechanistic details of this process are unclear. Additional studies aimed at resolving the molecular requirements for the final steps of R2 retrotransposition will be crucial for engineering R2 to perform transgene insertion in cells. Furthermore, systematic approaches for profiling the accuracy of TPRT have not been reported. The development of such methods will be critical for evaluating the safety of retroelement-mediated genome editing strategies.
A vast repertoire of retroelements exists in nature, yet only a few have been investigated for their genome editing potential. Certain RT enzymes with as-yet-unknown properties might be inherently well-suited for genome editing tasks. Systematic mining of retroelement diversity through bioinformatic analyses and pooled screening will be invaluable for the identification of RT enzymes with desirable characteristics (see the figure). This strategy is already bearing fruit for the development of compact prime editors (15). It is also worth noting that retroelements have already played an outsized role in shaping the human genome, roughly half of which is derived from MGEs (13). Thus, in developing retroelement-based DNA editing tools, humans are repurposing nature’s original genome engineers. ■
A path to retroelement-based programmable gene insertion.
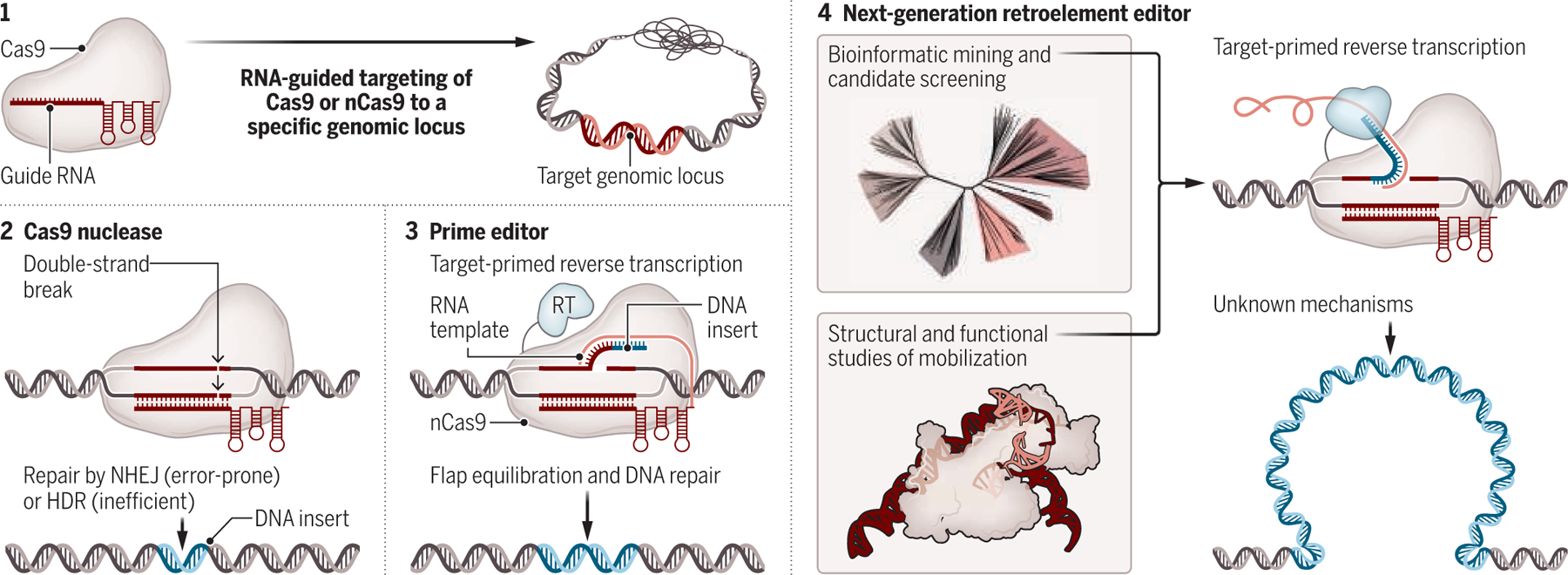
Genome editing tools commonly use Cas9 directed by a guide RNA to recognize the target site (1) but employ distinct strategies for the DNA editing step. In conventional nuclease-mediated editing (2), a DNA double-strand break is made by Cas9 and repaired by nonhomologous end joining (NHEJ) or homology-directed repair (HDR). In prime editing (3), nickase Cas9 (nCas9) generates a free DNA end from which a reverse transcriptase (RT) writes a new RNA-templated DNA sequence. Further exploration of retroelements could enable RNA-templated integration of larger DNA payloads (4).
ACKNOWLEDGMENTS
The authors thank D. Zhang and T. Wiegand for assistance with the figure, as well as A. Chen for insightful comments on the manuscript. S.T. is supported by a Medical Scientist Training Program grant (5T32GM145440-02) from the National Institutes of Health (NIH). S.H.S. is supported by NIH grant DP2HG011650, a Pew Biomedical Scholarship, a Sloan Research Fellowship, an Irma T. Hirschl Career Scientist Award, and a generous startup package from the Columbia University Irving Medical Center Dean’s Office and the Vagelos Precision Medicine Fund. S.H.S. is an inventor on patents and patent applications related to CRISPR–Cas systems and uses thereof, a cofounder of and scientific adviser to Dahlia Biosciences, a scientific adviser to Prime Medicine and CrisprBits, and an equity holder in Dahlia Biosciences and CrisprBits.
REFERENCES AND NOTES
- 1.Wilkinson ME, Frangieh CJ, Macrae RK, Zhang F, Science 380, 301 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng P et al. , Cell 186, 2865 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Wang JY, Doudna JA, Science 379, eadd8643 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Strecker J et al. , Science 365, 48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klompe SE, Vo PLH, Halpin-Healy TS, Sternberg SH, Nature 571, 219 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Lampe GD et al. , Nat. Biotechnol 10.1038/s41587-023-01748-1 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Raguram A, Banskota S, Liu DR, Cell 185, 2806 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H et al. , Science 289, 452 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Sharon E et al. , Cell 175, 544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anzalone AV et al. , Nature 576, 149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen PJ, Liu DR, Nat. Rev. Genet 24, 161 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luan DD, Korman MH, Jakubczak JL, Eickbush TH, Cell 72, 595 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Payer LM, Burns KH, Nat. Rev. Genet 20, 760 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Eickbush TH, Eickbush DG, Microbiol. Spectr 3, 3.2.02 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doman JL et al. , Cell 186, 3983 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]


