Abstract
Expression of the Escherichia coli multiple antibiotic resistance marA gene cloned in Mycobacterium smegmatis produced increased resistance to multiple antimicrobial agents, including rifampin, isoniazid, ethambutol, tetracycline, and chloramphenicol. Cloned marR or marA cloned in the antisense direction had no effect. Resistance changes were lost with spontaneous loss of the plasmid bearing marA. A MarA mutant protein, having an insertional mutation within either of its two alpha-helices of the first putative helix-turn-helix domain, failed to produce the multiresistance phenotype in E. coli and M. smegmatis, indicating that this region is critical for MarA function. These results strongly suggest that E. coli marA functions in M. smegmatis and that a mar-like regulatory system exists in this organism.
Multidrug resistance in Mycobacterium is presumed to occur via the accumulation of independent chromosomal mutations which affect susceptibility to individual drugs. In Escherichia coli and other members of the family Enterobacteriaceae, multidrug resistance is generally attributed to plasmids and transposons. Still, multidrug resistance can arise via derepression of the E. coli mar (multiple antibiotic resistance) operon, either by mutation or exposure to inducing compounds (4). In Mycobacterium, the observed relatively high frequency of multidrug resistance and the suggested relationship of inadequate treatment to the emergence of resistance (2) fit with the selection of E. coli Mar mutants. We looked for the possible existence of a mar-like regulatory drug resistance response in Mycobacterium smegmatis by examining antimicrobial susceptibility in cells expressing the cloned E. coli marA gene.
PCR oligonucleotide primers were used to prepare a wild-type marA amplicon from E. coli AG100 (9) chromosomal DNA, based on the annotated sequence (4). The oligonucleotide primers corresponded with nucleotide positions 1893 to 1910 and 2265 to 2282 and contained terminal EcoRI restriction enzyme sites to allow insertion of marA in frame with the hsp60 mycobacterial heat shock promoter resident on the E. coli-Mycobacterium shuttle plasmid pMV261 (11) (Table 1). We also used the “megaprimer” PCR method (12) to create insertional mutants of marA in the center of each alpha-helical region of the putative helix-turn-helix (HTH) domain of MarA (Fig. 1). These mutant marA genes were ligated to pMV261 and pET13a for testing in M. smegmatis and E. coli, respectively. Plasmids were introduced into E. coli and M. smegmatis mc2155 by electroporation with a Gene Pulser transfection apparatus (Bio-Rad, Richmond, Calif.) and selected on kanamycin (10 or 25 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli AG100 | Wild type | 9 |
| E. coli AG102 | Mar mutant of AG100 | 9 |
| E. coli BL21 | Expression strain for pET vectors | Novagen |
| M. smegmatis mc2155 | Electroporation competent | 18 |
| Plasmids | ||
| pMV261 | Mycobacterium-E. coli shuttle vector | 19 |
| pPM10 | pMV261::marA | This study |
| pPM10R | pMV261::marA in antisense orientation | This study |
| pPM11 | pMV261::marR | This study |
| pPM1989R | pPM10 insertional mutant of helix A | This study (Fig. 1) |
| pPM2016A | pPM10 insertional mutant of helix B | This study (Fig. 1) |
| pET13a | T7 expression vector | 20 |
| pEC10 | pET13a::marA | This study |
| pEC1989R | pEC10 insertional mutant of helix A | This study (Fig. 1) |
| pEC2016A | pEC10 insertional mutant of helix B | This study (Fig. 1) |
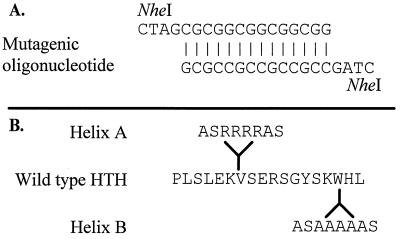
FIG. 1.
MarA mutants. (A) Nucleotide sequence of the mutagenic oligonucleotide. (B) Wild-type MarA amino acid residues 27 to 44, the putative HTH domain of MarA, based on the sequence (4). Mutants contained the amino acids shown at the indicated sites of insertion in helix A and helix B. Mutants differ in amino acid composition due to the mutagenic oligonucleotide inserting in opposite orientations.
Cultures of M. smegmatis mc2155 with and without plasmids were grown at 30 or 37°C by using 7H9 or 7H10 Middlebrook medium (Difco) enriched with Middlebrook albumin-dextrose complex (ADC) supplement and oleic albumin-dextrose complex supplement (OADC), respectively, supplemented with 0.05% Tween 80 and with kanamycin (10 μg/ml) where appropriate to maintain the Kanr plasmids. Antimicrobial susceptibilities were tested without kanamycin in 7H10-OADC antibiotic gradient plates (5) at 30 and 37°C. Tetracycline, chloramphenicol, norfloxacin, and phenazine methosulfate were purchased from Sigma Chemical Co. (St. Louis, Mo.). Isoniazid, rifampin, streptomycin sulfate, and ethambutol were kindly provided by J. Crawford (Centers for Disease Control and Prevention, Atlanta, Ga.), and sparfloxacin was received from Rhone-Poulenc (Paris, France).
M. smegmatis mc2155 bearing pMV261::marA showed increased resistance to multiple antimicrobial agents, including rifampin, isoniazid, ethambutol, chloramphenicol, and tetracycline, compared to the organism with vector alone (Table 2) when grown at 37°C but not at 30°C. Increased resistance to rifampin, however, was also noted at 30°C. Rifampin resistance also increased in the presence of vector alone at both temperatures, although this finding was variable. When it did occur, this was the only drug to which the vector appeared to affect parental susceptibility levels. Resistance of M. smegmatis to chloramphenicol increased twofold and resistance to tetracycline increased nearly fivefold in the presence of marA. Ethambutol and isoniazid resistance increased 1.5- and 2.5-fold at 37°C. Little if any change in susceptibility to nalidixic acid, phenazine methosulfate, or sparfloxacin occurred; some increased susceptibility was observed for norfloxacin and streptomycin (data not shown).
TABLE 2.
Antibiotic susceptibilities of M. smegmatis mc2155 organisms with and without plasmids bearing different E. coli mar genes
| M. smegmatis organism | % Growth in antibiotic gradienta
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RIF
|
INH
|
CML
|
TET
|
ETM
|
||||||
| 30°C | 37°C | 30°C | 37°C | 30°C | 37°C | 30°C | 37°C | 30°C | 37°C | |
| Wild type | 11 ± 1.2 | 11 ± 1.1 | 21 ± 1.2 | 20 ± 1.0 | 20 ± 0.8 | 22 ± 0.9 | 20 ± 1.9 | 19 ± 1.2 | 40 ± 2.2 | 44 ± 2.1 |
| Transformants bearing plasmids: | ||||||||||
| pMV261 | 22 ± 1.1 | 45 ± 2.6 | 22 ± 1.3 | 20 ± 1.0 | 22 ± 0.9 | 23 ± 1.1 | 20 ± 1.8 | 20 ± 1.3 | 42 ± 2.4 | 44 ± 1.5 |
| pPM10 | 68 ± 1.6 | 90 ± 2.1 | 15 ± 1.5 | 45 ± 2.2 | 13 ± 1.1 | 46 ± 1.6 | 15 ± 2.1 | 95 ± 3.1 | 38 ± 1.8 | 63 ± 2.7 |
| pPM10R | 24 ± 1.0 | 47 ± 2.2 | 20 ± 0.9 | 24 ± 1.7 | 22 ± 0.9 | 27 ± 1.1 | 21 ± 2.0 | 20 ± 1.0 | 38 ± 2.0 | 45 ± 2.4 |
| pPM11 | 24 ± 1.0 | 45 ± 2.1 | 22 ± 1.0 | 23 ± 1.2 | 22 ± 1.0 | 27 ± 1.0 | 22 ± 2.2 | 20 ± 1.3 | 40 ± 2.0 | 46 ± 2.4 |
| pPM1989R | ND | 10 ± 1.0 | ND | 18 ± 2.1 | ND | 26 ± 1.4 | ND | 20 ± 2.0 | ND | 48 ± 2.2 |
| pPM2016A | ND | 13 ± 0.8 | ND | 22 ± 1.0 | ND | 27 ± 2.2 | ND | 22 ± 1.7 | ND | 50 ± 2.4 |
The antibiotics were used at various concentrations (in micrograms per milliliter) as follows: rifampin (RIF), 150; isoniazid (INH), 3.5; chloramphenicol (CML), 30; tetracycline (TET), 0.3; and ethambutol (ETM), 2.0. Values are means ± standard deviations of experiments performed in triplicate. ND, not determined.
These changes in drug susceptibility were not seen with the marA gene cloned in the reverse orientation relative to the mycobacterial hsp60 promoter. Also, introduction of marR cloned with the same vector by PCR methods (primer nucleotide positions 1446 to 1462 and 1864 to 1879) caused no changes in susceptibility of M. smegmatis to any of the compounds tested (Table 2). Strains selected for spontaneous loss of plasmids by growth in the absence of kanamycin showed a return of the wild-type susceptibility phenotype (data not shown). While multidrug resistance was clearly temperature dependent and correlated with the presence of marA behind the heat shock promoter, it could reflect a resistance mechanism(s) per se which functions better at 37°C than at 30°C regardless of MarA expression. Of note, however, no temperature-dependent differences in susceptibility of wild-type cells were observed with any of the agents tested (Table 2).
To obtain support for the notion that the resistance phenotype was a direct result of MarA activity in the cell, insertional mutants targeted to the predicted HTH domain (1) of the MarA protein were constructed. Megaprimer PCR (12) was used to introduce an NheI restriction site into the centers of both the helix A (position 1989) and helix B (position 2016) regions of marA. A double-stranded synthetic oligonucleotide with compatible ends was ligated to the NheI sites to produce two distinct insertional mutants interrupting each of the two putative alpha-helical regions (Fig. 1). These mutated genes were cloned into pMV261, as described above, for susceptibility testing in M. smegmatis at 37°C. They were also expressed from the isopropyl-β-d-thiogalactopyranoside (IPTG)-regulated T7 promoter resident on plasmid pET13a (20) for testing in E. coli BL21. Insertional inactivation of MarA at either helix A or helix B abolished the multidrug resistance phenotype in both E. coli and M. smegmatis (Tables 2 and 3).
TABLE 3.
Antibiotic susceptibilities of E. coli organisms with plasmids bearing different E. coli mar genes
| Plasmid borne by E. coli transformant | % Growth in antibiotic gradienta
|
|||
|---|---|---|---|---|
| CML | TET | AMP | NAL | |
| pET13a | 20 ± 1.1 | 33 ± 1.0 | 2 ± 1.1 | 17 ± 0.8 |
| pEC10 | 100 | 100 | 11 ± 2.0 | 100 |
| pEC1989R | 20 ± 1.6 | 33 ± 1.4 | 2 ± 1.0 | 17 ± 1.2 |
| pEC2016A | 20 ± 1.3 | 33 ± 1.0 | 2 ± 1.7 | 17 ± 1.1 |
The antibiotics were used at various concentrations (in micrograms per milliliter) as follows: chloramphenicol (CML), 2.0; tetracycline (TET), 1.0; ampicillin (AMP), 3.0; and nalidixic acid (NAL), 1.0. Values are means ± standard deviations of experiments performed in triplicate.
To confirm marA expression, Northern blotting (10) was performed with total cellular RNA isolated by the TRIzol method (Gibco BRL, Gaithersburg, Md.) from mid-log-phase cells grown at 30 and 37°C in Middlebrook 7H9-ADC medium following 1 h of pretreatment with lysozyme (4 mg of Tris-EDTA [pH 8.0] per ml) at 30°C. Equal amounts of RNA, separated electrophoretically in 20 mM guanidine isothiocyanate, were probed with a radiolabeled marA PCR product. Hybridization signals were visualized with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
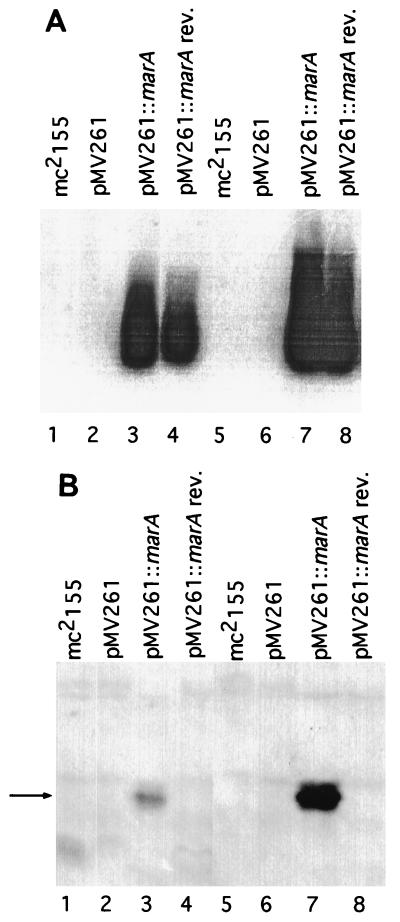
marA expression was observed in cells carrying the wild-type E. coli marA gene but not in the host carrying vector alone (Fig. 2A). The intensity of the marA hybridization signal was approximately fivefold higher in cells grown at 37 than at 30°C. As expected, a hybridization signal was detected in cells bearing the vector carrying marA in the reverse orientation, since a double-stranded marA probe was used.
FIG. 2.
Expression of the E. coli marA gene in M. smegmatis mc2155. Lanes 1 to 4, samples from M. smegmatis cells grown at 30°C. Lanes 5 to 8, samples from cells grown at 37°C. (A) Northern analysis with PCR-amplified marA probe (nucleotide primers 1910 to 1893 and 2265 to 2282). Each lane has 30 μg of total RNA from M. smegmatis mc2155. (B) Western analysis with anti-MarA rabbit antiserum. Each lane contains 7.5 μg of mycobacterial protein from supernatant fractions. The arrow indicates MarA protein.
Production of MarA protein was also examined. Anti-MarA antiserum was prepared with MarA purified from BL2(DE3)pLysS cells (20) bearing marA (1) cloned under the control of the T7 RNA polymerase initiation signals of pET13a. After induction with IPTG for 30 min, rifampin was added to maximize MarA synthesis. MarA was purified by a combination of the procedures of Li and Demple (14) and Langley et al. (13). Anti-MarA rabbit antiserum was generated with purified MarA by Biodesign International (Kennebunk, Maine).
For Western analysis, cell lysates were prepared from mid-log-phase M. smegmatis or E. coli cultures by sonication in buffer (10 mM Tris-HCl [pH 8.0], 3% sodium dodecyl sulfate) on ice. Prior to electrophoresis, samples were treated by boiling for 5 min in sample buffer (125 mM Tris-HCl [pH 6.8], 20% glycerol, 6 mM β-mercaptoethanol, 0.05% bromphenol blue), and equivalent amounts of total protein were resolved by electrophoresis in a sodium dodecyl sulfate–17.5% polyacrylamide electrophoresis gel. Proteins from E. coli AG100 and AG102 were used as negative and positive controls. Proteins were transferred to Immobilon-P membranes (Amersham) and analyzed by using rabbit anti-MarA antiserum and chemiluminescence detection (with a kit from New England Biolabs, Beverly, Mass.).
A protein band migrating to the same place as purified MarA and having the expected molecular mass (14.3 kDa) was detected in marA-containing cells grown at both temperatures (Fig. 2B); however, considerably more MarA was produced in cells incubated at 37°C. Since small amounts of MarA were detected at 30°C (Fig. 2B), the variable resistance to rifampin and the increased susceptibility phenotypes at 30°C may have been produced by relatively low cytoplasmic levels of MarA protein. By the same Western analysis, MarA was easily detected in E. coli and M. smegmatis lysates containing the mutant marA genes (data not shown).
The mechanism of MarA-mediated multidrug resistance in Mycobacterium is unknown. The lack of a resistance phenotype mediated by the two different expressed mutant MarA proteins suggests that the multidrug resistance we observed resulted from direct transcriptional activation of cognate promoters by MarA in M. smegmatis. Alternatively, MarA may have acted indirectly through induction of or interaction with endogenous proteins that mediate the mycobacterial Mar phenotype. In both instances, the multidrug resistance phenotype would have resulted from a mar-like regulatory system operating on other genes in this organism. The effect, as with MarA in E. coli, may be linked to activation of a yet-to-be-discovered multidrug efflux system. The presence of efflux-like proteins (6, 15), along with the earlier report of the existence of mycobacterial porin proteins (16, 21), indicates that, as in E. coli, effector proteins for mar-like multidrug resistance are present in Mycobacterium. The recently completed Mycobacterium tuberculosis genome sequencing project (17) identified at least two proteins similar to MarA. Determination of whether these elements represent an endogenous mar-like system in this species awaits further study.
In addition to defining a function for MarA in a heterologous genus, our results are the first direct evidence of structurally important regions of MarA. The HTH region we targeted in site-directed mutagenesis corresponds to regions in the homologous proteins AraC and XylS (8), which are involved in DNA binding and transcriptional activation (3, 7). Although the insertional mutations reported here involve significant changes to the wild-type protein, they point to the predicted HTH domain as critical for protein function.
Acknowledgments
We thank Bill Jacobs for providing plasmid pMV261 and M. smegmatis mc2155, and we thank Jack Crawford for providing antimicrobial agents and for his suggestions. We are also grateful to Mark Nelson and Laura McMurry for technical advice.
This work was supported in part by USPHS grant GM51661.
REFERENCES
- 1.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloom B R, Murray C J. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 3.Brunelle A, Schleif R F. Determining residue-base interaction between AraC protein and araI DNA. J Mol Biol. 1989;209:607–622. doi: 10.1016/0022-2836(89)90598-6. [DOI] [PubMed] [Google Scholar]
- 4.Cohen S P, Hachler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curiale M S, Levy S B. Two complementation groups mediate tetracycline resistance determined by Tn10. J Bacteriol. 1982;151:209–215. doi: 10.1128/jb.151.1.209-215.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doran J L, Pang Y, Mdluli K E, Moran A J, Victor T C, Stokes R W, Mahenthiralingam E, Kreiswirth B N, Butt J L, Baron G S, Treit J D, Kerr V J, Van Helden P D, Roberts M C, Nano F E. Mycobacterium tuberculosis efpA encodes an efflux protein of the QacA transporter family. Clin Diagn Lab Immunol. 1997;4:23–32. doi: 10.1128/cdli.4.1.23-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallegos M-T, Marqués S, Ramos J L. The TACAN4TGCA motif upstream from the −35 region in the ς70−ςs-dependent Pm promoter of the TOL plasmid is the minimum DNA segment required for transcription stimulation by XylS regulators. J Bacteriol. 1996;178:6427–6434. doi: 10.1128/jb.178.22.6427-6434.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George A M, Levy S B. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goda S K, Minton N P. A simple procedure for gel electrophoresis and Northern blotting of RNA. Nucleic Acids Res. 1995;23:3357–3358. doi: 10.1093/nar/23.16.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs W R, Jr, Snapper S B, Lugosi L, Bloom B R. Development of BCG as a recombinant vaccine vehicle. Curr Top Microbiol Immunol. 1990;155:153–160. doi: 10.1007/978-3-642-74983-4_11. [DOI] [PubMed] [Google Scholar]
- 12.Landt O, Grunert H P, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 13.Langley K E, Berg T F, Strickland T W, Fenton D M, Boone T C, Wypych J. Recombinant-DNA-derived bovine growth hormone from Escherichia coli. Eur J Biochem. 1987;163:313–321. doi: 10.1111/j.1432-1033.1987.tb10802.x. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Demple B. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J Biol Chem. 1994;269:18371–18377. [PubMed] [Google Scholar]
- 15.Liu J, Takiff H E, Nikaido H. Active efflux of fluoroquinolones in Mycobacterium smegmatis mediated by LfrA, a multidrug efflux pump. J Bacteriol. 1996;178:3791–3795. doi: 10.1128/jb.178.13.3791-3795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukhopadhyay S, Basu D, Chakrabarti P. Characterization of a porin from Mycobacterium smegmatis. J Bacteriol. 1997;179:6205–6207. doi: 10.1128/jb.179.19.6205-6207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B B R, Jacobs W R J, Cole S T. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 19.Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Benson L A, Bennett L T, Bansal G P, Young J F, Lee M H, Hatfull G F, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 20.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 21.Trias J, Jarlier V, Benz R. Porins in the cell wall of mycobacteria. Science. 1992;258:1479–1481. doi: 10.1126/science.1279810. [DOI] [PubMed] [Google Scholar]