Abstract
A minor population of wild strains of Escherichia coli contains a retron, a retroelement responsible for the synthesis of multicopy single-stranded DNA (msDNA). The retron is a genetic element consisting of the gene for reverse transcriptase (RT) and the msr-msd region under a single promoter. A single RNA transcript from the msr-msd region serves not only as a template but also as a primer for msDNA synthesis. Here, using a cell-free system with purified RT from retron Ec73, we examined whether the reaction can occur in a bimolecular reaction with use of separately expressed msr and msd transcripts. DNA sequencing of the cell-free product revealed that the sequence of the 5′-end region was identical to that of msDNA-Ec73, indicating that the cDNA synthesis was primed from the 2′-OH group of the specific internal G residue of the primer RNA, identical to the branching G residue in the RNA molecule of msDNA-Ec73. The present results raise an intriguing possibility for a role of bacterial retrons in vivo, the possibility that cellular mRNAs can be converted into cDNAs in retron-harboring cells if the mRNAs contain a sequence complementary to the sequence directly upstream of the branching G residue of the msr RNA transcript.
Bacterial reverse transcriptases (RTs) are unique among all RTs in terms of the priming reaction for cDNA synthesis (9). Using a single RNA molecule not only as a template but also as a primer, the RT forms an unusual 2′,5′-phosphodiester linkage between an internal G residue of the RNA molecule and the 5′ end of the cDNA to initiate cDNA synthesis (see references 1 to 3 for reviews). In this fashion, bacterial RTs synthesize an unusual type of single-stranded DNA in the cell, called multicopy single-stranded DNA (msDNA). msDNA is a DNA-RNA complex consisting of a single-stranded DNA molecule branching out from a guanosine residue (branching G) of an RNA molecule via a 2′,5′-phosphodiester linkage. A genetic element required for the msDNA synthesis is a retron, a novel prokaryotic retroelement which is integrated directly or indirectly through a prophage genome into a bacterial genome. The retrons so far identified are 1.3 to 3.0 kb in length and consist of at least three genetic components: msr, msd, and a gene for RT under a single promoter.
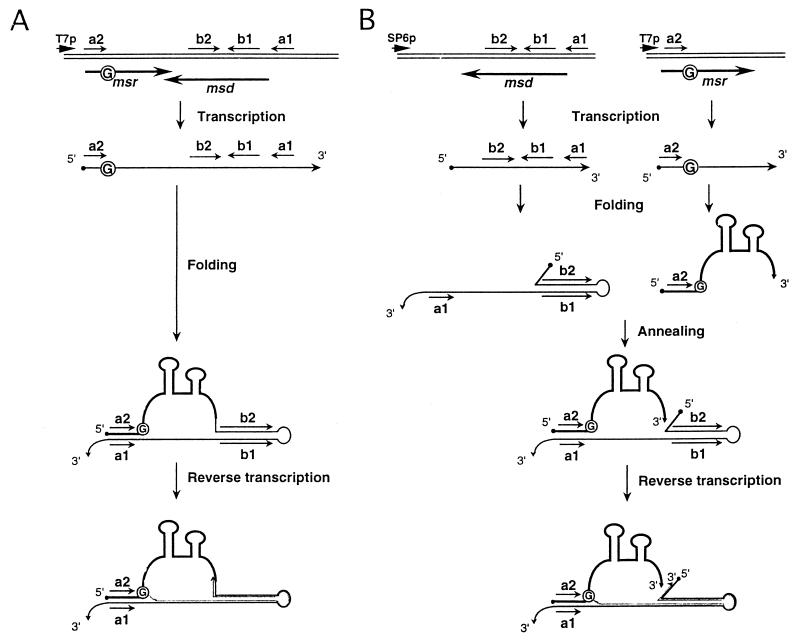
Retrons are thus transcribed as a single transcript, and the msr-msd region of the transcript serves as a template as well as a primer for the RT reaction. In the transcript, there are two inverted repeats (a1 and a2 in Fig. 1A) which form a stem. As a result, a G residue located at the 3′ end of the a2 sequence serves as a primer to initiate cDNA synthesis by forming a 2′,5′-phosphodiester linkage (Fig. 1A). The msr region of the RNA transcript is unique for each retron and is specifically recognized by RT from the same retron but not by that from other retrons (7). RT from retron Ec73 (RT-Ec73) was purified to homogeneity, and a cell-free system was established with purified RT and the RNA transcript corresponding to the msr-msd region from retron Ec73, which was synthesized in vitro by T7 RNA polymerase (8). The bacterial RT without any other factors was indeed able to initiate cDNA synthesis from the 2′-OH group of an internal G residue forming a 2′,5′-phosphodiester linkage. Figure 1A shows a schematic diagram for in vitro cDNA synthesis by bacterial RT.
FIG. 1.
Models for cDNA synthesis by bacterial RT using different templates. Wild-type msr-msd (A) and msr and msd (B) RNAs are transcribed separately under different promoters as shown. cDNA synthesis is carried out by mixing two RNA transcripts. Short, thin arrows represent the inverted repeats (a1-a2 and b1-b2). Thick arrows represent the genes for multicopy single-stranded RNA (msr) and msDNA (msd). The branching G residue is circled. Long, solid lines represent the transcripts from the msr region required for specific recognition by RT (7), and dotted lines correspond to cDNA.
As shown in Fig. 1A, the msr-msd transcript forms a stable duplex between the a2 and a1 sequences. As a result, the template RNA forms a loop between the a1 and a2 sequences. Therefore, cDNA synthesis initiated from the branching G residue (circled in Fig. 1A) cannot extend any further than the branching G residue, leaving the a2 sequence unused as template. In order to eliminate this blockage of cDNA synthesis by the branching G residue, the msr region and the msd region may be expressed under separate promoters to produce their transcripts separately, as shown in Fig. 1B. If the msr transcript thus produced is able to anneal to the msd transcript as shown in Fig. 1B, cDNA synthesis may be achieved from the branching G residue and may continue to the 5′ end of the template RNA encoded by msd.
In the present paper, we attempted to examine this possible scheme of cDNA synthesis by using retron Ec73 (10) as a model system. The results suggest that the msr transcript (primer RNA), even if produced separately from the msd region, is able to anneal to the msd transcript (template RNA). Thus, the cDNA synthesis is initiated from the 2′-OH group of the internal branching G residue in the msr transcript. This raises an interesting possible role for retrons in prokaryotes in producing cDNAs for cellular mRNAs in vivo.
msDNA synthesis using separate primer and template RNA molecules.
As shown in Fig. 1B, the transcript from the msr region is expected to hybridize with the transcript from the msd region, since they contain mutually complementary sequences, a1 and a2, which are derived from the msd and msr transcripts, respectively. The formation of the a1-a2 stem becomes a bimolecular reaction in this experiment, in contrast to the normal msDNA-priming transcript, which is able to form the stem in a unimolecular reaction (Fig. 1A). However, if a stem structure identical to that formed in the normal retron transcript is formed, the msr transcript may be able to function as a primer for cDNA synthesis.
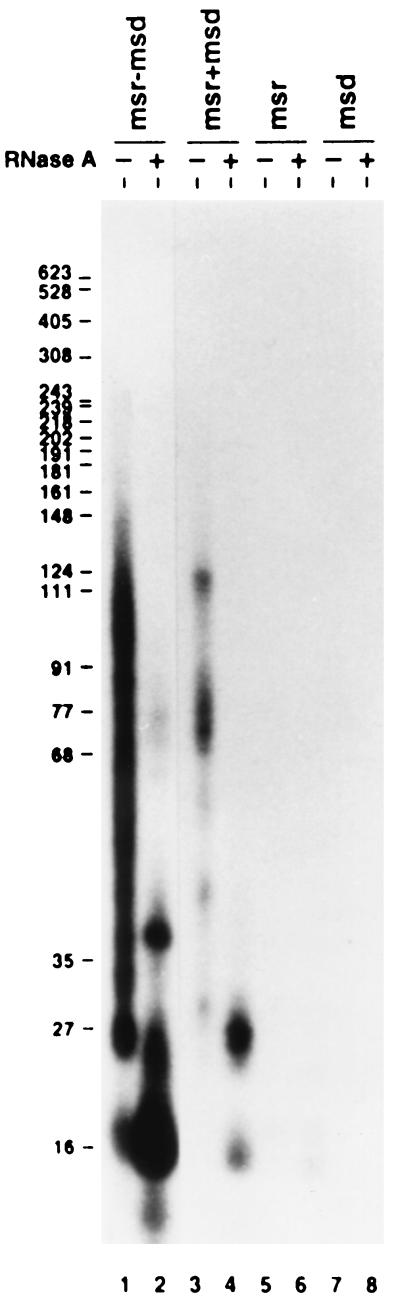
These two RNA transcripts were prepared with a T7 polymerase system as described in the legend to Fig. 2. cDNA synthesis was then tested in a cell-free system as described previously (8) with purified RT-Ec73 (His), a six-histidine tagged RT-Ec73 (8). Results are shown in lanes 3 and 4 of Fig. 2; clearly, cDNAs were synthesized as evident from the incorporation of [α-32P]dTTP into a number of bands in lane 3. The sizes of these bands were reduced to about 27 and 16 bases when the products were treated with RNase A (lane 4). These two major products after RNase A treatment appear to have sizes very similar to those of two major products from the single msr-msd transcript (lane 2). It appears that there are strong stop signals, probably due to the formation of stable secondary structures, which block further extension of cDNA in the present system. It is interesting to note that, in a preliminary experiment in vivo, cDNA synthesis could be extended much further, indicating that the other factors such as single-stranded DNA binding proteins may enhance cDNA synthesis in vivo (8a). In control experiments with either the msr transcript (lanes 5 and 6, Fig. 2) or the msd transcript (lanes 7 and 8), no cDNA was produced, or very little if any. This result indicates that cDNA production as shown in lanes 3 and 4 indeed results from a bimolecular reaction depending upon both msr and msd transcripts. The lower yield of cDNA in this system in comparison with that from the normal msDNA synthesis (compare lane 1 with lane 3 in Fig. 2) is likely due to the requirement of the formation of a hybrid between two separate RNA molecules for cDNA synthesis.
FIG. 2.
In vitro cDNA synthesis using different template RNAs. cDNAs were synthesized and labeled with [α-32P]dTTP as described below and separated on a 6% polyacrylamide–8 M urea gel before (lanes 1, 3, 5, and 7) or after (lanes 2, 4, 6, and 8) RNase A treatment. Lanes 1 and 2, cDNAs synthesized from the wild-type msr-msd transcript (Fig. 1A); lanes 3 and 4, cDNAs synthesized from the bimolecular reaction with the msr transcript as a primer and the msd transcript as a template (Fig. 1B); lanes 5 and 6, cDNAs synthesized only from the msr transcript; lanes 7 and 8, cDNAs synthesized only from the msd transcript. pBR322 digested with MspI and labeled with Klenow fragment and [α-32P]dCTP was used for molecular weight markers. The wild-type msr-msd RNA from retron Ec73 was synthesized in vitro with T7 RNA polymerase (Boehringer Mannheim) and pUCT7MS73 digested with BamHI as described previously (8). The msr RNA was made with T7 RNA polymerase with pSPdr73 digested with EcoRI. In order to get the transcript from the msd region, pSPdr73 was digested with MvnI and the 226-bp fragment containing the SP6 promoter and the msd region was purified by polyacrylamide gel electrophoresis. The purified DNA fragment was then used for in vitro transcription with SP6 RNA polymerase. All template DNAs were digested by adding RNase-free DNase (Boehringer Mannheim) to a final concentration of 0.2 U/ml and incubating the DNAs at 37°C for 15 min after the in vitro transcription reaction. pSPdr73 was constructed as follows. First, the BglII-BamHI fragment including the T7 promoter and the msr-msd region of retron Ec73 was isolated from pUCT7MS73 (8). This DNA fragment was further digested with AluI between msr and msd, and both fragments were ligated into pUC19 digested with SmaI. The plasmid in which the msd and msr regions were inserted in the reverse order (i.e., msd-msr instead of msr-msd) under the lac promoter was selected and designated pUCdr73. The msd-msr fragment was isolated from pUCdr73 with XbaI and EcoRI and ligated into the XbaI and EcoRI sites of pSP64 (Promega). The resulting plasmid was designated pSPdr73. E. coli CL83 (4) was used as a host strain for isolation of plasmids.
Analysis of cDNA products.
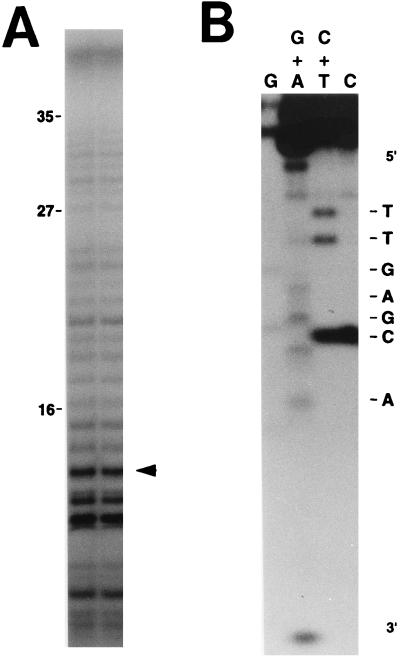
In order to unambiguously demonstrate that cDNA products from the bimolecular reaction with separate primer (msr transcript) and template (msd transcript) molecules were initiated from the same branching G residue as the products in the unimolecular reaction with the single msr-msd transcript (8), the DNA sequences of the cDNA products were determined by the method of Maxam and Gilbert (6). First, the cDNA products were labeled with [α-32P]ddATP and terminal deoxynucleotidyl transferase at their 3′ ends, and the labeled products were separated on a urea-polyacrylamide sequencing gel (Fig. 3A). Although the sizes of the cDNA products were extended as far as about 32 bases, the major products were about 11 to 15 bases in size. One of the major bands, indicated by an arrowhead in Fig. 3A, was extracted from the gel, and its DNA sequence was determined to be 5′ TTGAGCA…3′, as shown in Fig. 3B. Note that the product with only the msr transcript did not contain the sequences corresponding to the msd region (data not shown). The sequence determined in Fig. 3B is identical to the 5′-end sequence of msDNA-Ec73 synthesized in vivo as well as in vitro (8, 10), clearly demonstrating that the separately produced primer molecule, the msr transcript, functions as a primer to initiate cDNA synthesis. Thus, this priming reaction is likely initiated from the 2′-OH group of the branching G residue in the primer molecule with the msd RNA as a template.
FIG. 3.
Analysis of cDNA products synthesized from the msr-msd bimolecular reaction. (A) The cDNA products from the bimolecular reaction using the msr transcript and the msd transcript (Fig. 2, lane 4) were labeled at their 3′ ends with [α-32P]ddATP as described previously (8). The labeled sample was run in two lanes as shown. The band marked by an arrowhead was extracted from the gel, purified, and subjected to sequence analysis. Molecular weight markers were the same as those described in the legend to Fig. 2. (B) The cDNA product was extracted from the band shown by an arrowhead in panel A. The DNA sequence was determined by the method of Maxam and Gilbert (6).
Conclusion.
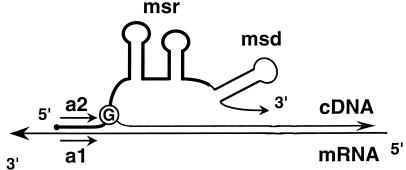
The present finding that the primer for msDNA synthesis can bind to a separate template RNA molecule suggests that such cDNA synthesis may occur in vivo for a specific mRNA if the mRNA contains a sequence complementary to the sequence 5′ to the branching G residue, namely, the a1 sequence, as shown in Fig. 4. Interestingly, the a1 sequence of retron Ec73 (12 matches of 13 bases) was found in an open reading frame at 7.2 min on the Escherichia coli chromosome. However, its orientation is opposite to that of the mRNA (the a1 sequence on its antisense RNA). The significance of this homology is unknown at present. Although bacterial retrons have been proposed to be the oldest retroelements in living organisms (3), their biological functions have not yet been identified, except for the production of msDNA. The present results imply a possible role for retrons in cDNA synthesis in bacteria. In addition to the possible cellular cDNA synthesis initiating at the 2′-OH group of the G residue at the end of the a2 sequence, retron RTs may be able to switch templates during cDNA synthesis. Thus, retrons might have participated in gene duplication in the prokaryotic genomes.
FIG. 4.
A model for in vivo cDNA synthesis from an mRNA containing the a1 sequence. The msr, msd, a1, and a2 regions are defined in Fig. 1. cDNA is initiated from the 2′-OH group of the branching G residue (circled) in the primer RNA.
With msDNA as a vehicle, artificial oligodeoxyribonucleotides have been synthesized in vivo and shown to effectively work as antisense DNA to block specific gene expression in E. coli (5). On the basis of the present results, it is possible to design a primer RNA for cDNA synthesis against any mRNAs by altering the a2 sequence of a primer molecule to a sequence complementary to that in a specific mRNA. In this fashion, one may be able to block the translation of the mRNA.
Acknowledgments
We thank Mei-Yin Hsu for purification of RT-Ec73(His).
This work was supported by a grant to S.I. from the National Institutes of Health (GM44012). This work was also supported in part by a grant-in-aid for scientific research to T.S. from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Inouye S, Inouye M. The retron: a bacterial retroelement required for synthesis of msDNA. Curr Opin Genet Dev. 1993;3:713–718. doi: 10.1016/s0959-437x(05)80088-7. [DOI] [PubMed] [Google Scholar]
- 2.Inouye S, Inouye M. Bacterial reverse transcriptase. In: Goff S, Skalka S, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 391–410. [Google Scholar]
- 3.Inouye S, Inouye M. Structure, function and evolution of bacterial reverse transcriptase. Virus Genes. 1996;11:81–94. doi: 10.1007/BF01728650. [DOI] [PubMed] [Google Scholar]
- 4.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao J-R, Shimada M, Inouye S, Inouye M. Gene regulation by antisense DNA produced in vivo. J Biol Chem. 1995;270:19684–19687. doi: 10.1074/jbc.270.34.19684. [DOI] [PubMed] [Google Scholar]
- 6.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavage. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 7.Shimamoto T, Hsu M-Y, Inouye S, Inouye M. Reverse transcriptases from bacterial retrons require specific secondary structures at the 5′-end of the template for the cDNA priming reaction. J Biol Chem. 1993;268:2684–2692. [PubMed] [Google Scholar]
- 8.Shimamoto T, Inouye M, Inouye S. The formation of the 2′,5′-phosphodiester linkage in the cDNA priming reaction by bacterial reverse transcriptases in a cell-free system. J Biol Chem. 1995;270:581–588. doi: 10.1074/jbc.270.2.581. [DOI] [PubMed] [Google Scholar]
- 8a.Shimamoto, T., R. Namba, and M. Miyawaki. Unpublished results.
- 9.Singer M F. Unusual reverse transcriptases. J Biol Chem. 1995;270:24623–24626. doi: 10.1074/jbc.270.42.24623. [DOI] [PubMed] [Google Scholar]
- 10.Sun J, Inouye M, Inouye S. Association of a retroelement with a P4-like cryptic prophage (retronphage φR73) integrated into the selenocystyl tRNA gene of Escherichia coli. J Bacteriol. 1991;173:4171–4181. doi: 10.1128/jb.173.13.4171-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]