Abstract
bfpA expression in enteropathogenic Escherichia coli is regulated by growth medium, temperature, and ammonium concentration and requires the BfpT protein (also called PerA), a member of the AraC family of transcriptional activators. Site-directed and PCR random mutagenesis, as well as deletion analysis of the bfpA upstream regulatory region, supported assignment of the promoter elements and demonstrated that the cis-acting elements that mediate BfpT-dependent regulation of bfpA are located between positions −85 and −46. Interestingly, this region shares 73% identity with a 40-bp-long AT-rich tract located upstream of the bfpT gene, which is essential for bfpT autoregulation.
Enteropathogenic Escherichia coli (EPEC) is a common cause of diarrhea, particularly among children under 6 months of age living in developing countries (16). Recently, a three-stage model by which EPEC infections proceed has been proposed (6). The initial stage involves the generalized nonintimate interaction of bacterial microcolonies with the surface of epithelial cells, in a pattern known as the localized adherence phenotype (5). This pattern of attachment requires the 80-kb EPEC adherence factor (EAF) plasmid, which contains a cluster of 14 tandemly arrayed genes; this cluster is sufficient to direct the production of the bundle-forming pilus (BFP), a type IV fimbria associated with microcolony formation and bacterial autoaggregation (8, 21, 23–25).
The expression of bfpA, the gene coding for the structural subunit of BFP (23), occurs during the exponential phase of growth, when it is modulated by the growth medium, ammonium concentration, and temperature (19). Our previous studies revealed that bfpA regulation is under the control of a regulatory region that extends further upstream from the putative −35 and −10 promoter sequences, which seems to determine the coordinate regulation of genes located downstream of bfpA (19, 21). This expression is regulated at the transcriptional level and requires the product of the bfpT gene, which is the first gene of the bfpTVW locus, localized 6.7 kb downstream of the bfp gene cluster on the EAF plasmid (27). bfpT encodes a 274-amino-acid protein, which belongs to the XylS-AraC family of transcriptional regulators (27). The bfpTVW locus, previously identified as per, has also been involved in the regulation of the eaeA and esp genes, whose products mediate the second and third stages of EPEC interactions with the host cells (6, 9, 12).
Interactions of BfpT with its target sites have been difficult to study in vitro, since different attempts to overproduce and purify it have been unsuccessful. We previously showed that a DNA fragment containing the sequence between nucleotides −94 and −55 of the bfpA regulatory region was bound by a T7-tagged BfpT fusion protein immobilized on Dynabeads; however, attempts to perform footprinting experiments with this fusion were unsuccessful (27). Thus, an alternative route was to genetically analyze the bfpA regulatory region, as presented here.
Deletion analysis of the bfpA regulatory region.
A series of 5′ upstream deletions of the bfpA-cat fusion carried on plasmid pCAT232 (19), containing all of the required elements for expression, were constructed by PCR amplification of the corresponding fragments and cloned into vector pKK232-8, which contains a promoterless cat gene (2). The nucleotide sequence of all cloned inserts was determined to confirm the precise positions of the deletions and to ensure that no mutations were introduced by the amplification reaction. The chloramphenicol acetyltransferase (CAT) activity directed by plasmids carrying these bfpA-cat deletions was tested in EPEC B171-8 grown in Dulbecco modified Eagle (DME) medium at 37°C, which are the optimal conditions for bfpA expression, and under conditions that are known to regulate bfpA expression, such as growth in Luria-Bertani (LB) medium at 37°C, DME medium at 25 and 39°C, and DME medium containing 15 mM ammonium sulfate at 37°C, as described before (19).
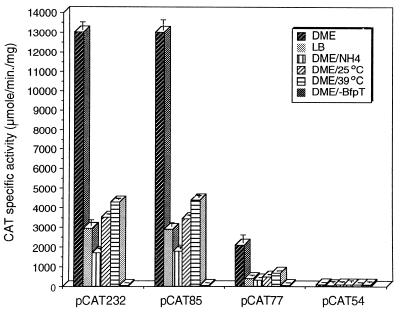
This analysis (Fig. 1) showed that a bfpA-cat deletion down to position −85 (pCAT85) had similar levels of expression and the same regulatory pattern in response to environmental cues as other fusions containing further upstream sequences. Also, a deletion to position −77 (pCAT77) showed an 84% reduction of the BfpT-dependent expression, although, interestingly, it still responded to regulatory signals to the same extent as the wild type. In contrast, only background activity was detected for deletions to position −54 or −40 (pCAT54 or pCAT40), both of which still contain the promoter (Fig. 1 and data not shown). These results indicated that the sequences required for BfpT-dependent expression of bfpA are located upstream of the −35 region and up to position −85.
FIG. 1.
Regulation of the bfpA 5′ regulatory region deletion fusions in response to environmental cues. The activities of plasmids pCAT232, pCAT85, pCAT77, and pCAT54 were tested in EPEC strain B171-8 grown in DME medium at 37, 25, and 39°C, in LB medium at 37°C, or in DME medium containing 15 mM ammonium sulfate at 37°C or in EPEC strain T::Gmr, a bfpT mutant derived from strain B171-8 (27), grown in DME medium at 37°C. The graph shows the maximal CAT-specific activity reached late in growth. The data are representative of at least three different experiments.
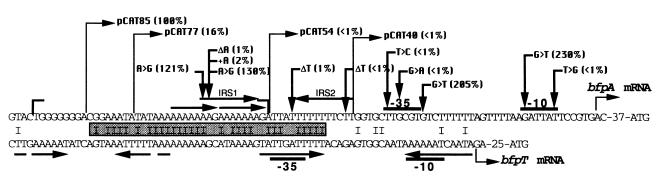
This AT-rich region contains two 8-bp-long direct-repeat elements, as well as two 10-bp-long inverted-repeat elements, which were designated IRS1 and IRS2 (Fig. 2). Although the precise role of these elements in BfpT binding remains unclear, since they are not sufficient for full activation (Fig. 2), it should be noticed that binding to tandem elements has been reported for other members of the AraC family, such as AraC, MelR, and VirF (3, 14, 28). Moreover, AT-rich sequences are necessary for the regulatory activity of other, closer homologs of BfpT, such as Rns and CfaD (regulation of the CS1 and CFA/I fimbrial operons in enterotoxigenic E. coli, respectively) (11, 18) and VirF (regulation of plasmid-encoded invasion proteins in Shigella species) (26). Interestingly, another common feature of Rns, CfaD, and VirF is that they seem to overcome the negative regulation by H-NS at their respective promoters, a mechanism that might also account for bfpA repression at temperatures below 37°C (11, 18, 26).
FIG. 2.
Nucleotide sequence alignment of the bfpA (upper line) and bfpT (lower line) 5′ regulatory regions. The upstream regulatory sequence up to position −85 contains all of the cis-acting elements required for the BfpT-dependent activation of bfpA (this work). The bfpA sequence between positions −85 and −46 shares a 73% identity over a 40-bp-long region (shaded bar) with the sequence between positions −65 and −26 of the bfpT regulatory region, which has been shown to be required for bfpT autoactivation (17). Brackets enclose the region bound by a T7-tag-BfpT fusion protein (27). Thin and thick broken arrows indicate the positions of each deletion and mutation, respectively, that affected CAT activity. The activities listed in parentheses are expressed as percentages of pCAT232 activity, which was assigned a value of 100% (see Fig. 1). Horizontal arrows above or below the sequences denote the inverted (IRS)- or direct-repeat elements. Promoter elements −35 and −10 and the transcription start sites are also indicated.
Mutational analysis of the bfpA regulatory region.
To pinpoint the position of cis-acting regulatory elements required for bfpA expression, random mutations were generated in the bfpA regulatory region contained in pCAT232, which was amplified under PCR conditions that enhance error-prone copying, as described previously (15). The PCR products were subcloned back into vector pKK232-8 (Ampr) and transformed into Escherichia coli HB101 carrying plasmid pBTA-BH1 (Kmr), which contains the bfpT regulatory locus (27). Mutations that reduced bfpA-cat expression were identified by selecting colonies that did not grow in concentrations of chloramphenicol noninhibitory for strains carrying the wild-type fusion (pCAT232), while transformants carrying mutations that improved bfpA-cat expression were screened for their ability to grow in a chloramphenicol concentration that inhibits the growth of a strain carrying the wild-type fusion. Candidates were assayed for CAT activity, as described before (19). Plasmid DNA from these clones was purified and the nucleotide sequence of the bfpA-cat regulatory region was determined, allowing the identification of two groups of mutations.
Promoter mutations.
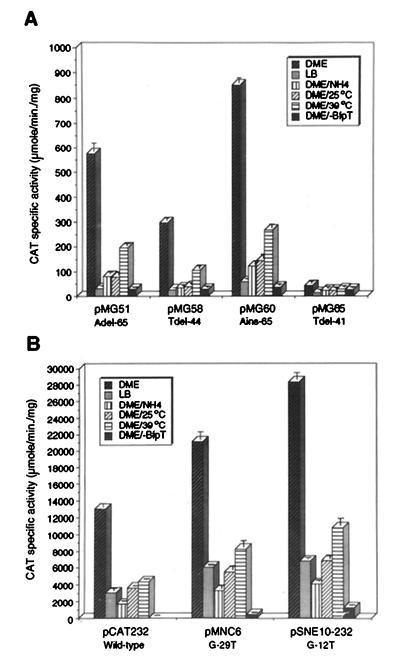
The sequence of the −35 promoter region of bfpA (TTGCGT) contains the most conserved residues of the consensus hexamer (TTGACA) at the first three positions. A T-to-C transition (T-35C) or a G-to-A transition (G-33A), at the first and third positions, respectively (Fig. 2), decreased expression of bfpA to the background level, showing that this sequence is critical for bfpA expression, in contrast to what is observed for the majority of the positively controlled promoters (20). Furthermore, a G-to-T transversion at position −29 (G-29T; 1 base downstream from the −35 hexamer) produced nearly a twofold increase in bfpA-cat expression under all growth conditions tested (Fig. 2 and 3B). This mutation did not modify the transcriptional start point (see below; Fig. 4B, lane 2) and caused a ninefold increase in the basal BfpT-independent expression levels (Fig. 3B), suggesting that it generated a stronger promoter. This is consistent with the fact that, in E. coli promoters, a T is the most frequently found residue 1 base downstream from the −35 hexamer (10).
FIG. 3.
Regulation of selected bfpA 5′ regulatory region mutants. The activities of bfpA-cat fusions containing down mutations (pMG51, pMG58, pMG60, and pMG65) (A) or promoter mutations (pMNC6 and pSNE10-232) (B) were tested EPEC strain B171-8 grown in DME medium at 37, 25, and 39°C, in LB medium at 37°C, or in DME medium containing 15 mM ammonium sulfate at 37°C or in strain T::Gmr, a bfpT mutant EPEC strain, grown in DME medium at 37°C. The graph shows the maximal CAT-specific activity reached late in growth. The data are representative of at least three different experiments.
FIG. 4.
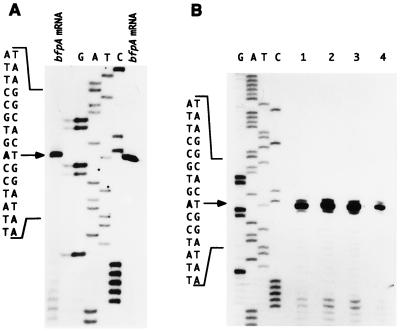
Primer extension analysis of bfpA and bfpA-cat transcripts. (A) Total RNA samples extracted from EPEC strain B171-8 were hybridized to a 5′-32P-end-labeled bfpA specific primer; primer extension was performed with avian myeloblastosis virus reverse transcriptase as described previously (19). Lanes G, A, T, and C correspond to the DNA sequence ladder obtained with the same primer. Arrow, the position of the extended products, which correspond to an A residue shown in boldface, 1 bp downstream from the G residue that was previously reported (19). The bfpA transcript is shown in lanes to the right and to the left of the sequence ladder. (B) Total RNA samples extracted from EPEC strain B171-8 carrying plasmid pCAT232 (wild type [lane 1]), pMNC6 (G-29T [lane 2]), or pSNE10-232 (G-12T [lane 3]) or from strain EPEC T::Gmr carrying pSNE10-232 (lane 4) were hybridized to a 5′-32P-end-labeled cat-specific primer.
Moreover, to better characterize the bfpA promoter, two site-directed mutations at the −10 hexamer were independently generated by PCR (1). A T-to-G transversion at position −7 (T-7G) that reduced the identity of the putative −10 region with the consensus sequence abolished bfpA-cat expression (Fig. 2), whereas a G-to-T transversion at position −12 (G-12T) (pSNE10-232), which brought the similarity of the putative −10 region closer to the consensus, caused more than a twofold increase in CAT activity, although its regulation in response to environmental cues was similar to that of the wild-type fusion (Fig. 2 and 3B). Interestingly, in the absence of BfpT, the G-12T mutation produced a 30-fold increase in the bfpA-cat basal level of expression (Fig. 3B). Primer extension analysis of this promoter mutant showed that transcription initiates at the wild-type position either in the wild-type EPEC strain or in its bfpT mutant derivative (Fig. 4A and B, lanes 1, 3, and 4), ruling out the possibility of having generated an alternative promoter. In summary, these results further support the assignment of the bfpA promoter (Fig. 2).
Mutations upstream of the promoter.
Further analysis of mutants with mutations randomly generated by PCR revealed that single deletions or a single insertion at different positions upstream from the promoter but downstream from the −85 position (e.g., an insertion or a deletion of one A at the 10-A tract between positions −65 and −74, a deletion of one T at the 8-T tract between positions −44 and −51, or a deletion of one T between positions −41 and −42 [plasmids pMG60, pMG51, pMG58, and pMG65, respectively]) decreased bfpA expression to less than 2% (Fig. 2 and 3A). Interestingly, this reduced level of expression still required BfpT and was regulated by the growth medium, temperature, and ammonium concentration (Fig. 3A).
This negative effect could have resulted from slight but significant local distortions in the DNA spatial structure, which would bring out of phase the BfpT-binding sites with respect to the promoter, probably altering its interactions with other molecules, i.e., RNA polymerase. In this regard, it has been observed for other regulatory proteins, such as CRP and FNR, that the exact spacing of their binding sites with respect to the promoter is crucial for activation (7, 29). Further analysis of site-directed mutants with full or half-turn insertions will be required to test this hypothesis. In contrast, two mutants with an A-to-G transition in the same region (A-65G or A-66G) rendered only a moderate positive effect on bfpA expression (Fig. 2).
The PCR random mutagenesis strategy did not render a wider variety of mutations, as illustrated by those that were generated by site-directed mutagenesis. In this respect, it is also possible that the effect of other mutations is not large enough to be detected by our screening procedure, contrasting with the larger effect caused by several single-base deletion or insertion mutants, which allowed their easy and recurrent isolation. In summary, new mutagenesis and screening schemes should be explored to exhaust all the possibilities.
The BfpT-independent expression of a bfpA promoter mutant is still repressed in LB medium.
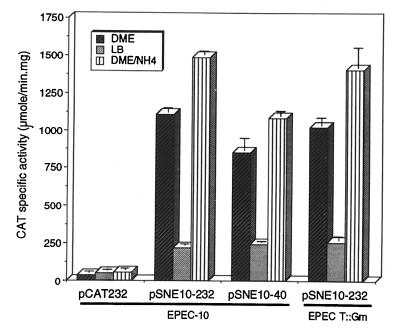
The BfpT-independent expression showed by pSNE10-232 (Fig. 5) was still repressed in LB medium but not by ammonium. Furthermore, a derivative of this mutant with a deletion to position −40 (pSNE10-40) behaved in the same manner (Fig. 5). These observations suggested that the different levels of bfpA expression in LB and DME medium could be mediated by a mechanism that acts directly on its promoter, while ammonium repression occurs through a different mechanism that requires BfpT and sequences upstream of the promoter. Moreover, since bfpA expression is selectively repressed upon entrance to stationary phase, we cannot exclude the possibility that the different levels of expression in DME and LB medium depend, at least partially, on how long the exponential phase of growth is sustained and that this phenomenon might be directly or indirectly mediated by RpoS (19, 30).
FIG. 5.
Regulation of the bfpA 5′ regulatory region mutant G-12T (contained in pSNE10-232) and its 5′ deletion derivative fused to the CAT reporter gene. The activities of pSNE10-232 and its 5′ deletion derivative pSNE10-40 were tested for EPEC strain B171-10 (EPEC-10), a spontaneous plasmid-cured derivative of B171-8 (this study), or for EPEC strain T::Gmr, a bfpT mutant strain, grown in DME or LB medium at 37°C or in DME medium containing 15 mM ammonium sulfate at 37°C. For comparison, expression directed by the wild-type fusion pCAT232 in B171-10 is also shown. The graph shows the maximal CAT-specific activity reached during the culture period. The data are representative of at least three different experiments.
The bfpA and bfpT regulatory regions share a common motif.
Recently, we have observed that bfpT expression is autoregulated and also modulated by the growth medium, temperature, and ammonium concentration (17). Considering these observations, we expected that common elements could be present in the regulatory regions of bfpA and bfpT. The nucleotide sequence alignment of these regions revealed the presence, as part of the minimal regulatory region of bfpT, of a sequence that shares 73% identity with the region between residues −85 and −46, which was shown to mediate regulation and BfpT-dependent expression of bfpA (Fig. 2). In contrast, no significant sequence similarities could be found with the bfpA region upstream from position −84 or downstream from position −46 (Fig. 2). Interestingly, the sequence comprised between positions −84 and −65, which proved to be critical in bfpA activation and is part of the bfpA-bfpT homologous motif, is located two full turns further upstream in bfpA with respect to bfpT, suggesting that BfpT can activate transcription from different locations with respect to the promoter, as long as the correct phase is maintained (Fig. 2), as has been described for many regulatory proteins in E. coli (4, 7, 29).
Concluding remarks.
This study led us to determine that the sequence between positions −85 and −55 is essential for the BfpT-dependent activation and ammonium regulation of bfpA, probably constituting the BfpT-binding motif. The region between positions −55 and −35, which resembles an UP element (13, 22), probably accounts for the stronger promoter activity shown by bfpA in comparison with that of bfpT, which lacks this element (Fig. 2) (17), although this hypothesis remains untested. All of this provides the basis toward further understanding the molecular mechanisms that control the expression of BFP and possibly other virulence factors in EPEC.
Acknowledgments
We particularly thank Francisco Santana for excellent technical assistance. We thank Enrique Morett for critical reading of the manuscript. J.L.P. thanks Dave Bieber for his collaborative effort and many helpful discussions at the early stage of this work.
V.H.B. was supported by a Ph.D. fellowship from the Consejo Nacional de Ciencia y Tecnología (no. 90275). This work was supported by grants from the Universidad Nacional Autónoma de México (DGAPA IN208095) and from the Consejo Nacional de Ciencia y Tecnología (CONACyT 1027P-N).
REFERENCES
- 1.Ali S A, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques. 1995;18:746–750. [PubMed] [Google Scholar]
- 2.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 3.Caswell R, Webster C, Busby S. Studies on the binding of the Escherichia coli MelR transcription activator protein to operator sequences at the melAB promoter. Biochem J. 1992;287:501–508. doi: 10.1042/bj2870501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cravioto A, Gross R, Scotland S, Rowe B. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol. 1979;3:95–99. [Google Scholar]
- 6.Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/S0966-842X(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 7.Gaston K, Bell A, Kolb A, Buc H, Busby S. Stringent spacing requirements for transcription activation by CRP. Cell. 1990;62:733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- 8.Giron J A, Ho A S Y, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Duarte O G, Kaper J B. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harley C B, Reynolds R P. Analysis of E. coli promoters. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordi B J, Dagberg B, de Haan L A, Hamers A M, van der Zeijst B A, Gaastra W, Uhlin B E. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 1992;11:2627–2632. doi: 10.1002/j.1460-2075.1992.tb05328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenny B, Finlay B B. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc Natl Acad Sci USA. 1995;92:7991–7995. doi: 10.1073/pnas.92.17.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landini P, Volkert M R. RNA polymerase α subunit binding in positively controlled promoters: a new model for RNA polymerase-promoter interaction and transcriptional activation in the Escherichia coli ada and aidB genes. EMBO J. 1995;14:4329–4335. doi: 10.1002/j.1460-2075.1995.tb00107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee N, Franklyn C, Hamilton E. Arabinose-induced binding of AraC protein to araI2 activates the araBAD promoter. Proc Natl Acad Sci USA. 1987;84:8814–8818. doi: 10.1073/pnas.84.24.8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung D, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 16.Levine M M. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis. 1987;155:377–385. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- 17.Martínez-Laguna Y, Calva E, Puente J L. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Regulation of bfpT, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli, abstr. H-171; p. 313. [DOI] [PubMed] [Google Scholar]
- 18.Murphree D, Froehlich B, Scott J R. Transcriptional control of genes encoding CS1 pili: negative regulation by a silencer and positive regulation by Rns. J Bacteriol. 1997;179:5736–5743. doi: 10.1128/jb.179.18.5736-5743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puente J L, Bieber D, Ramer S W, Murray W, Schoolnik G K. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 20.Raibaud O, Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- 21.Ramer S W, Bieber D, Schoolnik G K. BfpB, an outer membrane lipoprotein required for the biogenesis of bundle-forming pili in enteropathogenic Escherichia coli. J Bacteriol. 1996;178:6555–6563. doi: 10.1128/jb.178.22.6555-6563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross W, Gosink K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 23.Sohel I, Puente J L, Murray W, Vuopio-Varkila J, Schoolnik G K. Cloning and characterization of the bundle-forming pilin gene of enteropathogenic Escherichia coli and its distribution in Salmonella serotypes. Mol Microbiol. 1993;7:563–575. doi: 10.1111/j.1365-2958.1993.tb01147.x. [DOI] [PubMed] [Google Scholar]
- 24.Sohel I, Puente J L, Ramer S W, Bieber D, Wu C-Y, Schoolnik G K. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J Bacteriol. 1996;178:2613–2628. doi: 10.1128/jb.178.9.2613-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone K D, Zhang H-Z, Carlson L K, Donnenberg M S. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol Microbiol. 1996;20:325–337. doi: 10.1111/j.1365-2958.1996.tb02620.x. [DOI] [PubMed] [Google Scholar]
- 26.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J Bacteriol. 1993;175:6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tobe T, Schoolnik G K, Sohel I, Bustamante V H, Puente J L. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:963–975. doi: 10.1046/j.1365-2958.1996.531415.x. [DOI] [PubMed] [Google Scholar]
- 28.Wattiau P, Cornelis G R. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J Bacteriol. 1994;176:3878–3884. doi: 10.1128/jb.176.13.3878-3884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wing H J, Williams S M, Busby S J W. Spacing requirements for transcription activation by E. coli FNR protein. J Bacteriol. 1995;177:6704–6710. doi: 10.1128/jb.177.23.6704-6710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Johnson R C. Identification of genes negatively regulated by Fis: Fis and RpoS comodulate growth-phase-dependent gene expression in Escherichia coli. J Bacteriol. 1995;177:938–947. doi: 10.1128/jb.177.4.938-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]