Highlights
-
•
Increase in relative abundances of beneficial gut microbes observed for SDB and BYB.
-
•
Acetate and butyrate levels were higher for SDB after 24-h faecal fermentation.
-
•
Inflammatory cytokines were downregulated by SDB fermentation supernatants.
-
•
Spontaneous sourdough fermentation influences gut health at the metabolite level.
Keywords: Tritordeum, Sourdough fermentation, Simulated in vitro digestion, Gut microbiota, Epithelial integrity
Abstract
Spontaneous fermentation of Tritordeum flour enhances the nutritional potential of this hybrid cereal. However, the effect of consumption of Tritordeum sourdough bread (SDB) on gut health remains to be elucidated. This study investigated the effect of in vitro digestion and faecal fermentation of SDB compared to that of traditional baker's yeast (BYB) Tritordeum bread. After 24-h anaerobic faecal fermentation, both SDB and BYB (1% w/v) induced an increase in the relative abundances of Bifidobacterium, Megasphaera, Mitsuokella, and Phascolarctobacterium genera compared to baseline, while concentrations of acetate and butyrate were significantly higher at 24 h for SDB compared to those for BYB. Integrity of intestinal epithelium, as assessed through in vitro trans-epithelial electrical resistance (TEER) assay, was slightly increased after incubation with SDB fermentation supernatants, but not after incubation with BYB fermentation supernatants. The SDB stimulated in vitro mucosal immune response by inducing early secretion of inflammatory cytokines, IL-6 and TNF-α, followed by downregulation of the inflammatory trigger through induction of anti-inflammatory IL-10 expression. Overall, our findings suggest that Tritordeum sourdough can modulate gut microbiota fermentation activity and positively impact the gut health.
Graphical abstract
Introduction
Gut microbiota regulates immune homeostasis in the intestine by maintaining epithelial integrity lining the intestine and prevents immunogenic molecules from disrupting this intestinal barrier. The intestinal barrier comprises a single layer of epithelial cells connected to each other by tight junctions and is selectively permeable to nutrients, water, and beneficial microbial metabolites (Kunzelmann and Mall, 2002). The integrity of intestinal barrier is determined quantitatively by transepithelial electrical resistance (TEER) assay that measures electrical resistance across the epithelial monolayer using an epithelial volt-ohm-meter (EVOM) (Srinivasan et al., 2015). The TEER measurements have been previously defined for intestinal epithelial cell lines (e.g., Caco-2 cells) and therefore are commonly used in the in vitro co-culture model to study the barrier function of the gastrointestinal tract (Artursson et al., 2001).
Furthermore, gut microbiota-derived metabolites (e.g., short chain fatty acids (SCFAs), amino acids and their derivatives, etc.) regulate physiological functions in the gut through specific signalling pathways (Gasaly et al., 2021). These metabolites are produced by intestinal microorganisms during the digestion of nutrients, such as dietary fibres, proteins, and peptides, which usually escape the digestion in the upper gut. Primarily, the most common SCFAs released by the intestinal microbiota during digestion are acetate, propionate, and butyrate in the molar ratio of 3:1:1. The proteins are also hydrolysed by proteases releasing amino acids, which are either absorbed by the host or metabolized by the intestinal microbiota. Moreover, alterations in the gut microbiota, known as dysbiosis, can damage the intestinal epithelial integrity (termed as “leaky gut”) triggering intestinal inflammation or releasing harmful metabolites, which increases the susceptibility of host to various diseases (Cani, 2019; Shen et al., 2021).
The simulation of gastrointestinal digestion using an in vitro model is being widely used for studying the digestibility of foods to overcome the limitations of in vivo experiments (Minekus et al., 2014). The influence of these digested foods on the gut microbiota and its metabolome can be investigated using an in vitro batch culture fermentation model using fresh faecal samples (Rumney and Rowland, 1992). Although the gastrointestinal function depends immensely on the dietary habits of the faecal donors, it is possible to observe some significant differences in the digestibility and absorption of nutrients in the gut (Yang and Rose, 2016).
Previously, fermentation during sourdough bread preparation for a longer time (8 h) significantly decreased the relative abundance of δ-Proteobacteria and reduced gas production when consumed by patients with inflammatory bowel syndrome (IBS) (Costabile et al., 2014). A decrease in the alpha diversity of fungal microbiota and enhanced gastric emptying was also demonstrated after the consumption of sourdough pasta by overweight and obese adults (Shah et al., 2020). Only a few studies have investigated the in vitro faecal fermentation of sourdough bread, which is the focus of our study. These studies suggested that the differences in the gut microbiota were minimal after 24 h of faecal fermentation and the impact of sourdough was mostly evident in the metabolites released by the gut microbiota (Da Ros et al., 2021). The in vitro faecal fermentation of whole grains processed using different methods classified the faecal microbiome of sourdough bread into high carbohydrate utilization (CU), with increased butyrate production (Smith et al., 2020). In another study, sourdough and baker's yeast breads were fermented using two faecal inocula from individuals following a Mediterranean diet, using the Twin Mucosal-SHIME® (Simulator of Human Intestinal Microbial Ecosystem) for two weeks. Faecal fermentation of both the breads did not show any significant changes in the luminal and mucosal microbiota. However, the metabolomic study revealed a profound increase in the concentrations of SCFAs (acetate, propionate, butyrate), isovaleric and 2-methylbutyric acids as well as amino acids [aspartate, threonine, glutamate, and γ-aminobutyric acid (GABA)] with sourdough bread (Da Ros et al., 2021). An in vivo challenge to healthy adults consuming sourdough bread demonstrated faster gastric emptying and lower post-prandial glycaemic responses (Rizzello et al., 2019). A combined approach of metagenomic and metaproteomic analyses revealed that the consumption of sourdough Carasau bread by rats increased the abundance of gut microbiota belonging to the following families, Verrucomicrobiaceae, Bacillaceae, Moraxellaceae, Nitrospinaceae, and Thermaceae. At the genus level, a decrease in Alistipes and Mycoplasma was observed, which are considered as the intestinal pathobionts associated with low protein diets (Abbondio et al., 2019). Moreover, the abundance of protein families responsible for sporulation indicated the metabolic dormancy of Clostridium after sourdough bread consumption (Abbondio et al., 2019).
Furthermore, the metabolites released during the microbial fermentation in the gastrointestinal tract modulate immune responses during inflammatory diseases by inhibiting the expression of inflammatory and proinflammatory cytokines (Shen et al., 2021). The inhibition of inflammatory response by SCFAs was shown to be mediated through mechanisms like activation of GPCRs (G protein-coupled receptors) or inhibition of histone deacetylases that recruit leukocytes at the inflamed site to combat the inflammation (Vinolo et al., 2011). Moreover, the transformation of gut microbiota by antibiotic treatment in mice demonstrated the role of gut microbiota in controlling metabolic endotoxemia and the consequent inflammation in diet-induced obesity and type 2 diabetes (Cani et al., 2008). To our knowledge, there is limited information available on the microbial metabolites and intestinal inflammation in response to changes in gut microbiota on exposure to sourdough or its fermentation supernatants.
The research work presented in the current study is in continuation with our previous findings on the dynamics of microbial ecology, biochemical and nutritional characteristics during Tritordeum sourdough preparation (Arora et al., 2022). Tritordeum is an amphidiploid cereal resulting from the cross between wild barley (Hordeum chilense) and durum wheat (Triticum turgidum ssp. durum). This golden cereal is regarded as one of the most sustainable food ingredients combining the nutritional and functional properties of the two cereals. In this study, we aim to investigate and compare the in vitro faecal fermentation of Tritordeum breads prepared by the addition of spontaneously fermented Tritordeum sourdough (SDB) or commercial baker's yeast (BYB) by studying their impact on the human gut microbiota and the subsequent metabolites released during the fermentation. Further, we explored the potential of these test breads to improve intestinal permeability and immune function, the two key components of gut health.
Materials and methods
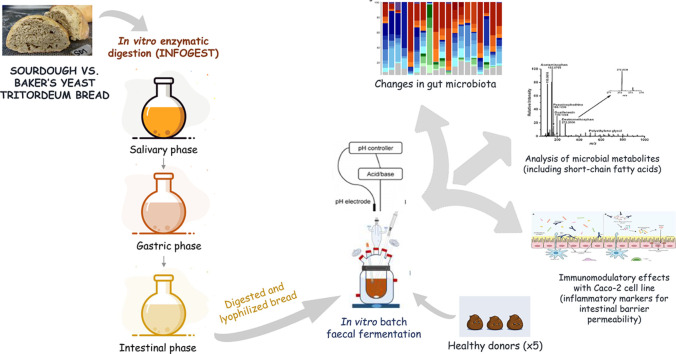
Preparation of tritordeum breads
Breads were prepared according to typical Italian bread recipe having dough yield of 160 at the Bakery Insperience pilot plant of the Micro4Food lab (Libera Universitá di Bolzano, Italy). The bread formulas were as follows: (1) Tritordeum baker's yeast bread (BYB) made with 250 g Tritordeum flour, 150 g tap water and 1.5 % (w/w) of commercial baker's yeast; and (2) Tritordeum sourdough bread (SDB) made with 175 g Tritordeum flour, 105 g tap water and 120 g Tritordeum sourdough (30 %, [w/w] of total dough). A continuous high-speed mixer (60 × g, dough mixing time 5 min) was used to prepare the doughs. Fermentation of doughs was allowed at 30 °C for 2.5 h. All breads were baked at 230 °C for 35 min (Omega 2, Bongard, Italy).
In vitro digestion of breads
The Tritordeum breads (BYB and SDB) were subjected to simulate the human digestion using the in vitro digestion protocol as standardized by (Minekus et al., 2014) with slight modifications. Inulin, a readily fermented fibre, was employed as control and underwent the same in vitro digestion. Firstly, the oral phase was simulated by the mixing of 30 g of bread with 21 mL simulated salivary fluid (SSF) [0.5 M KCl, 0.5 M KH2PO4, 1 M NaHCO3, 0.15 M MgCl2 and 0.5 M (NH4)2CO3] in a stomacher bag for 90 s. The contents were transferred to a 250-mL glass bottle followed by the addition of 0.15 mL 0.3 M CaCl2(H2O)2, 3 mL 1500 U/mL α-amylase (Sigma-Aldrich) and 5.85 mL mQ H2O and incubated for 2 min at 37 °C with shaking at 150 rpm. Then the oral bolus was mixed with 45 mL simulated gastric fluid (SGF) [0.5 M KCl, 0.5 M KH2PO4, 1 M NaHCO3, 2 M NaCl, 0.15 M MgCl2 and 0.5 M (NH4)2CO3] containing 10 mg/mL lipid vesicles, 9.6 mL 25,000 U/mL pepsin (Sigma Aldrich), 30 µL 0.3 M CaCl2(H2O)2 and 4.17 mL mQ H2O. This gastric phase was adjusted to pH 3.0 with 1 M HCl and incubated for 2 h at 37 °C with shaking at 150 rpm. At the end of incubation (or gastric phase), the intestinal phase was simulated by the addition of 66 mL simulated intestinal fluid (SIF) [0.5 M KCl, 0.5 M KH2PO4, 1 M NaHCO3, 2 M NaCl and 0.15 M MgCl2], 30 mL 800 U/mL pancreatin, 4.8 g porcine bile extract, 240 µL 0.3 M CaCl2(H2O)2 and 7.86 mL mQ H2O. The intestinal phase was adjusted to pH 7.0 with 1 M NaOH and incubated for 2 h at 37 °C with shaking at 150 rpm. Additionally, passive intestinal absorption of water and other hydrolytic by-products resulting from digestion in the small intestine was simulated using Spectra/Por® Dialysis Membrane MWCO 1000 KDa (Repligen, Waltham, MA, USA). The digested breads after the intestinal phase were tightly sealed using a strong thread at both ends and incubated overnight at 4 °C with magnetic stirring in a tank containing 10 mM NaCl with a volume equal to 100 × of total intestinal phase in the membrane. The samples were then frozen at −80 °C overnight, then freeze-dried and stored at −80 °C as 2 g aliquots in 50 mL tubes, until further use.
In vitro batch-culture fermentation of substrates using human faeces
Fresh faecal samples were collected from four healthy male and female volunteers aged between 30 and 50 years with no history of antibiotic treatment or probiotics in the three months preceding the experiment. The faecal samples were kept in the anaerobic chamber and inoculated within 1 h of collection. For each fermentation, the faecal samples from one volunteer were used in four fermentation vessels with negative control (blank), positive control (inulin), Tritordeum baker's yeast bread (BYB) and Tritordeum sourdough bread (SDB) as substrates. Faecal slurry (10% w/v of faeces) was prepared in sterile and pre-reduced 1 × phosphate buffered saline (PBS) solution and homogenized in a stomacher bag for 2 min. The fermentation vessels were temperature-controlled at 37 °C with a circulating water bath and 20 mL of faecal slurry was added to each fermentation vessel containing 180 mL of basal nutrient medium. The substrates (2 g each) were added to the respective vessels and fermentation was carried out for 24 h. Anaerobiosis was guaranteed by continuous nitrogen flow through the fermentation medium. The samples (5 mL each) were collected at 0, 5, 10, and 24 h and centrifuged at 13,000 rpm for 5 min. at 4 °C and stored at −80 °C immediately. The pellets were analysed for gut microbiota composition and the supernatants were analysed for microbial metabolites, as well as employed for in vitro cell culture experiments.
Analysis of gut microbiota by 16S rRNA gene sequencing
Total DNA was extracted from faecal fermentation pellets using MP Biomedicals™ FastDNA™ SPIN DNA Isolation Kit for Feces (Thermo Fisher Scientific, Waltham, MA, USA) following manufacturer's instructions, and the quality and quantity of DNA was measured using NanoDrop® 8000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). PCR amplification of V3–V4 regions of 16S rRNA gene was performed with the primer pair 341F (5′-CCTACGGGNGGCWGCAG-3′) and 806R (5′-GACTACNVGGGTWTCTAATCC-3′) (Klindworth et al., 2013) with a reaction volume of 25 µL. The reaction mix per sample consisted of 9 µL PCR H2O, 12.5 µL 2 × KAPA Hifi HotStart Ready Mix (Kapa Biosystems Ltd., UK), 0.5 μL of each primer (1 M) and 2.5 μL DNA (5 ng/μL). All PCR reactions were performed in the Verity™ 96-well Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA) with the following amplification protocol: 5 min at 95 °C (initialization), 30 cycles of 30 s at 95 °C (denaturation), 30 s at 55 °C (annealing), and 30 s at 72 °C (extension), followed by a final extension of 5 min at 72 °C. The amplified PCR products were examined using 1.5 % (w/v) agarose gel electrophoresis and purified using an Agencourt AMPure XP system (Beckman Coulter, Brea, CA, USA). Illumina library preparation was executed according to 16S Metagenomic Sequencing Library Preparation (Illumina) and adapters (Illumina Nextera XT Index Primer) were attached after seven PCR cycles. Libraries were purified using Agencourt AMPure XP (Beckman) and sequenced on an Illumina® MiSeq (PE300) platform (MiSeq Control Software 2.0.5 and Real-Time Analysis software 1.16.18, Illumina, San Diego, CA, USA). Raw sequences obtained from Illumina sequencing platform were analysed using the Quantitative Insights Into Microbial Ecology (QIIME) 2.0 pipeline (Bolyen et al., 2019) and the taxonomic classification was identified using Silva database Version 138 (Quast et al., 2013).
Nucleotide sequence accession number
The sequences are available in the Sequence Read Archive of NCBI (accession number PRJNA952160).
Targeted metabolite analysis using NMR spectroscopy
The fermentation supernatants from −80 °C were thawed overnight at 4 °C for quantification of metabolites using targeted approach through NMR analysis. Briefly, 900 µL of each supernatant was mixed with 100 µL deuterium oxide (D2O), vortexed vigorously for 15 s and filtered with Sartorius 0.22 μm PVDF syringe filters (Thermo Fisher Scientific, Waltham, MA, USA). Each filtered mix (600 µL) was transferred in 5-mm NMR tube and the spectra were recorded on Bruker Avance Neo 600 (base frequency 600 MHz for 1H nuclei), equipped with a broadband Z-gradient probe (5 mm sample tubes) and SampleCase 24-position autosampler. The spectra were acquired and processed using Topspin 4.1.1 software in the automation mode with Icon NMR 5.2.1. The deuterium lock signal was optimized for the 9:1 mixture of H2O and D2O (v/v). All proton NMR spectra were recorded using the noesygppr1d pulse sequence with automatic adjustment of water signal suppression frequency (o1p) and power level utilized for pulse was 47.10 dB (25 Hz suppression window). The size of the spectrum (sweep width, SW) was 20.8 ppm, time domain (TD) consisted of 65,536 (64 K) data points, number of scans (NS) was 64 and the number of dummy scans (DS) was 4, the time for relaxation delay (D1) was 10 s, receiver gain (RG) for all spectra was fixed at 101, and baseopt digitization mode was used. Acquisition of each spectrum was preceded by automatic adjustment of the probe (ATMA routine) and automatic shimming (TOPSHIM). Spectra were processed in the TopSpin software with the size of real spectrum (SI) set to 131,072 (128 K, 2xTD) data points, and apk0.noe phase correction au program was applied automatically to each spectrum. Quantitative analyses were performed using AssureNMR software with the external standard technique (ERETIC or Electronic REference To access In vivo Concentrations) (Hong et al., 2013), with the 2 mM sucrose reference solution in 9:1 mixture of H2O and D2O (v/v) used as the external standard.
Analysis of intestinal epithelium integrity and mucosal immunomodulation using in vitro cell culture models
Caco-2 cell culture
Human epithelial colorectal adenocarcinoma Caco-2 cell line (ATCC® HTB-37™, passage number between 50 and 60) were used for cell culture experiments to determine the intestinal membrane integrity and co-culture experiments with peripheral blood mononuclear cells (PBMCs) for pro- and anti-inflammatory cytokine analyses on exposure to faecal fermentation supernatants. The Caco-2 cell line was maintained in Dulbecco's modified Eagle's medium (DMEM) with high glucose (4.5 g/L) (Lonza, Switzerland) supplemented with 20 % decomplemented (56 °C, 60 min) fetal bovine serum (Lonza, Switzerland), 100 U/mL penicillin (Biological Industries, Israel), 100 µg/mL streptomycin (Biological Industries, Israel), 1 % non-essential amino acids (Euroclone, Milan), 2 mM glutamine and 0,25 µg/mL amphotericin B (Biological Industries, Israel) in humidified CO2 (5 %) incubator at 37 °C. Prior to the experiment, Caco-2 cells were maintained in T-75 cm2 flasks (Sarstedt, Nümbrecht, Germany) and passaged after 70 % confluency using 0.05 % trypsin–0.5 mM EDTA (Lonza, Switzerland) for detachment of cells from the flask wall. Medium was refreshed every two days until 70 % confluency was achieved. The transwell inserts with membrane filters of pore size 0.1 µm (BD Falcon, Sacco s.r.l, Cadorago, Como, Italy) were coated with rat tail collagen Type I (Sigma Aldrich) and kept to dry overnight under sterile conditions. Caco-2 cells were harvested to obtain a cell suspension of 1 × 105 cells/cm2 and 2.5 mL was added to each transwell insert. The cells were allowed to multiply and create a monolayer for 13 days. Uniformity of the monolayer was tested by measuring the passage of current through the monolayer during consecutive days, by employing a volt-ohm-meter, as described below. Briefly, 1.5 mL of DMEM medium was added to the basolateral chamber of the 6-well plate with inserts. Faecal fermentation supernatants collected at t = 24 h were pooled from all the donors for the respective substrates (BYB or SDB) for the experiments. All control and test substrates were prepared on the day of the experiment. Propionic acid (10 mM) and ethanol (7 %) were used as positive and negative controls, respectively, and each pooled test substrate was used at 10 % (v/v) in DMEM culture medium. All the controls and substrates were added in replicates (n = 3) to the apical chamber with a final volume of 2.5 mL.
Trans epithelial electrical resistance (TEER) assay
TEER measurements were performed using an epithelial volt-ohm-meter (EVOM, World Precision Instruments Inc., Sarasota, FL, USA). Plates were left at room temperature for 30 min prior to TEER measurements to stabilize the temperature in the wells. The membrane integrity of Caco-2 cell monolayers was assessed before the addition of test substrates (t0). The media was then removed from basolateral chambers followed by the removal from apical chamber. The control or test treatments were added to the apical layer and cell culture medium in the basolateral chamber. The plates were incubated at 37 °C in the humidified 5 % CO2 incubator and resistance was measured again after 24 h (t24). The TEER was calculated using the following equation (Srinivasan et al., 2015):
Where, area of the semipermeable membrane= 9.6 cm2. The change in TEER for each insert was calculated using the following formula:
where,
TEERt24=TEER measurement after 24 h,
TEERt0 = TEER measurement before the addition of control or test substrates.
Isolation of PBMCs from human blood for Caco-2/PBMC co-culture model
The peripheral blood mononuclear cells (PBMCs) were used in the in vitro co-culture Caco-2-PBMC as a model of intestinal mucosa. Isolation of PBMCs was performed from whole blood donations from healthy humans (n = 3), received from the Transfusion Unit of Santa Chiara Hospital, Trento, Italy. PBMCs were obtained by density gradient centrifugation method using Lymphoprep™ (Thermo Fisher Scientific, Waltham, MA, USA). The experimental plan was approved by the local Ethical Committee of Azienda Provinciale dei Servizi Sanitari (APSS, Santa Chiara Hospital, Italy; approval document n. 401/2015). The study was designed in conformity with the international recommendation (Dir. EU 2001/20/EC) and its Italian counterpart (DM 15 Luglio 1997; D.Lvo 211/2003; D.L.vo 200/2007) for clinical trial and following the Declaration of Helsinki, to assure protection and care of subjects involved. The human blood was diluted in 1 × PBS (1:1) followed by its addition to 50 mL tubes containing Lymphoprep™ reagent in the ratio of 2:1 (blood: Lymphoprep™). The tubes were then centrifuged at 400 × g for 20 min at room temperature without break to avoid the disturbance caused in the layer separation. PBMCs were carefully collected from the ring surrounding the Lymphoprep™ using a 1 mL pipette in a 50 mL tube containing the complete RPMI 1640 medium to preserve the cell viability. The PBMCs were washed twice in RPMI 1640 medium and finally in 1 × PBS to determine the cell concentration using trypan blue assay. The PBMCs were again resuspended in complete RPMI 1640 medium (Sigma-Aldrich), supplemented with 10 % decomplemented (56 °C, 60 min) fetal bovine serum (Lonza, Switzerland), 1 % penicillin (100 U)–streptomycin (100 μg/mL), 1 % 200 mM l-glutamine, and 1 % 100 mM sodium pyruvate (Biological Industries, Israel), at a cell concentration of 2 × 106 cells/mL and 1.5 mL of this cell suspension was added in the basolateral chamber. The plates were incubated at 37 °C in the humidified 5 % CO2 incubator overnight for co-culture experiment.
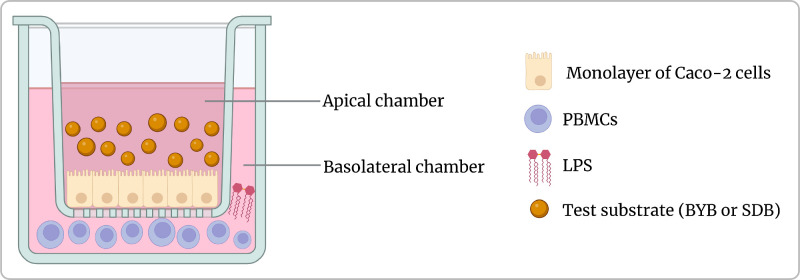
In vitro Caco-2/PBMC co-culture model of intestinal mucosa
The Caco-2/PBMC co-culture system (Fig. 1) was developed to investigate the anti-inflammatory effects of faecal fermentation supernatants collected at time 24 h of fermentation using the test substrates, after an inflammatory stimulus represented by lipopolysaccharide (LPS) (Borsci et al., 2020; Kämpfer et al., 2017). The transwell inserts containing the Caco-2 cells at a concentration of 1 × 105 cells/cm2 were transferred to the plate containing PBMCs incubated overnight at a concentration of 2 × 106 cells/mL. The PBMCs in the basolateral chamber were then refreshed with complete RPMI 1640 medium and the apical chamber was replaced by fresh DMEM medium. Then the LPS was added to the co-culture system at 10 ng/mL and incubated for 2 h at 37 °C in humidified 5 % CO2 incubator. The pooled fermented supernatants of test substrates (BYB or SDB) were then added (10 % v/v) to the apical chamber (n = 3) as mentioned previously in section 2.6.1. The samples were collected at different time points (0, 3, 6 and 24 h) for the analysis of inflammatory cytokines released by cells to the culture medium and tight junction proteins isolated from Caco-2 cells.
Fig. 1.
Illustration of in vitro co-culture experimental setup with human adenocarcinoma cells (Caco-2 cells) in the apical chamber and peripheral blood mononuclear cells (PBMCs) isolated from human blood in the basolateral chamber. The inflammatory response was stimulated with the addition of lipopolysaccharide (LPS) in the basolateral chamber and the fermentation supernatants of Tritordeum breads BYB and SDB), represented here as test substrates, were incubated on the apical side containing Caco-2 monolayer.
Quantification of cytokines using Magpix®
The supernatants from apical and basolateral chambers in the Caco-2/PBMCs co-culture system at 0 and 24 h were subjected to cytokine analysis using Magpix®. The supernatants were centrifuged at 13,000 × g for 5 min at 4 °C to separate the cell debris. IL-10, IL-1β, IL-6 and TNF-α were quantified using a cytokine magnetic bead-based panel (Milliplex MAP kit, Millipore Corp., Billerica, MA, USA) and measured by a Magpix® instrument (Luminex, Texas, USA) and Xponent software (version 4.2, Luminex Corp, Austin, Texas, US) according to manufacturer's instructions. Blanks and standard curves were included for each plate with the overall precision (2–19 %) and accuracy (87–107 %) based on the calibration range (3.2–10,000 pg/mL) of the standards (Millipore).
RNA isolation and gene expression analysis of cytokines
Caco-2 cells and PBMCs collected after 0-, 3- and 6-h incubation with faecal fermentation supernatants were resuspended in 500 μL TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and stored at −80 °C until total RNA extraction according to manufacturer's instructions. The total RNA was quantified using Nanodrop 8000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The RNA quality was examined using 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA) and the samples with RNA Integrity Number (RIN) > 8 were subjected to reverse transcription using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems™, Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer's instructions with reaction volume of 20 µL containing 2 µL 10 × RT buffer, 0.8 μL 25 × dNTP Mix (100 mM), 2.0 μL 10 × RT Random Primers, 1.0 μL MultiScribe™ Reverse Transcriptase, 4.2 μL DEPC-treated water and 10 μL template RNA (5 ng/μL). The cDNA was stored at −80 °C until further use. The expression level of inflammatory genes (IL-10, IL-1β, IL-6 and TNF-α) was determined by quantitative Real Time-PCR (qRT-PCR) using ViiA™ 7 System (Thermo Fisher Scientific, Waltham, MA, USA). The reaction volume of 20 µL was prepared for each primer pair containing 10 μL of KAPA Probe FAST qPCR Master Mix 2 × Universal (Kapa Biosystems, Sigma-Aldrich, Germany), 0.4 μL 50 × Rox Low (10 ng/μL), 1 μL TaqMan Assay (TaqMan® Gene Expression Assays, Applied Biosystems, USA), 2 μL of cDNA (10 ng/μL) and 6.6 μL H2O. The Cycle threshold (Ct) values were normalized for each sample against their respective geometric mean for housekeeping genes (18S and GAPDH). Gene expression was calculated as the relative fold change (2−ΔΔCt), wherein ΔΔCt was obtained by subtracting ΔCt (mean Ct of housekeeping genes subtracted from the tested gene) of test substrates from ΔCt of negative control (culture medium).
Quantification of tight junction proteins by western blot analysis
The Caco-2 cells were collected after 0 and 24 h of incubation with faecal fermentation supernatants in co-culture with PBMCs. Analysis of tight junction proteins was carried out to assess the membrane integrity (Bianchi et al., 2019). The cells were rinsed twice with ice-cold 1 × PBS followed by the addition of 350 µL/well cell Lysis buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 % Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM NaF, 2 mM imidazole) supplemented with a protease inhibitor cocktail (Complete, Mini, EDTA-free, Roche, Monza, Italy). The cell lysates were centrifuged at 14,000 × g for 10 min at 4 °C and the supernatant was used for determining the protein concentration using Pierce™ BCA Protein Assay Kit (Thermo Fisher, Waltham, MA, USA). The proteins were precipitated with acetone (Sigma-Aldrich, Germany) overnight at 4 °C in the ratio of 4:1. The precipitated samples were centrifuged at 18, 000 × g for 10 min and the pellet was dried for 5–10 min to evaporate acetone. The pellets were resuspended in 15 μL Loading buffer 1 × (63 mM Tris–HCl, pH 6.8, 2 % SDS, 10 % glycerol, 0.1 % 2-mercaptoethanol and 0.005 % bromophenol blue) to a final concentration of 35 µg protein/15 μL. The mixture was heated at 70 °C for 5–10 min for multi-pass membrane proteins and then loaded on 12 % SDS-polyacrylamide gel (35 μg proteins/well). The gels were run at 100 V for 1.5 h after which the gels were prepared for western blotting procedure. The SDS-PAGE gel was blotted on PVDF membrane (Immobilon-P, Millipore, Millipore Merck Corporation, Burlington, MA, USA) for 1 h at 100 V, at 4 °C. The PVDF membrane was then rinsed with TRIS-Buffered Saline (TBS) and then incubated in TBS 0.05 % Tween (TBS-T) with 5 % skim milk powder (Oxoid, Milan, Italy) solution for 1 h at room temperature (RT). This was followed by washing three times with TBS-T. The membranes were then immersed overnight with primary monoclonal antibodies (Santa Cruz Biotechnology, Texas, USA) (initial concentration 200 μg/mL) diluted 1:1000 in TBS-T with a 1 % skim milk powder solution at 4 °C. Next day, the membrane is washed (3 ×) with TBS-T and then exposed to HRP-conjugated secondary antibodies in TBS-T with 0.5 % skim milk powder solution for 1 h at room temperature. The membrane was again washed (3 ×) with TBS-T, 10 min each and only 1 × wash with PBS-T. The protein bands were then visualized by developing solution of 10 × CN/DAB Concentrate with the Stable Peroxide Substrate (1:10 ratio). The reaction was stopped by the removal of solution and washing with distilled water once. High quality images were obtained for the membranes using a digital camera. The protein bands were quantified using ImageJ Software (Java-based software designed by National Institute of Health, https://imagej.nih.gov/ij) by subtracting the background and then expressed as the ratio of test protein over control protein (β-actin) for each lane, according to the instructions by (Gallo-Oller et al., 2018). The change in protein expression was quantified as fold-change normalized with β-actin (positive control) and medium (negative control).
Statistical analysis
The statistical analysis for microbial diversity (α- and β-diversities) was performed using QIIME 2.0 pipeline and statistically significant differences (P < 0.05) within each substrate were calculated by Tukey's Honestly Significant Difference (HSD). In addition, the relative abundances obtained using QIIME 2.0 were analysed by non-parametric Wilcoxon test using R studio version 3.4.1. The P-values for the statistical significance in all the experiments were corrected using Benjamini–Hochberg false discovery rate (FDR) with P < 0.05 as statistically significant. The significant differences in metabolite concentrations between different substrates were estimated by Wilcoxon test using R studio version 3.4.1. The statistical significance (P < 0.05) of TEER measurements, cytokine quantification, gene expression and western blot analysis was calculated using unpaired t-test in Microsoft Excel. The correlation between relative abundances of bacterial genera and metabolites released during faecal fermentation was calculated by Spearman's correlation analysis. All the results are expressed as mean ± SD.
Results
Gut microbial ecology
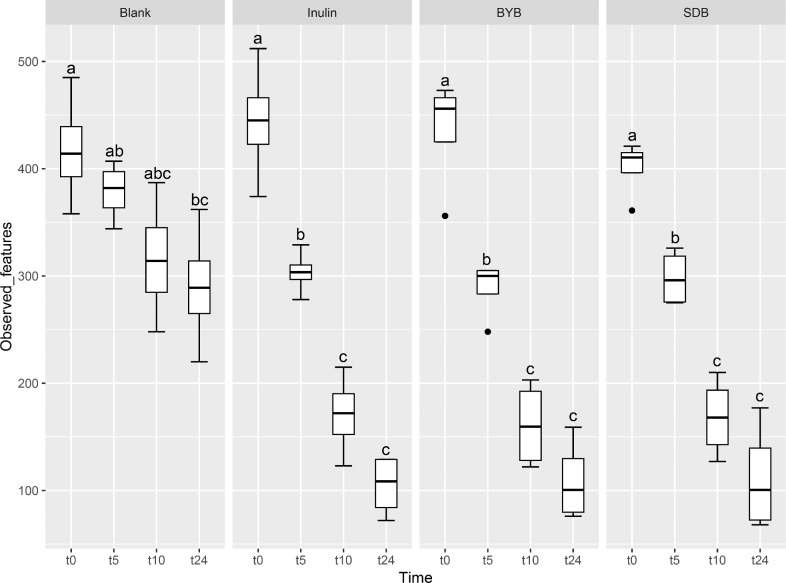
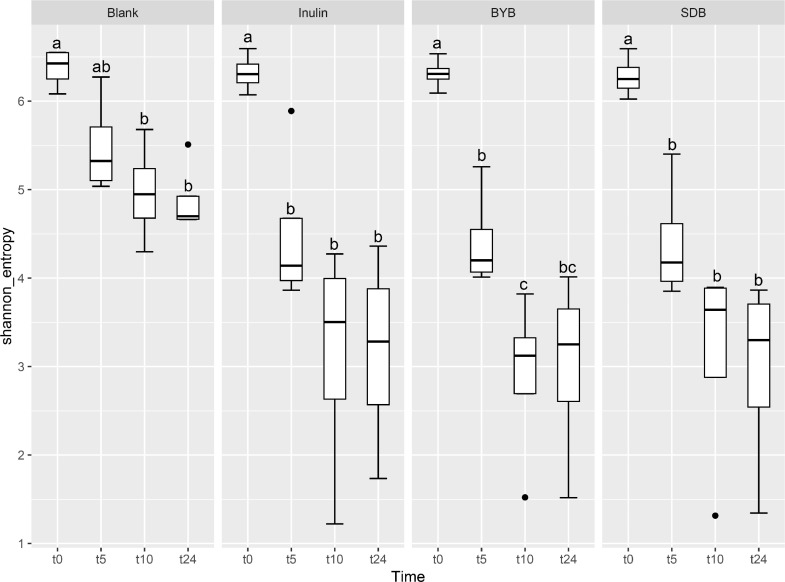
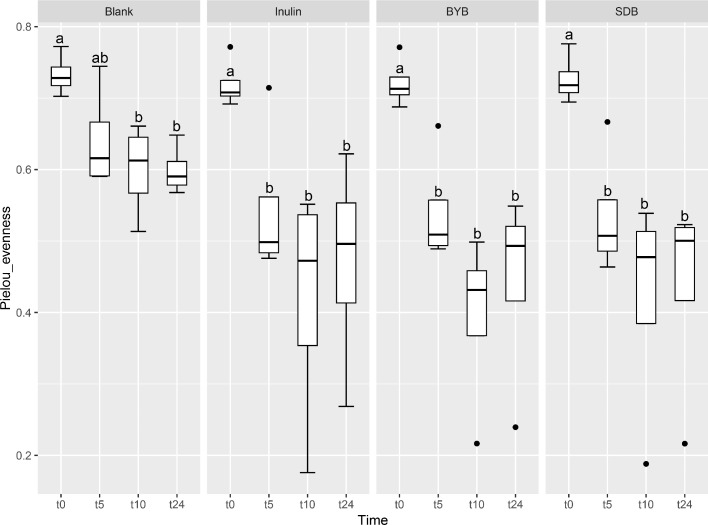
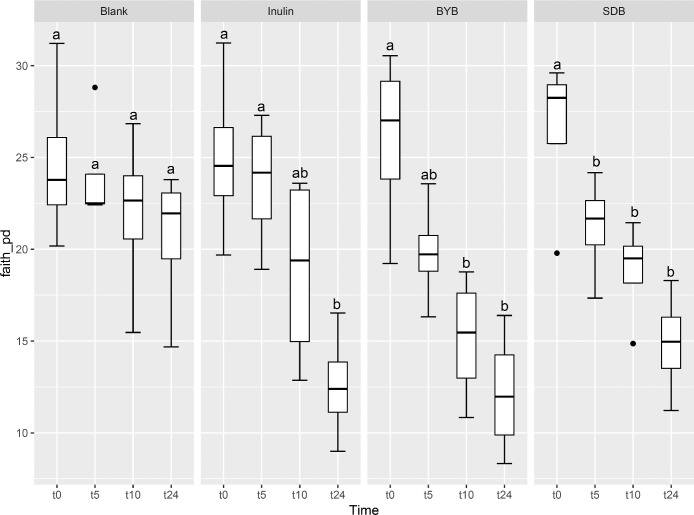
Illumina® MiSeq (PE300) sequencing of gut microbial 16S rRNA gene amplicons yielded 5823,585 reads in total, with 90, 993.52 ± 10,604.09 raw reads per sample. These raw reads were processed using QIIME 2.0 pipeline to remove chimeras, low quality sequences, and Cyanobacteria sequences derived from the flour origin, resulting in 5821,089 total reads with 90,954.52 ± 10,593.21 of raw reads per sample. Bacterial α-diversity was analysed through richness, evenness and phylogenetic indices expressed as observed number of features (or OTUs) (Fig. 2A), the Shannon entropy index (Fig. 2B), Pielou's evenness (Fig. 2C) and the Faith Phylogenetic Diversity (PD) index (Fig. 2D). The faecal-fermented substrates [Inulin and Tritordeum breads with commercial baker's yeast (BYB) and type I sourdough (SDB)] demonstrated different impacts on bacterial α-diversity when compared to negative control (Blank) at baseline (t0) and after 5 (t5), 10 (t10), and 24 (t24) h of anaerobic fermentation. No significant differences (P > 0.05) were observed between all the treatments at the time of inoculation (t0). The bacterial richness (number of observed features) decreased with time for all substrates when compared to that of blank from t5 onwards (P < 0.05) (Fig. 2A and Table 1). In addition, the evenness (indicated by Shannon index and Pielou's evenness) and phylogenetic diversity (Faith PD index) decreased from 10 h when compared to blank, which was not significant after FDR correction. Moreover, only the bacterial richness showed significant decline between t0 and t24 (P < 0.05) for all the substrates indicating minimal differences from t5 to t24.
Fig. 2.
Bacterial α-diversity using observed features (OTUs) (A), Shannon index (B), Pielou's evenness (C), and Faith Phylogenetic Diversity (PD) index (D) at t = 0 h (t0), t = 5 h (t5), t = 10 h (t10) and t = 24 h (t24) of faecal fermentation with the following substrates: Blank (no substrate), Inulin (as control), Tritordeum breads with commercial baker's yeast (BYB) and spontaneous sourdough (SDB). Line inside the box represents the median (n = 4), whiskers from either side of the box represent the first and the third quartiles, respectively. The letters indicate statistically significant differences at P < 0.05 within each substrate calculated by Tukey's Honestly Significant Difference (HSD).
Table 1.
P-values as a measure of significant differences in α-diversity indices (Observed OTUs, Shannon, Pielou, and Faith PD indices) at 0 (t0), 5 (t5), 10 (t10), and 24 (t24) h of faecal fermentation of Blank, Inulin, and Tritordeum breads prepared with baker's yeast (BYB) or sourdough (SDB). The significance (P-values) was calculated using Kruskal–Wallis and FDR correction.
| P-value (α-Diversity Indices) | ||||
|---|---|---|---|---|
| Comparison at t = 0 h | Observed OTUs | Shannon | Pielou | Faith PD |
| Blank vs Inulin | 0.577 | 0.932 | 0.634 | 0.932 |
| Blank vs BYB | 0.916 | 0.799 | 0.634 | 0.799 |
| Blank vs SDB | 0.916 | 0.932 | 0.916 | 0.932 |
| Inulin vs BYB | 1.000 | 1.000 | 0.916 | 1.000 |
| Inulin vs SDB | 0.245 | 0.799 | 0.634 | 0.799 |
| BYB vs SDB | 0.411 | 0.932 | 0.776 | 0.932 |
| Comparison at t = 5 h | ||||
| Blank vs Inulin | 0.046 | 0.270 | 0.333 | 0.270 |
| Blank vs BYB | 0.046 | 0.167 | 0.333 | 0.167 |
| Blank vs SDB | 0.046 | 0.167 | 0.333 | 0.167 |
| Inulin vs BYB | 0.916 | 0.799 | 0.776 | 0.799 |
| Inulin vs SDB | 0.769 | 0.932 | 1.000 | 0.932 |
| BYB vs SDB | 0.769 | 0.932 | 0.916 | 0.932 |
| Comparison at t = 10 h | ||||
| Blank vs Inulin | 0.046 | 0.052 | 0.221 | 0.052 |
| Blank vs BYB | 0.046 | 0.052 | 0.067 | 0.052 |
| Blank vs SDB | 0.046 | 0.052 | 0.127 | 0.052 |
| Inulin vs BYB | 0.916 | 0.799 | 0.916 | 0.799 |
| Inulin vs SDB | 1.000 | 0.932 | 1.000 | 0.932 |
| BYB vs SDB | 0.916 | 0.607 | 0.634 | 0.607 |
| Comparison at t = 24 h | ||||
| Blank vs Inulin | 0.046 | 0.052 | 0.333 | 0.052 |
| Blank vs BYB | 0.046 | 0.052 | 0.067 | 0.052 |
| Blank vs SDB | 0.046 | 0.052 | 0.067 | 0.052 |
| Inulin vs BYB | 1.000 | 0.932 | 0.333 | 0.932 |
| Inulin vs SDB | 0.916 | 0.932 | 0.776 | 0.932 |
| BYB vs SDB | 0.916 | 0.932 | 1.000 | 0.932 |
| Comparison of t = 0 vs t = 24 h | ||||
| Blank_t0 vs Blank_t24 | 0.081 | 0.052 | 0.067 | 0.052 |
| Inulin_t0 vs Inulin_t24 | 0.046 | 0.052 | 0.067 | 0.052 |
| BYB_t0 vs BYB_t24 | 0.046 | 0.052 | 0.067 | 0.052 |
| SDB_t0 vs SDB_t24 | 0.046 | 0.052 | 0.067 | 0.052 |
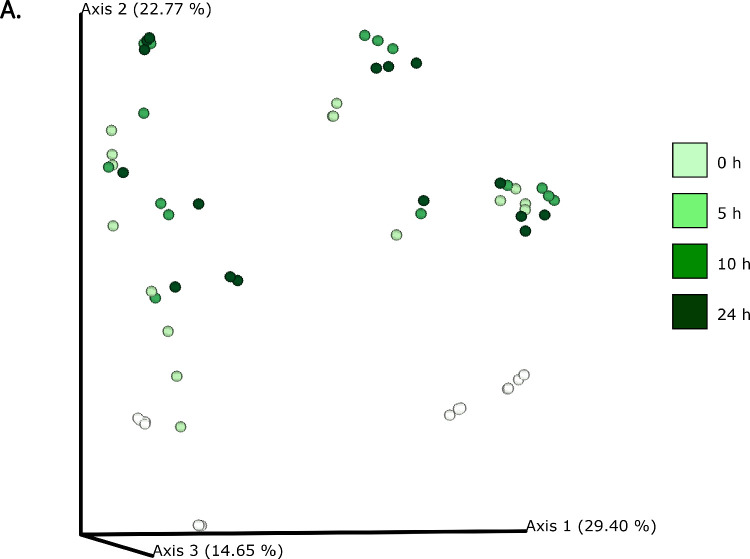
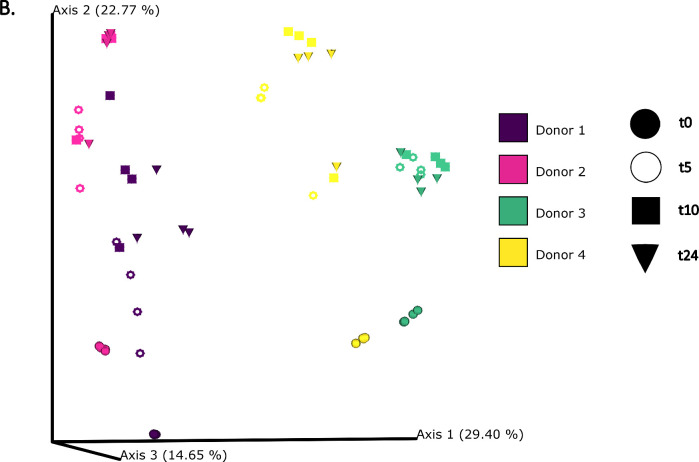
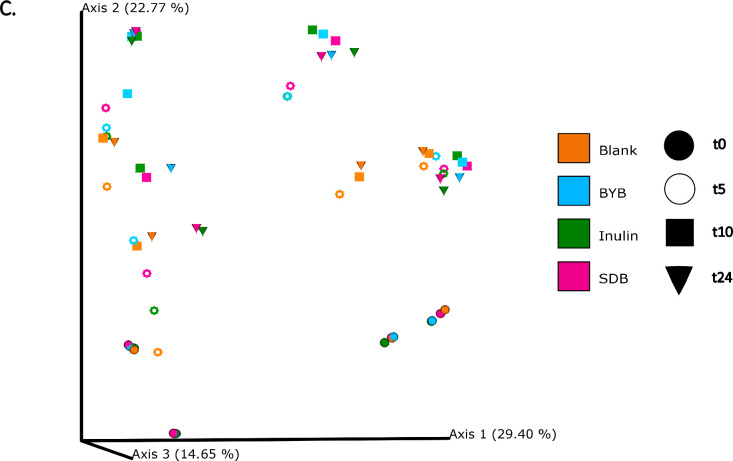
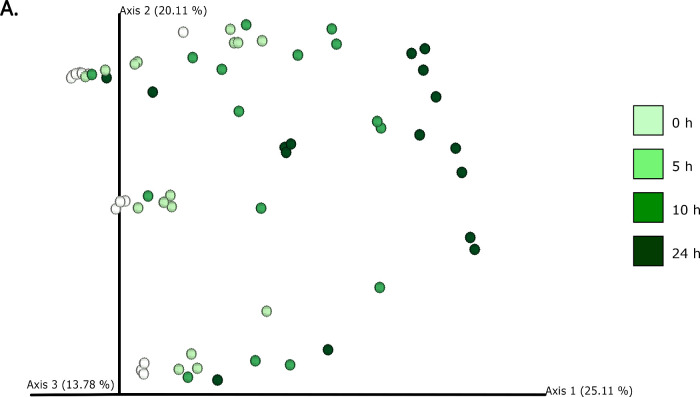
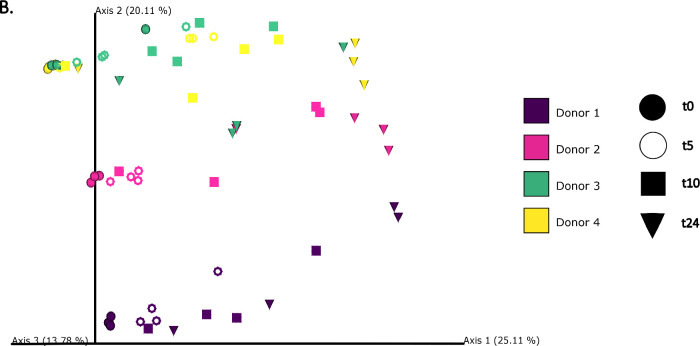
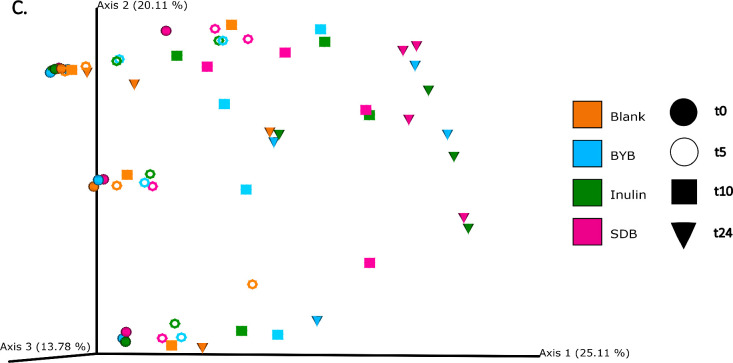
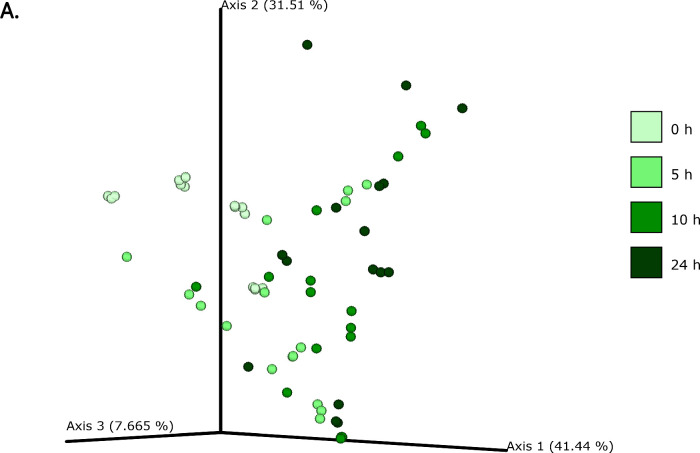
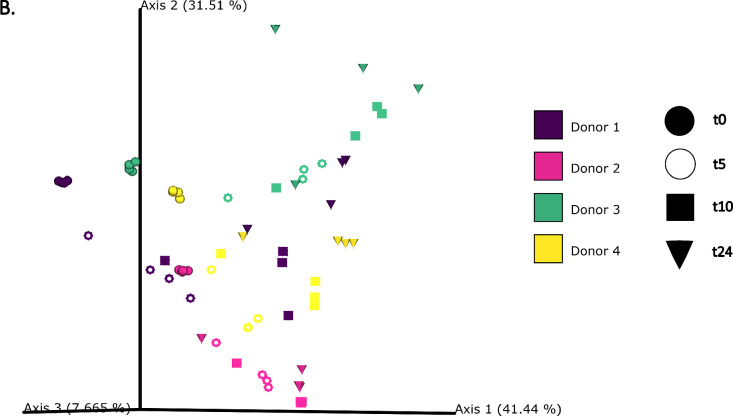
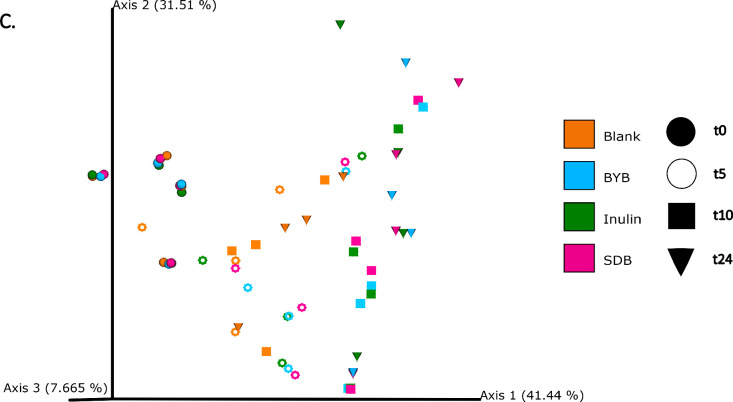
The differences in bacterial composition following fermentation of different substrates were assessed as β-diversity using Bray–Curtis dissimilarity matrix (Fig. 3A‒C), Unweighted (Fig. 4A‒C) and Weighted (Fig. 5A‒C) UniFrac distances. As compared to Unifrac PCoA plots, Bray–Curtis plots demonstrated differences arising at the donor and substrate levels. The baseline fermentation (t0) segregated from t5, t10, and t24 time points of faecal fermentation for all substrates in all the PCoA plots. No statistically significant differences were observed in β-diversity indices when comparing substrates (SDB, BYB, or Inulin) or control (blank) within the same time point (t0, t5, t10, or t24). The statistical significance between time points was highlighted for the blank as well as for each fermentation substrate (Table 3). The negative control (blank) showed significant increase in Weighted Unifrac distances only at t10 (P = 0.033) and t24 (P = 0.033) when compared to the baseline (t0). The other fermentation substrates (Inulin, BYB and SDB) were statistically significant for all the β-diversity metrics when baseline (t0) was compared to t10 and t24 (Table 2). Therefore, the faecal fermentation with different substrates showed a significant impact based on α- and β-diversities of gut microbiota composition when compared to the negative control.
Fig. 3.
Bacterial β-diversity represented as Principal Component Analysis (PCoA) using Bray–Curtis dissimilarity index (n = 4 for each fermentation substrate at each time point). Different shades of green represent time points (0, 5, 10, and 24 h) of faecal fermentation (A). Different colours highlight different faecal donors (n = 4) with time points indicated as different shapes (B). Different colours highlight different substrates (n = 4) with time points indicated as different shapes (C).
Fig. 4.
Bacterial β-diversity represented as Principal Component Analysis (PCoA) using Unweighted Unifrac distances (n = 4 for each fermentation substrate at each time point). Different shades of green represent time points (0, 5, 10, and 24 h) of faecal fermentation (A). Different colours highlight different faecal donors (n = 4) with time points indicated as different shapes (B). Different colours highlight different substrates (n = 4) with time points indicated as different shapes (C).
Fig. 5.
Bacterial β-diversity represented as Principal Component Analysis (PCoA) using Weighted Unifrac distances (n = 4 for each fermentation substrate at each time point). Different shades of green represent time points (0, 5, 10, and 24 h) of faecal fermentation (A). Different colours highlight different faecal donors (n = 4) with time points indicated as different shapes (B). Different colours highlight different substrates (n = 4) with time points indicated as different shapes (C).
Table 3.
P-values as a measure of significant differences in the concentrations of IL-10, IL-1β, IL-6 and TNF-α under LPS stimulation upon exposure to control Medium or test substrates (BYB and SDB). The statistical differences were determined as the change in cytokine concentrations from 0 to 6 (Δt6–t0), 6 to 24 (Δt24–t6) and 0 to 24 (Δt24–t0) hours of incubation. The significance (P-values) was calculated using pairwise t-test.
| APICAL SIDE (P-values) | |||||
|---|---|---|---|---|---|
| IL-10 | IL-1β | IL-6 | TNF-α | ||
| Δt6–t0 | BYB vs Medium | 0.205 | 0.500 | 0.252 | 0.877 |
| SDB vs Medium | 1.000 | 1.000 | 0.223 | 0.459 | |
| BYB vs SDB | 0.205 | 0.500 | 0.493 | 0.494 | |
| Δt24–t6 | BYB vs Medium | 0.586 | 0.545 | 0.587 | 0.506 |
| SDB vs Medium | 0.970 | 0.505 | 0.527 | 0.744 | |
| BYB vs SDB | 0.680 | 0.565 | 0.617 | 0.662 | |
| Δt24–t0 | BYB vs Medium | 0.583 | 0.472 | 0.519 | 0.412 |
| SDB vs Medium | 0.954 | 0.432 | 0.455 | 0.658 | |
| BYB vs SDB | 0.682 | 0.510 | 0.573 | 0.627 | |
| BASOLATERAL SIDE (P-values) | |||||
| IL-10 | IL-1β | IL-6 | TNF-α | ||
| Δt6–t0 | BYB vs Medium | 0.169 | 0.859 | 0.024 | 0.115 |
| SDB vs Medium | 0.060 | 0.668 | 0.042 | 0.050 | |
| BYB vs SDB | 0.568 | 0.753 | 0.751 | 0.798 | |
| Δt24–t6 | BYB vs Medium | 0.312 | 0.850 | 0.575 | 0.338 |
| SDB vs Medium | 0.687 | 0.464 | 0.122 | 0.031 | |
| BYB vs SDB | 0.144 | 0.506 | 0.232 | 0.387 | |
| Δt24–t0 | BYB vs Medium | 0.234 | 0.847 | 0.457 | 0.050 |
| SDB vs Medium | 0.403 | 0.495 | 0.633 | 0.162 | |
| BYB vs SDB | 0.211 | 0.545 | 0.373 | 0.161 | |
Table 2.
P-values of statistical differences for each faecal fermentation substrate (Blank, Inulin, BYB, and SDB after pairwise comparisons of bacterial β-diversity indices (Permanova with 999 permutations) between time points 0 (t0), 5 (t5), 10 (t10), and 24 (t24) h.
| P-value (β-Diversity Indices) | |||
|---|---|---|---|
| BLANK | Bray-Curtis | Unweighted Unifrac | Weighted Unifrac |
| t0 vs t5 | 0.551 | 0.673 | 0.120 |
| t0 vs t10 | 0.052 | 0.667 | 0.033 |
| t0 vs t24 | 0.053 | 0.548 | 0.033 |
| t5 vs t10 | 0.869 | 0.901 | 0.803 |
| t5 vs t24 | 0.789 | 0.961 | 0.281 |
| t10 vs t24 | 0.766 | 0.910 | 0.835 |
| INULIN | |||
| t0 vs t5 | 0.165 | 0.709 | 0.034 |
| t0 vs t10 | 0.025 | 0.037 | 0.03 |
| t0 vs t24 | 0.034 | 0.021 | 0.026 |
| t5 vs t10 | 0.785 | 0.774 | 0.353 |
| t5 vs t24 | 0.687 | 0.035 | 0.217 |
| t10 vs t24 | 0.811 | 0.285 | 0.479 |
| BYB | |||
| t0 vs t5 | 0.085 | 0.644 | 0.021 |
| t0 vs t10 | 0.024 | 0.029 | 0.029 |
| t0 vs t24 | 0.028 | 0.023 | 0.028 |
| t5 vs t10 | 0.777 | 0.635 | 0.311 |
| t5 vs t24 | 0.702 | 0.034 | 0.228 |
| t10 vs t24 | 0.769 | 0.854 | 0.555 |
| SDB | |||
| t0 vs t5 | 0.059 | 0.701 | 0.041 |
| t0 vs t10 | 0.031 | 0.035 | 0.038 |
| t0 vs t24 | 0.026 | 0.031 | 0.033 |
| t5 vs t10 | 0.799 | 0.753 | 0.347 |
| t5 vs t24 | 0.640 | 0.022 | 0.177 |
| t10 vs t24 | 0.793 | 0.236 | 0.767 |
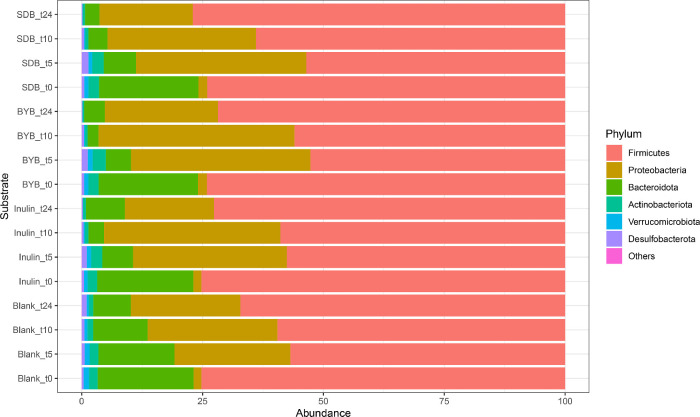
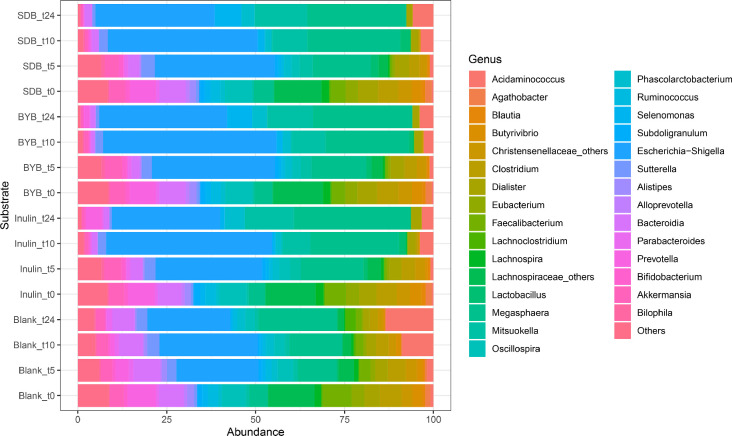
The gut microbiota composition comprised of mainly of the phyla Firmicutes, Bacteroidota, Proteobacteria, Actinobacteriota, Verrucomicrobiota, and Desulfobacterota (Fig. 6A). No significant differences were observed in the relative abundances at the baseline (t0) at all taxonomy levels between the test substrates while some differences were observed at the donor level (Supplementary Table 1). Firmicutes constituted 50–75 % of phyla throughout the fermentation period in all the substrates. Furthermore, Proteobacteria increased significantly from ∼2 % up to 30–50 % for all substrates while Bacteriodota declined from 20 % to ∼8 % (Blank) or ∼2 % (Inulin, BYB, and SDB) (Supplementary Table 2).
Fig. 6.
Bacterial relative abundance (%) at phylum (A) and genus (B) levels for all the substrates (Blank, Inulin, Tritordeum baker's yeast (BYB) and sourdough (SDB) breads) inoculated for in vitro batch culture fermentation and analysed at time points 0 (t0), 5 (t5), 10 (t10) and 24 (t24) h. The abundances are represented as mean percentage with n = 4. Less abundant phyla include bacteria with a relative abundance less than 0.01 % in fewer than 25 % of samples; less abundant genera include bacteria with a relative abundance less than 0.5 % in fewer than 25 % of samples.
The average relative abundances of gut microbiota at the genus level are depicted in Fig. 6B. The test substrates (BYB and SDB) showed significant differences (P < 0.05) for selected genera as compared to Blank. In contrast, no significant differences (P > 0.05) were observed between test substrates and Inulin at the genus level. The relative abundances of Bacteroides were reduced significantly from t5 onwards with BYB (P = 0.015), SDB (P = 0.022) and Inulin (P = 0.019) as compared to Blank which showed no change in this genus. In contrast, Bifidobacterium abundance was increased significantly at t24 with BYB (P = 0.031), SDB (P = 0.032) and Inulin (P = 0.023) whereas it was significantly reduced in the Blank (P = 0.029). Moreover, the genera Butyrivibrio, Christensenellaceae, Clostridium, Faecalibacterium, Oscillospiraceae, Lachnospiraceae, and Ruminococcus disappeared almost completely after 24 h of faecal fermentation in BYB, SDB, and Inulin as compared to Blank. Furthermore, a significant increase was observed for relative abundances of Escherichia-Shigella (BYB, P = 0.010; SDB, P = 0.027; Inulin, P = 0.015), Megasphaera (BYB, P = 0.047; SDB, P = 0.044; Inulin, P = 0.024), Mitsuokella (BYB, P = 0.030; SDB, P = 0.041; Inulin, P = 0.033) and Phascolarctobacterium (BYB, P = 0.035; SDB, P = 0.048; Inulin, P = 0.035) (Supplementary Table 3).
Targeted metabolite analysis by NMR spectroscopy
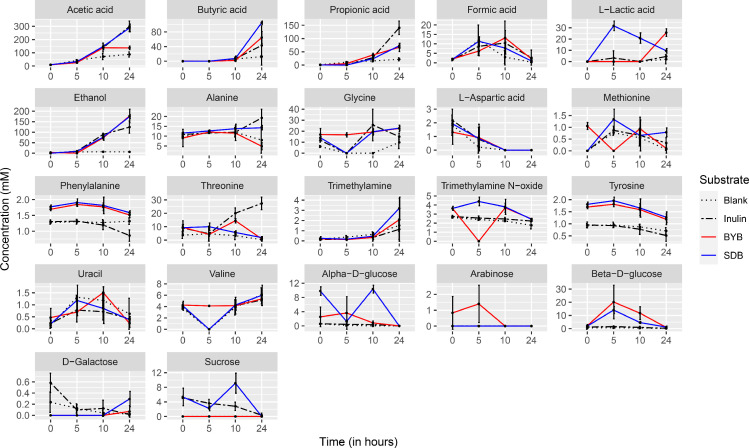
The changes in microbial metabolites during batch culture faecal fermentation of controls (Blank and Inulin) and test substrates (BYB and SDB) were analysed by Targeted Metabolomics using NMR Spectroscopy. The concentration of different metabolites estimated during 0 (t0), 5 (t5), 10 (t10), and 24 (t24) h of faecal fermentation are depicted as a line graph in Fig. 7 and categorized into short chain fatty acids (SCFA), amino acids and their derivatives, sugars, and alcohol. Average concentrations of all the metabolites quantified in the faecal supernatants are mentioned in the Supplementary Table 3. The main SCFAs, i.e., acetic, butyric, and propionic acids, increased over time for all with a total concentration of 121.74 ± 73.52 mM (Blank), 485 ± 73.52 mM (Inulin), 270.71 ± 46.71 mM (BYB), and 467.96 ± 43.18 mM (SDB), with acetic acid being the highest SCFA in all. Acetic acid and butyric acid concentrations were significantly higher (P < 0.05) with SDB when compared to BYB whereas propionic acid was highest (P < 0.05) with inulin. Formic acid, another by-product of anaerobic microbial fermentation, increased up to 10 h, followed by its decrease at 24 h of faecal fermentation with Inulin, BYB and SDB without any significant differences, whereas no significant changes were observed in the negative control (blank) throughout. Lactic acid concentrations were initially higher in SDB (31.70 ± 4.20 mM), as anticipated due to the presence of sourdough which further decreased by the end of fermentation (9.51 ± 2.04 mM), indicating its metabolism to SCFAs, as discussed further. On the contrary, BYB substrate had increased concentrations after 24 h of faecal fermentation (25.68 ± 3.42 mM). Ethanol production increased continuously for all the substrates when compared to blank, while it was significantly lower (P < 0.05) with inulin (124.37 ± 28.94 mM) as substrate than BYB (177.14 ± 32.70 mM) and SDB (173.98 ± 9.36 mM). Release of amino acids by proteolysis and their utilization are common metabolic activities in the gut influenced by the microbiota. In this context, amino acid concentrations were fluctuating (no defined pattern) throughout the faecal fermentation with all the substrates (Inulin, BYB, and SDB), except l-aspartic acid and tyrosine which exhibited continuous decrease by the end of 24 h of faecal fermentation. Trimethylamine slightly increased during the faecal fermentation (P > 0.05) and its derivative trimethylamine-N-oxide did not show any significant changes after 24 h. The utilization of sugars for survival by the gut bacteria was apparent in the fermentation supernatants which were negligible or below the detection limit by the end of fermentation.
Fig. 7.
Line graphs depicting the changes in concentrations of different metabolites reported during the in vitro batch culture faecal fermentation of substrates (Blank, Inulin, Tritordeum breads prepared with baker's yeast (BYB) or sourdough (SDB)) at time points 0, 5, 10 and 24 h. Data are represented as a mean of 4 independent faecal fermentations. Vertical bars represent the standard deviation.
Effect of substrates on the teer measurements
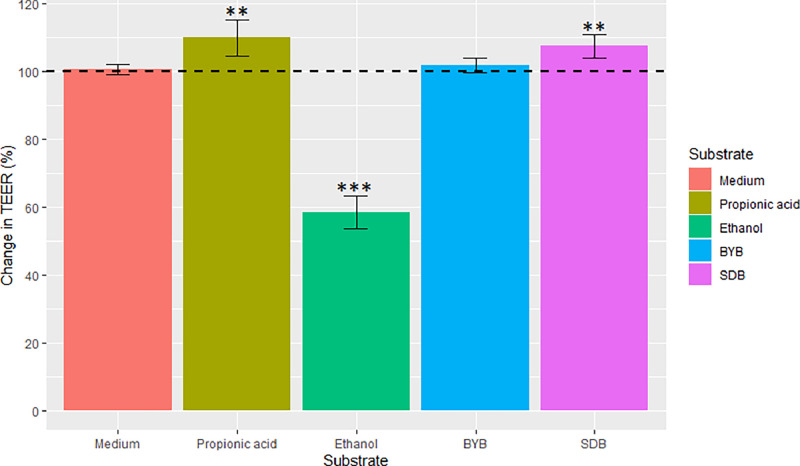
Changes in epithelial gut permeability were reported as percentage increase or decrease TEER measurement (Ω cm2) after 24 h incubation with test substrates (BYB and SDB faecal fermentation supernatants) and controls (Propionic acid and Ethanol) and these changes were then compared to control medium, the (Fig. 8). The exposure to propionic acid (positive control) increased the% change TEER significantly after 24 h by 9.19 ± 3.67 % (P < 0.01), whereas ethanol (negative control) significantly reduced it by 41.94 ± 4.93 % (P < 0.0001) when compared to the control medium. The exposure to fermentation supernatants from two types of Tritordeum breads showed a significant (P < 0.05) increase only with SDB (6.71 ± 3.36 %) while BYB exhibited a similar behaviour as the control medium (1.02 ± 0.71 %). These results indicate that Tritordeum sourdough can influence the epithelial barrier integrity in the gastrointestinal tract.
Fig. 8.
Changes in trans-epithelial electrical resistance (TEER) across the differentiated Caco-2 monolayers when exposed to different test substrates. The change in TEER is the percentage (%) change compared to the initial TEER (horizontal dotted line) for each substrate. The values are plotted as mean ± SD with experimental (n = 3) and technical replicates (n = 3). Statistical significance compared to control medium is indicated by ** P < 0.01 or *** P < 0.001.
Quantification of tight junction proteins of Caco-2 cells by western blot analysis
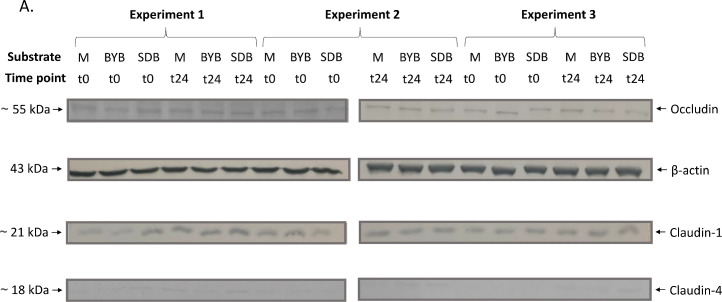
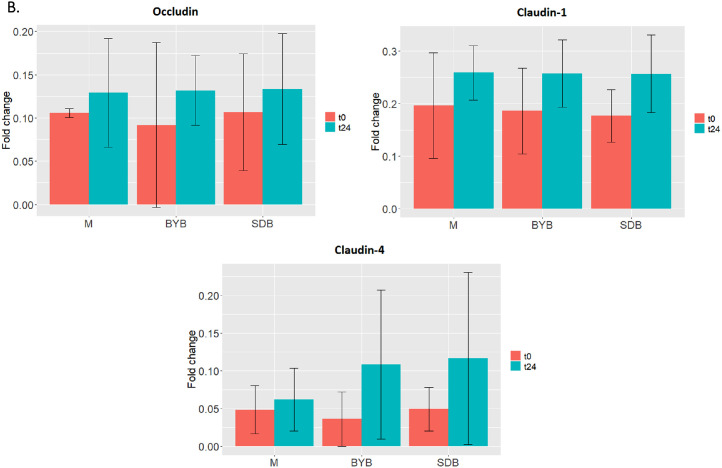
In order to investigate the intestinal permeability, tight junction proteins (TJP) of Caco-2 cells maintaining the integrity of the gut membrane were subjected to western blot analysis after the co-culture experiment with PBMCs. The Caco-2 TJPs (Claudin-1, Occludin and Claudin-4) showed an increase (P > 0.05), after 24 h with the control medium as well as the fermentation supernatants of test substrates (BYB and SDB) (Fig. 9). Although not statistically significant (P > 0.05), Claudin-4 was more expressed after incubation both with BYB and with SDB faecal fermentation supernatants as opposed to control medium.
Fig. 9.
(A) Western blot of tight junction proteins expressed by Caco-2 cells at baseline (t0) and after 24 h (t24) incubation with fermentation supernatants of BYB and SDB), or with control medium (M) (n = 3). Molecular weight (kDa) is indicative of each protein. (B) Proteins’ quantification through densitometric analysis with ImageJ software, using β-actin as reference protein. No significant differences were observed between different substrates and between different timepoints within each substrate.
Quantification of inflammatory and anti-inflammatory cytokines by Magpix®
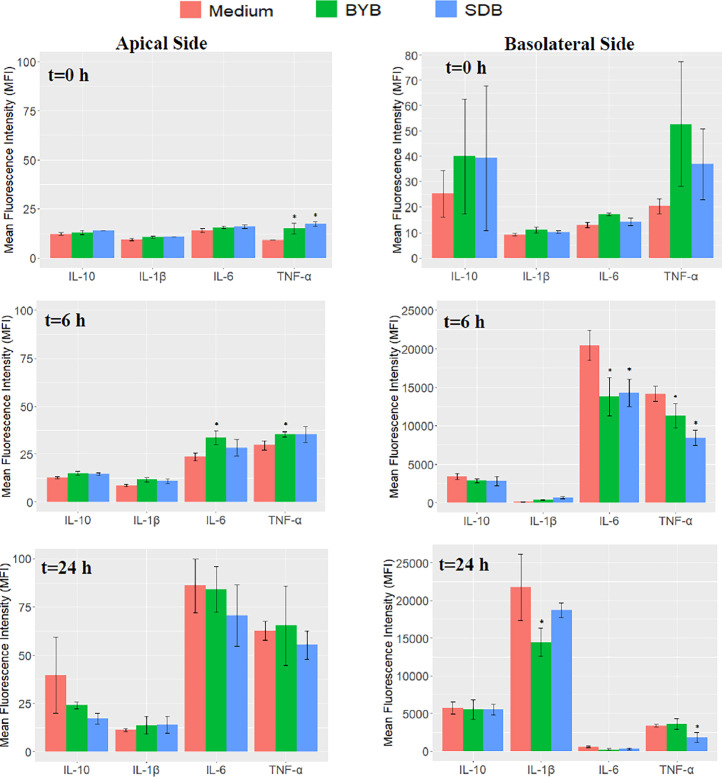
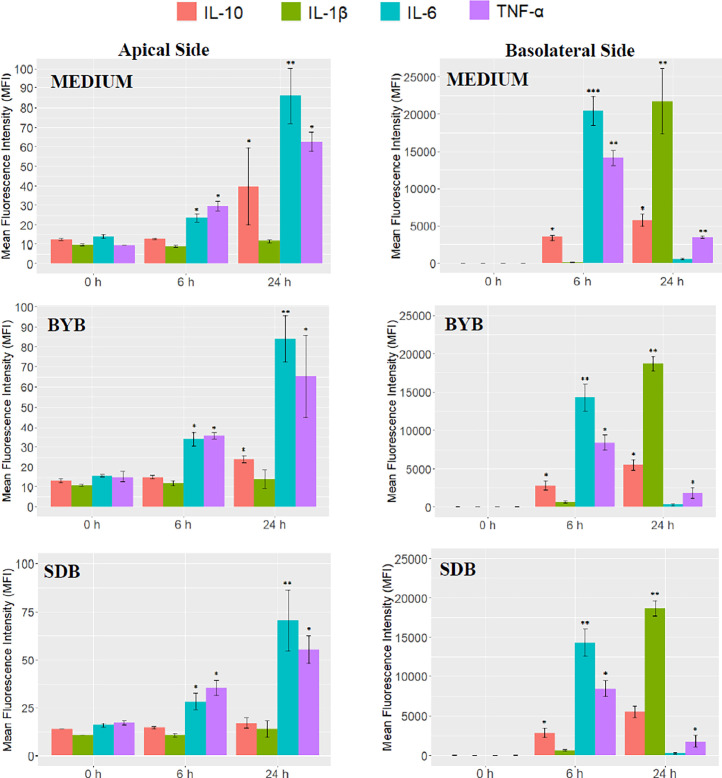
Secretion of cytokines during the Caco-2-PBMCs co-culture experiments (n = 3) after an initial inflammatory stimulus (i.e., incubation with bacterial LPS) followed by exposure to test substrates (BYB and SDB 24-h fermentation supernatants) or control medium, was quantified in the apical and basolateral supernatants at 0, 6 and 24 h (Fig. 10). Some variability was observed between test substrates and control medium at baseline (t0). Therefore, comparisons between test substrates and control medium were carried out by comparing the Delta values (i.e., the change in cytokine quantification between t0 and t6, between t6 and t24 and between t0 and t24), as well as by comparing the quantification of cytokines at each timepoint. The Luminex Mean Fluorescence Intensity (MFI) indicated an increase in secretion of inflammatory cytokines (IL-6 and TNF-α) in the apical as well as basolateral side at 6 h incubation with both test substrates and also with control medium (Fig. 11). However, while both BYB and SDB induced a significantly lower PBMCs inflammatory response compared to control medium, as demonstrated by increased IL-6 and TNF-α secretion in the basolateral side, the inflammatory response of Caco-2 was higher after incubation with BYB than SDB (i.e., significantly higher IL-6 and TNF-α compared to control medium). IL-6 and TNF-α secretion further increased in the apical side at 24 h incubation, whereas it decreased on the basolateral side. However, no significant differences in cytokine release were observed between the test substrates and the medium at t24 h. The decreased inflammatory cytokines in the basolateral chamber can be linked to their downregulation by the anti-inflammatory cytokine IL-10, which was elevated at t6 and at t24. Both BYB and SDB induced statistically lower inflammatory cytokines IL-6 and TNF-α on the basolateral after 6 h incubation when compared to the control medium. Inflammatory IL-1β showed a delayed secretion by PBMCs, since its concentration appeared low at t0 and t6, but it increased at t24 on the basolateral side. Similar to IL-6 and TNF-α, secretion of IL-1β was lower for SDB and BYB compared to that for control medium, and this difference was significant only for BYB. Moreover, the delta values in Table 3 indicated a significant increase (P < 0.05) in IL-6 and TNF-α between t0 and t6 for the control medium as compared to BYB and SDB substrates on the apical as well as basolateral side. Besides, the increase in IL-6 between t0 and t6 was significantly higher (P < 0.05) with BYB substrate as compared to that with SDB. To support these results obtained for cytokine quantification in cell culture supernatants, we also investigated gene expression of the same cytokines, as discussed in the next section and Fig. 12.
Fig. 10.
Secretion of IL-10, IL-1β, IL-6, and TNF-α by co-cultured Caco-2 (apical side) and PBMCs (basolateral side) under LPS stimulation at baseline (t = 0 h) and after 6 and 24 h of incubation with fermentation supernatants of test substrates (SDB or BYB, pool of 4 faecal fermentations) or with medium as control. Bar plots represent median values of Luminex Mean Fluorescent Intensity (MFI), and the error bars represent standard deviation. Note that the limits of y-axis are different for both apical and basolateral sides, and the initial time point (t = 0 h) of basolateral side has different y-axis limits than 6 and 24 h. Significant differences compared to control medium within each cytokine are indicated by the asterisk (*) (P < 0.05).
Fig. 11.
Secretion of IL-10, IL-1β, IL-6, and TNF-α by co-cultured Caco-2 (apical side) and PBMCs (basolateral side) under LPS stimulation at baseline (t = 0 h) and after 6 and 24 h of incubation with fermentation supernatants of test substrates (BYB or SDB, pool of 4 faecal fermentations) or with medium as control. Bar plots represent median values of Luminex Mean Fluorescent Intensity (MFI), and the error bars represent standard deviation. Significant differences compared to t = 0 h within each cytokine for each substrate are indicated by * P < 0.05) or ** P < 0.01.
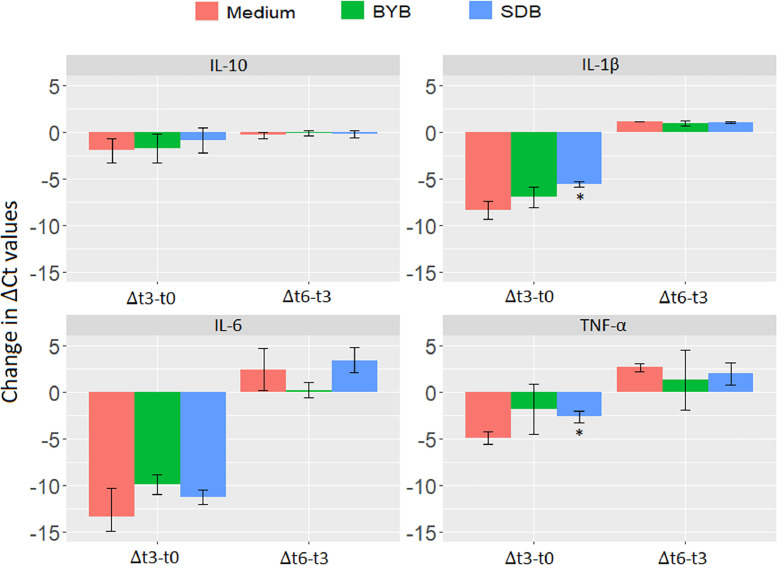
Fig. 12.
Gene expression analysis of cytokines IL-10, IL-1β, IL-6, and TNF-α in PBMCs during co-culture experiment with Caco-2 cells. The results are expressed as change in ΔCt values from baseline (t0) to 3 h (t3) and then up to 6 (t6) h incubation with fermentation supernatants of negative control Medium and test substrates (BYB or SDB, pool of 4 faecal fermentations) represented as Δt3-t0 and Δt6-t3, respectively. Error bars represent the standard deviation (n = 3). The change in ΔCt values was significantly reduced (P < 0.05) for IL-1β and TNF-α when incubated with SDB after 3 h of incubation as compared to control medium. Statistical significance compared to control Medium is indicated with an asterisk (*).
Quantification of inflammatory and anti-inflammatory cytokines by gene expression analysis
The effect of fermentation supernatants of Tritordeum breads (BYB and SDB) was also investigated by the quantification of gene expression of inflammatory (IL-1β, IL-6 and TNF-α) and anti-inflammatory (IL-10) cytokines when induced by LPS (Fig. 12). The ΔCt values were calculated by subtracting the mean Ct of reference genes (18S and GAPDH) from the mean Ct of test gene (or cytokine). These values were then used for determining the change in gene expression from 0 (t0) to 3 (t3) and 3 (t3) to 6 (t6) h of incubation with control medium and test substrates (BYB and SDB). The test substrates were compared with the control medium for determining the relative gene expression of all the cytokines. The inflammatory cytokines (IL-1β, IL-6 and TNF-α) exhibited reduction in ΔCt after 3 h of incubation with control medium and both the test substrates. The ΔCt values for IL-1β and TNF-α were significantly reduced (P < 0.05) for SDB when compared to control medium after 3 h of incubation as observed in Δt3−t0 (Fig. 12). On the contrary, ΔCt values for these inflammatory cytokines were slightly increased from 3 to 6 h of incubation with control medium as well as test substrates, except no change in IL-6 with BYB substrate. However, no significant changes were observed when test substrates were compared to control medium. On the contrary, the ΔCt values of anti-inflammatory cytokine, IL-10, showed minor decrease from 0 to 3 h and no change from 3 to 6 h for control medium or test substrates (BYB and SDB).
Correlation of faecal fermentation metabolites with gut microbiota, TEER measurements and inflammatory and anti-inflammatory cytokines
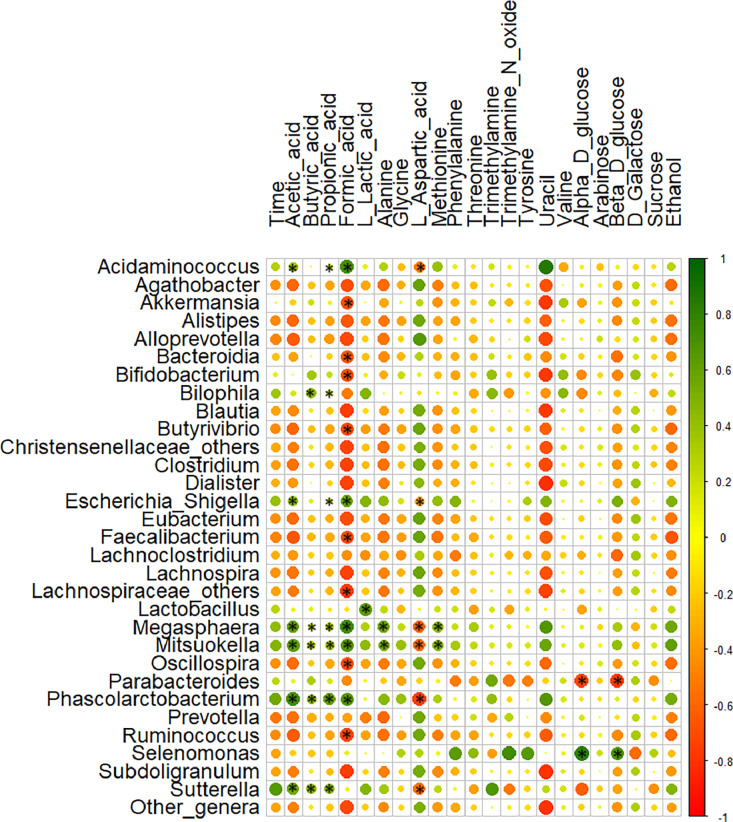
The role of intestinal gut microbiota in influencing the metabolite concentrations during faecal fermentation is well established. In this context, Spearman's correlation matrix between the relative abundances of gut microbiota genera and the metabolites identified over time during the faecal fermentation with different substrates was generated (Fig. 13). SCFAs comprising of acetic, butyric and propionic acids, showed a positive correlation (P < 0.05) with Acidaminococcus, Bilophila, Escherichia-Shigella, Megasphaera, Mitsuokella, Phascolarctobacterium and Sutterella. Formic acid exhibited a very strong negative correlation with most of the identified genera except Acidaminococcus (P = 0.002), Escherichia-Shigella (P = 0.021), Megasphaera (P = 0.001), Mitsuokella (P = 0.006) and Phascolarctobacterium (P = 0.007). Lactic acid was positively correlated with Lactobacillus (P = 0.015). Alanine and methionine were positively correlated with Megasphaera (P = 0.029) and Mitsuokella (P = 0.036). l-aspartic acid showed a negative correlation with Acidaminococcus (P = 0.041), Escherichia-Shigella (n.s., P = 0.153), Megasphaera (P = 0.018), Mitsuokella (P = 0.021), Phascolarctobacterium (P = 0.002) and Sutterella (P = 0.027). Both α- and β-d-glucose showed a positive correlation with Selenomonas (P = 0.031) and a negative correlation with Parabacteroides (P = 0.025).
Fig. 13.
Spearman correlation (n = 4) between the relative abundances of genera of gut microbiota and metabolites produced during the faecal fermentation with different substrates (Inulin, BYB and SDB). The colour scale bar indicates a positive (green) or negative (red) correlation (*P < 0.05).
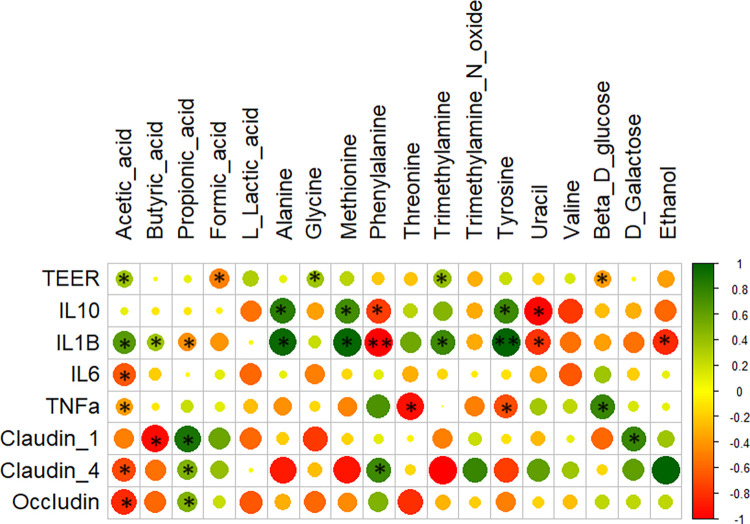
Additionally, the Spearman's correlation matrix was also generated between the metabolites and factors influencing the intestinal membrane permeability and inflammation as measured during the in vitro faecal fermentation of BYB and SDB (Fig. 14). Acetic acid and butyric acid showed a positive correlation with IL-1β (P = 0.041) and TEER measurements and a negative correlation with IL-6 (P = 0.038), TNF-α (P = 0.045) and TJP (Claudin-1, Claudin-4, and Occludin). On the other hand, propionic acid is positively correlated with the TJP (P < 0.05). The amino acids, namely, alanine, methionine and tyrosine showed a strong positive correlation (P < 0.05) with IL-1β and anti-inflammatory cytokine IL-10 whereas phenylalanine was negatively correlated (P < 0.05) with these cytokines. Threonine and tyrosine were negatively correlated with the inflammatory cytokine, TNF-α, while β-d-glucose is positively correlated with this cytokine (P < 0.05). Trimethylamine was positively correlated with the TEER, IL-10 and IL-1β (P < 0.05) whereas its derivative, trimethylamine-N-oxide, was negatively correlated (P > 0.05). As observed during the TEER measurements, ethanol is negatively correlated with the intestinal permeability (TEER) and the anti-inflammatory cytokines (P < 0.05). It is difficult to precisely define the strong correlations between these faecal fermentation metabolites and biomarkers of epithelial permeability and inflammation due to high inter-individual variation of the donors. Further studies using animal models should be conducted to reduce such variations for easy traceability of the impact of sourdough.
Fig. 14.
Spearman correlation (n = 3) of the in vitro faecal fermentation metabolites with TEER measurements, anti-inflammatory cytokine IL-10, inflammatory cytokines IL-1β, IL-6, and TNF-α and tight junction proteins (Claudin-1, Claudin-4 and Occludin), as measured during the co-culture experiment of Caco-2/PBMCs on exposure to faecal fermentation supernatants with test substrates (BYB and SDB). The colour scale bar indicates a positive (green) or negative (red) correlation (*P < 0.05, **P < 0.01).
Discussion
The impact of fermented foods, such as sourdough, on the gut microbiota and their influence on the gastrointestinal health has been recently reviewed (Dimidi et al., 2019). Improvement in the nutritional value by hydrolysis of complex dietary fibres, reduction in fat rancidity, and enhancement of starch and protein digestibility along with vitamins and minerals bioavailability are some of the beneficial outcomes of sourdough fermentation (Fernández-Peláez et al., 2020). Besides, the prebiotic effect of sourdough by modulating the gut microbiota leading to the production of fermentation-derived bioactive peptides, biogenic amines and other health-promoting metabolites has also been discussed previously (Dimidi et al., 2019). In this prospect, we studied the digestibility and immunomodulatory effects of Tritordeum breads prepared by the addition of Tritordeum sourdough (SDB) vs. commercial baker's yeast (BYB).
Initially, the Tritordeum breads were subjected to in vitro digestion (Minekus et al., 2014) followed by the batch culture faecal fermentation for 24 h to analyse the changes in gut microbiota and microbial metabolites. Based on the α-diversity indices, gut microbiota composition showed a marked decrease in microbial diversity during the faecal fermentation of positive control (Inulin) and test substrates (BYB and SDB) as compared to negative control (Blank) at 5, 10 and 24 h (Fig. 1). This shift in diversity indicates the substrate-dependent fermentation allowing the growth of selective gut microorganisms. The β-diversity analyses demonstrated differences in the gut microbiota at the donor level indicating variability due to inter-individual differences. At the phylum level, gut microbiota comprised the highest relative abundance of Firmicutes followed by Proteobacteria, Bacteroidota, Actinobacteriota, Verrucomicrobiota and Desulfobacterota, throughout the faecal fermentation for all the substrates and the blank. A decrease in Bacteriodota was observed up to 8 % (Blank) and 2 % (Inulin, BYB and SDB) after 24 h which can be attributed to the limitations of in vitro faecal fermentation model (Payne et al., 2012). At the genus level, the relative abundance of Escherichia-Shigella and Megasphaera dominated and increased significantly (P < 0.05) with the controls (Inulin and Blank) as well as the test substrates (BYB and SDB) at 5,10, and 24 h of faecal fermentation. The microorganisms belonging to these genera can utilize carbohydrates as their energy source, which were readily available with Tritordeum breads (SDB and BYB) as well as inulin. Besides, Escherichia-Shigella is usually outnumbered by other anaerobic bacteria in the gut, however, this increase during fermentation can be attributed to poor anaerobic conditions which allowed the multiplication of these facultative anaerobic bacteria (Russell and Jarvis, 2001). Furthermore, the abundance of Mitsuokella and Phascolarctobacterium was also significantly elevated after 24 h of faecal fermentation with BYB, SDB, and inulin substrates without any significant differences amongst them. In contrast, these genera did not show any significant changes with the Blank. The genus Bacteroides was significantly decreased with BYB, SDB and inulin substrates at 24 h whereas no change was observed with the Blank. On the other hand, the relative abundances of Bifidobacterium increased with BYB, SDB, and inulin fermentation which was slightly higher (P > 0.05) with inulin, whereas this genus was significantly reduced with the negative control (Blank). Many genera disappeared after 24 h fermentation with BYB, SDB and inulin substrates which included Butyrivibrio, Christensenellaceae, Clostridium, Faecalibacterium, Oscillospiraceae, Lachnospiraceae and Ruminococcus. These genera did not show any significant changes in the fermentation vessel of Blank. The grains of Tritordeum contain 43 % higher dietary fibres than durum wheat (Suchowilska et al., 2021) which explains the increase in relative abundances of Bifidobacterium, Megasphaera (Wu et al., 2021), Mitsuokella (Healey et al., 2017) and Phascolarctobacterium (Wu et al., 2021) in Tritordeum breads (BYB and SDB). Some studies reported the increase in Mitsuokella as a result of enteric dysfunction in children (Ordiz et al., 2017), obesity (Palmas et al., 2021), or in faeces of marathon runners (Zhao et al., 2018). Moreover, Phascolarctobacterium can utilize succinate secreted by Bacteroides (Ikeyama et al., 2020) limiting the availability to Clostridium which essentially requires succinate for its growth (Nagao-Kitamoto et al., 2020). Additionally, increased concentrations of succinate have also been associated with intestinal inflammation (Connors et al., 2018). Therefore, the increased abundance of Phascolarctobacterium can help in regulating the intestinal inflammation.
Furthermore, changes in microbial metabolites examined by targeted metabolite analysis (Fig. 7) revealed the increased production of total SCFAs with BYB (270.71 ± 46.71 mM), SDB (467.96 ± 43.18 mM) and inulin (485 ± 73.52 mM) as opposed to the negative control (121.74 ± 73.52 mM). Acetic and butyric acid concentrations were significantly higher with SDB as compared to BYB and inulin, which did not differ in the concentration of these SCFAs. Propionic acid, on the other hand, was highest with inulin as compared to SDB and BYB. A plausible explanation for significant increase in SCFAs is the saccharolytic fermentation by gut microbiota (Rowland et al., 2018). Furthermore, a recent study by (Smith et al., 2020) reported high-butyrate production during in vitro faecal fermentation of sourdough bread as reported in our results obtained with sourdough bread (SDB). While both the Tritordeum breads (BYB and SDB) were prepared with the same type of flour, the differences lie in the addition of baker's yeast or sourdough and hence differ in the bacterial composition of breads during fermentation. Sourdough microbiota was dominated by lactic acid bacteria which increased the total dietary fibre content in SDB (data under publication). Since the sourdough lactic acid bacteria can secrete exopolysaccharides (EPS) (Gänzle and Gobbetti, 2013), these EPS can be made available to the gut microbiota during digestion leading to the increased production of SCFAs as opposed to baker's yeast bread. Other organic acids, such as lactic and formic acids, are also considered as SCFAs which showed a significant increase up to 5 and 10 h, respectively, followed by gradual decrease during fermentation of Inulin, BYB and SDB (Fig. 7). Formic acid production is not well-studied in the human gut; however, studies have demonstrated its higher concentrations during high-fibre intake in pigs (Tao et al., 2019). The decreasing lactic acid concentrations can be associated with the increasing abundance of Megasphaera, which converts lactic acid to butyric and propionic acids (Shetty et al., 2013) and considered as a potential microorganism to control ruminal acidosis by degradation of lactic acid (Chen et al., 2019; Henning et al., 2010). Studies have indicated that cross-feeding of lactate to gut microbiota often leads to an increased production of butyrate due to the preference of lactate as a carbon source by some gut microbes (Duncan et al., 2004; Muñoz-Tamayo et al., 2011). Moreover, researchers have reported the anti-inflammatory potential of butyrate in intestinal homeostasis (Liu et al., 2018) and SCFAs (acetate, butyrate and propionate) were also reported to be therapeutic for inflammatory disorders, including IBS (Tedelind et al., 2007). The increasing concentration of ethanol was observed with all the substrates indicating the continuous fermentation of substrates and metabolism of gut microbiota (Fodje et al., 2009; Han et al., 2013). Sugars were also degraded and/or utilized by the intestinal microbiota during faecal fermentation with all the substrates as demonstrated in previous studies (Kaoutari et al., 2013). Additionally, the changes in concentration of amino acids during in vitro faecal fermentation of all substrates (inulin, BYB and SDB) demonstrated the proteolytic action of gut microbiota and their ability to utilize these amino acids for the production of SCFAs (Lin et al., 2017; Neis et al., 2015). Spearman's correlation matrix demonstrated a strong influence or correlation of the gut microbiota mainly on SCFAs, formic acid, alanine, methionine, L-aspartic acid and glucose. Acidaminococcus, Bilophila, Escherichia-Shigella, Megasphaera, Mitsuokella, Phascalarctobacterium and Sutterella were found to be positively correlated (P < 0.05) with acetic, butyric, and propionic acids as demonstrated previously (Han et al., 2013) whereas negatively correlated with formic acid. Among amino acids, alanine and methionine were positively correlated (P < 0.05) with Megasphaera and Mitsuokella whereas l-aspartic acid showed a negative correlation (P < 0.05) with Acidaminococcus, Megasphaera, Mitsuokella, Phascolarctobacterium and Sutterella. Both α- and β-D-glucose showed a significantly positive and negative correlation with Selenomonas and Parabacteroides, respectively. Previous research studies have indicated similar correlation of metabolites with Acidaminococcus, Escherichia-Shigella and Megasphaera genera (Lin et al., 2017).
Furthermore, the impact of Tritordeum breads after fermentation was examined on the gut health. The intestinal epithelium acts as a barrier between invading pathogens or inflammatory molecules to prevent inflammation or any other systemic diseases. Moreover, the integrity of epithelial barrier is also maintained by the TJPs that are selectively permeable to nutrients. The function of intestinal epithelial barrier can be compromised leading to a condition called leaky gut. This is a common clinical diagnosis in individuals suffering from CD, IBS or other diseases leading to gut dysbiosis (Odenwald and Turner, 2013; Teshima and Meddings, 2008). The intestinal permeability can be assessed by Transepithelial/transendothelial electrical resistance (TEER) to determine the integrity of tight junctions in the intestinal epithelium by measuring the ohmic resistance across the epithelium monolayer (Srinivasan et al., 2015). Hence, we determined the TEER measurements of intestinal Caco-2 epithelial cells on exposure to fermentation supernatants of BYB and SDB collected after 24 h of faecal fermentation with these substrates. The permeability of intestinal epithelium in response to fermentation supernatants demonstrated an increase (P < 0.05) in the TEER measurements with SDB, which was slightly lower than the positive control (propionic acid). BYB showed same results as the control medium while ethanol reduced the TEER measurements by 41.94 ± 4.93 %. Additionally, the TJP of Caco-2 cells were subjected to western blot analysis for Claudin-1, Claudin-4 and Occludin. The western blot results indicated no differences between the control medium, BYB and SDB, although some increase in Claudin-4 was observed for BYB and SDB when compared to control medium.
Furthermore, the inflammatory cytokines studied using in vitro co-culture model of Caco-2 cells and PBMCs demonstrated an increase in IL-6 and TNF-α on the apical and basolateral sides after 6 h of incubation which was lower in BYB and SDB than control medium only on the basolateral side (P < 0.05). These inflammatory responses were regulated by the increase in IL-10 at 24 h with lower MFI values observed only for IL-1β with BYB supernatant as compared to control medium and SDB. However, the quantitative gene expression analysis of these cytokines indicated lower expression of IL-1β, IL-6 and TNF-α after 3 and 6 h of incubation with both BYB and SDB substrates. The decrease in expression of IL-1β was significantly higher with SDB as compared to BYB. While the potential of sourdough in reducing the inflammatory response has been previously established by decreased synthesis of IFN-γ (Calasso et al., 2012) or decreased release of monocyte chemoattractant protein-1 (MCP-1) and TNF-α (Huang et al., 2020), its functional role in intestinal permeability and inflammation still needs to be elaborated. Our results demonstrated the early secretion of inflammatory cytokines, IL-6 and TNF-α, which was downregulated by the increase in anti-inflammatory cytokine (IL-10) with no statistically significant differences between SDB and BYB. However, on the contrary, significantly increased TEER measurements were observed with SDB supernatants as compared to those with BYB supernatant. Previously, the generation of antibodies against baker's yeast have been demonstrated in response to Crohn's disease or Inflammatory Bowel Disease (IBD) (Main et al., 1988; Mitsuyama et al., 2016). While no significant differences were detected between SDB and BYB in the secretion of cytokines, significantly higher TEER measurements for SDB might suggest an increased inflammatory response with BYB than with SDB.
The metabolites secreted by gut microbiota can also have an impact on the epithelial permeability and inflammation. Therefore, we generated a Spearman's correlation matrix between metabolites and the markers of intestinal barrier (TEER and TJP) and inflammation (pro- and anti-inflammatory cytokines). A positive correlation was found between acetic acid and TEER measurements while propionic acid was positively correlated with TJP (Claudin-1, Claudin-4 and Occludin). Acetic acid was negatively correlated with inflammatory cytokines IL-6 and TNF-α. While many other correlations were established, it is difficult to support our results with this data due to high inter-individual variability.
Conclusively, our findings suggest that Tritordeum breads prepared with the addition of sourdough or baker's yeast did not show significant differences in the gut microbiota composition during in vitro faecal fermentation with these breads for 24 h. However, the differences were observed in the metabolic output, in particular, increased concentrations of SCFAs with SDB substrate as compared to BYB. Moreover, the epithelial membrane integrity of intestine was improved with the downregulation of inflammatory response. These results indicate that sourdough has the potential to increase the functional value of baked goods by improving the digestibility and human gut health. However, our findings should be further confirmed in laboratory animals and eventually in human studies, to provide better insights into the mechanisms of human gut physiology.
Funding
This work was supported by CRC 2018 PRO4HEALTHFOOD funding. Support was also provided by the project SYSTEMIC “An integrated approach to the challenge of sustainable food systems: adaptive and mitigatory strategies to address climate change and malnutrition”, Knowledge hub on Nutrition and Food Security, which has received funding from national research funding parties in Belgium (FWO), France (INRA), Germany (BLE), Italy (MIPAAF), Latvia (IZM), Norway (RCN), Portugal (FCT), and Spain (AEI) in a joint action of JPI HDHL, JPI-OCEANS and FACCE-JPI launched in 2019 under the ERA-NET ERA-HDHL (n 696295). This study has also received funding from the Autonomous Province of Trento, with EU co-financing (FRUITOMICS, FESR 2014–2020 Program of the Autonomous Province of Trento, Italy).
CRediT authorship contribution statement
Kashika Arora: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. Giulia Gaudioso: Data curation, Investigation, Methodology. Pavel Solovyev: Data curation, Formal analysis, Methodology, Funding acquisition, Writing – review & editing. Kieran Tuohy: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. Raffaella Di Cagno: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. Marco Gobbetti: Funding acquisition, Resources. Francesca Fava: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2023.100214.
Contributor Information
Raffaella Di Cagno, Email: raffaella.dicagno@unibz.it.
Francesca Fava, Email: francesca.fava1978@gmail.com.
Appendix. Supplementary materials
Data availability
No data was used for the research described in the article.
References
- Abbondio M., Palomba A., Tanca A., Fraumene C., Pagnozzi D., Serra M., Marongiu F., Laconi E., Uzzau S. Fecal metaproteomic analysis reveals unique changes of the gut microbiome functions after consumption of sourdough Carasau bread. Front. Microbiol. 2019;10:1733. doi: 10.3389/fmicb.2019.01733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora K., Carafa I., Fava F., Tuohy K.M., Nikoloudaki O., Gobbetti M., Cagno R.D. Sourdough performances of the golden cereal Tritordeum: dynamics of microbial ecology, biochemical and nutritional features. Int. J. Food Microbiol. 2022;374 doi: 10.1016/j.ijfoodmicro.2022.109725. [DOI] [PubMed] [Google Scholar]
- Artursson P., Palm K., Luthman K. Caco-2 monolayers in experimental and theoretical predictions of drug transport. Adv. Drug Deliv. Rev. 2001;46:27–43. doi: 10.1016/s0169-409x(00)00128-9. [DOI] [PubMed] [Google Scholar]
- Bianchi M.G., Chiu M., Taurino G., Brighenti F., Del Rio D., Mena P., Bussolati O. Catechin and procyanidin B2 modulate the expression of tight junction proteins but do not protect from inflammation-induced changes in permeability in human intestinal cell monolayers. Nutrients. 2019;11:E2271. doi: 10.3390/nu11102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., Bai Y., Bisanz J.E., Bittinger K., Brejnrod A., Brislawn C.J., Brown C.T., Callahan B.J., Caraballo-Rodríguez A.M., Chase J., Caporaso J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsci G., Barbieri S., Guardamagna I., Lonati L., Ottolenghi A., Ivaldi G.B., Liotta M., Tabarelli de Fatis P., Baiocco G., Savio M. Immunophenotyping reveals no significant perturbation to pbmc subsets when co-cultured with colorectal adenocarcinoma Caco-2 cells exposed to X-rays. Front. Immunol. 2020;11:1077. doi: 10.3389/fimmu.2020.01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calasso M., Vincentini O., Valitutti F., Felli C., Gobbetti M., Di Cagno R. The sourdough fermentation may enhance the recovery from intestinal inflammation of coeliac patients at the early stage of the gluten-free diet. Eur. J. Nutr. 2012;51:507–512. doi: 10.1007/s00394-012-0303-y. [DOI] [PubMed] [Google Scholar]
- Cani P.D. Microbiota and metabolites in metabolic diseases. Nat. Rev. Endocrinol. 2019;15:69–70. doi: 10.1038/s41574-018-0143-9. [DOI] [PubMed] [Google Scholar]
- Cani P.D., Bibiloni R., Knauf C., Waget A., Neyrinck A.M., Delzenne N.M., Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- Chen L., Shen Y., Wang C., Ding L., Zhao F., Wang M., Fu J., Wang H. Megasphaera elsdenii lactate degradation pattern shifts in rumen acidosis models. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00162. https://www.frontiersin.org/article/10.3389/fmicb.2019.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors J., Dawe N., Van Limbergen J. The role of succinate in the regulation of intestinal inflammation. Nutrients. 2018;11:25. doi: 10.3390/nu11010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costabile A., Santarelli S., Claus S.P., Sanderson J., Hudspith B.N., Brostoff J., Ward J.L., Lovegrove A., Shewry P.R., Jones H.E., Whitley A.M., Gibson G.R. Effect of breadmaking process on in vitro gut microbiota parameters in irritable bowel syndrome. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0111225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ros A., Polo A., Rizzello C.G., Acin-Albiac M., Montemurro M., Di Cagno R., Gobbetti M. Feeding with sustainably sourdough bread has the potential to promote the healthy microbiota metabolism at the colon level. Microbiol. Spectr. 2021;9 doi: 10.1128/Spectrum.00494-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimidi E., Cox S.R., Rossi M., Whelan K. Fermented foods: definitions and characteristics, impact on the gut microbiota and effects on gastrointestinal health and disease. Nutrients. 2019;11:1806. doi: 10.3390/nu11081806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S.H., Louis P., Flint H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Peláez J., Paesani C., Gómez M. Sourdough technology as a tool for the development of healthier grain-based products: an update. Agronomy. 2020;10:1962. doi: 10.3390/agronomy10121962. [DOI] [Google Scholar]
- Fodje A.M.L., Chang P.R., Leterme P. In vitro bile acid binding and short-chain fatty acid profile of flax fiber and ethanol co-products. J. Med. Food. 2009;12:1065–1073. doi: 10.1089/jmf.2008.0242. [DOI] [PubMed] [Google Scholar]
- Gallo-Oller G., Ordoñez R., Dotor J. A new background subtraction method for western blot densitometry band quantification through image analysis software. J. Immunol. Methods. 2018;457:1–5. doi: 10.1016/j.jim.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Gänzle M., Gobbetti M. In: Handbook on Sourdough Biotechnology. Gobbetti M., Gänzle M., editors. Springer US; 2013. Physiology and biochemistry of lactic acid bacteria; pp. 183–216. [DOI] [Google Scholar]
- Gasaly N., de Vos P., Hermoso M.A. Impact of bacterial metabolites on gut barrier function and host immunity: a focus on bacterial metabolism and its relevance for intestinal inflammation. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.658354. https://www.frontiersin.org/article/10.3389/fimmu.2021.658354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K.J., Pitman W.D., Kim M., Day D.F., Alison M.W., McCormick M.E., Aita G. Ethanol production potential of sweet sorghum assessed using forage fiber analysis procedures. Glob. Change Biol. Bioenergy. 2013;5:358–366. doi: 10.1111/j.1757-1707.2012.01203.x. [DOI] [Google Scholar]
- Healey G., Murphy R., Butts C., Brough L., Rosendale D., Blatchford P., Stoklosinski H., Coad J. Variability in gut microbiota response to an inulin-type fructan prebiotic within an in vitro three-stage continuous colonic model system. Bioact. Carbohydr. Diet. Fibre. 2017;11:26–37. doi: 10.1016/j.bcdf.2017.07.001. [DOI] [Google Scholar]
- Henning P.H., Horn C.H., Steyn D.G., Meissner H.H., Hagg F.M. The potential of Megasphaera elsdenii isolates to control ruminal acidosis. Anim. Feed Sci. Technol. 2010;157:13–19. doi: 10.1016/j.anifeedsci.2009.12.011. [DOI] [Google Scholar]
- Hong R.S., Hwang K., Cho H.E., Lee H.J., Hong J.T., Moon D.C. Survey of ERETIC2 NMR for quantification. J. Korean Magn. Reason. Soc. 2013;17:98–104. doi: 10.6564/JKMRS.2013.17.2.098. [DOI] [Google Scholar]
- Huang X., Schuppan D., Rojas Tovar L.E., Zevallos V.F., Loponen J., Gänzle M. Sourdough fermentation degrades wheat alpha-amylase/trypsin inhibitor (ATI) and reduces pro-inflammatory activity. Foods. 2020;9:943. doi: 10.3390/foods9070943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeyama N., Murakami T., Toyoda A., Mori H., Iino T., Ohkuma M., Sakamoto M. Vol. 9. MicrobiologyOpen; 2020. p. e1111. (Microbial Interaction Between the Succinate-Utilizing Bacterium Phascolarctobacterium faecium and the Gut Commensal Bacteroides thetaiotaomicron). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpfer A.A.M., Urbán P., Gioria S., Kanase N., Stone V., Kinsner-Ovaskainen A. Development of an in vitro co-culture model to mimic the human intestine in healthy and diseased state. Toxicol. In Vitro. 2017;45:31–43. doi: 10.1016/j.tiv.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaoutari A.E., Armougom F., Gordon J.I., Raoult D., Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzelmann K., Mall M. Electrolyte transport in the mammalian colon: mechanisms and implications for disease. Physiol. Rev. 2002;82:245–289. doi: 10.1152/physrev.00026.2001. [DOI] [PubMed] [Google Scholar]
- Lin R., Liu W., Piao M., Zhu H. A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids. 2017;49:2083–2090. doi: 10.1007/s00726-017-2493-3. [DOI] [PubMed] [Google Scholar]
- Liu H., Wang J., He T., Becker S., Zhang G., Li D., Ma X. Butyrate: a double-edged sword for health? Adv. Nutr. 2018;9:21–29. doi: 10.1093/advances/nmx009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main J., McKenzie H., Yeaman G.R., Kerr M.A., Robson D., Pennington C.R., Parratt D. Antibody to Saccharomyces cerevisiae (bakers’ yeast) in Crohn's disease. Br. Med. J. 1988;297:1105–1106. doi: 10.1136/bmj.297.6656.1105. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1834894/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minekus M., Alminger M., Alvito P., Ballance S., Bohn T., Bourlieu C., Carrière F., Boutrou R., Corredig M., Dupont D., Dufour C., Egger L., Golding M., Karakaya S., Kirkhus B., Le Feunteun S., Lesmes U., Macierzanka A., Mackie A., Marze S., McClements D.J., Ménard O., Recio I., Santos C.N., Singh R.P., Vegarud G.E., Wickham M.S.J., Weitschies W., Brodkorb A. A standardised static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014;5:1113–1124. doi: 10.1039/c3fo60702j. [DOI] [PubMed] [Google Scholar]
- Mitsuyama K., Niwa M., Takedatsu H., Yamasaki H., Kuwaki K., Yoshioka S., Yamauchi R., Fukunaga S., Torimura T. Antibody markers in the diagnosis of inflammatory bowel disease. World J. Gastroenterol. 2016;22:1304–1310. doi: 10.3748/wjg.v22.i3.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Tamayo R., Laroche B., Walter E., Doré J., Duncan S.H., Flint H.J., Leclerc M. Kinetic modelling of lactate utilization and butyrate production by key human colonic bacterial species. FEMS Microbiol. Ecol. 2011;76:615–624. doi: 10.1111/j.1574-6941.2011.01085.x. [DOI] [PubMed] [Google Scholar]
- Nagao-Kitamoto H., Leslie J.L., Kitamoto S., Jin C., Thomsson K.A., Gillilland M.G., Kuffa P., Goto Y., Jenq R.R., Ishii C., Hirayama A., Seekatz A.M., Martens E.C., Eaton K.A., Kao J.Y., Fukuda S., Higgins P.D.R., Karlsson N.G., Young V.B., Kamada N. Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat. Med. 2020;26:608–617. doi: 10.1038/s41591-020-0764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neis E.P.J.G., Dejong C.H.C., Rensen S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients. 2015;7:2930–2946. doi: 10.3390/nu7042930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald M.A., Turner J.R. Intestinal permeability defects: is it time to treat? Clin. Gastroenterol. Hepatol. 2013;11:1075–1083. doi: 10.1016/j.cgh.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordiz M.I., Stephenson K., Agapova S., Wylie K.M., Maleta K., Martin J., Trehan I., Tarr P.I., Manary M.J. Environmental enteric dysfunction and the fecal microbiota in Malawian children. Am. J. Trop. Med. Hyg. 2017;96:473–476. doi: 10.4269/ajtmh.16-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmas V., Pisanu S., Madau V., Casula E., Deledda A., Cusano R., Uva P., Vascellari S., Loviselli A., Manzin A., Velluzzi F. Gut microbiota markers associated with obesity and overweight in Italian adults. Sci. Rep. 2021;11:5532. doi: 10.1038/s41598-021-84928-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne A.N., Zihler A., Chassard C., Lacroix C. Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol. 2012;30:17–25. doi: 10.1016/j.tibtech.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzello C.G., Portincasa P., Montemurro M., Di Palo D.M., Lorusso M.P., De Angelis M., Bonfrate L., Genot B., Gobbetti M. Sourdough fermented breads are more digestible than those started with baker's yeast alone: an in vivo challenge dissecting distinct gastrointestinal responses. Nutrients. 2019;11:2954. doi: 10.3390/nu11122954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland I., Gibson G., Heinken A., Scott K., Swann J., Thiele I., Tuohy K. Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 2018;57:1–24. doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumney C.J., Rowland I.R. In vivo and in vitro models of the human colonic flora. Crit. Rev. Food Sci. Nutr. 1992;31:299–331. doi: 10.1080/10408399209527575. [DOI] [PubMed] [Google Scholar]
- Russell J.B., Jarvis G.N. Practical mechanisms for interrupting the oral-fecal lifecycle of Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001;3:265–272. [PubMed] [Google Scholar]
- Shah S., Brown P.D.S., Mayengbam S., Gänzle M.G., Wang W., Mu C., Lettrari S., Bertagnolli C., Shearer J. Metabolic and gut microbiota responses to sourdough pasta consumption in overweight and obese adults. Front. Nutr. 2020;7 doi: 10.3389/fnut.2020.615003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G., Wu J., Ye B.C., Qi N. Gut microbiota-derived metabolites in the development of diseases. Can. J. Infect. Dis. Med. Microbiol. 2021 doi: 10.1155/2021/6658674. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty S.A., Marathe N.P., Lanjekar V., Ranade D., Shouche Y.S. Comparative genome analysis of Megasphaera sp. reveals niche specialization and its potential role in the human gut. PLoS ONE. 2013;8:e79353. doi: 10.1371/journal.pone.0079353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C., Van Haute M.J., Rose D.J. Processing has differential effects on microbiota-accessible carbohydrates in whole grains during in vitro fermentation. Appl. Environ. Microbiol. 2020;86:e01705–e01720. doi: 10.1128/AEM.01705-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan B., Kolli A.R., Esch M.B., Abaci H.E., Shuler M.L., Hickman J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015;20:107–126. doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchowilska E., Wiwart M., Przybylska-Balcerek A., Stuper-Szablewska K. The profile of bioactive compounds in the grain of various x Tritordeum genotypes. J. Cereal Sci. 2021;102 doi: 10.1016/j.jcs.2021.103352. [DOI] [Google Scholar]
- Tao S., Bai Y., Zhou X., Zhao J., Yang H., Zhang S., Wang J. In vitro fermentation characteristics for different ratios of soluble to insoluble dietary fiber by fresh fecal microbiota from growing pigs. ACS Omega. 2019;4:15158–15167. doi: 10.1021/acsomega.9b01849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedelind S., Westberg F., Kjerrulf M., Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J. Gastroenterol. 2007;13:2826–2832. doi: 10.3748/wjg.v13.i20.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima C.W., Meddings J.B. The measurement and clinical significance of intestinal permeability. Curr. Gastroenterol. Rep. 2008;10:443–449. doi: 10.1007/s11894-008-0083-y. [DOI] [PubMed] [Google Scholar]
- Vinolo M.A.R., Rodrigues H.G., Nachbar R.T., Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3:858–876. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D.T., Fu Y., Guo H., Yuan Q., Nie X.R., Wang S.P., Gan R.Y. In vitro simulated digestion and fecal fermentation of polysaccharides from loquat leaves: dynamic changes in physicochemical properties and impacts on human gut microbiota. Int. J. Biol. Macromol. 2021;168:733–742. doi: 10.1016/j.ijbiomac.2020.11.130. [DOI] [PubMed] [Google Scholar]
- Yang J., Rose D.J. The impact of long-term dietary pattern of fecal donor on in vitro fecal fermentation properties of inulin. Food Funct. 2016;7:1805–1813. doi: 10.1039/c5fo00987a. [DOI] [PubMed] [Google Scholar]
- Zhao X., Zhang Z., Hu B., Huang W., Yuan C., Zou L. Response of gut microbiota to metabolite changes induced by endurance exercise. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00765. https://www.frontiersin.org/article/10.3389/fmicb.2018.00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.