Abstract
mRNA pseudoknots have a stimulatory function in programmed −1 ribosomal frameshifting (−1 PRF). Though we previously presented a model for how mRNA pseudoknots might activate the mechanism for −1 PRF, it did not address the question of the role that they may play in positioning the mRNA relative to the ribosome in this process [E. P. Plant, K. L. M. Jacobs, J. W. Harger, A. Meskauskas, J. L. Jacobs, J. L. Baxter, A. N. Petrov and J. D. Dinman (2003) RNA, 9, 168–174]. A separate ‘torsional restraint’ model suggests that mRNA pseudoknots act to increase the fraction of ribosomes directed to pause with the upstream heptameric slippery site positioned at the ribosome's A- and P-decoding sites [J. D. Dinman (1995) Yeast, 11, 1115–1127]. Here, experiments using a series of ‘pseudo-pseudoknots’ having different degrees of rotational freedom were used to test this model. The results of this study support the mechanistic hypothesis that −1 ribosomal frameshifting is enhanced by torsional resistance of the mRNA pseudoknot.
INTRODUCTION
The structure of an RNA molecule is widely recognized to play a role in many processes, including structurally organizing complex RNAs, the assembly of ribonucleoprotein complexes, and in translational recoding and regulation [reviewed in (1)]. One common RNA folding motifs is a pseudoknot, the folding back of a single-stranded RNA onto itself to form two helical structures with single-stranded loops joining them (2). Many such structures can be inferred from RNA sequences and frameshifting function has been demonstrated for some of these [reviewed in (3–5)]. However, though much theoretical progress has been made in understanding how mRNA pseudoknots promote efficient −1 ribosomal frameshifting (6), a complete understanding of this mechanism remains untested.
Programmed −1 ribosomal frameshift signals are typically divided into three components. From 5′ to 3′ these are (i) a ‘slippery site’ in the form N NNW WWH, where N must be a stretch of any three identical nucleotides, where W is either three A or U residues, and H is A, C or U (spacing indicates the unshifted zero frame), (ii) a spacer region and (iii) an mRNA structural element, most often a pseudoknot. The general model posits that upon encountering the mRNA pseudoknot, an elongating ribosome is forced to pause such that the anticodons of its A- and P-site tRNAs are base-paired with the zero-frame codons of the slippery site. The nature of the tRNA–mRNA interactions is such that a relative slip of −1 nucleotide still allows base-pairing in the non-wobble positions. The slippage occurs during the ribosomal pause, and it has been shown that changes affecting ribosome pause times affect frameshift efficiencies [reviewed in (7)]. An important observation is that even though mRNA pseudoknots and energetically equivalent stem–loop structures appear to promote ribosome pausing with equal effectiveness, mRNA pseudoknots are more efficient at promoting −1 PRF (8). Our ‘9 Å’ model (6) provided a refinement of the original ‘simultaneous slippage’ (9,10) model of frameshifting by suggesting that rather than the entire ribosome having to slip one base in the 5′ direction, slippage could be accomplished by moving the small section of mRNA in the downstream tunnel by one base in the 3′ direction. We have proposed that this is accomplished by the bulky and difficult to unwind mRNA pseudoknot structures becoming wedged in the downstream entrance tunnel of the ribosome, preventing the downstream region of the mRNA from being pulled into the ribosome by the equivalent of one base during the accommodation step of elongation. This blockage would introduce tension into the spacer region, which could be resolved by unpairing the mRNA from the tRNAs, allowing the mRNA to slip 1 nt backwards, resulting in a net shift of reading frame by −1 base.
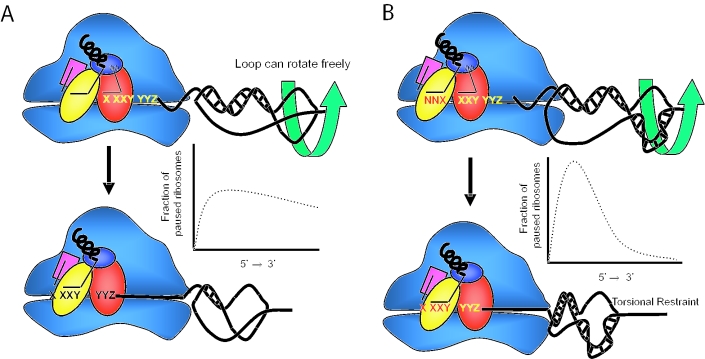
Though the 9 Å model provides a partial explanation for why mRNA pseudoknots promote programmed −1 ribosomal frameshifting (−1 PRF) more efficiently than simple stem–loop structures, it does not answer the question of how the mRNA pseudoknot directs the ribosome to pause at the correct position along the mRNA. A complementary ‘torsional restraint’ model addresses this issue (11). When a stem–loop structure is unwound by an elongating ribosome, unwinding of the stem forces the loop to rotate. Since a simple stem–loop is not restrained, the loop can rotate freely and only the base pairs within the stem resist ribosomal movement, and thus the potential energy of unwinding should be distributed along the length of Stem 1 (Figure 1A). However, if the loop is anchored or restrained, as it is in a pseudoknot by Stem 2, since the intrinsic ribosomal helicase is processive (12), Stem 1 cannot be fully unwound until Stem 2 is first denatured. Mechanically, as the ribosome begins to unwind the base of Stem 1, Stem 2 forces the supercoiling in the remainder of Stem 1, providing extra resistance to ribosome movement. At some specific point, the resistance to ribosome movement provided by the supercoiling counteracts the forward movement of the ribosome, increasing the likelihood that the ribosome will stop at a precise point along the mRNA. Energetically, since full unwinding of Stem 1 is dependent on complete denaturation of Stem 2, the potential energy of unwinding of the pseudoknot structure should similarly be directed toward one point. Viewed either mechanically or energetically, this point is where ribosomes will be directed to specifically pause on the mRNA. If it occurs with the tRNAs in ribosomal A- and P-sites positioned at the slippery site, then frameshifting is stimulated. This is summarized in Figure 1B. The efficiency of −1 PRF can thus be viewed as a function of (i) the fraction of ribosomes paused over the slippery site and (ii) the rate at which the structure can be denatured. There is increasing evidence from single molecule experiments that unfolding occurs in quick ‘rips’ at a particular force (13), suggesting that in the case of unfolding pseudoknots, frameshifting efficiency is related to both the energy barriers to unfolding the pseudoknot structure and the resistance of the structure against the force of the ribosome. In the context of the torsional restraint model, this resistance is dependent on the ability of Stem 2 to remain intact while Stem 1 is being unwound.
Figure 1.
The torsional resistance model. (A) An elongating ribosome can easily unwind a simple stem–loop because there is no resistance to the rotation of the loop, even with a long stem. Thus, the energy required to unwind the structure is broadly distributed along its length. (B) The ribosome meets added resistance to unwinding Stem 1 of a pseudoknot because Loop 1 cannot easily rotate. The energy of unwinding is narrowly focused at one point in the structure. If the pseudoknot is properly placed, the ribosome will be directed to pause with the slippery site positioned in the A- and P-site, setting up the frameshift. The theoretical ‘graphs’ are inserted to further illustrate this point, though they are not based on any actual measurements.
There is experimental data that indirectly support this model: (i) disruption of the first 3 bp of Stem 1, which would displace the ribosome's pause site to a point 3′ of the slippery site, has been shown to eliminate frameshifting (14); (ii) destabilizing Stem 2, which would allow it to be unwound more readily, has been shown to result in decreased frameshifting efficiency (15–17); (iii) replacing bulges in Stem 1 with base pairs would increase the energy required to unwind the first three bases, and a longer ribosomal pause over the slippery site would follow, yielding increased efficiencies in −1 frameshifting (15,18); (iv) destabilizing the base of Stem 1 by replacing G:C base pairs with A:U pairs decreases −1 frameshifting efficiencies (19,20); (v) the model eliminates the need for a ‘pseudoknot recognizing factor’, the evidence of which has not been forthcoming in either competition assays in in vitro translation systems (21) or by gel retardation assays (J. D. Dinman, unpublished data); and (vi) elimination of a potential torsion-restraining Stem 2, but not of a non-torsion-restraining Stem 2 in HIV-1, resulted in decreased −1 PRF efficiencies (22).
Though all of the cited studies support the torsional restraint model, none has directly addressed it. In the experiments presented in this study, a series of ‘pseudo-pseudoknot’ containing reporter constructs were used to test the torsional restraint hypothesis. In vitro frameshifting assays show that frameshifting can be significantly stimulated by limiting the rotational freedom of the loop region of a stem–loop structure, and that the degree of rotational freedom of Stem 1 is important in determining the extent of −1 PRF. Furthermore, mRNA toeprint analyses reveal a pseudo-pseudoknot-specific strong stop 16 nt 3′ of the slippery site, consistent with this structure being able to direct ribosomes to pause with their A- and P-sites positioned at the slippery site.
MATERIALS AND METHODS
Construction of plasmids
All synthetic DNA oligonucleotides were purchased by IDT (Coralville, IA). The modified L-A viral −1 PRF signal containing the G GGU UUA slippery site followed by a simple stem–loop was amplified from pJD18 (23) using the primers luc5′b (5′-CCCCAAGCTTATGACTTCTAGGCAGGGTTTAGG-3′) and luc3′b (5′-CCCCCCATGGGACGTTGTAAAAACGACGGGATC-3′). These were digested with HindIII and NcoI (restriction sites are underlined) and cloned into the firefly luciferase reporter plasmid pT7-LUC minus 3′-untranslated region-A50 (24). In the resulting reporter construct (pJD214-18), expression of firefly luciferase requires a −1 frameshift, and the 5′ sequence of the Stem 2 is not able to base pair with the 3′ sequence, so that only a stem–loop rather than a pseudoknot is able to form. The same primers were used to amplify DNA from pJDRΨ (23) to make pJD214-Rψ. In this construct, complementary mutations (5′-GCUGGC-3′ to 5′-CGACCG-3′) in the 3′ acceptor sequence of the pseudoknot-forming region of Stem 2 allow the formation of an mRNA pseudoknot that has previously been shown to promote frameshifting at the same frequency as the wild type (23). The primer luc5′CON (5′-CCCCAAGCTTATGACTTCTAGGCAAGGGTTTAGG-3′) contains an additional A nucleotide upstream of the slippery site and was used to make pJD214-0, the zero-frame control. To eliminate the possibility of internal initiation occurring at the luciferase initiation codon downstream of the frameshift signals, the AUG codon was changed to AUA. The Stratagene QuikChange kit was used to mutate pJD214-18 and pJD214-Rψ into pJD336-18 and pJD366-Rψ, respectively, using the oligonucleotides 5′-GGCGTTCTTCTATGGGACGTTGTAAAAACGGATC-3′ and 5′-GATCCGTCGTTTTTACAACGTCCCATAGAAGACGCC-3′ (the mutated codon is underlined). pJD366-18 was further mutated to make a zero-frame control by placing an A upstream of the slippery site using the oligonucleotides 5′-TGACTTCTAGGCAAGGGTTTAGGAGTG and 5′-CACTCCTAAACCCTTGCCTAGAAGTCA (the inserted base is underlined).
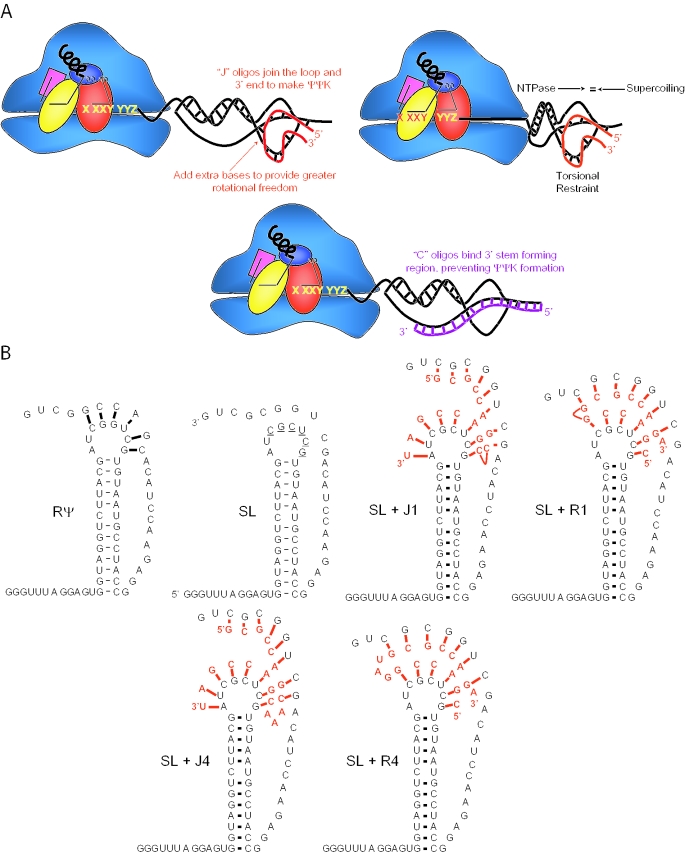
A series of synthetic DNA oligonucleotides were designed to join the loop acceptor region of mRNA transcribed from pJD366-18 to the downstream region that forms the pseudoknot in the wild-type L-A −1 PRF signal. In the J-oligos, the 3′ sequences base pair with the loop of the mRNA transcribed from pJD366-18, and the 5′ regions of these oligos base pair with the downstream sequence. This orientation is reversed for the R-oligos. These general orientations are shown in Figure 3B. The naming of the oligonucleotide names refers to the number of additional residues placed between the regions of complementarity. The bases complementary to the pJD366-18 sequence are underlined.
J1 5′-GCGCCAGCCGACCGAT-3′
J2 5′-GCGCCAGCACGACCGAT-3′
J3 5′-GCGCCAGCAACGACCGAT-3′
J4 5′-GCGCCAGCAAACGACCGAT-3′
R1 5′-CGACCGGCCAGCTGA-3′
R2 5′-CGACCGGGCCAGCTGA-3′
R3 5′-CGACCGGAGCCAGCTGA-3′
R4 5′-CGACCGGATGCCAGCTGA-3′
C 5′-CAGCGCCAGCTGTAG-3′
Figure 3.
Cartoons of pseudo-pseudoknot constructs. (A) Addition of J-oligo (R-oligos not shown) to a stem–loop containing mRNA to form a pseudo-pseudoknot mimics the activity of a pseudoknot (top and middle). C-oligos out compete J- or R-oligos, reverting the structure to a stem–loop. (B) Structures of the pJD366-RΨ pseudoknot (RΨ), of the pJD366-18 stem–loop (SL) and of pseudo-pseudoknots formed by annealing SL with J1-, R1-, J4- and R4-oligos.
Preparation of plasmids and mRNA synthesis
Plasmid DNAs were prepared using the Qiagen mini-prep kits and were linearized with DraI in a total volume of 20 μl. Proteins were eliminated by the addition of 2 μl of 1 mg/ml proteinase K and SDS to a final concentration of 0.5% followed by digestion at 50°C for 30 min. Volumes were then increased to 100 μl, extracted twice with phenol/chloroform, and DNA was precipitated with 10 μl NH4Ac and 250 μl ethanol. The purified DNA was resuspended in DEPC-treated H2O. To prepare synthetic mRNAs, 2 μl of purified linear DNAs were used for in vitro transcription using the Ambion T7 mMachine mMessage kit. RNAs were precipitated using 30 μl DEPC H2O and 25 μl LiAc. The RNA was resuspended in 11 μl DEPC H2O (1 μl in 500 would give an OD260 of 0.05–00.1; 1–2 μg/μl).
In vitro translation and frameshifting assays
To anneal the oligonucleotides with the mRNA, J-oligos, R-oligos or the equivalent volumes of dilution buffer alone (20 mM Tris, pH 7.4, 2 mM MgCl2 and 50 mM EDTA final concentration) were added to synthetic mRNA (0.5 μg), and the mixtures were first incubated in a 70°C heating block for 10 min; the block was then removed and allowed to cool to 37°C (30 min), after which they were briefly spun down and incubated on ice. In all experiments, the molar ratios of J- and R-oligos to synthetic mRNAs were 100:1. In experiments using the competing oligonucleotide (C-oligo), this was added to either 0.5:1 or 1:1 molar ratios with either J- or R-oligonucleotides. Reticulocyte lysates were thawed on ice, 15 μl of −met and 15 μl of −leu master mixes plus 20 μl of H2O were added to 400 μl of lysate, and 19 μl of this was added to each annealed reaction to start the in vitro translation reactions. These were incubated at 30°C for 60 min (the reaction reached a plateau after 30–35 min where the greatest difference was seen between the zero-frame controls and the frameshifting plasmids) (data not shown), and the reactions were then placed on ice. An aliquot of 7.5 μl from each in vitro translation reaction was added to 50 μl of the prewarmed luciferase reagent, and luminescence readings were taken after a 3 s delay for 15 s in triplicate using a Turner 20/20 Luminometer.
RNase assay
Synthetic transcripts generated from DraI-digested pJD366-18 (∼1.7 kb) were 5′ end labeled using [γ-32P]CTP. These RNAs (4 μl) were incubated with 1 μl of annealing buffer and either 1 μl of H2O or 1 μl of an oligo (0.25 ng) at 70°C. The heating block was allowed to cool at room temperature for 40 min before 8 μl of RNaseH buffer was added (20 mM HEPES, 50 mM KCl, 10 mM MgCl2 and 1 mM DTT). An aliquot of 1 μl of enzyme was added (mung bean nuclease, RNaseH or RNaseT1) and the reactions incubated at 37°C for 1 h. The reactions were stopped by adding 4 μl of stop solution, the products separated through a 6% polyacrylamide–urea denaturing gel and visualized by autoradiography.
mRNA toeprinting
JD366-18 mRNA (1 μg in 8 μl) was annealed with 2 μl of 3′ end-labeled toeprinting primer (5′-CGTACGTGATCTTCACC-3′, complementary to sequence 240 bp 3′ of the slippery site) as described above. This was added to 15 μl of lysate (200 μl Ambion retic lysate, 7.5 μl of each master mix [−leu and −met] and 70 μl of 250 mM KCl), except for 2 μl, which was added to 15 μl of RT buffer [50 mM Tris–HCl (25), 40 mM KCl, 6 mM MgCl2, 5 mM DTT and 575 μM dNTPs] to be used as a no-ribosome control. In vitro translation reactions were incubated at room temperature for 10 min, which was empirically determined to provide the optimum amount of time to allow ribosomes to initiate translation and pause at the frameshift signal. Subsequently, 15 μl of RT buffer containing RNasin inhibitor and cycloheximide (to a final concentration of 100 ng/μl) was added to stop translation. To this, 2 μl of Superscript II (Invitrogen) was added and the reaction incubated at room temperature for 10 min. Reactions were terminated by phenol:chloroform extraction and 15 μl of stop solution added. The toeprinting primer was also used in conjunction with pJD366-18 to produce sequencing ladders by standard dideoxynucleotide chain termination methods using Sequenase (USB). Products were separated though 6% polyacrylamide–urea denaturing gels and visualized using a Storm phosphorImager (Pharmacia).
RESULTS
Pseudo-pseudoknots stimulate frameshifting, and frameshifting efficiency changes with the degree of pseudo-pseudoknot rotational freedom
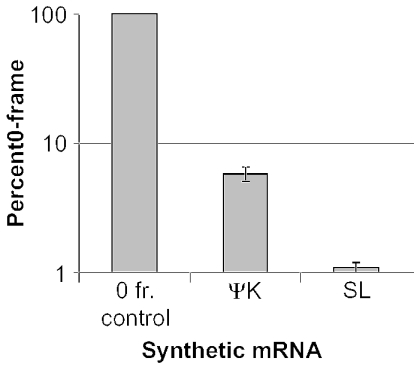
We previously showed in intact yeast cells that the pseudoknot containing mRNA produced from pJDRΨ was able to promote efficient −1 PRF, whereas one in which only a stem–loop can form, transcribed from pJD18, could not (23). As a first step in this study, we tested the ability of synthetic mRNAs produced from pJD366-RΨ and from pJD366-18, two plasmids derived from these parental constructs, to promote −1 PRF. Total luciferase activities produced from these synthetic mRNAs were divided by the luciferase activity produced from the zero-frame control plasmid, pJD366-0, and multiplied by 100% to determine −1 PRF efficiencies. The results show that the trends observed in yeast were replicated in vitro, i.e. JD366-RΨ mRNA promoted ∼8% efficiency of −1 PRF as compared with ∼1.1% promoted by JD366-18 mRNA (Figure 2).
Figure 2.
A pseudoknot promotes more efficient frameshifting than a stem–loop in vitro. mRNAs generated from pJD366-0 (0), pJD366-RΨ (RΨ) and pJD366-18 (SL) were used as substrates for in vitro translation using rabbit reticulocyte lysates. Results are shown as percent of the zero-frame control. Assays were performed three times in triplicate and error bars represent standard deviations.
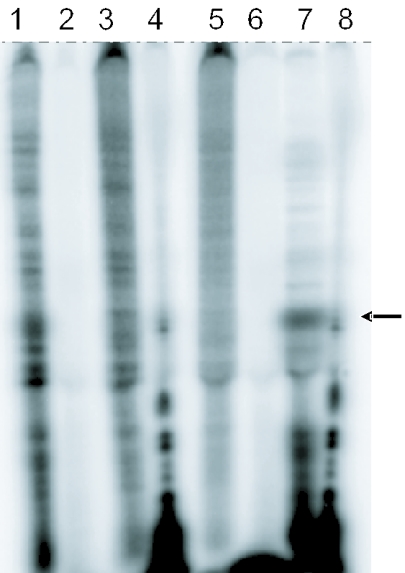
The ‘torsional restraint’ model predicts that conditions that would inhibit the rotational freedom of the loop region of the pJD366-18-derived mRNA should result in enhanced −1 PRF efficiency. The strategy used in this study was to anneal this mRNA with synthetic oligonucleotides complementary to both the loop region and to the sequence downstream that is normally involved in pseudoknot formation. These ‘pseudo-pseudoknots’ would be predicted to restore a pseudoknot-like structure to the mRNA. This is diagrammed in Figure 3A. Two different classes of oligonucleotides having different orientations relative to the mRNA were used to this end: ‘joining’ (J-) and ‘reverse’ (R-) oligos. The orientation of the J-oligos promotes the formation of a structure containing the equivalent of a Loop 2 region, while that of the R-oligos promotes a Loop 1 equivalent. The model also predicts that pseudo-pseudoknots having different degrees of rotational freedom should promote different frequencies of ribosome pausing over the slippery site, resulting in different efficiencies of −1 PRF. In order to control this parameter, increasing numbers of nucleotides were inserted between the mRNA hybridizing regions of the J- and R-oligos. The additional non-complementary bases in the J-oligos are 3′ to the stem–loop residues involved in Stem 2, thus effectively increasing Loop 2. Similarly, the additional non-complementary bases in the R-oligos are 5′ to the loop acceptor residues and correspond to an increased Loop 1. The structure of the stem–loop of pJD366-18 and its maximum base-paired interactions with representative J- and R-oligos are shown in Figure 3B. To demonstrate that an oligonucleotide–mRNA hybrid was capable of forming under the assay conditions, the J1-oligo was incubated with 5′ [32P]labeled JD366-18 mRNA and subjected to RNaseH digestion. Digestion of the RNA–DNA hybrid resulted in a labeled 110 nt fragment, demonstrating that the oligonucleotide bound to the mRNA at the position of the pseudoknot (Figure 4).
Figure 4.
Short oligonucleotides bind to mRNA. mRNA from pJD366-18 T7 run offs was incubated with the joining oligonucleotide J1 and subjected to nuclease treatment. Lanes 1–4 contain mRNA without the oligonucleotide and lanes 5–8 mRNA with oligonucleotide. Lanes 2 and 6 were treated with Mung Bean nuclease; lanes 3 and 7 with RNaseH; lanes 4 and 8 with RNaseT1; and lanes 1 and 5 untreated. The arrow indicates the 5′ 110 nt fragment after incubation with J1 and RNaseH (lane 7).
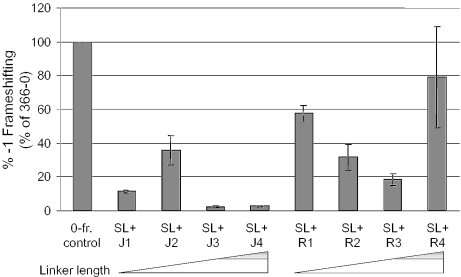
Having demonstrated the utility of the in vitro frameshifting assay and that the J- and R-series of oligonucleotides were able to hybridize with synthetic mRNA produced from pJD366-18, the next step was to monitor frameshifting efficiencies promoted by these hybrid species. Significant increases in frameshifting were observed with the incubation of pJD366-18 mRNA with oligonucleotides J1 (∼10%) and J2 (∼35%), while only modest increases were seen with J3 and J4 (Figure 5). These findings are consistent with the notion that changes in the degree of rotational freedom of the structure would affect the distribution of paused ribosomes in the vicinity of the slippery site.
Figure 5.
Efficient frameshifting is stimulated by pseudo-pseudoknots. In vitro translation assays were performed in retic lysates with mRNAs derived from pJD366-18 (SL) to which J- or R-oligos were annealed. Luciferase activities were divided by those obtained using mRNAs generated from pJD366-0, and the resulting ratios were multiplied by 100 to calculate percent frameshifting. The averages of three independent experiments performed in triplicate are shown. Error bars denote standard deviation.
One potential complication with the J-oligos is the possibility that they could interact with the Loop 2–Stem 1 region. In the R-oligos, the additional bases are distal to any possible Loop 2–Stem 1 interactions and would be more analogous to increasing Loop 1. The R-oligos stimulated −1 PRF to an even higher extent than the J-oligos (Figure 5). Importantly, increasing the length of the bridging regions in these oligonucleotides (R1 to R3), which is predicted to increase the rotational freedom of the stem–loop, resulted in decreased frameshifting activity as predicted by the torsional resistance model. However, addition of three residues between the two binding regions of the R-oligo (R4) resulted in an unexpected increase in frameshifting with a very large amount of variation.
In a series of control experiments, 8 nt oligos complementary to the 5′ (Loop 1) and 3′ Stem 2 forming regions of the pseudo-pseudoknot were hybridized to the SL mRNA and −1 PRF assays were performed. Neither of these were able to stimulate −1 PRF, even at concentrations in 100-fold molar excess to the mRNA template (data not shown). Though supportive of our central hypothesis, it is also possible that these results were due to the thermodynamic instability of the RNA:DNA duplexes through the course of the experimental protocol.
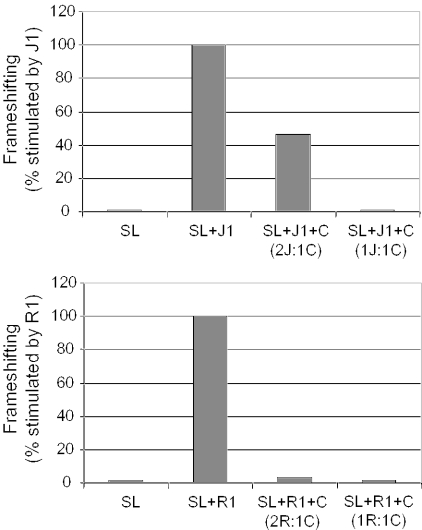
Pseudo-pseudoknot structure and function can be competed away
To determine whether the stimulation of frameshifting was specifically due to the bridging of the stem–loop with downstream sequence (the pseudo-pseudoknot), as opposed to the nonspecific presence of an RNA:DNA hybrid, the competing oligonucleotide (C-oligo) was designed to form a 15 bp duplex with JD366-18 mRNA, including the 3′ Stem 2 forming region, which was expected to significantly out compete either the J- or R-oligos from binding to this site, thus disrupting formation of the pseudo-pseudoknot (see Figure 3A). Additionally, in the presence of the C-oligo, the J- and R-oligos were still predicted to hybridize with the 5′ Stem 2 forming region, enabling us to address the question of whether this interaction alone was able to stimulate frameshifting. The results demonstrate that the addition of the C-oligo severely inhibited the abilities of both the J- and R-oligos to promote efficient frameshifting (Figure 6). These findings demonstrate that (i) frameshifting was specifically stimulated by bridging of the 5′ and 3′ Stem 2 forming regions by the J- and R-oligos, and (ii) that the presence of an RNA:DNA hybrid at the 5′ Stem 2 forming region was not sufficient to stimulate frameshifting by itself.
Figure 6.
Competition for J- or R-oligo binding sites inhibits its ability to promote efficient frameshifting. mRNA transcribed from pJD366-18 (SL) was annealed with either J- or R-oligos alone, or in combination with different concentrations of competing (C-) oligos (in ratios of 2:1 or equimolar as indicated). Sample marked SL is mRNA alone. Luciferase activities generated from in vitro translation reactions in rabbit reticulocyte lysates were divided by those obtained using mRNAs generated from pJD366-0, and the resulting ratios were multiplied by 100 to calculate percent frameshifting.
Pseudo-pseudoknots direct ribosomes to pause over the slippery site
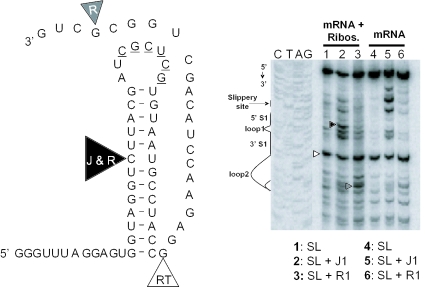
The torsional restraint model predicts that pseudoknots should direct elongating ribosomes to pause at one specific location on the mRNA, rather than being distributed along Stem 1. We used mRNA toeprint assays to test this hypothesis. In mRNA toeprint reactions, the movement of reverse transcriptase is blocked by paused ribosomes, resulting in a strong stop positioned ∼16–18 nt 3′ of the P-site of eukaryotic ribosomes (25). Synthetic JD366-18 mRNAs were annealed with the sequencing oligonucleotide and either J1, R1 or no second oligo, and these were then used for in vitro translation reactions. After a period of time (10 min were empirically determined to be optimal), elongation reactions were stopped by the addition of cycloheximide, and reverse transcription reactions were initiated on the sequencing oligonucleotides. In parallel, control reverse transcription reactions were carried out using synthetic JD366-18 mRNA and oligonucleotides, but without in vitro translation. The results are consistent with the model, showing that the J1- and R1-oligos specifically promoted one strong reverse transcriptase stop 16 nt 3′ of the P-site of the slippery site only the in the in vitro translation reactions (Figure 7). As further predicted by the model, a broad distribution of stops of equal intensities was observed in this region with JD366-18 mRNA alone (Figure 7, lane 1). Importantly, the +16 stop was not observed when toeprint reactions were carried in the absence of ribosomes. Additional strong stops were also of interest. One corresponding to the 3′ end of the base of Stem 1 was observed in all samples, consistent with the presence of this structure. Both J- and R-oligo-specific pauses were also observed. The reason for the strong pause in the J-oligo is unknown. The R-oligo-specific pause is perhaps more revealing. It occurs at the 3′ end of the RNA:DNA hybrid formed by this oligo and the mRNA, a structure that should also promote pausing of reverse transcriptase.
Figure 7.
Pseudo-pseudoknots direct ribosomes to pause over the slippery site. mRNAs generated from pJD366-18 (SL) were annealed with the sequencing oligonucleotide and either J1-, R1- or no oligo (lanes 1, 2 and 3, respectively), and these were then used for in vitro translation reactions. Reactions were stopped after 10 min by the addition of cycloheximide, and reverse transcription reactions were initiated on the sequencing oligonucleotides. In parallel, control reverse transcription reactions were carried out using synthetic JD366-18 mRNA and oligonucleotides, but without in vitro translation (lanes 4–6). The positions of the slippery site, loops and stems of the pseudo-pseudoknots are indicated next to a sequencing reaction. Arrowheads indicate positions of reverse transcriptase strong stops and these are mapped to a representation of the stem–loop structure of pJD366-18.
DISCUSSION
The results presented in this study provide strong support for the torsional restraint model of programmed −1 frameshifting. Specifically, we demonstrated that RNA:DNA hybrids that mimic mRNA pseudoknots can significantly stimulate frameshifting. As predicted by the model, changing the rotational freedom of the structure by altering the lengths in the J1- and R1-oligos between the 5′ and 3′ mRNA hybridizing regions resulted in changes in their abilities to stimulate −1 frameshifting. The demonstration that these ‘pseudo-pseudoknot’ structures cause elongating ribosomes to specifically pause with their A- and P-sites positioned at the slippery site provides independent evidence in support of the model.
In the case of the J-oligo series, frameshifting was best stimulated by J2, suggesting the structure created and the rotational freedom allowed by it was optimal for −1 PRF. The experimental design is such that we assume a similar rate of unfolding for each oligo as the predicted maximum base pairing is the same for them all. However, we do note that the type of nucleotides separating the two, separately paired regions of the oligos, and their presentation, may play a role in −1 PRF efficiency. The recent NMR structural solution of the SRV-1 pseudoknot revealed a highly structured Loop 2–Stem 1 interface including base triples involving an A residue at the 3′ end of Loop 2 (26). The additional base in the J2-oligonucleotide is also an A. Mutagenesis experiments in this region by other groups showed, for example, that replacing the 3′ base in Loop 2 of IBV with an A residue promoted a significant increase in frameshifting efficiency (27), and mutation or removal of the A residue at the 3′ base in Loop 2 of the BWYV pseudoknot reduced frameshifting levels (17). This part of the pseudoknot has been proposed to be important in a frameshifting model where differential transition state energy barriers (due to small differences in local structure, stability or dynamics) are the primary determinant of frameshifting efficiency (3). Indeed, a Loop 2–Stem 1 triplex interaction seen in smaller frameshifting pseudoknots from luteoviruses has been shown to be critical for −1 PRF, and that similar pseudoknots lacking the triplex are less efficient at frameshifting [(28) and references therein]. This extra structural feature would limit the rate of unfolding and provide extra anchoring of Stem 2 as the ribosome attempts to unwind Stem 1, i.e. it too would help to provide additional torsional restraint. It is also possible that although the J3- and J4-oligonucleotides also help to form a pseudo-pseudoknot, the additional bases may interfere with the stabilization of Stem 1.
With the R-oligos, a general correlation was observed between minimization of rotational freedom and frameshifting efficiency, though this was not the case of the R4-oligo. Since the stability of the pseudo-pseudoknot generated with R4 should be similar to that of the other oligonucleotides based on the base-pairing, this result suggests that there are additional considerations to be uncovered with regard to the pseudoknot structure influencing frameshifting. Addition of residues in the R-oligos was analogous to lengthening Loop 1, which is typically short in −1 frameshifting pseudoknots. Limited and conflicting data are available on the importance of Loop 1 in −1 frameshifting pseudoknots. In one study, addition of three A bases to Loop 1 did not affect frameshifting efficiency (15), while in another all the mutations made in this region were detrimental to frameshifting efficiency (17). Given the complex interactions occurring between the helices and loops in this region, we cannot yet account for why the R4-oligo stimulated frameshifting so efficiently and with such variable results.
Examination of the RNA toeprint data presented here reveals that both of the pseudo-pseudoknot structures formed by the J1- and R1-oligos promoted strong stops of the reverse transcriptase ∼16 nt 3′ of the P-site codon of the slippery site, consistent with the hypothesis that the presence of Stem 2 forces ribosomes to pause with their A- and P-sites positioned over the slippery site. Previous studies mapping the lagging edge of paused ribosomes, i.e. mRNA heelprint studies, did not reveal any striking differences between the effects of pseudoknots versus stem–loops (8,16). Interestingly, using this method, the ribosomal pauses appeared distributed over a broader stretch of mRNA (∼4 nt) than observed here. It is possible that some critical level of resolution is lost in the requirement for many additional manipulations of substrates using the mRNA heelprint as compared with the toeprint methods.
A remaining question centers on whether the role of the RNA pseudoknot in −1 PRF is passive or active. In the ‘9 Å solution’ (6), the frameshift mechanism is activated by movement of the A-site codon–anticodon complex by 1 base in the 5′ direction upon accommodation. As currently described, the mRNA pseudoknot merely passively blocks entry of the downstream message into the ribosome, resulting in stretching of the segment of mRNA located between the codon–anticodon complex and the pseudoknot. By this model, all of the energetic input for the frameshift is derived from hydrolysis of GTP by eEF1A. However, it is possible that the pseudoknot may also actively contribute to the frameshift mechanism. Specifically, pulling the downstream message into the ribosome at accommodation could result in unwinding of Stem 1 of the pseudoknot by one additional base pair. The energetic cost of so doing would be to introduce an equivalent amount of torsional resistance into Stem 2. If Stem 2 were to release this resistance by ‘pulling back’, the base pair in Stem 1 would be re-formed, which in turn would contribute to the energy required to dissociate the A- and P-site codon–anticodon complexes from the zero-frame. This would be followed by slippage of the mRNA by 1 base in the 3′ direction relative to the ribosome, followed by the formation of −1 frame codon–anticodon complexes. As such, the proposed active role for the mRNA pseudoknot would further reduce the energetic barrier to −1 PRF. In sum, we suggest that the ‘torsional restraint model’ can be combined with the ‘9 Å solution’ to mechanistically explain the original ‘simultaneous slippage’ model of −1 PRF (9,10). In other words, the 9 Å solution + torsional restraint = simultaneous slippage.
Two recent publications have also shown that oligonucleotide:mRNA duplexes can stimulate efficient −1 ribosomal frameshifting (29,30). These studies differed from the present one in a number of ways, particularly insofar as they examined the effects duplex structures immediately 3′ of the slippery site rather than addressing mRNA pseudoknot related questions. The findings support the notion that the specific location of ribosome pausing on the mRNA plays a critical role in determining frameshifting, though they do come with caveats, e.g. neither study directly mapped ribosomal pausing, and the use of different slippery sites and downstream contexts likely contributed to disparate findings for the optimal distances between the 3′ ends of slippery sites and 5′ ends of frameshift-stimulating oligonucleotides. Although potentially useful therapeutically there are no known natural examples of frameshifting stimulated in this manner, and thus these results do not affect the hypothesis presented here. However, these studies are important in that they raise the possibility for a new role for micro-RNAs in regulating gene expression, and for therapeutic approaches to correcting inborn errors of metabolism due to the presence of frameshift mutations.
Acknowledgments
We thank Reed Wickner, who first conceived the torsional restraint model, and Ignacio Tinoco, who originally challenged us to design an experimental approach. This work was supported by a grant to J.D.D. from the National Institutes of Health (GM 58859). Funding to pay the Open Access publication charges for this article was also provided by the NIH (GM 58859).
Conflict of interest statement. None declared.
REFERENCES
- 1.Gesteland R.F., Atkins J.F. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 2.Puglisi J.D., Wyatt J.R., Tinoco I., Jr A pseudoknotted RNA oligonucleotide. Nature. 1988;331:283–286. doi: 10.1038/331283a0. [DOI] [PubMed] [Google Scholar]
- 3.Giedroc D.P., Theimer C.A., Nixon P.L. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farabaugh P.J. Programmed Alternative Reading of the Genetic Code. Austin TX: R.G. Landes Company; 1997. [Google Scholar]
- 5.Brierley I. Ribosomal frameshifting on viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 6.Plant E.P., Jacobs K.L.M., Harger J.W., Meskauskas A., Jacobs J.L., Baxter J.L., Petrov A.N., Dinman J.D. The 9-angstrom solution: how mRNA pseudoknots promote efficient programmed −1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harger J.W., Meskauskas A., Dinman J.D. An “integrated model” of programmed ribosomal frameshifting and post-transcriptional surveillance. Trends Biochem. Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- 8.Kontos H., Napthine S., Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacks T., Madhani H.D., Masiraz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacks T., Power M.D., Masiarz F.R., Luciw P.A., Barr P.J., Varmus H.E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 11.Dinman J.D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takyar S., Hickerson R.P., Noller H.F. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 13.Onoa B., Tinoco I., Jr RNA folding and unfolding. Curr. Opin. Struct. Biol. 2004;14:374–379. doi: 10.1016/j.sbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Morikawa S., Bishop D.H.L. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brierley I.A., Rolley N.J., Jenner A.J., Inglis S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu C., Tzeng T.-H., Bruenn J.A. Ribosomal movement impeded at a pseudoknot required for ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y.G., Su L., Maas S., O'Neill A., Rich A. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc. Natl Acad. Sci. USA. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinman J.D., Wickner R.B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bekaert M., Bidou L., Denise A., Duchateau-Nguyen G., Forest J.P., Froidevaux C., Hatin I., Rousset J.P., Termier M. Towards a computational model for −1 eukaryotic frameshifting sites. Bioinformatics. 2003;19:327–335. doi: 10.1093/bioinformatics/btf868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Napthine S., Liphardt J., Bloys A., Routledge S., Brierley I. The role of RNA pseudoknot stem 1 length in the promotion of efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ten Dam E., Brierley I., Inglis S., Pleij C. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucleic Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinman J.D., Richter S., Plant E.P., Taylor R.C., Hammell A.B., Rana T.M. The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl Acad. Sci. USA. 2002;99:5331–5336. doi: 10.1073/pnas.082102199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinman J.D., Icho T., Wickner R.B. A −1 ribosomal frameshift in a double-stranded RNA virus forms a gag–pol fusion protein. Proc. Natl Acad. Sci. USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 25.Dmitriev S.E., Pisarev A.V., Rubtsova M.P., Dunaevsky Y.E., Shatsky I.N. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 2003;533:99–104. doi: 10.1016/s0014-5793(02)03776-6. [DOI] [PubMed] [Google Scholar]
- 26.Michiels P.J., Versleijen A.A., Verlaan P.W., Pleij C.W., Hilbers C.W., Heus H.A. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liphardt J., Napthine S., Kontos H., Brierley I. Evidence for an RNA pseudoknot loop–helix interaction essential for efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nixon P.L., Cornish P.V., Suram S.V., Giedroc D.P. Thermodynamic analysis of conserved loop–stem interactions in P1-P2 frameshifting RNA pseudoknots from plant Luteoviridae. Biochemistry. 2002;41:10665–10674. doi: 10.1021/bi025843c. [DOI] [PubMed] [Google Scholar]
- 29.Olsthoorn R.C., Laurs M., Sohet F., Hilbers C.W., Heus H.A., Pleij C.W. Novel application of sRNA: stimulation of ribosomal frameshifting. RNA. 2004;10:1702–1703. doi: 10.1261/rna.7139704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard M.T., Gesteland R.F., Atkins J.F. Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides. RNA. 2004;10:1653–1661. doi: 10.1261/rna.7810204. [DOI] [PMC free article] [PubMed] [Google Scholar]