Abstract
We have previously characterized transcription factor LZIP to be a growth suppressor targeted by hepatitis C virus oncoprotein. In search of proteins closely related to LZIP, we have identified a liver-enriched transcription factor CREB-H. LZIP and CREB-H represent a new subfamily of bZIP factors. CREB-H activates transcription by binding to cAMP responsive element, box B, and ATF6-binding element. Interestingly, CREB-H has a putative transmembrane (TM) domain and it localizes ambiently to the endoplasmic reticulum. Proteolytic cleavage that removes the TM domain leads to nuclear translocation and activation of CREB-H. CREB-H activates the promoter of hepatic gluconeogenic enzyme phosphoenolpyruvate carboxykinase. This activation can be further stimulated by cAMP and protein kinase A. CREB-H transcript is exclusively abundant in adult liver. In contrast, the expression of CREB-H mRNA is aberrantly reduced in hepatoma tissues and cells. The enforced expression of CREB-H suppresses the proliferation of cultured hepatoma cells. Taken together, our findings suggest that the liver-enriched bZIP transcription factor CREB-H is a growth suppressor that plays a role in hepatic physiology and pathology.
INTRODUCTION
The establishment and maintenance of a differentiated phenotype in a given tissue requires specific gene expression program. In liver, this is accomplished by the coordinated action of a group of liver-enriched transcription factors (LETFs). Well-characterized LETFs include hepatocyte nuclear factors-1 (HNF1), HNF3, HNF4, HNF6, CAATT enhancer-binding proteins (C/EBPs) and D-box binding protein. These factors are activated in a precise temporal and spatial order during liver development (1,2), and they form a regulatory network that controls the expression of a repertoire of liver-specific genes (3). Thus, HNF3 is required for inducing the endoderm to adopt a hepatic fate (4). Once the pre-hepatic cells are committed, HNF4α is essential for complete differentiation of hepatocytes (5) and C/EBPα regulates a number of genes required for energy metabolism in adult liver (6).
Dysregulation of liver-specific transcription is a hallmark of hepatocellular carcinoma (HCC) (7). Indeed, most of the dysregulated genes identified in an HCC expression profiling study are known to be regulated by LETFs (8). In addition, among the LETFs, HNF1, HNF3β, HNF4α and HNF4γ have been shown to be up-regulated in HCC samples whereas the expression of C/EBPα is reduced (8). In this regard, C/EBPα may serve as a growth suppressor through stabilization of p21 (9,10), inhibition of cyclin-dependent kinases 2 and 4 (11,12), and repression of E2F-dependent transcription (13,14).
bZIP transcription factors are a large family of sequence-specific DNA-binding proteins characterized by a basic domain, which interacts with DNA, and a leucine zipper or coiled coil region critical for dimerization (15,16). C/EBPα and CREB are two prototypic bZIP proteins that have been extensively studied (6,17). In the CREB family of bZIP transcription factors, box B-binding factor 2 (BBF2)/dCREB-A (18,19) has been identified in a screen for proteins that specifically bind to the box B element in the fat-body-specific enhancer of the Drosophila alcohol dehydrogenase gene. The fat body, a functional counterpart of the mammalian liver, is a major organ for energy metabolism in Drosophila. It is noteworthy that BBF2 also binds to and activates CRE and box B-like element located in the promoter of human alcohol dehydrogenase as well as rat tyrosine aminotransferase genes (18). Thus, box B-like elements and their binding proteins likely represent an important and evolutionarily conserved component in the liver-specific regulatory circuit. Interestingly, a human liver-specific bZIP protein called CREB-H has also been described. CREB-H specifically binds to box B and activates box B-dependent transcription (20), suggesting that it may be functionally equivalent to BBF2.
A mammalian homolog of BBF2 termed LZIP/CREB3 has previously been identified and characterized (21–23). LZIP interacts with host cell factor-1 and host cell factor-like protein 1 (21,23,24). The ubiquitously expressed LZIP protein binds to canonical CRE and regulates cell proliferation. It is also a binding partner and transformation cofactor of hepatitis C virus core protein. Loss of LZIP function in NIH3T3 cells induces morphological transformation and anchorage-independent growth (22). In searching for additional factors closely related to LZIP, we identified a mouse bZIP protein orthologous to human CREB-H. LZIP and CREB-H represent a new subfamily of bZIP proteins with a unique putative transmembrane (TM) domain lined to the bZIP region. Removal of TM domain and the remaining C-terminal sequences translocates CREB-H from endoplasmic reticulum (ER) to the nucleus and renders it a potent transactivator. CREB-H activates transcription by binding not only to CRE and box B but also to ATF6 element. CREB-H transcript is exclusively abundant in adult liver. In contrast, the expression of CREB-H mRNA is aberrantly reduced in hepatoma tissues and cells. Overexpression of the full-length CREB-H and its active form suppress proliferation of HepG2 hepatoma cells. Our findings indicate that CREB-H is a liver-enriched bZIP transcription factor critically involved in hepatic physiology and hepatocarcinogenesis.
MATERIALS AND METHODS
Plasmids
The mouse CREB-H cDNA clone (IMAGE 1887290) was obtained from Invitrogen. CREB-H (amino acids 1–479) and CREB-HΔTC (amino acids 1–318) cDNA fragments were PCR-amplified and subcloned into expression vector pcDNA3.1/V5-His (Invitrogen) via restriction sites XhoI and XbaI, into pCMV-FLAG (Kodak) via restriction sites HindIII and SalI, and into pEGFP-C (Clontech) via restriction sites HindIII and XhoI. Mammalian expression plasmids containing the complete coding region of human CREB and ATF4, and the active form of human ATF6 (amino acids 1–373) were constructed by PCR amplification of the corresponding cDNA fragments and inserted into pCMV-FLAG via restriction sites HindIII and XhoI. Plasmids expressing human C/EBPα (9) and ATF6 (25) were kindly provided by Dr Gretchen Darlington and Dr Ron Prywes, respectively.
Plasmids expressing the fusion protein GAL4BD-CREB-H and its deletion mutants were constructed by in-frame insertion into plasmid pM (Clontech) via restriction sites (5′ and 3′): SalI and XbaI for BD-FL and BD-(1–318); EcoRI and SalI for BD-(63–318), BD-(154–479) and BD-(154–318); SalI and PstI for BD-(1–62) and BD-(1–280); SalI and HindIII for BD-(1–120), BD-(1–180) and BD-(1–240). Plasmid expressing the bZIP region of CREB-H (CREB-HZ; amino acids 206–318 plus C-terminal polyhistidine tag) was constructed by PCR amplification and in-frame insertion to the bacterial expression plasmid pET32c (Novagen) via restriction sites NdeI and XhoI.
Reporter plasmid pCRE-Luc and expression plasmid for the catalytic subunit of protein kinase A (PKA) were from Stratagene. pATF6-Luc was derived from pGL3-basic (Promega) and contains six copies of canonical ATF6-binding sites (26). pPEPCK-Luc containing −490 to +73 of rat phosphoenolpyruvate carboxykinase (PEPCK) promoter (27) was a kind gift from Dr Marc Montminy. pGAL4-Luc was a gift from Dr Karen Kibler. pSV-βGal was purchased from Promega. Lentiviral expression vector for CREB-H was derived from pLenti6/V5-D-TOPO (Invitrogen).
Antibodies
Polyclonal antiserum α-CH8 and α-CH9 were raised in two rabbits, respectively, against a purified GST-CREB-H (1–84) fusion protein, which contains the N-terminal 84 amino acids of mouse CREB-H. The crude antiserum was further purified by pre-adsorbing the anti-glutathione S-transferase (GST) antibodies onto a GST–Sepharose column. Rabbit polyclonal anti-V5 and mouse monoclonal anti-FLAG were from Sigma. Mouse monoclonal anti-BrdU conjugated to fluorescein was from Roche. Mouse monoclonal antibodies against green fluorescent protein (GFP), V5, calnexin and GM130 were from Santa-Cruz, Invitrogen, Affinity Bioreagents and BD Biosciences, respectively.
Northern blotting
Northern blot analysis was performed using 32P-labeled 661 bp random-primed fragment generated by restriction digestion of mouse CREB-H cDNA (corresponding to nucleotides 258–918) using EcoRI and PstI. Blots of poly(A)+ RNAs from mouse tissues and cancer cell lines were purchased from Clontech and were probed as recommended by the manufacturer.
Electrophoretic mobility shift assay (EMSA)
Recombinant polyhistidine-tagged protein of CREB-H bZIP region (CREB-HZ; amino acids 206–318) was expressed and purified from Escherichia coli according to Novagen's instructions. Probe labeling and EMSA were performed as described previously (28). Canonical sequences for CRE, AP1, Sp1 and κB motifs have also been described previously (22). Sequences of other sense-strand oligonucleotides used are as follows: box B in fruitfly alcohol dehydrogenase-1 gene (29), 5′-GATCTTGTACACGTAATCGTAGGATCC-3′; C/EBP in human albumin gene (30), 5′-GATCTTGATTTTGTAATGGGGGGATCC-3′; CRE in rat PEPCK gene (31), 5′-GATCTCCCTTACGTCAGAGGAGGATCC-3′; ERSE-I element in human GRP78 (an ER chaperone) gene (32), 5′-GATCTCCAATCGGCGGCCTCCACGGGATCC-3′; and ERSE-II element in human Herp (a TM protein of ER) gene (33), 5′-ATTGGGCCACG-3′.
Western blotting
Harvested cells were lysed either by repeated freezing and thawing or in RIPA buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate) supplemented with 2 mM phenylmethylsulfonyl fluoride, 2 μg/ml pepstatin A, 2 μg/ml aprotinin, 2 μg/ml leupeptin and 2 μg/ml soybean trypsin inhibitor. Samples containing equal amounts of protein were separated by SDS–PAGE and electroblotted onto Immobilon-P membranes (Millipore) using Hoefer SemiPhor semi-dry blotting apparatus (Amersham). Blots were blocked with 5% skim milk, followed by incubation with primary antibodies. Blots were then incubated with goat anti-rabbit or anti-mouse secondary antibody conjugated to horseradish peroxidase (Amersham) and visualized by enhanced chemiluminescence (Amersham).
Cell cultures, transient transfection, lentiviral transduction and luciferase assays
Human hepatoma cell lines HepG2 and Hep3B, human cervical carcinoma cell line HeLa and mouse hepatoma cell line Hepa1-6 were grown in DMEM containing 10% fetal bovine serum, 100 U/ml penicillin-G and 100 μg/ml streptomycin. Cells were maintained at 37°C in a humidified atmosphere at 5% CO2. For luciferase reporter assays, cells were cultured in 24-well plates for transient transfection. HeLa cells were transfected using LipofectAMINE 2000™ reagent (Invitrogen). HepG2 cells were transfected by calcium phosphate co-precipitation method. All cells were transfected for at least 16 h before harvest. For forskolin (Fsk) treatment, cells were incubated in the presence of 10 μM Fsk for 12 h before harvest. Reporter assays were performed using the Dual-Luciferase™ kit (Promega). Luminescence was measured with a LB9570 luminometer (EG&G).
Preparation of lentiviral stock and lentiviral transduction of HepG2 cells were performed using the manufacturer's protocols (Invitrogen).
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed as detailed elsewhere (34), except that the cross-linking step was performed by adding 1% formaldehyde to HepG2 or Hepa1-6 cells for 5 min. The primers used for PCR amplification of the endogenous human PEPCK promoters are forward primer 1, 5′-GACTGTGACCTTTGACTATGGGGTGACATC-3′ (−454 to −424); reverse primer 1, 5′-CTGGATCACGGCCAGGGTCAGTTATGC-3′ (−150 to −123); forward primer 2, 5′-GTTGAGGGCTCGAAGTCTCCCAGCATTC-3′ (−339 to −311); reverse primer 2, 5′-GGCACGAATGTGAGGTCACCTGAAATAG-3′ (+206 to +234). The primers for PCR amplification of rat PEPCK promoter are forward primer, 5′-GTGCAGCCAGCAGCATATG-3′ (−378 to −360); reverse primer, 5′-CTTGGTAGCTAGCCCTCCTC-3′ (−21 to −2). DNA was PCR-amplified from the immunoprecipitated chromatin (number of cycles: 30; annealing temperature: 65°C; extension time: 25 s). Amplification of human MAD1 promoter was performed as described previously (34). The number of cycles for all PCR was optimized to ensure that the amplification is within a quantitative linear range.
Confocal microscopy
MCF-7 and HepG2 were cultured on cover-slips that are situated inside the wells of a 6-well plate. Transfected or lentivirus-transduced cells were washed with phosphate-buffered saline (PBS) and then fixed in ice-cold 50% acetone/50% methanol for 10 min, followed by three washes with PBS. Cells were co-stained for CREB-H with calnexin/GM130. The cover-slips containing cells were mounted on glass slide using VECTORSHIELD agent (Vector). Confocal immunofluorescence microscopy was then performed on BioRad MRC1024 system and the images were captured at 63× magnification with the help of the LaserSharp software.
Patients and tumor samples
Sampling of tumor tissues was performed as detailed elsewhere (35). In short, 26 Chinese patients (18 males and 8 females) who had had surgical resection of the tumors at the University of Hong Kong Medical Center were randomly selected for study. Paired samples of their HCCs and corresponding non-tumorous liver tissues were used. The patients' ages ranged from 29 to 74 years (mean: 56.85 years). The tumor size ranged from 2.3 to 27 cm (mean: 8.86 cm), with 20 (76.9%) of them being ≥5 cm in diameter. Eight (30.8%) of the tumors were at earlier stage (tumor, node, and metastasis stages I and II), and the remaining 69.2% (18 cases) were at more advanced stages (stages III and IV). The sera were positive for hepatitis B surface antigen in 21 (80.77%) patients. All specimens were obtained immediately after surgical resection, snap-frozen in liquid nitrogen and kept at −70°C. Frozen sections were cut from the tumor and non-tumorous liver blocks separately and stained for histological examination to ensure homogeneous cell populations of tissues.
RNA extraction and RT–PCR analysis
Total RNA was extracted from human HCC and their corresponding non-tumorous livers with TRIzol reagent as described by the manufacturer (Invitrogen). cDNA was synthesized from 1 μg of total RNA by using oligo(dT)16 as primer and with Gene-Amp RNA PCR kit (Applied Biosystems). PCR amplification using a set of primers (forward: 5′-AGTGTTCTCCAGAACTTTGC-3′; reverse: 5′-TGCACGTCCTGAGCCAGT-3′) was performed to give a product of 286 bp (from nucleotide 1187–1472). A fragment of β-actin was amplified as a control. The PCR was performed in a 9700 thermocycler (PerkinElmer). PCR was stopped at the exponential phases, 28 cycles for CREB-H and 28 cycles for β-actin, with one cycle of hot-start at 95°C for 12 min, followed by amplification at 94°C for 30 s, 53°C for 30 s, 72°C for 30 s and a final elongation at 72°C for 5 min. The PCR products were analyzed by electrophoresis on 1.5% agarose gels, and their signal intensities were measured using GelWorks ED Intermediate Software (UVP).
Growth suppression assays
For β-galactosidase (β-gal) staining experiments, 5 × 105 HepG2 cells were cultured in each well of a 6-well plate, and co-transfected at day 0 with pSV-βgal (2 μg) and a CREB-H-expressing plasmid (0.2 μg; either pcDNA3.1/V5-CREB-H or pcDNA3.1/V5-CREB-HΔTC) at a ratio of 10:1. On day 1 and day 5 post-transfection, cells were harvested and stained with X-gal for β-gal activity. Briefly, the cells were washed three times with PBS and then fixed by adding 0.2% glutaraldehyde in PBS for 5 min at room temperature. Cells were washed again three times with PBS and then X-gal/PBS was added. β-gal staining was performed at 37°C in a humidified atmosphere at 5% CO2 for at least 2 h. Cell growth was calculated by counting the number of blue cell clusters (more than four cells) in five different fields at 10× and 40× magnification under microscope.
For BrdU staining experiments, 1 × 105 HepG2 cells were cultured on cover-slips in 6-well plate. Cells were transfected with either a control plasmid pcDNA3.1/V5-β-gal or a CREB-H-expressing plasmid for 16 h. Cells were then incorporated with BrdU (10 μM) for 6 h, washed three times with PBS and fixed with ice-cold methanol/acetone (1/1, v/v). Cells were subsequently stained with rabbit anti-V5 and fluorescein-conjugated mouse anti-BrdU. A Texas red-conjugated secondary antibody against rabbit IgG (Cappel) was used to visualize the V5 signals. For quantitative analysis, 200 transfected cells from each transfection were examined. Cells were counted as BrdU-positive if the intensity of BrdU staining was increased more than 5-fold compared with adjacent untransfected cells on the same focal plane. Three independent transfections were analyzed for each group of cells. The percentage of blue cells reported represents the average of three transfections.
RESULTS
CREB-H belongs to a new subfamily of bZIP transcription factors
In search of proteins closely related to LZIP, we identified several mouse expressed sequence tag clones that might encode novel bZIP proteins homologous to LZIP. DNA sequencing of one clone (IMAGE 1887290) confirmed that this cDNA codes for a protein of 479 amino acids harboring a characteristic bZIP domain (GenBank accession no. AF392874). The coding sequence is reasoned to be complete in light of the existence of an in-frame termination codon immediately upstream of the putative initiating ATG. More than 75% of the residues in this novel protein are similar to those in LZIP.
At the time of initial identification, this mouse LZIP-like protein had no existing counterparts in the databases, except for a paralogous protein termed OASIS/CREB3L1 (36). Subsequently, a highly homologous human protein was identified in a search for liver-specific transcription factor through random sequencing of cDNAs in libraries derived from human liver. This protein designated CREB-H is specifically expressed in hepatocytes (20). In light of the high homology between the two proteins (67% identity and 73% similarity), the LZIP-related protein we identified likely represents the mouse ortholog of human CREB-H. Notably, the official name for CREB-H as approved by Mouse Genome Informatics is cAMP responsive element binding protein 3-like 3 protein (CREB3L3; http://www.informatics.jax.org). For simplicity and consistency, hereafter we will call it mouse CREB-H. The mouse CREB-H locus was found to locate in chromosome 10. The gene is 8.5 kb in length and contains 12 exons.
Interestingly, several additional mammalian bZIP proteins significantly homologous to LZIP and CREB-H have been identified more recently. These include AIBZIP/Atce1/CREB4/CREB3L4 (37,38) and human BBF2H7/CREB3L2 (39). These proteins contain a short stretch of hydrophobic amino acids, which was predicted to be a TM domain by two programs HMMTOP (Hungarian Academy of Sciences; http://www.enzim.hu/hmmtop) and PredictProtein (Columbia University; http://cubic.bioc.columbia.edu/predictprotein). The C-terminal TM domain immediately adjacent to the bZIP region is highly conserved in this unique group of bZIP proteins. Indeed, phylogenetic analysis based on protein sequences segregates these bZIP proteins into a new subfamily distinct from CREB and ATF6.
CREB-H preferentially recognizes CRE, ATF6 and box B elements
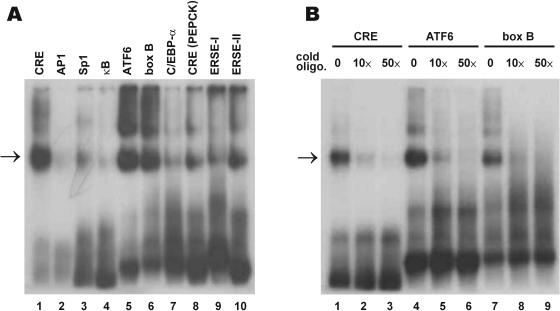
Human CREB-H binds to CRE and box B (20). However, mouse Atce1 closely related to CREB-H has been shown to recognize κB motif instead of CRE (38). To clarify this issue, recombinant polyhistidine-tagged mouse CREB-HZ protein, which contains the bZIP region (amino acids 206–318), was expressed and purified from E.coli. This protein was then tested for its ability to bind various 32P-labeled DNA elements using the EMSA assay. In addition to CRE, box B and κB site, canonical ATF6-binding element (26) was also examined because ATF6 has a TM domain (40) and shares some similarity to CREB-H subfamily proteins. ERSE-I and ERSE-II are two different ATF6-responsive elements found in the promoter of GRP78 (32) and Herp (33) genes, respectively. CREB-H was found to bind strongly to canonical CRE, ATF6 and box B elements (Figure 1A, lanes 1, 5 and 6), but not to AP-1 or κB motif (lanes 2 and 4). CREB-H also bound less potently to CRE in the rat PEPCK gene, ERSE-I and ERSE-II (lanes 8–10), but it hardly recognized Sp1 and C/EBPα motifs (lanes 3 and 7). Results from the competition EMSA assay using 10- and 50-fold cold oligonucleotides verified the specific interaction of CREB-H with CRE, ATF6 and box B elements (Figure 1B).
Figure 1.
CREB-H preferentially recognizes CRE, ATF6 and box B elements. (A) EMSA assay. EMSA was performed using 0.2 μg of purified CREB-HZ protein (amino acids 206–318), 3 pmol of 32P-labeled oligonucleotides representing the indicated DNA binding motifs, and 0.5 μg poly(dI–dC). (B) Competition assay. A 10- and 50-fold excess (10× and 50×) of cold oligonucleotides (cold oligo.) were added to the reactions in lanes 2, 3, 5, 6, 8 and 9. The reactions in lanes 1, 4 and 7 received 50-fold excess of cold irrelevant oligonucleotides corresponding to GAL4-binding motif. The retarded bands containing the CREB-H-DNA complex are highlighted by arrows.
Proteolytic processing of CREB-H in cultured cells
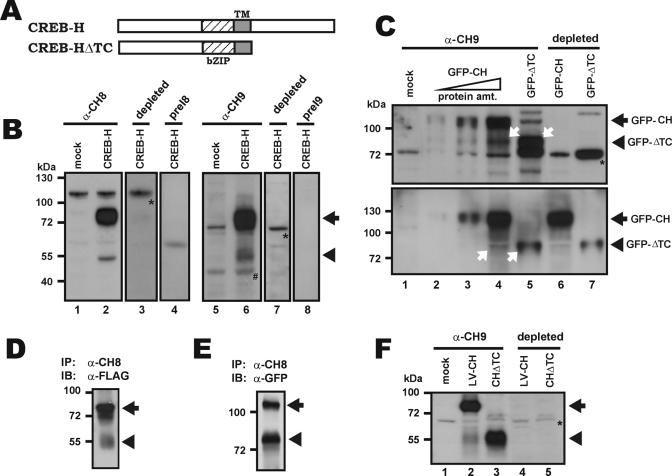
We raised two polyclonal antisera (α-CH8 and α-CH9) in rabbits against the N-terminal 84 amino acids of CREB-H. We then performed western blotting with these antibodies to probe CREB-H protein in Hepa1-6 hepatoma cells that were either mock-transfected (Figure 2B, lanes 1 and 5), transfected with an untagged CREB-H-expressing plasmid (Figure 2B, lanes 2–4) or transfected with a FLAG-tagged CREB-H expressing plasmid (Figure 2B, lanes 6–8). Consistent with our results from northern blot analysis (see below in Figure 6), the amount of endogenous CREB-H protein in Hepa1-6 cells was very low (Figure 2B, lanes 1 and 5). In contrast, several additional α-CH8/9-reactive protein bands were observed in CREB-H-overexpressing Hepa1-6 cells (Figure 2B, lanes 2 and 6).
Figure 2.
Proteolytic processing of CREB-H in hepatoma cells. (A) Schematic representation of the structure of CREB-H and CREB-HΔTC. (B) Western blot analysis. Empty vector (mock; lanes 1 and 5), pFLAG-CREB-H (lanes 2–4) and pcDNA-CREB-H (lanes 6–8) were transfected separately into mouse hepatoma cell line Hepa1-6 for 24 h. Cells were then harvested and lysed. The whole cell extracts thus obtained were analyzed by 10% SDS–PAGE followed by western blotting using α-CH8 (lanes 1 and 2), α-CH9 (lanes 5 and 6) and their respective pre-immune sera (preI8 and preI9; lanes 4 and 8). Antibody depletion was performed (lanes 3 and 7) by pre-incubating α-CH8/9 with Sepharose beads coupled to GST-CREB-H (1–84) fusion protein (10 μg) containing the N-terminal 84 amino acids of CREB-H. The full-length CREB-H protein and the 50 kDa cleaved product of CREB-H are indicated with arrow and arrowhead, respectively. The 110- and 72 kDa cross-reactive bands are highlighted with asterisk. Trace amount of a 45 kDa band (marked with ‘#’) was seen in mock- and pFLAG-CREB-H-transfected Hepa1-6 cells (lanes 5 and 6), but it could not be detected with depleted α-CH9 serum (lane 7). This species recognized by α-CH9 but not by α-CH8 likely represents an endogenous cross-reactive protein (C) Western blot analysis was performed as in B, except that GFP-CREB-H (GFP-CH) and GFP-CREB-HΔTC (GFP-ΔTC) constructs were used. Lanes 2–4 received increasing amounts of protein (protein amt.). The blot in the upper panel was probed with α-CH9 (lanes 5–6) or α-CH9 depleted with an excess (10 μg) of GST-CREB-H (1–84) (lanes 6 and 7). The same blot was stripped and re-probed with a mouse monoclonal antibody to GFP (lower panel). The GFP-CREB-H and GFP-CREB-HΔTC bands are indicated. White arrows highlight the GFP-CREB-HΔTC band (lane 5) and the cleavage product of GFP-CREB-H of the same size (lane 4). A 72 kDa cross-reactive band is indicated with an asterisk. (D and E) Hepa1-6 cells were transfected with pFLAG-CREB-H and the pGFP-CREB-H. CREB-H protein was immunoprecipitated (IP) with α-CH8 antibody and the precipitates were immunoblotted (IB) with α-FLAG and α-GFP antibodies, respectively. (F) Western blot analysis was performed with extracts of HepG2 cells transduced with a lentivirus expressing CREB-H (LV-CH). Protein extract of HepG2 cells transfected with CREB-HΔTC (CHΔTC) plasmid (lanes 3 and 5) was used as a size reference for the proteolytic fragment.
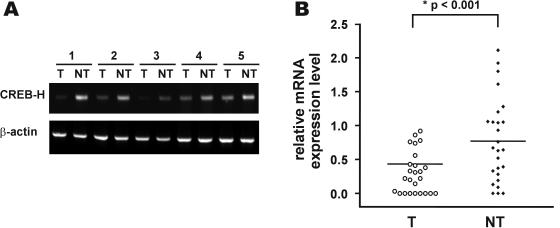
Figure 6.
CREB-H is underexpressed in human HCC tissues. Surgically resected HCC (T) and adjacent non-cancerous liver tissues (NT) from 26 patients were subjected to RNA extraction and subsequent RT–PCR using primers that amplify 286 bp cDNA fragment of human CREB-H, as well as a fragment of β-actin. Representative RT–PCR results were shown in (A). The normalized values of the relative quantities of the CREB-H PCR products as compared with the respective amount of β-actin were shown in a scatterplot (B). The lines indicate the median.
To verify the identity of these reactive bands, we tested the pre-immune sera (Figure 2B, lanes 4 and 8) and performed protein blocking assay by pre-incubating α-CH8/9 with an excess amount of recombinant CREB-H protein (Figure 2B, lane 3 and 7). Because neither pre-immune nor depleted α-CH8/9 recognized the bands of ∼80 and 50 kDa in size (Figure 2B, lanes 3, 4, 7 and 9), these bands should represent specific CREB-H-derived species. While the ∼80 kDa proteins (Figure 2B, lanes 2 and 6, bands with an arrow) plausibly represent the full-length CREB-H and FLAG-CREB-H, the 50 kDa species (bands with an arrowhead) might be a cleaved product (Figure 2B, compare lane 2 to 3, and lane 6 to 7). Both forms of CREB-H might have extensive post-translational modifications because their sizes are larger than predicted.
Since α-CH8/9 recognizes the N-terminus of CREB-H, the appearance of 50 kDa α-CH-reactive species suggests site-specific proteolysis of CREB-H at the C-terminus. To verify the specificity of α-CH8/9 and to further characterize the proteolytic processing of CREB-H, we expressed two versions of CREB-H proteins (Figure 2A, full-length CREB-H and truncated CREB-HΔTC) fused to GFP. In CREB-HΔTC, a C-terminal part including the TM domain was removed (Figure 2A). Comparison of this artificially truncated protein with the proteolytic fragments of CREB-H might shed light on the cleavage site in CREB-H. To this end, Hepa1-6 cells were transfected with GFP-CREB-H and GFP-CREB-HΔTC plasmids and the cell extracts were examined by western blotting (Figure 2C).
With increasing amounts of protein loaded, α-CH9 was able to detect a band of ∼110 kDa in size that corresponds to GFP-CREB-H (Figure 2C, upper panel, lanes 2–4). The blot was re-probed with a monoclonal anti-GFP antibody (α-GFP; Figure 2C, lower panel, lanes 3 and 4). The reaction of the 110 kDa band with both α-CH9 and α-GFP verified the identity of GFP-CREB-H and it also lent further support to the specificity of α-CH9.
Likewise, both α-CH9 and α-GFP recognized the GFP-CREB-HΔTC protein of 75 kDa (Figure 2C, lane 5). Interestingly, in cells that had been transfected with a GFP-CREB-H plasmid but not a GFP-CREB-HΔTC construct, an α-CH9- and α-GFP-reactive band of about the same size was also observed (Figure 2C, compare lane 4 with lane 5, bands with arrows). This species plausibly represents a processed form of CREB-H that had been cleaved at a specific site. Because the size of this cleavage product is almost identical to that of GFP-CREB-HΔTC, the actual cleavage site was predicted to be very close to the junction between bZIP and TM regions (Figure 2A). It is noteworthy that this cleaved species of 77 kDa shown in Figure 2C corresponds to the 50 kDa band in Figure 2B. The difference in molecular mass (∼27 kDa) was accounted for by GFP. In addition, when we performed immunoprecipitation with α-CH8 and extracts of cells transfected with either FLAG-CREB-H or GFP-CREB-H plasmid, two major protein species corresponding to the full-length and cleaved forms of CREB-H, respectively, were found in the precipitates (Figure 2D and E).
Next, we asked whether proteolysis also occurs in HepG2 cells stably transduced with a lentivirus expressing CREB-H. In keeping with results obtained from transfected cells, a strong ∼80 kDa band and a weaker ∼50 kDa species reactive to α-CH9 were observed in extracts of lentivirus-transduced cells (cf. Figure 2F, lane 2 with Figure 2B, lane 5). These two protein bands were not seen when α-CH9 was depleted with recombinant CREB-H (Figure 2F, lane 4). As a size reference for the ∼50 kDa band observed in lentivirus-transduced cells, protein extract of HepG2 cells expressing CREB-HΔTC, which did not react to depleted α-CH9 (Figure 2F, lane 5), was also examined (cf. lane 3 with lane 2).
CREB-H activates CRE- and ATF6 element-dependent transcription
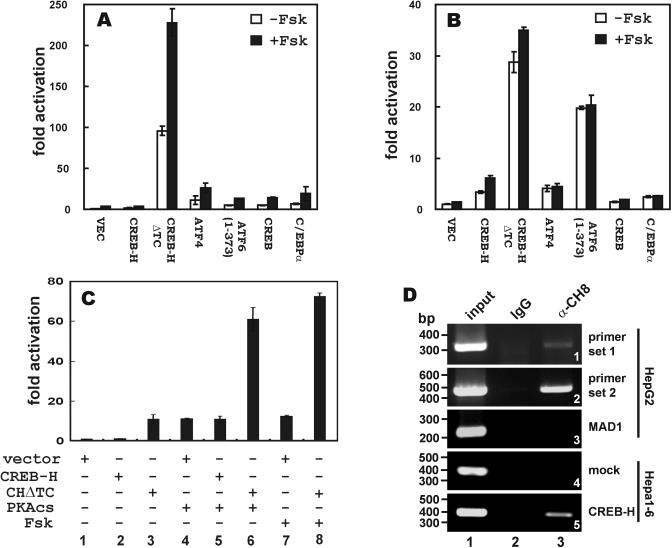
Human CREB-H has been shown to activate box B-dependent transcription (20). Box B is a fat-body-specific enhancer in Drosophila and box B-like sequences in mammalian cells have not been characterized (18). In light of the strong binding of CREB-H to CRE and ATF6 element (Figure 1), we next asked whether CREB-H was able to drive transcription from CRE or ATF6 element. To address this, we co-transfected a CREB-H expressing plasmid and either pCRE-Luc or pATF6-Luc reporter plasmid into HepG2 hepatoma cells and assayed for luciferase activity. For comparison, we also tested the activities of ATF4, ATF6, CREB and C/EBPα on CRE and ATF6 enhancers (Figure 3A and B). While the full-length CREB-H weakly activated CRE-dependent transcription (Figure 3A, 1.6-fold activation), it was capable of stimulating the ATF6 enhancer to 4.9-fold (Figure 3B). This potency is comparable with that of ATF4, CREB and C/EBPα (11.3-, 3.9- and 6.9-fold activation, respectively).
Figure 3.
Transcriptional activity of CREB-H and potentiation by cAMP. Empty vector (VEC) and plasmids (0.3 μg of each) expressing CREB-H, CREB-HΔTC (CHΔTC), ATF4, ATF6, CREB and C/EBPα were individually co-transfected with pCRE-Luc (A), pATF6-Luc (B) or pPEPCK-Luc (C) plasmid (0.2 μg) into HepG2 (A and B) and Hep3B (C) cells with or without the addition of Fsk or catalytic subunit of PKA (PKAcs). An SV40 promoter-driven Renilla luciferase plasmid was also transfected into the cells. The cell lysates were measured for Firefly luciferase luminescence and the readings were normalized by the luminescence of Renilla luciferase. (D) ChIP assay was performed with α-CH8 and extracts of HepG2 cells transfected with pcDNA-CREB-H. One control group received 10% of input chromatin (group 1). The other control group was given pre-immune IgG (group 2). Primer sets 1 and 2 were used to amplify sequences in the proximal promoter of human PEPCK gene (panels 1 and 2). Experiments were also carried out with primers for amplifying human MAD1 promoter (panel 3). Similar ChIP assays were performed in mock-transfected (panel 4) and CREB-H-expressing (panel 5) Hepa1-6 cells carrying a rat PEPCK promoter. Enforced expression of CREB-H was achieved by co-transfection with pcDNA-CREB-H. No CREB-H-bound DNA could be detected when input chromatin was omitted (data not down).
It is generally accepted that removal of the TM domain from ATF6 and LZIP leads to the activation of these transcription factors (40,41). The proteolytic cleavage of ATF6 can be induced in vivo in response to ER stress (40), but the physiological stimuli for the activation of LZIP remain elusive. CREB-H also contains a TM domain, which is probably removed through site-specific proteolytic cleavage (Figure 2) as exemplified in the cases of ATF6 (42) and SREBPs (43,44). To test the hypothesis that CREB-H could also be activated through regulated proteolysis, we asked whether the truncated CREB-HΔTC devoid of the TM and C-terminal domains (Figure 2A) might have a higher transcriptional activity. Indeed, CREB-HΔTC acted as a transcriptional activator on both CRE and ATF6 element, resulting in ∼100- and 30-fold increase of luciferase activity (Figure 3A and B). In this setting, CREB-HΔTC is more potent than CREB, ATF4, ATF6 and C/EBPα. This raised the possibility that the truncated CREB-HΔTC might be functionally equivalent to the physiologically active form of CREB-H generated through site-specific proteolysis in vivo (see Figure 2).
CREB-H is activated in response to cAMP stimulation
cAMP is a ubiquitous second messenger and regulator of gene transcription (45,46). In one well-characterized pathway, cAMP activates PKA, which phosphorylates and activates CREB (17). In addition, C/EBPs are also responsive to cAMP stimulation (6). This prompted us to investigate the regulation of CREB-H by cAMP. We measured the activities of CREB-H and CREB-HΔTC on the pCRE-Luc and pATF6-Luc reporters with or without the addition of Fsk, which stimulates adenylate cyclase leading to increased cAMP levels (Figure 3A and B). Fsk did not activate ATF6 element-dependent transcription significantly with the exception of CREB-H, which was stimulated mildly by Fsk (Figure 3B, ∼80% increase in luciferase activity). As to CRE-dependent transcription, both CREB-H and CREB-HΔTC were modestly responsive to Fsk stimulation, resulting in up to 100% increase in reporter activity (Figure 3A, 3.7- and 227.8-fold activation, respectively). However, in the control experiment in which the empty vector alone was co-transfected, Fsk also stimulated CRE-dependent transcription (Figure 3A, 3.7-fold activation). Thus, while it remains unclear as to whether Fsk specifically stimulates CREB-H, Fsk activation of endogenous transcription factors alone unlikely accounted for the increase of CRE-dependent luciferase activity observed in the presence of CREB-HΔTC and Fsk. Thus, cAMP might be directly or indirectly involved in regulating CREB-H and CREB-HΔTC.
CREB-H activates PEPCK promoter
While the DNA-binding and gene-activating abilities of LZIP, CREB-H and other transcription factors in the same subfamily have been well documented (20,22,23,37,38,47,48), their physiological targets remain to be identified. As a first step toward identifying the gene targets of CREB-H (20), we asked whether it might bind and activate CRE-dependent transcription in liver. In this regard, the gene encoding gluconeogenic enzyme PEKCK has been extensively used as a model for studying the regulation of CREB- and C/EBP-dependent transcription in liver (49–51). Based on this reasoning, we have demonstrated that CREB-H binds modestly to the CRE in rat PEPCK promoter (Figure1A, lane 8). Because CREB-H binds moderately to the rat PEPCK CRE, here we set out to investigate whether it might activate the PEPCK promoter. Indeed, CREB-HΔTC but not the full-length CREB-H stimulated the PEPCK promoter activity when overexpressed in hepatoma cell line Hep3B (Figure 3C, lane 3). PEPCK is abundantly expressed in liver and is an important metabolic enzyme that controls gluconeogenesis in both liver and adipose tissues. It is well known that cAMP/PKA can activate the PEPCK gene transcription through the cAMP response unit in the PEPCK promoter (6,50). As such, when the cAMP pathway is stimulated by either treatment of Fsk or the expression of PKA catalytic subunit in Hep3B cells, the PEPCK promoter is stimulated (Figure 3C, lanes 4 and 7) (52). Consistent with our finding that Fsk was able to further enhance CREB-HΔTC activation of CRE (Figure 3A), we observed that the expression of CREB-HΔTC cooperated with either PKA or Fsk in the activation of PEPCK promoter (Figure 3C, lanes 6 and 8).
Because the reporter assays were performed in cells transfected with PEPCK-Luc plasmid, we next investigated the binding of CREB-H to genomic PEPCK promoter in CREB-H-expressing cells using ChIP assay (Figure 3D). We observed that α-CH8 but not pre-immune IgG precipitated a CREB-H–DNA complex containing endogenous PEPCK promoter (Figure 3D, panels 1 and 2, lane 3 compared with lane 2). In a control experiment, α-CH8 did not pull down an irrelevant genomic sequence in human MAD1 promoter (Figure 3D, panel 3, lane 3). In further support of the specificity of the interaction between CREB-H and the PEPCK promoter, ChIP assays performed in non-CREB-H-expressing Hepa1-6 cells (see Figure 5) yielded no CREB-H–DNA complex (Figure 3D, panel 4), whereas the PEPCK promoter was amplified from the CREB-H-containing immunoprecipitate prepared from CREB-H-expressing Hepa1-6 cells. Thus, our results consistently revealed that CREB-H specifically associates with PEPCK promoter in vivo.
Figure 5.
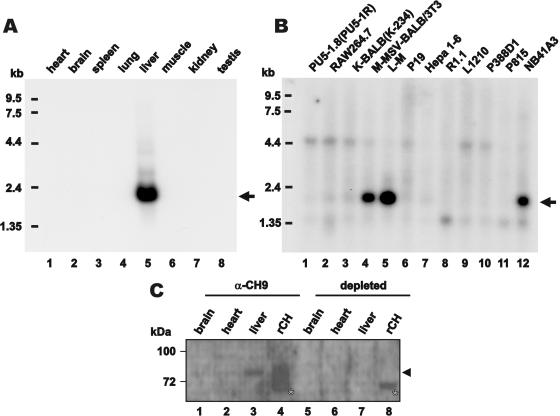
CREB-H is exclusively expressed in the liver of adult mouse and is differentially expressed in mouse cancer cell lines. Northern blot analysis was performed on mouse tissues (A) and cancer cell lines (B). CREB-H transcripts are arrowed. Western blotting was also performed on protein extracts from mouse tissues (C). Mouse brain, heart and liver tissues were probed with α-CH9 (lanes 1–4) and depleted α-CH9 (lanes 5–8). Recombinant CREB-H (rCH) prepared from transfected Hepa1-6 cells was also loaded as a positive control (lanes 4 and 8). CREB-H protein is highlighted with an arrowhead. The 72 kDa cross-reactive bands are indicated with an asterisk.
Subcellular localization of CREB-H
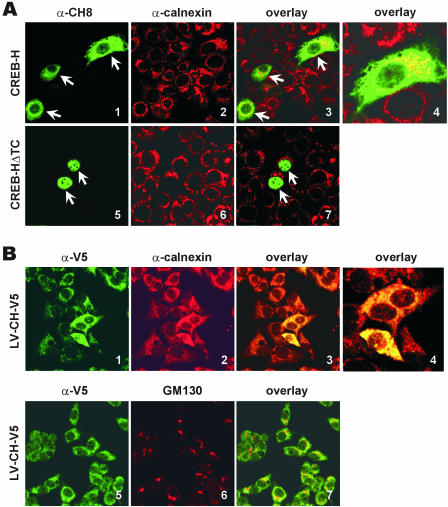
We expressed CREB-H and CREB-HΔTC proteins in MCF-7 cells and determined their subcellular localization. CREB-H localized predominantly in the cytoplasm (Figure 4A, panel 1), whereas CREB-HΔTC was exclusively in the nucleus (panel 5). Because ATF6 with a TM domain localizes to ER membrane and is activated by intramembrane proteolysis (40), we investigated the association of CREB-H with ER by co-staining cells for CREB-H and calnexin, an ER marker. As shown in Figure 4A, full-length CREB-H co-localized with calnexin (panels 3 and 4 compared with panel 7). These results suggest that CREB-H, similar to ATF6, localizes to the ER.
Figure 4.
Subcellular localizations of CREB-H and CREB-HΔTC. (A) pcDNA-CREB-H (panels 1–4) and pcDNA-CREB-HΔTC (panels 5–7) plasmids were separately transfected into MCF-7 cells. Cells were fixed and co-stained with α-CH8 (green, panels 1 and 5) and α-calnexin (red, panels 2 and 6). Green (CREB-H) and red (calnexin) fluorescent signals were overlaid by computer assistance (panels 3, 4 and 7). Panel 4 is a high magnification image of a portion of panel 3. Co-localizations are shown in yellow. The same fields are shown in panels 1–3 and 5–7. Arrows indicate transfected cells. (B) Similar co-localization studies were performed with HepG2 cells transduced with a lentivirus expressing V5-tagged CREB-H (LV-CH-V5). Green (CREB-H) and red (calnexin or GM130) fluorescent signals were overlaid in panels 3, 4 and 7. Panel 4 is a high magnification image of a portion of panel 3. Results are representative of three independent experiments.
Next, we examined the localization of CREB-H protein in HepG2 cells homogenously expressing V5-tagged CREB-H from a lentiviral vector. Again, a typical cytoplasmic localization pattern was observed when we stained the lentivirus-transduced cells with anti-V5 (Figure 4B, panels 1 and 5). We noticed a significant co-localization of CREB-H and calnexin (Figure 4B, panels 1–4). While the staining patterns of CREB-H and the Golgi marker GM130 were distinct, a fraction of CREB-H was also found in the Golgi apparatus (Figure 4B, panels 5–7). Thus, the full-length but inactive CREB-H (Figure 3) is retained predominantly in the ER. On the contrary, CREB-HΔTC acts as a potent transactivator plausibly owing to constitutive nuclear localization.
CREB-H expression is abundant in adult liver but reduced in hepatoma cells
We performed northern blot analysis of CREB-H mRNA in mouse tissues and cells. Consistent with previous findings in human (20), CREB-H mRNA was exclusively and abundantly detected in adult mouse liver (Figure 5A, lane 5). On the other hand, the CREB-H signal was not observed in a mouse hepatoma cell line Hepa1-6 (Figure 5B, lane 7). In addition, we also found that human CREB-H transcript is underexpressed in most hepatoma cell lines tested (data not shown). It is, however, noteworthy that mouse CREB-H transcript is highly expressed in 3T3 fibroblasts immortalized by mammary sarcoma virus (M-MSV-BALB/3T3), cancerous subcutaneous adipocytes (L-M) and NB41A3 cancer cells derived from neuroblastoma (Figure 5B, lanes 4, 5 and 12).
Consistent with results from northern blot analysis, CREB-H protein was detected in mouse liver, but not in brain or heart (Figure 5C, lanes 1–3). The identity of the endogenous CREB-H protein in mouse liver was verified by comparing with untagged recombinant CREB-H (Figure 5C, lane 4) and by antibody depletion experiments (Figure 5C, lanes 5–8). Notably, the truncated CREBΔTC form was undetectable in the blot, implicating that CREB-H is not constitutively active in mouse liver.
Because CREB-H is abundantly expressed in normal adult liver but underexpressed in liver cancer cell lines, we speculated that CREB-H might be critically involved in hepatocarcinogenesis. With this hypothesis in mind, we performed RT–PCR screening of CREB-H transcripts in human HCC tissues. The CREB-H mRNA was significantly underexpressed in HCC samples when compared with corresponding non-tumorous liver (Figure 6, P < 0.001 by Wilcoxon test). A more than 2-fold reduction in steady-state amounts of CREB-H mRNA was observed in 11 of the 26 HCC tissues (see Figure 6A for representative RT–PCR results and Figure 6B for a scatterplot).
Growth suppressive activity of CREB-H in hepatoma cells
The underexpression of CREB-H in HCC cells and tissues (Figures 5 and 6) suggests that it might play a role in hepatocarcinogenesis. Considered together with the tumor suppressive activities of other bZIP proteins, such as C/EBPα (11,12), C/EBPβ (53) and LZIP (22), we queried whether CREB-H might also serve a growth suppressive function in the liver. We carried out a growth assay in HepG2 cells using transient co-transfection of pSV-βgal, a β-gal expression vector driven by SV40 promoter, plus excess amount (10×) of CREB-H expressing plasmids. This ensures that the blue cells are the CREB-H transfected cells when staining with X-gal. This method has previously been used to study the growth suppressive activity of various proteins, including C/EBPα and C/EBPβ (9,12,54).
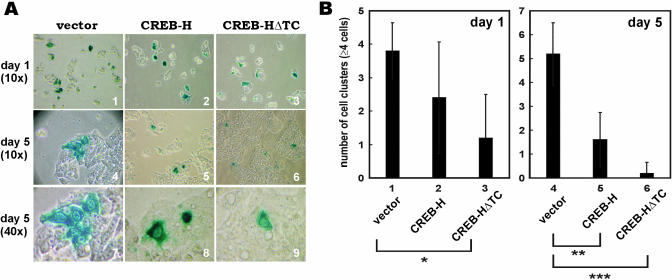
On day 1 after co-transfection, slight differences were seen in the number of blue cell clusters among the groups that had received vector alone, full-length CREB-H and CREB-HΔTC (Figure 7A, panels 1–3). On average, four cell clusters (of ≥4 cells) were seen in vector-transfected cells, whereas two and one clusters were evident in CREB-H- and CREB-HΔTC-overexpressing cells, respectively (Figure 7B, columns 1–3). However, on day 5 after transfection, significant differences were observed among vectors, CREB-H and CREB-HΔTC groups (Figure 7A, panels 4–9; Figure 7B, columns 4–6). On average, four cell clusters increased to seven cell clusters in vector-transfected cells, whereas only one or less than one cell cluster was found in CREB-H- and CREB-HΔTC-overexpressing cells (Figure 7B, columns 4–6). Western blot analysis confirmed that the protein expression levels of CREB-H and CREB-HΔTC in HepG2 cells on day 1 and day 5 post-transfection were similar (data not shown). Thus, CREB-H and CREB-HΔTC inhibited the proliferation of HepG2 cells.
Figure 7.
Overexpression of CREB-H suppresses growth in cultured human cells. (A) pcDNA3.1/V5 vector, pcDNA3.1/V5-CREB-H and pcDNA3.1/V5-CREB-HΔTC were co-transfected with pSV-βgal at a ratio of 10:1 into HepG2 cells. Cells were harvested and stained with X-gal for β-gal activity on day 1 and day 5 post-transfection. Images of the blue-stained cells were taken at 10× magnification for cells harvested on day 1, and at both 10× and 40× magnifications for cells harvested on day 5. (B) Cell clusters containing more than four cells were counted in five different fields at 10× and 40× magnifications and scored in a bar graph. Results represent the average of five independent experiments. Error bars indicate the SE. On day 1, the number of blue cells in the CREB-HΔTC group was significantly lower than in the group that had received vector alone (P < 0.01 by t-test; marked with *). On day 5, the numbers of blue cells in CREB-H and CREB-HΔTC groups were also significantly lower than the group with vector alone (P < 0.002 and P < 0.0001, respectively; marked with ** and ***).
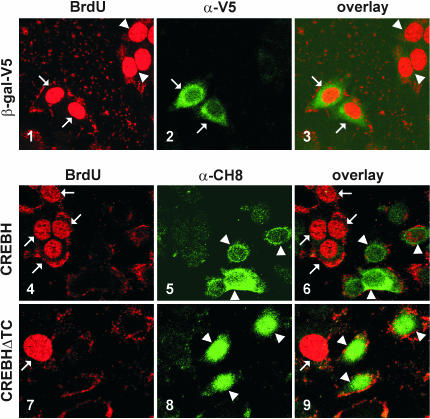
To further characterize the growth inhibitory activity of CREB-H, we measured 5-bromo-2′-deoxyuridine (BrdU) incorporation in CREB-H- and CREB-HΔTC-expressing HepG2 cells using confocal microscopy. BrdU incorporation indicates the cellular activity of DNA synthesis that dictates competency of cell proliferation. This method has been widely used to study cell growth and cell cycle progression. In particular, it has been adopted to demonstrate the growth suppressive role of transiently expressed C/EBPα in cultured cells (12). In our control experiment, the expression of β-gal did not affect BrdU incorporation (Figure 8, panels 1–3). In stark contrast, BrdU staining was undetectable in either CREB-H- or CREB-HΔTC-expressing HepG2 cells (Figure 8, panels 4–9; compare transfected cells with arrowheads to surrounding untransfected cells with arrows). Quantitative analysis by cell counting indicates that none of the CREB-H- or CREB-H-positive cells was incorporating BrdU, whereas ∼50% (47.6 ± 5.0%) of control cells expressing β-gal were positive for BrdU. In another word, BrdU incorporation was significantly reduced in CREB-H- or CREB-HΔTC-expressing cells compared with control cells (P < 0.0001 by t-test). Thus, the activation of CREB-H can inhibit S-phase entry and cell proliferation.
Figure 8.
Overexpression of CREB-H inhibits S-phase entry. (A) pcDNA3.1/V5-β-gal, pcDNA3.1-CREB-H and pcDNA3.1-CREB-HΔTC were transfected into HepG2 cells for 16 H. Cells were then treated with 10 μM BrdU for 6 h, and fixed. Cells were subsequently stained with α-BrdU-fluorescein and α-V5 or α-CH8 antibodies. Red (BrdU) and green (β-gal, CREB-H or CREB-HΔTC) fluorescent signals were overlaid in panels 3, 6 and 9. The same fields are shown in panels 1–3, 4–6 and 7–9. The arrows indicate transfected cells. Similar results were obtained from three transfections.
DISCUSSION
In this study, we characterized a newly identified liver-enriched transcription factor called CREB-H. CREB-H contains a TM domain and represents a new subfamily of bZIP proteins that have not been extensively studied. CREB-H binds to CRE, box B and ATF6 enhancer elements (Figure 1) and activates transcription from CRE, ATF6 site and PEPCK promoter in response to cAMP/PKA (Figure 3). CREB-H associated with the ER in the cytoplasm (Figure 4) is proteolytically processed in hepatoma cells to generate an active form devoid of the repressive TM domain (Figure 2). This active form of CREB-H constitutively localizes to the nucleus (Figure 4). CREB-H is exclusively expressed in adult liver and is significantly underexpressed in HCC tissues and cells (Figures 5 and 6). Finally, CREB-H functions as a growth suppressor in cultured cells (Figures 7 and 8). Thus, CREB-H serves regulatory roles in liver-specific transcription and the growth of hepatocytes.
Collectively, our data derive mechanistic insight into the proteolytic processing, nuclear translocation and regulated activation of CREB-H. In addition, we also provide the first evidence for the growth suppressive activity of CREB-H. Our findings have implications in liver physiology and carcinogenesis.
A new subfamily of bZIP proteins
Compared with prototypic bZIP proteins, such as CREB and C/EBPα, CREB-H is unique in bearing a hydrophobic TM domain immediately adjacent to the dimerization and DNA-binding domains situated in the middle of the protein (Figure 2A). Additional bZIP proteins that share a domain architecture and high homologies with CREB-H can be classified into a new subfamily structurally and functionally different from CREB and ATF6.
In this expanding subfamily, LZIP/CREB3, OASIS and BBF2H7 exhibit broad expression profiles, while CREB-H and AIBZIP are specifically expressed in liver and prostate/testis, respectively. LZIP/CREB3 has been implicated in the suppression of herpes simplex virus replication and in the establishment of viral latency in trigeminal ganglia (55) through association with HCF-1 (21,23). OASIS has initially been isolated from astrocytes in long-term culture, which serve as an in vitro gliosis model (36), implicating a role in inflammation and stress responses. Detailed developmental expression profiling has revealed that OASIS is involved in the late phase of osteoblast differentiation (56). BBF2H7 has recently been retrieved from database as a novel human protein whose C-terminal part was fused to the FUS genes in low grade fibromyxoid sarcoma as a result of chromosomal translocation (39). Human AIBZIP has been found to be expressed abundantly and specifically in prostate tissues and tumors under the control of androgens, suggesting a role in prostate development and carcinogenesis (37). Interestingly, the mouse ortholog of AIBZIP termed Atce1 has also been found in reproductive tissues exhibiting a restricted post-meiotic expression pattern in spermatids in mouse testis (38). Apparently, bZIP proteins in this emerging subfamily serve both general and tissue-specific regulatory functions in various biological processes.
bZIP proteins form homo- and heterodimers through their leucine zipper domains. Selective dimerization is an additional determinant for the functional diversity and specificity of bZIP factors. In this regard, one recent study based on protein arrays has raised the possibility of heterodimerization between bZIP proteins within the emerging LZIP subfamily (57). Hence, it would be of great interest to investigate whether and how CREB-H might interact with other members of this subfamily, such as LZIP and OASIS.
TM domain and activation of CREB-H
Although CREB-H and ATF6 are in phylogenetically separate groups, both proteins have a TM domain immediately downstream of the bZIP region. ATF6 ambiently localizes to the ER and functions as a proximal sensor of ER stress (58,59). In response to the accumulation of unfolded proteins in the ER, ATF6 is activated by regulated intramembrane proteolysis to release its active N-terminal fragment, which translocates into the nucleus to activate the transcription of ER-resident chaperones and folding enzymes (40). This active form of ATF6 binds to the consensus ER stress elements (ERSEs) constitutively pre-occupied by NF-Y (32,33,60). CREB-H binds modestly to ERSEs (Figure 1); however, it neither activates the genes coding for ER chaperones, such as GRP78 and GRP94, nor responds to the ER stress-inducing agents, such as tunicamycin (K.-T. Chin and D.-Y.Jin, unpublished data). Thus, it remains to be understood whether CREB-H might regulate gene transcription through the ERSEs.
The ATF6 site (5′-TGACGTGG-3′) is another ER stress responsive element identified through binding site screening experiments using the bZIP domain of ATF6 (26). While it remains to be seen that ATF6 can directly bind to the ATF6 site (32), XBP1, another bZIP protein which is the downstream target of ATF6 (60), has been demonstrated to recognize the ATF6 element (61). Noteworthily, both OASIS (47) and CREB-H (Figure 2) interacted strongly with the ATF6 element. Taken together with the fact that both CREB-H and ATF6 have similar domain structure (data not shown) and localize to ER (Figure 4), CREB-H and ATF6 are likely regulated through distinct but related mechanisms. In this scenario, CREB-H might activate unidentified genes that contain recognition sites similar to the ATF6 element but distinct to the ERSEs. Because ATF6 and XBP1 have been shown to affect only a subset of ER stress targets (62), CREB-H might target a different subset of genes. Meanwhile, it might also cooperate with ATF6 and XBP1 to regulate some common targets.
ATF6 is an archetype of TM domain-containing bZIP factors and it is cleaved by site-1 and site-2 proteases in response to ER stress to release an active N-terminal fragment (42,63). Interestingly, LZIP is also cleaved by the same proteases that act on ATF6 and the activated N-terminal form of LZIP likely translocates into the nucleus to activate transcription (41). However, the physiological stimuli that trigger LZIP cleavage and activation are not understood. In particular, unlike ATF6 or OASIS (64) but similar to CREB-H, LZIP does not respond to tunicamycin stimulation (K.-T. Chin and D.-Y. Jin, unpublished data) (41). Currently, we have no evidence of CREB-H involvement in ER stress response.
In this study, we obtained several lines of data that strongly support the regulated activation of CREB-H. First, we detected a proteolytic fragment of CREB-H in hepatoma cells, which is almost identical to CREB-HΔTC in size (Figure 2). Second, we demonstrated the ER and nuclear localizations of CREB-H and CREB-HΔTC, respectively (Figure 4). Finally, we documented that CREB-HΔTC activates transcription more potently than CREB-H (Figure 3). These findings strongly support the notion that CREB-H could be activated through intramembrane cleavage and subsequent nuclear translocation of an active form equivalent to CREB-HΔTC. In this regard, it will be of interest to elucidate the upstream signals and molecular events that trigger the cleavage of CREB-H.
CREB-H activation of PEPCK promoter
We showed that the promoter of hepatic gluconeogenic enzyme PEPCK is activated by overexpression of CREB-HΔTC, but not the full-length CREB-H (Figure 3C). The cAMP response unit, which mediates the activation of PEPCK promoter by the cAMP/PKA pathway, has been shown to bind a number of bZIP transcription factors, including CREB, CREM, C/EBPα, C/EBPβ, ATF2, ATF3 and AP-1 (49,50,52,65,66). As CREB-H binds to PEPCK-CRE in vitro (Figure 1) and to endogenous PEPCK promoter in vivo (Figure 3D), CREB-H likely activates PEPCK transcription through direct binding to the CRE element (−91 to −84) in its promoter. Plausibly, the regulation of PEPCK by the liver-specific transcription factor CREB-H would be physiologically relevant.
Our results indicate that CREB-HΔTC and cAMP/PKA synergistically activate the PEPCK promoter (Figure 3C). This raises the possibility that post-cleavage activation of CREB-H by cAMP/PKA-dependent phosphorylation might be responsible for regulating PEPCK expression and hence glucose homeostasis in the liver. However, we could not identify potential PKA phosphorylation sites in CREB-HΔTC (data not shown). It remains to be elucidated as to whether CREB-H is phosphorylated by PKA on a non-consensus site. Alternatively, CREB-H might cooperate with other cAMP-responsive CRE-binding factors or transcriptional co-regulators to activate PEPCK expression. In this regard, CREB has been shown to synergize with other transcription factors and co-activators to mediate cAMP responsiveness (67,68). Nevertheless, it is of interest to see whether cAMP and PKA are required for CREB-H activation of PEPCK promoter. PKA inhibitors, such as H89, could be used to address whether the kinase is dispensable for the activity of CREB-HΔTC on the PEPCK promoter.
Growth suppressive function of CREB-H in liver
LETFs are known to be required for liver development and for maintaining its differentiated functions (1,2,4). In this study, we characterized CREB-H, a novel LETF whose expression is exclusively abundant in adult liver (Figure 5A). Notably, CREB-H is underexpressed in HCC tissues and cell lines (Figures 5B and 6). Our findings suggest that CREB-H might play a pivotal role in hepatocyte growth and differentiation. In this context, the loss of CREB-H function in HCC might contribute to the initiation and/or progression of cancer. In support of this model, overexpression of CREB-H sufficiently induced growth suppression in cultured cells (Figure 8).
With respect to the liver-specific and growth suppressor functions, we noticed a strong resemblance of CREB-H to C/EBPα, another LETF of the bZIP family. C/EBPα is involved in regulating hepatic growth and differentiation (69). C/EBPα is also a multifaceted growth suppressor that interferes with various growth regulatory pathways (9–12,14,70). The mechanisms through which CREB-H induces growth arrest in liver remain to be elucidated. In this regard, we noted that forced overexpression of CREB-H in hepatoma cells might trigger apoptosis because the number of CREB-H transfected cell clusters (blue cells in Figure 7A) decreased during prolonged culture. In addition, CREB-HΔTC could not be stably expressed in HepG2 or Hepa1-6 cells transduced with a lentivirus carrying a CREB-HΔTC expression cassette. In fact, the cells were all dead within the first day of lentiviral transduction (K.-T. Chin and D.-Y. Jin, unpublished data).
Since hepatitis C virus core protein targets LZIP (22) which is structurally related to the liver-specific CREB-H, we reasoned that CREB-H might be a more physiologically relevant target of hepatitis C virus core protein. Our preliminary work suggests that CREB-H does not interact directly with hepatitis C virus core protein (data not shown). However, CREB-H could also be targeted indirectly through an interaction with LZIP. Additionally, LZIP is a binding partner of transcriptional co-activator HCF1 (71,72) as well as its related protein HCLP-1 (24), both of which are implicated in cell proliferation (71). It remains to be seen whether CREB-H might interact with HCF-1 and/or HCLP-1. The relationship of CREB-H with LZIP and partners of LZIP merits further study.
Acknowledgments
The authors thank Y.P. Ching for helpful discussions; M. Montminy, K. Mori, K.V. Kibler, G.J. Darlington and R. Prywes for reagents; and Y.P. Ching, J.W.P.Yam, C.M. Wong, A.C.S. Chun, Y.T. Siu and K.L. Siu for critical reading of the manuscript. D.-Y.J. is a Leukemia and Lymphoma Society Scholar. This work was supported by grants to D.-Y.J. from the Hong Kong Research Grants Council (Project HKU 7294/02M and Project N-HKU015/00). The National Natural Science Foundation (Project 3001161945) and the Ministry of Science and Technology of China (Projects 2001AA221041 and G1998051002 under the National Program for Key Basic Research Projects) also provided support to B.-Q.Q. and J.-G.Y. in Beijing. Funding to pay the Open Access publication charges for this article was provided by the Hong Kong Research Grants Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Costa R.H., Kalinichenko V.V., Holterman A.X., Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 2.Duncan S.A. Transcriptional regulation of liver development. Dev. Dyn. 2000;219:131–142. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1051>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- 3.Schrem H., Klempnauer J., Borlak J. Liver-enriched transcription factors in liver function and development. Part I: The hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol. Rev. 2002;54:129–158. doi: 10.1124/pr.54.1.129. [DOI] [PubMed] [Google Scholar]
- 4.Zaret K.S. Liver specification and early morphogenesis. Mech. Dev. 2000;92:83–88. doi: 10.1016/s0925-4773(99)00326-3. [DOI] [PubMed] [Google Scholar]
- 5.Watt A.J., Garrison W.D., Duncan S.A. HNF4: a central regulator of hepatocyte differentiation and function. Hepatology. 2003;37:1249–1253. doi: 10.1053/jhep.2003.50273. [DOI] [PubMed] [Google Scholar]
- 6.Roesler W.J. The role of C/EBP in nutrient and hormonal regulation of gene expression. Annu. Rev. Nutr. 2001;21:141–165. doi: 10.1146/annurev.nutr.21.1.141. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi Y., Wang W., Ninomiya T., Nagano H., Ohta K., Itoh H. Liver enriched transcription factors and differentiation of hepatocellular carcinoma. Mol. Pathol. 1999;52:19–24. doi: 10.1136/mp.52.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L., Hui L., Wang S., Gong J., Jin Y., Wang Y., Ji Y., Wu X., Han Z., Hu G. Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res. 2001;61:3176–3181. [PubMed] [Google Scholar]
- 9.Timchenko N.A., Wilde M., Nakanishi M., Smith J.R., Darlington G.J. CCAAT/enhancer-binding protein α (C/EBPα) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 10.Timchenko N.A., Harris T.E., Wilde M., Bilyeu T.A., Burgess-Beusse B.L., Finegold M.J., Darlington G.J. CCAAT/enhancer binding protein α regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol. 1997;17:7353–7361. doi: 10.1128/mcb.17.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris T.E., Albrecht J.H., Nakanishi M., Darlington G.J. CCAAT/enhancer-binding protein-α cooperates with p21 to inhibit cyclin-dependent kinase-2 activity and induces growth arrest independent of DNA binding. J. Biol. Chem. 2001;276:29200–29209. doi: 10.1074/jbc.M011587200. [DOI] [PubMed] [Google Scholar]
- 12.Wang H., Iakova P., Wilde M., Welm A., Goode T., Roesler W.J., Timchenko N.A. C/EBPα arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell. 2001;8:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 13.Timchenko N.A., Wilde M., Iakova P., Albrecht J.H., Darlington G.J. E2F/p107 and E2F/p130 complexes are regulated by C/EBPα in 3T3-L1 adipocytes. Nucleic Acids Res. 1999;27:3621–3630. doi: 10.1093/nar/27.17.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timchenko N.A., Wilde M., Darlington G.J. C/EBPα regulates formation of S-phase-specific E2F-p107 complexes in livers of newborn mice. Mol. Cell. Biol. 1999;19:2936–2945. doi: 10.1128/mcb.19.4.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinson C., Myakishev M., Acharya A., Mir A.A., Moll J.R., Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell. Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hai T., Hartman M.G. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 17.Shaywitz A.J., Greenberg M.E. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 18.Abel T., Bhatt R., Maniatis T. A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev. 1992;6:466–480. doi: 10.1101/gad.6.3.466. [DOI] [PubMed] [Google Scholar]
- 19.Smolik S.M., Rose R.E., Goodman R.H. A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol. Cell. Biol. 1992;12:4123–4131. doi: 10.1128/mcb.12.9.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omori Y., Imai J., Watanabe M., Komatsu T., Suzuki Y., Kataoka K., Watanabe S., Tanigami A., Sugano S. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 2001;29:2154–2162. doi: 10.1093/nar/29.10.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freiman R.N., Herr W. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin D.Y., Wang H.L., Zhou Y., Chun A.C., Kibler K.V., Hou Y.D., Kung H., Jeang K.T. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 2000;19:729–740. doi: 10.1093/emboj/19.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R., Yang P., O'Hare P., Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H.J., Wong C.M., Chen J.H., Qiang B.Q., Yuan J.G., Jin D.Y. Inhibition of LZIP-mediated transcription through direct interaction with a novel host cell factor-like protein. J. Biol. Chem. 2001;276:28933–28938. doi: 10.1074/jbc.M103893200. [DOI] [PubMed] [Google Scholar]
- 25.Zhu C., Johansen F.E., Prywes R. Interaction of ATF6 and serum response factor. Mol. Cell. Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Shen J., Arenzana N., Tirasophon W., Kaufman R.J., Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 27.Herzig S., Long F., Jhala U.S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 28.Jin D.Y., Jeang K.T. HTLV-I Tax self-association in optimal trans-activation function. Nucleic Acids Res. 1997;25:379–387. doi: 10.1093/nar/25.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer J.A., Maniatis T. Drosophila Adh: a promoter element expands the tissue specificity of an enhancer. Cell. 1988;53:451–461. doi: 10.1016/0092-8674(88)90165-1. [DOI] [PubMed] [Google Scholar]
- 30.Williams S.C., Cantwell C.A., Johnson P.F. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 31.Wynshaw-Boris A., Lugo T.G., Short J.M., Fournier R.E., Hanson R.W. Identification of a cAMP regulatory region in the gene for rat cytosolic phosphoenolpyruvate carboxykinase (GTP): use of chimeric genes transfected into hepatoma cells. J. Biol. Chem. 1984;259:12161–12169. [PubMed] [Google Scholar]
- 32.Yoshida H., Haze K., Yanagi H., Yura T., Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins: Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 33.Kokame K., Kato H., Miyata T. Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. J. Biol. Chem. 2001;276:9199–9205. doi: 10.1074/jbc.M010486200. [DOI] [PubMed] [Google Scholar]
- 34.Chun A.C., Jin D.Y. Transcriptional regulation of mitotic checkpoint gene MAD1 by p53. J. Biol. Chem. 2003;278:37439–37450. doi: 10.1074/jbc.M307185200. [DOI] [PubMed] [Google Scholar]
- 35.Ching Y.P., Wong C.M., Chan S.F., Leung T.H., Ng D.C., Jin D.Y., Ng I.O. Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. J. Biol. Chem. 2003;278:10824–10830. doi: 10.1074/jbc.M208310200. [DOI] [PubMed] [Google Scholar]
- 36.Honma Y., Kanazawa K., Mori T., Tanno Y., Tojo M., Kiyosawa H., Takeda J., Nikaido T., Tsukamoto T., Yokoya S., et al. Identification of a novel gene, OASIS, which encodes for a putative CREB/ATF family transcription factor in the long-term cultured astrocytes and gliotic tissue. Brain Res. Mol. Brain Res. 1999;69:93–103. doi: 10.1016/s0169-328x(99)00102-3. [DOI] [PubMed] [Google Scholar]
- 37.Qi H., Fillion C., Labrie Y., Grenier J., Fournier A., Berger L., El-Alfy M., Labrie C. AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up-regulated by androgens in LNCaP human prostate cancer cells. Cancer Res. 2002;62:721–733. [PubMed] [Google Scholar]
- 38.Stelzer G., Don J. Atce1: a novel mouse cyclic adenosine 3′,5′-monophosphate-responsive element-binding protein-like gene exclusively expressed in postmeiotic spermatids. Endocrinology. 2002;143:1578–1588. doi: 10.1210/endo.143.5.8822. [DOI] [PubMed] [Google Scholar]
- 39.Storlazzi C.T., Mertens F., Nascimento A., Isaksson M., Wejde J., Brosjo O., Mandahl N., Panagopoulos I. Fusion of the FUS and BBF2H7 genes in low grade fibromyxoid sarcoma. Hum. Mol. Genet. 2003;12:2349–2358. doi: 10.1093/hmg/ddg237. [DOI] [PubMed] [Google Scholar]
- 40.Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raggo C., Rapin N., Stirling J., Gobeil P., Smith-Windsor E., O'Hare P., Misra V. Luman, the cellular counterpart of herpes simplex virus VP16, is processed by regulated intramembrane proteolysis. Mol. Cell. Biol. 2002;22:5639–5649. doi: 10.1128/MCB.22.16.5639-5649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye J., Rawson R.B., Komuro R., Chen X., Dave U.P., Prywes R., Brown M.S., Goldstein J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 43.Sakai J., Rawson R.B., Espenshade P.J., Cheng D., Seegmiller A.C., Goldstein J.L., Brown M.S. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 44.Rawson R.B., Zelenski N.G., Nijhawan D., Ye J., Sakai J., Hasan M.T., Chang T.Y., Brown M.S., Goldstein J.L. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 45.Servillo G., Della Fazia M.A., Sassone-Corsi P. Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM. Exp. Cell Res. 2002;275:143–154. doi: 10.1006/excr.2002.5491. [DOI] [PubMed] [Google Scholar]
- 46.Roesler W.J. What is a cAMP response unit? Mol. Cell. Endocrinol. 2000;162:1–7. doi: 10.1016/s0303-7207(00)00198-2. [DOI] [PubMed] [Google Scholar]
- 47.Omori Y., Imai J., Suzuki Y., Watanabe S., Tanigami A., Sugano S. OASIS is a transcriptional activator of CREB/ATF family with a transmembrane domain. Biochem. Biophys. Res. Commun. 2002;293:470–477. doi: 10.1016/S0006-291X(02)00253-X. [DOI] [PubMed] [Google Scholar]
- 48.Nikaido T., Iseki K., Mori T., Takaki H., Yokoya S., Hagino S., Takeda J., Zhang Y., Takeuchi M., Kikuchi S., et al. Expression of OASIS, a CREB/ATF family transcription factor, in CNS lesion and its transcriptional activity. Brain Res. Mol. Brain Res. 2002;108:129–138. doi: 10.1016/s0169-328x(02)00521-1. [DOI] [PubMed] [Google Scholar]
- 49.Wilson H.L., McFie P.J., Roesler W.J. Different transcription factor binding arrays modulate the cAMP responsivity of the phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 2002;277:43895–43902. doi: 10.1074/jbc.M203169200. [DOI] [PubMed] [Google Scholar]
- 50.Croniger C., Leahy P., Reshef L., Hanson R.W. C/EBP and the control of phosphoenolpyruvate carboxykinase gene transcription in the liver. J. Biol. Chem. 1998;273:31629–31632. doi: 10.1074/jbc.273.48.31629. [DOI] [PubMed] [Google Scholar]
- 51.Routes J.M., Colton L.A., Ryan S., Klemm D.J. CREB (cAMP response element binding protein) and C/EBPα (CCAAT/enhancer binding protein) are required for the superstimulation of phosphoenolpyruvate carboxykinase gene transcription by adenoviral E1a and cAMP. Biochem. J. 2000;352:335–342. [PMC free article] [PubMed] [Google Scholar]
- 52.Lamers W.H., Hanson R.W., Meisner H.M. cAMP stimulates transcription of the gene for cytosolic phosphoenolpyruvate carboxykinase in rat liver nuclei. Proc. Natl Acad. Sci. USA. 1982;79:5137–5141. doi: 10.1073/pnas.79.17.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buck M., Chojkier M. Signal transduction in the liver: C/EBPβ modulates cell proliferation and survival. Hepatology. 2003;37:731–738. doi: 10.1053/jhep.2003.50155. [DOI] [PubMed] [Google Scholar]
- 54.Buck M., Poli V., van der Geer P., Chojkier M., Hunter T. Phosphorylation of rat serine 105 or mouse threonine 217 in C/EBPβ is required for hepatocyte proliferation induced by TGF α. Mol. Cell. 1999;4:1087–1092. doi: 10.1016/s1097-2765(00)80237-3. [DOI] [PubMed] [Google Scholar]
- 55.Lu R., Misra V. Potential role for luman, the cellular homologue of herpes simplex virus VP16 (α gene trans-inducing factor), in herpesvirus latency. J. Virol. 2000;74:934–943. doi: 10.1128/jvi.74.2.934-943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikaido T., Yokoya S., Mori T., Hagino S., Iseki K., Zhang Y., Takeuchi M., Takaki H., Kikuchi S., Wanaka A. Expression of the novel transcription factor OASIS, which belongs to the CREB/ATF family, in mouse embryo with special reference to bone development. Histochem. Cell Biol. 2001;116:141–148. doi: 10.1007/s004180100279. [DOI] [PubMed] [Google Scholar]
- 57.Newman J.R., Keating A.E. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 58.Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- 59.Kaufman R.J. Orchestrating the unfolded protein response in health and disease. J. Clin. Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M., Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 62.Lee A.H., Iwakoshi N.N., Glimcher L.H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R.J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kondo S., Murakami T., Tatsumi K., Ogata M., Kanemoto S., Otori K., Iseki K., Wanaka A., Imaizumi K. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nature Cell Biol. 2005;7:186–194. doi: 10.1038/ncb1213. [DOI] [PubMed] [Google Scholar]
- 65.Roesler W.J., Vandenbark G.R., Hanson R.W. Identification of multiple protein binding domains in the promoter-regulatory region of the phosphoenolpyruvate carboxykinase (GTP) gene. J. Biol. Chem. 1989;264:9657–9664. [PubMed] [Google Scholar]
- 66.Allen-Jennings A.E., Hartman M.G., Kociba G.J., Hai T. The roles of ATF3 in liver dysfunction and the regulation of phosphoenolpyruvate carboxykinase gene expression. J. Biol. Chem. 2002;277:20020–20025. doi: 10.1074/jbc.M200727200. [DOI] [PubMed] [Google Scholar]
- 67.Roesler W.J., Graham J.G., Kolen R., Klemm D.J., McFie P.J. The cAMP response element binding protein synergizes with other transcription factors to mediate cAMP responsiveness. J. Biol. Chem. 1995;270:8225–8232. doi: 10.1074/jbc.270.14.8225. [DOI] [PubMed] [Google Scholar]
- 68.Screaton R.A., Conkright M.D., Katoh Y., Best J.L., Canettieri G., Jeffries S., Guzman E., Niessen S., Yates J.R., III, Takemori H., et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 69.Wang N.D., Finegold M.J., Bradley A., Ou C.N., Abdelsayed S.V., Wilde M.D., Taylor L.R., Wilson D.R., Darlington G.J. Impaired energy homeostasis in C/EBP α knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 70.Wang H., Goode T., Iakova P., Albrecht J.H., Timchenko N.A. C/EBPα triggers proteasome-dependent degradation of cdk4 during growth arrest. EMBO J. 2002;21:930–941. doi: 10.1093/emboj/21.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Julien E., Herr W. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 2003;22:2360–2369. doi: 10.1093/emboj/cdg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goto H., Motomura S., Wilson A.C., Freiman R.N., Nakabeppu Y., Fukushima K., Fujishima M., Herr W., Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]