Abstract
Aberrant mRNAs whose open reading frame (ORF) is truncated by the presence of a premature translation-termination codon (PTC) are recognized and degraded in eukaryotic cells by a process called nonsense-mediated mRNA decay (NMD). Here, we report the development of a reporter system that allows monitoring of NMD in mammalian cells by measuring the fluorescence of green fluorescent protein (GFP). The NMD reporter gene consists of a T-cell receptor-β minigene construct, in which the GFP-ORF was inserted such that the stop codon of GFP is recognized as PTC. The reporter mRNA is therefore subjected to NMD, resulting in a low steady-state mRNA level, an accordingly low protein level and hence a very low green fluorescence in normal, NMD-competent cells that express this reporter gene. We show that the inactivation of NMD by RNAi-mediated knockdown of the essential NMD factor hUpf1 or hSmg6 increases the NMD reporter mRNA level, resulting in a proportional increase of the green fluorescence that can be detected by flow cytometry, spectrofluorometry and fluorescence microscopy. With these properties, our GFP-based NMD reporter system could be used for large-scale screenings to identify NMD-inhibiting drugs or NMD-deficient mutant cells.
INTRODUCTION
Quality control checkpoints at different stages during gene expression are necessary to keep production of aberrant gene products low and thus to allow proper functioning of the cells. On the post-transcriptional level, a process called nonsense-mediated mRNA decay (NMD) has evolved in all eukaryotes examined so far to recognize and specifically degrade aberrant mRNAs, in which the open reading frame (ORF) is truncated by the presence of a premature translation-termination codon (PTC) (1). Two important sources giving rise to PTC-containing mRNAs (PTC+ mRNAs) in metazoans are mutations in the DNA and pre-mRNA splicing. It is estimated that about one-third of all inherited genetic disorders and many forms of cancer are caused by mutations that result in the generation of PTC+ mRNA (2). In humans, more than half of all pre-mRNAs undergo alternative splicing (3), and about one-third of these alternative transcripts contain a PTC (4). This demonstrates that NMD is an important cellular process that prevents (or at least reduces) the production of potentially deleterious truncated proteins and thereby modulates the clinical manifestations of many genetic diseases (2,5,6). In addition, immunoglobulin and T-cell receptor (TCR) genes more often than not acquire PTCs during the programmed V(D)J rearrangements which take place during lymphocyte maturation (7).
The molecular mechanisms underlying NMD are not well understood. Seven different NMD factors have been genetically identified in Caenorhabditis elegans (8,9), and the orthologues of three of them have also been genetically identified in Saccharomyces cerevisiae (10,11). The orthologue genes in humans, mice and Drosophila have all been found by homology searches (12–20). Owing to the lack of functional screens, putative mammalian NMD factors without orthologues in a genetically amenable organism are therefore still awaiting their discovery. It seems likely that additional, yet unknown, NMD factors might exist in vertebrates, considering that NMD appears to be more regulated and more sophisticated in more complex organisms. For example, hUpf1 activity appears to be regulated by a cycle of phosphorylation and dephosphorylation in metazoans, while S.cerevisiae lacks the orthologues involved in regulating hUpf1's phosphorylation state (1). Of interest for this study, the human orthologue of C.elegans SMG6, hSmg6, functions in dephosphorylation of hUpf1 (20) and is required for NMD (19). In addition, hSmg6 was also found to associate with telomerase and its overexpression uncaps telomeres (18). Also consistent with the idea of more factors being involved in NMD of more complex organisms, several factors of the exon junction complex were found to play a role in human NMD (21–25), while in Drosophila, the exon junction complex is dispensable for NMD (19).
Although technically quite demanding, mutagenesis screens in diploid human cells have been shown to be feasible (26,27), provided a suitable reporter system is available. To study mammalian NMD, several genes with PTCs at various positions were established as ‘model systems’ (28–32). But since the analysis of NMD of these genes requires analysis of mRNA, they are not suitable as reporters for screens. We, therefore, decided to develop a NMD reporter system where the readout was based on fluorescence in intact cells or whole cell extracts. We figured that such a NMD reporter system would not only be a valuable tool for basic research but could also be useful for drug screening. Given the large number of genetic diseases originating from PTC-causing mutations, there is also a considerable pharmaceutical interest to develop substances that interfere with NMD. Here, we describe the construction and characterization of a green fluorescent protein (GFP)-based NMD reporter system that allows detection of NMD-deficient cells by the detection of increased green fluorescence. We show that this NMD reporter system can be used in conjunction with flow cytometry, spectrofluorometry and fluorescence microscopy.
MATERIAL AND METHODS
Plasmids
The parental vector expressing the TCR-β minigene (pβ433) has been described previously (33). pβ510 was generated from pβ433 by replacing the NotI–SalI fragment with a double-stranded oligo (5′-gcGGCCGCGGCGGCGCCCTATAAAACCCAGCGGCGCGACGCGCCACCACCGCCGAGACCGGTACCGtcgac-3′; sequence of forward oligo is in upper case, NotI and SalI sites in italics, transcription start site underlined), which creates an intronless 5′-untranslated region of 62 bp. The EGFP ORF was PCR amplified using primers 5′-ATCTATAGCATATGCCATGGTGAGCAAGGGCGAG-3′ and 5′-ATCGACACCATATGCTCAGTACTTGTACAGCTCGTCCATGC-3′ (NdeI site in italics, stop codon underlined), digested with NdeI and inserted into the NdeI site of pβ510.
Finally, the leader exon sequence was exchanged for a sequence encoding the haemagglutinin A epitope by replacing the SalI–EcoRI fragment with a double-stranded oligo (5′-gTCGACACCATGGCCTACCCATATGATGTTCCAGATTACGCTTCACTCGAACGCGTAAGTGAGTGCTGGTCAGGCCACTGGTGTGCTTTCTTTTTTTAGaattc-3′; sequence of forward oligo is in upper case, SalI and EcoRI sites in italics, translation start site underlined). The resulting plasmid is called pβ510-HA-TCRβ-GFP PTC+. The plasmid pβ510-HA-TCRβ-GFP ΔJCin was generated by deleting the JC intron using fusion PCR. Detailed cloning strategy and primer sequences are available upon request. The HA-TCRβ-GFP gene of both plasmids was sequenced.
pSUPERpuro vector was created by inserting a puromycin-resistance cassette lacking the HindIII site of the puromycin ORF (from pTRE2pur, Clontech) into the XhoI site of pSUPER (34). Sequences coding for short hairpin RNAs (shRNAs) were inserted as double-stranded oligos into pSUPERpuro between the BglII and HindIII as described previously (34). The two target sequences of hUpf1 were5′-GAGAATCGCCTACTTCACT-3′ (pSUPERpuro-hUpf1/I) and 5′-GATGCAGTTCCGCTCCATT-3′ (pSUPERpuro-hUpf1/II) and the two target sequences of hSmg6 were 5′-GGGTCACAGTGCTGAAGTA-3′ (pSUPERpuro-hSmg6/I) and 5′-GCTGCAGGTTACTTACAAG-3′ (pSUPERpuro-hSmg6/II).
Cell culture and transfection
HeLa cells were grown in DMEM (Invitrogen) supplemented with 10% heat inactivated fetal calf serum (FCS), 100 U/ml penicillin and 100 μg/ml streptomycin. LipofectAmine (Invitrogen) was used for all transient transfections according to the manufacturer's protocol. For RNAi experiments, cells were seeded in 8 cm dishes and transfected with 2 μg empty pSUPERpuro (mock), a mixture of 1 μg pSUPERpuro-hUpf1/I and 1 μg pSUPERpuro-hUpf1/II, or a mixture of 1 μg pSUPERpuro-hSmg6/I and 1 μg pSUPERpuro-hSmg6/II using 8 μl LipofectAmine (Invitrogen). An aliquot of 1.5 μg/ml puromycin (Calbiochem) was added to the cells from 24 h post-transfection until the evening before analysis (4 days post-transfection) to eliminate the untransfected cells. Before analysis, the cells were cultured without puromycin for at least 16 h to avoid potential effects of this translation inhibitor on NMD. In cells transfected with the empty pSUPERpuro, relative levels of NMD substrates and mRNAs without a PTC were identical irrespective of whether the cells were treated with puromycin or not (data not shown), confirming that the puromycin treatment did not interfere with NMD. For microscopy, 10 mm glass coverslips were placed in the culture dish before seeding the cells.
To establish NMD reporter-expressing HeLa cell lines, pβ510-HA-TCRβ-GFP PTC+ was transfected with Gene Jammer Transfection Reagent (Stratagene) according to the manufacturer's instructions, and stable transfectants were selected by cultivating the cells in the presence of 500 μg/ml of G418 (Geneticin, Invitrogen) for 2 weeks, before single cell clones were isolated and expanded. As a control, stably transfected clonal cell lines were also established using a reporter construct that lacks the JC intron (pβ510-HA-TCRβ-GFP ΔJCin).
The monoclonal cell lines were treated with 100 μg/ml cycloheximide (CHX) for 3 h to analyse the response of NMD reporter mRNA or mRNA of the ΔJCin control construct to inhibition of NMD.
RNA analysis
Total cellular RNA was purified using the Absolutely RNA RT–PCR Miniprep Kit (Stratagene) according to the manufacturer's protocol. For real-time PCR analysis, 1 μg was reverse transcribed in 50 μl Stratascript buffer in the presence of 200 ng random hexamers, 0.4 mM dNTP mix, 40 U RNasIn (Promega) and 50 U Stratascript (Stratagene) according to the manufacturer's protocol. Reverse transcribed material corresponding to 40 ng RNA was amplified in 25 μl Universal PCR MasterMix, no AmpErase (Applied Biosystems) using the following primers and TaqMan probes. TCR-β mRNA was measured over the junction between the VDJ exon and the C1 exon using 400 nM forward primer om59 (5′-GCGGTGCAGAAACGCTGTA-3′), 400 nM reverse primer om60 (5′-TGGCTCAAACAAGGAGACCTT-3′) and 200 nM TaqMan probe TM59/60 (5′-FAM-CTCGAGGATCTGAGAAATGTGACTCCACC-TAMRA-3′). β-Globin mRNA was measured over the junction between exons 2 and 3 using 800 nM forward primer om111 (5′-GCTGCACTGTGACAAGCTGC-3′), 800 nM reverse primer om112 (5′-AAAGTGATGGGCCAGCACAC-3′) and 200 nM TaqMan probe TM111/112 (5′-FAM-TCCTGAGAACTTCAGGCTCCTGGGCAAC-TAMRA-3′). hUpf1 mRNA, GAPDH mRNA and 18S rRNA were measured using pre-developed assay reagents from Applied Biosystems. For 18S rRNA measurements, cDNA corresponding to 400 pg RNA instead of 40 ng was used. Real-time PCR was run on the GeneAmp 5700 and 7000 Sequence Detection Systems (Applied Biosystems) using the standard thermal profile.
The conditions for the traditional endpoint RT–PCR shown in Figure 1B were as follows: RNA was reverse transcribed using an anchored primer against the poly(A) tail (5′-N30-VN-3′) and Stratascript reverse transcriptase in Stratascript first strand buffer according to the manufacturer's protocol (Stratagene). Subsequent PCR was performed with a primer binding to the HA-tag (om153; 5′-TCCAGATTACGCTTCACTCGAAC-3′) and a reverse primer binding to exon C1 (om96; 5′-GATGGCTCAAACAAGGAGACCTT-3′). An aliquot of the PCR was analysed on a 1.5% agarose gel and stained with ethidium bromide.
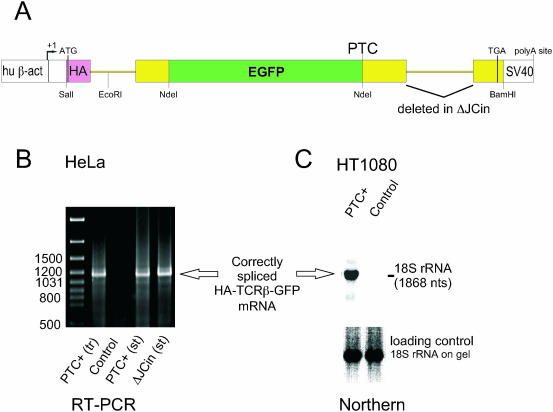
Figure 1.
Expression of the HA-TCRβ-GFP reporter gene yields correctly spliced mRNA. (A) Schematic representation of the NMD reporter gene. Sequences derived from human β-actin, mouse TCR-β and EGFP are shown in white, yellow and green, respectively. The haemagglutinin epitope (HA) is shown in purple. The positions of the transcription start site (+1), translation start codon (ATG), PTC and the normal stop codon (TGA) are depicted, and relevant restriction sites are indicated. The intron deleted in the control construct ΔJCin is marked. See text for details. (B) RNA from HeLa cells transiently (tr) or stably (st) expressing the HA-TCRβ-GFP reporter gene (PTC+) or the control construct with the deleted JC intron (ΔJCin) was reverse transcribed and amplified with primers annealing to the first and last exon. RNA from untransfected HeLa cells served as a negative control. The PCR products were resolved on a 1.5% agarose gel. (C) An aliquot of 10 μg RNA from HT1080 cells stably expressing the HA-TCRβ-GFP reporter gene (PTC+) and from untransfected HT1080 cells (control) was separated on a 1.2% agarose gel. After ethidium bromide staining of the gel to reveal the ribosomal RNAs (see 18S rRNA loading control), the RNA was transferred to a nylon membrane and hybridized with a 32P-labelled random-primed probe against TCRβ. The position on the membrane of the 18S rRNA is marked on the right.
For the northern blot shown in Figure 1C, 10 μg of total cellular RNA was separated on a 1.2% agarose gel containing 1% formaldehyde and 1× MOPS. A picture of the ethidium bromide stained gel before blotting was taken to use the 18S rRNA band as loading control. The RNA was transferred to positively charged nylon membrane (Roche Diagnostics) in 20× SSC by standard capillary blotting method. Probe labelling, hybridization, washing and signal detection was as described elsewhere (32). Approximately 50 ng of a purified SalI–BamHI fragment comprising the TCR-β minigene sequence was used as template for the 32P-labelled probe.
Flow cytometry analysis
About 106 cells were harvested and resuspended in phosphate-buffered saline (PBS)/1% FCS and then analysed using a FACScan (BD, San Jose, CA). GFP fluorescence was excited at 488 nm and emission was measured with a 530/30 nm bandpass filter. HeLa cells without the NMD reporter gene (plain HeLa) served as a control for autofluorescence and were used to set the gate for GFP positive cells. The data were processed using FlowJo software (Tree Star, Ashland, OR). The geometric mean values were used to compare the amount of fluorescence in the different samples.
Western blot analysis
Cells were lysed and denatured in SDS loading buffer and these whole cell lysates were separated on 10% SDS–PAGE. Proteins were then transferred to Optitran BA-S 85 reinforced nitrocellulose (Schleicher & Schuell) and probed with different antibodies. The monoclonal anti-HA mouse antibody (BabCO, Berkeley, CA) was used at a dilution of 1:1000, the anti-hUpf1 rabbit antiserum [(14), a kind gift from Jens Lykke-Andersen] was diluted 1:2500, and supernatant of the hybridoma cell line Y12 (producing the anti-Sm antibody) was diluted 1:400. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibody (Promega) was used as secondary antibodies at a dilution of 1:2500, and detected with the ECLplus Western Blotting Detection System (Amersham Pharmacia Biotech).
Dual scanning microplate spectrofluorometer
Proteins were extracted in 1% deoxycholic acid, 1% NP40, 25 mM Tris–HCl (pH 8), 50 mM NaCl and protease inhibitor cocktail (Complete Mini, EDTA-free; Roche). The cells were incubated at 4°C for 30 min on a shaker and then centrifuged at 16 000 g for 5 min. The supernatant was immediately frozen in liquid nitrogen and stored at −20°C. GFP fluorescence was measured on a SPECTRAmax Gemini (Molecular Devices) using 96-well black plates with flat, black bottom. GFP fluorescence was excited at 488 nm and measured at 512 nm, the cut-off was set at 495 nm. SOFTmaxPRO software was used for data analysis.
Confocal microscopy
Images were collected on a Leica TCS SP2 AOBS laser scanning confocal microscope equipped with a HCX PL APOlbd.BL 63.0× 1.2W objective (Leica Microsystems Inc., Exton, PA). The cells were grown on coverslips, washed once with PBS and then fixed in 4% paraformaldehyde and mounted on DAKO fluorescent anti-fading mounting medium (DAKO Corporations). For GFP detection, cells were scanned using the 488 nm laser line and emitted light between 500 and 600 nm was collected. The pinhole was opened to 418 μm and the photomultipliers were set to 580 V. The same settings for image acquisition and processing have been applied for all samples to allow comparison of the fluorescence intensities among the different samples.
RESULTS AND DISCUSSION
Design of the NMD reporter gene
Fluorescent proteins have become a valuable tool for innumerable applications in modern biology, because the light emitted by these proteins can be detected relatively easily by many different techniques. Therefore, we decided to develop a reporter system for NMD that is based on the enhanced green fluorescent protein (EGFP, Clontech). To this end, we inserted the EGFP ORF in frame into the middle exon of a TCR-β minigene (Figure 1A). In this position, more than 50 nt upstream of the 3′-most 5′-splice site, the stop codon of the EGFP ORF is recognized as a PTC and, thus, the mRNA is subjected to NMD. A TCR-β minigene was chosen for our reporter gene, (i) because NMD of TCR-β transcripts has been extensively studied and (ii) because PTCs in these TCR-β minigenes cause a dramatic reduction in the steady-state mRNA levels (35,36). The particular TCR-β minigene used here has fused the first 120 bp of exon C1 to base pair number 117 of exon C4 and, therefore, lacks part of the constant region. Since the signal peptide encoded by the Vleader exon is still present in this TCR-β minigene, while the region of the gene encoding the membrane-spanning domain is missing, it was likely that this TCRβ-GFP protein would be secreted from the cells. To prevent this potential problem, and to introduce a tag that allows easy detection of the TCRβ-GFP protein in western blots, we replaced the signal peptide encoding sequence in the Vleader exon with a sequence encoding for the haemagglutinin epitope (Figure 1A, HA-tag).
In addition, a control construct was derived from the NMD reporter gene by deleting the second intron (Figure 1A, ΔJCin). This ΔJCin construct encodes the same TCRβ-GFP fusion protein as the NMD reporter construct, but because the PTC in the ΔJCin construct is now located in the last exon, its mRNA is not expected to be a substrate for NMD.
In RNA samples from HeLa cells stably or transiently expressing the TCRβ-GFP reporter gene, we amplified by RT–PCR a product of the size expected for the correctly spliced mRNA (Figure 1B), indicating that insertion of the EGFP ORF into the VDJ exon, which results in an unusually long internal exon, did not induce alternative splicing of the pre-mRNA. Correct splicing was also confirmed for the ΔJCin construct stably expressed in HeLa cells (Figure 1B), and for the NMD reporter gene stably expressed in the diploid human cell line HT1080: northern blotting revealed a single and specific band for HA-TCRβ-GFP mRNA in the expected size range (Figure 1C). We conclude that the vast majority of HA-TCRβ-GFP transcripts are spliced correctly in human cells, both when expressed episomally (transient transfection) or from a genomic locus (stable transfection).
The GFP fluorescence is proportional to the mRNA level of the reporter gene
The concept of a GFP-based NMD reporter system is to use the GFP fluorescence as a measure for the steady-state mRNA level of the NMD reporter. Thus, the reporter system is only useful, if the intensity of the GFP fluorescence is proportional to the mRNA level. To test this, HeLa cells were transfected with increasing amounts of NMD reporter plasmid DNA. Seventy-two hours post-transfection, a fraction of the cells was analysed by flow cytometry (Figure 2A), and from the rest of the cells, RNA was isolated and relative reporter mRNA levels were measured by real-time RT–PCR. The comparison between the percentage of GFP-positive cells multiplied by the average intensity of GFP fluorescence (Figure 2B) and the relative reporter mRNA levels (Figure 2C) for the different plasmid DNA amounts shows that the GFP signal is indeed highly proportional to mRNA levels.
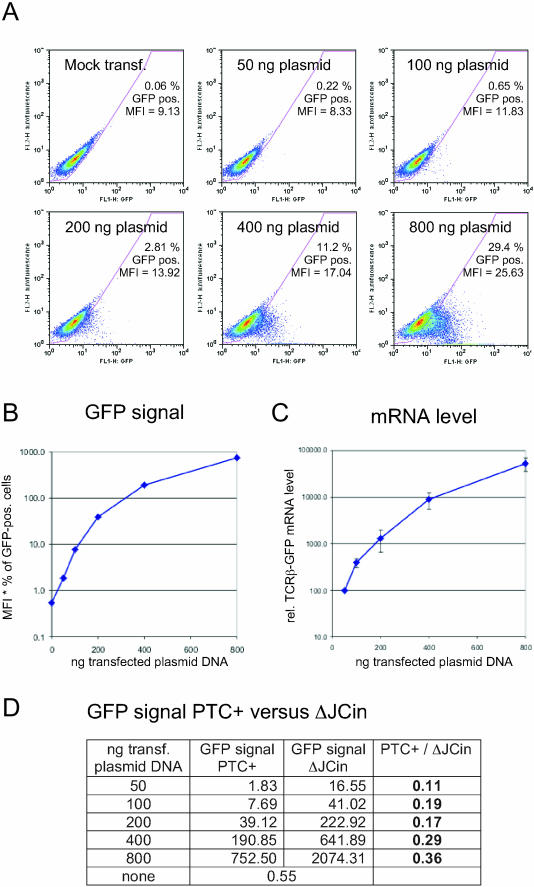
Figure 2.
The GFP fluorescence is proportional to the mRNA level of the reporter gene. (A) Forty-eight hours after transfection with the indicated amounts of the HA-TCRβ-GFP reporter gene together with a constant amount of a β-globin-expressing plasmid (41), the HeLa cells were analysed by flow cytometry. The gate for GFP positive cells was set so that the autofluorescence of the mock-transfected cells did not score as GFP positive. MFI, mean fluorescence intensity. (B) MFI values multiplied with the percentage of GFP positive cells from (A) were plotted against the amount of transfected NMD reporter plasmid DNA. (C) From an aliquot of the cells used in (A), RNA was isolated and relative NMD reporter mRNA and β-globin mRNA was measured by real-time RT–PCR. Relative reporter mRNA levels were normalized to relative β-globin mRNA levels, and average values and standard deviations of three real-time PCR runs are shown. (D) Analogous to the experiment shown in (A) and (B) for the PTC+ reporter gene, the GFP signals from cells transiently transfected with different amounts of the ΔJCin control construct were determined by flow cytometry. As in (B), the GFP signal is defined as MFI* % of GFP positive cells.
The same experiment was also carried out with the ΔJCin control construct, and the respective GFP signals (mean fluorescence intensity multiplied by percentage of GFP-positive cells) are shown in comparison with the GFP signals determined for the PTC+ construct (Figure 2D). With increasing amounts of transfected plasmid DNA, the GFP signal of the NMD reporter construct changes from 9-fold lower than the GFP signal of the NMD-resistant control construct to only 3-fold lower. The extent of NMD cannot be determined accurately from this experiment for two reasons. When low amounts of plasmid DNA is transfected, most of the resulting weak GFP signals will be hidden within the autofluorescence of the cells, and only a small percentage of the cells qualifies as ‘GFP positive’ (fluorescence signal distinct from the autofluorescence seen in the mock-transfected control cells) and contributes to the overall GFP signal. Because this effect is more pronounced for the PTC+ than for the ΔJCin, the extent of NMD will be overestimated with low plasmid DNA amounts. On the other hand, we know from experience with a number of different NMD reporter genes that NMD can easily be saturated by the expression of high amounts of NMD substrate (A. Paillusson and O. Mühlemann, unpublished data). This partially saturated NMD results in a decreased difference between the PTC+ and the ΔJCin GFP signal measured with high plasmid DNA amounts. Taking both effects into consideration, we estimate that non-saturated NMD reduces the fluorescence of our TCRβ-GFP fusion protein expressed from the NMD reporter construct ∼4- to 6-fold.
Establishing cell lines stably expressing the NMD reporter
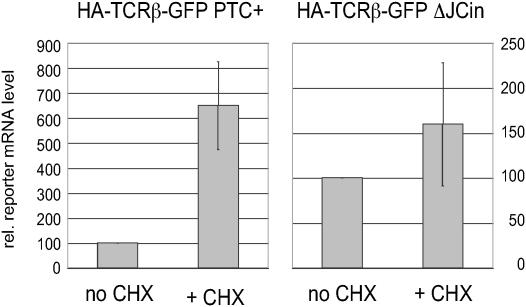
After having confirmed that the GFP signal proportionally reflects the mRNA level of the NMD reporter, we transfected HT1080 (data not shown) and HeLa cells with our NMD reporter plasmid and selected a number of single cell clones that had integrated the reporter gene into the genome. In order to select a suitable NMD reporter system, we first screened the monoclonal cell lines by real-time RT–PCR for lines expressing a clearly detectable level of the HA-TCRβ-GFP reporter mRNA, which allowed us to eliminate ‘false positives’ that did not express the reporter gene at all (data not shown). Cell lines expressing the reporter gene were then treated with the translation inhibitor CHX for 3 h, which is a standard way to inhibit NMD (31), and the increase in reporter mRNA levels was quantified by real-time RT–PCR. As a control, we also generated and analysed in parallel stably transfected clonal cell lines expressing the ΔJCin construct. Because this construct lacks an intron downstream of the PTC, its mRNA should not be subjected to NMD. Figure 3 shows the relative mRNA levels of a NMD reporter-expressing cell line (left panel) and of a cell line expressing the ΔJCin control construct (right panel) after the inhibition of NMD with CHX. The NMD reporter mRNA increased on average 6.5-fold upon CHX treatment, similar to the previously tested PTC-containing TCR-β minigenes (37), whereas the mRNA level of the ΔJCin control construct increased only slightly, confirming that this is not a substrate for NMD.
Figure 3.
NMD reporter mRNA stably expressed in a single-cell clone increases upon inhibition of translation by CHX treatment. After transfection of HeLa cells with the NMD reporter gene (HA-TCRβ-GFP PTC+) or with the control construct (HA-TCRβ-GFP ΔJCin) and selection for G418 resistance, cell lines derived from single-cell clones were established. RNA was isolated from these cell lines after treatment with 100 μg/ml cycloheximide for 3 h (+CHX) or without CHX treatment (no CHX). Relative NMD reporter mRNA was measured by real-time RT–PCR and normalized to relative GAPDH mRNA levels. For each cell line, the NMD reporter mRNA level of non-treated cells was set to 100 and the mRNA level of the CHX treated cells is shown relative to it. Average values and standard errors of five real-time PCR runs are shown.
RNAi-mediated depletion of hUpf1 and hSmg6 causes increased mRNA and protein levels of the NMD reporter gene
Treatment of cells with CHX is an easy and quick way to inhibit NMD, but because it blocks all cellular translation, unspecific effects influencing the level of NMD reporter mRNA could also occur. Therefore, we decided to interfere with NMD specifically by knocking down essential NMD factors using RNAi techniques. To this end, we used the pSUPER plasmid (34), from which RNAi-inducing shRNAs can be expressed. To be able to use such a transfection-based RNAi approach for subsequent biochemical analysis, an almost 100% transfection efficiency or a way to eliminate untransfected cells before analysis is required. Since the first option is not always feasible, we opted for the second and introduced into the pSUPER vector the puromycin resistance gene, which allows elimination of the untransfected cells during the course of the RNAi experiment (see Materials and Methods). The NMD reporter gene-expressing HeLa cell line was transfected with such pSUPERpuro plasmids encoding shRNAs against the two essential human NMD factors hUpf1 (12,13,38) or hSmg6 [also called hEST1A or Smg5/7a (18–20)], and the effect on the reporter mRNA level was analysed (Figure 4). Compared with cells transfected with the empty pSUPERpuro (Mock), mRNA levels of the NMD reporter increased between 6- and 11-fold when hUpf1 was knocked down by RNAi (Figure 4A, left panel, and Figure 4C), whereas the ΔJCin mRNA level in the control cell lines was not affected (Figure 4A, right panel). While RNAi against hUpf1 worked quite efficiently, resulting in a 10-fold or greater reduction of hUpf1 mRNA (Figure 4C, and data not shown) and barely detectable hUpf1 protein levels (Figure 4B) under our experimental conditions, RNAi against hSmg6 resulted only in a ∼3-fold reduction of hSmg6 mRNA in our hands (Figure 4C, and data not shown). Yet, this moderate reduction of hSmg6 levels caused the same increase of the NMD reporter mRNA as knockdown of hUpf1 (Figure 4C). This result suggests that hSmg6 might be a limiting factor for NMD function. Collectively, these results show that the expression of our NMD reporter gene increases as predicted when NMD deficiency is induced by RNAi.
Figure 4.
RNAi-mediated knockdown of essential NMD factors results in the expected increase of NMD reporter mRNA. (A) HeLa cell lines stably expressing the HA-TCRβ-GFP reporter construct (PTC+) or the control construct lacking the JC intron (ΔJCin) were transfected with pSUPERpuro plasmids expressing shRNAs against hUpf1, or with the empty pSUPERpuro (mock) as a control. After elimination of the non-transfected cells by treatment with puromycin, RNA was isolated 4 days post-transfection and relative NMD reporter mRNA was measured by real-time RT–PCR and normalized to relative GAPDH mRNA levels. Average values of three PCR runs from a typical experiment are shown. (B) The hUpf1 knockdown in the cells used in (A) was monitored by a western blot using hUpf1-specific polyclonal antisera. Cell extract of untransfected HeLa cells was used as a reference in the first lane. Detection of Sm proteins B/B′ with the monoclonal antibody Y12 served as loading control. (C) Average reporter mRNA levels determined from three independent knockdowns of hUpf1 or hSmg6 in the cell line expressing the NMD reporter construct (PTC+). The relative reporter mRNA levels were measured by real-time RT–PCR and normalized to relative GAPDH mRNA levels. Relative hUpf1 and hSmg6 mRNA levels were also determined to monitor the efficiency of the knockdowns. hUpf1 mRNA was ∼10-fold reduced compared with the mock, and hSmg6 mRNA ∼3-fold (data of one typical experiment is shown).
Detection of NMD deficiency by flow cytometry
In order to use the NMD reporter system as a tool for screening large number of cells for NMD deficiency, its readout (i.e. GFP fluorescence) should be detectable by techniques capable of analyzing many cells within a short time. To detect and isolate NMD-deficient cells in a large cell population, flow cytometry is today's method of choice. To test the suitability of our NMD reporter system for analysis by flow cytometry, we knocked down hUpf1 and hSmg6 in the reporter gene-expressing HeLa cell line as in the previous experiment and determined the fluorescence emitted by these cells using FACScan. As a reference signal for GFP in NMD-competent cells, the reporter cell line was transfected with the empty pSUPERpuro (Mock), and to control for autofluorescence of the cells, mock-transfected parental HeLa cells were also analysed. The two cell populations with RNAi-mediated NMD deficiency exhibited clearly more GFP fluorescence than the mock-transfected reporter cell line, demonstrating that flow cytometry can be used to analyse the NMD reporter system (Figure 5A). In the mock-transfected reporter cells, almost no GFP signal could be detected over the background caused by the autofluorescence, similar to the situation when small amounts of NMD reporter gene were transiently expressed (Figure 2A). Since almost no GFP signal over background could be detected in mock-transfected reporter cells, the fold increase upon knockdown of hUpf1 and hSmg6 cannot be calculated. As a control, NMD reporter mRNA levels were measured from the same cells analysed by flow cytometry. As in previous experiments (Figure 4C), the RNAi-mediated knockdown of hUpf1 and hSmg6 resulted in an ∼10-fold increase of reporter mRNA (Figure 5B). RNA from the mock-transfected parental HeLa cells gave no detectable signal in the real-time RT–PCR assay, confirming that the TaqMan probe for measuring reporter mRNA is specific (data not shown). In summary, this experiment shows that our NMD reporter system can be used in conjunction with flow cytometry to detect NMD-deficient cells.
Figure 5.
Detection of increased GFP fluorescence by flow cytometry analysis after RNAi-mediated knockdown of NMD factors. (A) hUpf1 or hSmg6 was knocked down in the NMD reporter gene-expressing HeLa cells as in Figure 4, and the cells were analysed by flow cytometry 4 days post-transfection. (B) From an aliquot of the same cells used for flow cytometry, relative reporter mRNA levels were determined by real-time RT–PCR. Average numbers of four PCR runs, normalized to endogenous GAPDH mRNA levels, are shown.
Detection of NMD deficiency using a microplate spectrofluorometer
For other types of automated large-scale screens, the preferred analysis method includes the use of a fluorescence plate reader. A typical example for such an application would be a screening of chemical libraries for compounds that inhibit NMD. Although the nature of the reporter system would prevent the identification of compounds that inhibit NMD by blocking translation, it might still be very useful to identify novel compounds that inhibit NMD through other mechanisms, such as binding to essential NMD factors. We, therefore, wanted to test whether our NMD reporter cells can be used to detect NMD deficiency by measuring the GFP fluorescence in whole cell extracts using a microplate spectrofluorometer (SpectraMAX Gemini). To this end, hUpf1 was again knocked down in the NMD reporter cell line, and mock-transfected reporter cells and mock-transfected plain HeLa cells were used as controls. Whole cell extracts of each cell population were prepared and the fluorescence at 512 nm (GFP) of different dilutions thereof was measured. The values obtained with the NMD reporter cells were normalized by subtracting the background values measured with plain HeLa (Figure 6A). Very similar to the result of the flow cytometry analysis, no GFP signal significantly over background could be detected in the NMD-competent, mock-transfected reporter cells, even with an amount of cell lysate corresponding to 106 cells. In contrast, a linear increase of GFP fluorescence, proportional to the amount of lysate used, was readily detected in reporter cells where hUpf1 was knocked down. This demonstrates that in general the NMD reporter system can also be used in combination with a fluorescence plate reader. However, the minimal amount of lysate required to detect a GFP signal clearly over background in the hUPF1-depleted cells corresponds to ∼4 × 105 cells, which is too much for a large-scale screening. Thus, in order to use the NMD reporter system for spectrofluorometer-based drug discovery, the sensitivity of the assay needs to be improved (see below).
Figure 6.
Detection of NMD deficiency by spectrofluorometry. (A) The GFP signal in extract corresponding to the indicated amount of cells from mock-transfected (black circles) and pSUPERpuro-hUpf1 transfected NMD reporter cells (grey squares), and from plain HeLa cells was measured with a dual scanning spectrofluorometer. The values measured in plain HeLa were used to subtract the background. RNAi was performed as in Figure 4. (B) Aliquots of the cells used in (A) were checked by western blotting using antibodies against hUPF1 (14) and the HA-tag to monitor the knockdown efficiency and the increase of the NMD reporter protein level, respectively. The two Sm proteins B and B′, detected with the monoclonal antibody Y12, served as loading control.
To assess the efficiency of the hUpf1 knockdown and its effect on the level of reporter protein, aliquots of the cells used in Figure 6A were analysed by western blot analysis (Figure 6B). Probing with the anti-hUPF1 antibody shows that in this experiment too, the expression of shRNAs targeting hUpf1 mRNA led to a strong reduction in hUPF1 protein. With the anti-HA antibody, NMD reporter protein could be detected in the hUpf1 knockdown cells, but not in the mock-transfected cells, consistent with result from the spectrofluorometer. The monoclonal antibody Y12, which detects the Sm proteins B, B′, D1 and D3 (39), was used as a loading control. Real-time RT–PCR showed that the NMD reporter mRNA levels increased 3.2-fold in this particular experiment (data not shown).
Detection of NMD-deficiency by fluorescence microscopy
The third detection method that we tested on our NMD reporter cells was fluorescence microscopy using a confocal laser scanning microscope. As previously, hUpf1 was knocked down in the NMD reporter-expressing HeLa cell line by transfection of hUpf1 targeting pSUPERpuro followed by selection of the puromycin-resistant cells. Four days after transfection, these cells were examined under the fluorescence microscope and compared with the mock-transfected, NMD-competent reporter cells and untransfected HeLa cells. Only weak green fluorescence could be detected mainly in the cytoplasm of mock-transfected reporter cells when the pinhole of the confocal microscope was opened to achieve maximal sensitivity (Figure 7D). The average fluorescent signal was nevertheless significantly higher than the autofluorescence seen in untransfected HeLa cells (Figure 7B), and we therefore conclude that in contrast to flow cytometry (Figure 5A), the low basal GFP level of the NMD reporter can be detected by laser scanning fluorescence microscopy. In contrast to this low basal level of GFP in the mock-transfected reporter cells, the green fluorescence, which is predominantly cytoplasmic, is much more intense of hUPF1-depleted reporter cells (compare Figure 7D and F). Differential interference contrast (DIC) images of the same cells showed no obvious morphological changes 4 days after transfection of pSUPERpuro plasmids (compare Figure 7C and E with Figure 7A), suggesting that at this time point, abrogation of NMD has not yet severely damaged the cells (see below). We conclude that our NMD reporter system can also be used to screen for individual cells by fluorescence microscopy.
Figure 7.
Detection of NMD deficiency by confocal laser scanning fluorescence microscopy. The NMD reporter cell line was grown on coverslips and transfected with the empty pSUPERpuro plasmid (mock; C and D) or with pSUPERpuro-Upf1 (E and F) as in Figure 4. Untransfected HeLa cells served as a control for background autofluorescence (A and B). After fixation, the cells were analysed using a confocal laser scanning fluorescence microscope. Differential interference contrast (DIC) images (A, C and E) and fluorescence images in the GFP channel (B, D and E) of the same cells are shown.
CONCLUSIONS
Here, we describe the construction of a GFP-based reporter system, in which increased green fluorescence of the cells signals NMD deficiency. We validate this reporter system by demonstrating that increased GFP signals are in fact detected by flow cytometry, spectrofluorometry and fluorescence microscopy in cells, where NMD has been abrogated by knocking down essential NMD factors using RNAi techniques. In all these tests, our NMD reporter system behaved as expected, proving its potential to become a valuable tool for future studies of NMD.
If the GFP-based NMD reporter system is used to screen for NMD-deficient cells, one must be aware that, for example, mutants that enhance transcription or translation, or that inhibit general mRNA turnover or protein degradation will also result in increased GFP fluorescence. To identify such false positives, samples with increased GFP signal must therefore be validated by further experiments. To this end, reporter mRNA levels and mRNA of an endogenous gene can be measured, and in addition, expression and analysis of another well-established NMD reporter mRNA can be used to confirm NMD deficiency.
For some automated large-scale screenings, the currently generally low GFP signal emitted by the reporter may limit its application. However, there are several ways to improve the sensitivity of the system if required. On the one hand, detection systems are becoming more and more sensitive, and on the other hand, it should be possible to generate cell lines that contain more copies of the HA-TCRβ-GFP gene and, thus, would be expected to generate more fluorescence per cell. For flow cytometry applications, substitution of GFP in our reporter gene by another fluorescent protein variant that emits light at a different wavelength might overcome the current problem of low GFP signals being hidden in the autofluorescence of the cells (Figures 2A and 5A). To increase the sensitivity of the reporter system in spectrofluorometric applications, GFP could be replaced by firefly luciferase. Because luciferase produces chemiluminescence in an enzymatic reaction by oxidation of luciferin, detection of more light per reporter molecule would be expected when using a microplate spectrofluorometer.
In parallel to the HeLa NMD reporter system mainly described here, we have also generated a diploid HT1080 cell line that expresses our NMD reporter gene (Figure 1C, and data not shown) and used these cells for a mutagenesis screen with the goal of obtaining NMD-deficient cell lines. The mutagenesis scheme and the selection of high GFP-expressing cells by flow cytometry was essentially as described for a similar screen, which yielded mutant cell clones deficient for rapid ARE-dependent IL-3 mRNA turnover (26). Although high GFP-expressing cells occurred at a very low frequency in our mutated cell population, we were never able to proliferate the high GFP-expressing cells after sorting (data not shown). This was not due to cell culturing problems, because from the mutated cell pool, cell lines from single, low GFP-expressing cells could be obtained with a reasonable plating efficiency (data not shown). Meanwhile, we believe that the reason for the failure of this mutagenesis screen might be that NMD could be essential for survival of mammalian cells, even in cell culture. In our RNAi experiments targeting hUpf1 and hSmg6, the cells die 7–8 days after transfection (A. Paillusson and O. Mühlemann, unpublished data). In addition, from all attempts to stably transfect pSUPERpuro targeting hUpf1, we only obtained cell clones in which the hUpf1 protein level was reduced 5-fold or less (data not shown), while hUpf1 is usually reduced ∼10-fold or more when cells transfected with the same pSUPERpuro plasmids are assayed 4 or 5 days after transfection (Figures 4B and 6B, and data not shown). Furthermore, Upf1 knock-out is lethal in mice, with embryos dying 6–7 days post coitus (40). All these observations are consistent with the suggestion that NMD might be essential for viability of mammalian cells. However, it is also possible that hUpf1 and hSmg6 play additional roles in other essential cellular processes.
In conclusion, the GFP-based NMD reporter gene system described here faithfully reports NMD deficiency in individual cells by increasing the fluorescence of these cells. Because the NMD reporter system functions in combination with flow cytometry, spectrofluorometry and fluorescence microscopy, it has the potential to be used for automated large-scale screening applications, although some modifications might be necessary for specific applications (see above). With these properties, this NMD reporter gene system represents a new and valuable tool in the toolbox of researchers investigating the molecular mechanisms of mammalian NMD.
Acknowledgments
The authors thank Jens Lykke-Andersen for providing anti-hUPF1 antibody and Andreas Gruber for help with the spectrofluorometer. This work was supported by the Kanton Bern and by grants to O.M. from the Swiss National Science Foundation (grants 31-61720.00 and 3100A0-102159), the Novartis Foundation for Biomedical Research, Basel, Switzerland, and the Helmut Horten Foundation, Agno, Switzerland. C.M.A. is supported by a grant from the Swiss National Science Foundation to Joachim Lingner. The Open Access publication charges for this article were paid by the Swiss National Science Foundation grant 3100A0-102159.
Conflict of interest statement. None declared.
REFERENCES
- 1.Maquat L.E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nature Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 2.Frischmeyer P.A., Dietz H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 3.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 4.Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendell J.T., Dietz H.C. When the message goes awry. disease-producing mutations that influence mRNA content and performance. Cell. 2001;107:411–414. doi: 10.1016/s0092-8674(01)00583-9. [DOI] [PubMed] [Google Scholar]
- 6.Holbrook J.A., Neu-Yilik G., Hentze M.W., Kulozik A.E. Nonsense-mediated decay approaches the clinic. Nature Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 7.Li S., Wilkinson M.F. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- 8.Cali B.M., Kuchma S.L., Latham J., Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pulak R., Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 10.Leeds P., Peltz S.W., Jacobson A., Culbertson M.R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 11.Leeds P., Wood J.M., Lee B.S., Culbertson M.R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Applequist S.E., Selg M., Raman C., Jack H.M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perlick H.A., Medghalchi S.M., Spencer F.A., Kendzior R.J., Jr, Dietz H.C. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc. Natl Acad. Sci. USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lykke-Andersen J., Shu M.D., Steitz J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 15.Serin G., Gersappe A., Black J.D., Aronoff R., Maquat L.E. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol. Cell. Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendell J.T., Medghalchi S.M., Lake R.G., Noensie E.N., Dietz H.C. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 2000;20:8944–8957. doi: 10.1128/mcb.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita A., Ohnishi T., Kashima I., Taya Y., Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichenbach P., Hoss M., Azzalin C.M., Nabholz M., Bucher P., Lingner J. A human homolog of yeast est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 2003;13:568–574. doi: 10.1016/s0960-9822(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 19.Gatfield D., Unterholzner L., Ciccarelli F.D., Bork P., Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiu S.Y., Serin G., Ohara O., Maquat L.E. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Hir H., Gatfield D., Izaurralde E., Moore M.J. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim V.N., Yong J., Kataoka N., Abel L., Diem M.D., Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J. 2001;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibuya T., Tange T.O., Sonenberg N., Moore M.J. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nature Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 24.Palacios I.M., Gatfield D., St Johnston D., Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 25.Lykke-Andersen J., Shu M.D., Steitz J.A. Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 26.Stoecklin G., Ming X.F., Looser R., Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 2000;20:3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKendry R., John J., Flavell D., Muller M., Kerr I.M., Stark G.R. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl Acad. Sci. USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng J., Fogel-Petrovic M., Maquat L.E. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol. Cell. Biol. 1990;10:5215–5225. doi: 10.1128/mcb.10.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baserga S.J., Benz E.J., Jr Nonsense mutations in the human beta-globin gene affect mRNA metabolism. Proc. Natl Acad. Sci. USA. 1988;85:2056–2060. doi: 10.1073/pnas.85.7.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belgrader P., Maquat L.E. Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations can facilitate exon skipping. Mol. Cell. Biol. 1994;14:6326–6336. doi: 10.1128/mcb.14.9.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter M.S., Doskow J., Morris P., Li S., Nhim R.P., Sandstedt S., Wilkinson M.F. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 32.Buhler M., Paillusson A., Muhlemann O. Efficient downregulation of immunoglobulin mu mRNA with premature translation-termination codons requires the 5′-half of the VDJ exon. Nucleic Acids Res. 2004;32:3304–3315. doi: 10.1093/nar/gkh651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoecklin G., Stoeckle P., Lu M., Muehlemann O., Moroni C. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA. 2001;7:1578–1588. [PMC free article] [PubMed] [Google Scholar]
- 34.Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 35.Gudikote J.P., Wilkinson M.F. T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts. EMBO J. 2002;21:125–134. doi: 10.1093/emboj/21.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muhlemann O., Mock-Casagrande C.S., Wang J., Li S., Custodio N., Carmo-Fonseca M., Wilkinson M.F., Moore M.J. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell. 2001;8:33–43. doi: 10.1016/s1097-2765(01)00288-x. [DOI] [PubMed] [Google Scholar]
- 37.Carter M.S., Li S., Wilkinson M.F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 38.Sun X., Perlick H.A., Dietz H.C., Maquat L.E. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lerner E.A., Lerner M.R., Janeway C.A., Jr, Steitz J.A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl Acad. Sci. USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medghalchi S.M., Frischmeyer P.A., Mendell J.T., Kelly A.G., Lawler A.M., Dietz H.C. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 41.Thermann R., Neu-Yilik G., Deters A., Frede U., Wehr K., Hagemeier C., Hentze M.W., Kulozik A.E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]