Abstract
Although efficient methods exist to assemble synthetic oligonucleotides into genes and genomes, these suffer from the presence of 1–3 random errors/kb of DNA. Here, we introduce a new method termed consensus shuffling and demonstrate its use to significantly reduce random errors in synthetic DNA. In this method, errors are revealed as mismatches by re-hybridization of the population. The DNA is fragmented, and mismatched fragments are removed upon binding to an immobilized mismatch binding protein (MutS). PCR assembly of the remaining fragments yields a new population of full-length sequences enriched for the consensus sequence of the input population. We show that two iterations of consensus shuffling improved a population of synthetic green fluorescent protein (GFPuv) clones from ∼60 to >90% fluorescent, and decreased errors 3.5- to 4.3-fold to final values of ∼1 error per 3500 bp. In addition, two iterations of consensus shuffling corrected a population of GFPuv clones where all members were non-functional, to a population where 82% of clones were fluorescent. Consensus shuffling should facilitate the rapid and accurate synthesis of long DNA sequences.
INTRODUCTION
Methods for the automated chemical synthesis of oligonucleotides (1,2) and their assembly into long double-stranded DNA (dsDNA) sequences by PCR (3,4) and LCR (5) have enabled the chemical synthesis of genes and even entire viral genomes (6,7). These technological advances have helped spur the formation of the new field of synthetic biology (8), which aims at defining the functional units of living organisms through the modular engineering of synthetic organisms. In addition, the demand for fully synthetic gene length DNA fragments of defined sequence has dramatically increased in recent years for use in applications such as codon optimization (9), construction of DNA vaccines (10), de novo synthesis of novel biopolymers (11), or simply to gain access to known DNA sequences when original templates are unavailable. The future demand for long synthetic DNA is likely to dramatically increase when it becomes cheaper/faster to synthesize a desired sequence than to obtain it by other means.
The assembly of DNA is currently limited by the presence of random sequence errors in synthetic oligonucleotides that arise from side reactions during synthesis (incomplete couplings, misincorporations, etc.) and resulting in 1–3 errors/kb (7,12,13). The deleterious impact of these errors becomes more significant as the desired lengths of synthetic DNA increase. Indeed, in the remarkable assembly of the PhiX 174 bacteriophage genome (5386 bp) using gel-purified, synthetic oligonucleotides, the products contained an average of ∼2 lethal errors/kb resulting in 1 plaque-forming genomes per 20 000 clones (7). A functional selection (plaque formation) was required in this study to identify a clone with the correct sequence. Thus, error reduction/correction is a requirement for the efficient production of long synthetic DNA of defined sequence. However, the process of sequencing multiple clones and manual correction of errors is both costly and time consuming.
Several methods have been reported for the removal of error-containing sequences in populations of DNA. These methods rely upon the selective destruction (14,15) or physical separation (16,17) of mismatch-containing heteroduplexes. Smith and Modrich (14) reported the selective destruction of error-containing sequences in PCR products by generating dsDNA breaks upon overdigestion with the Escherichia coli mismatch-specific endonuclease MutHLS (18). Gel purification and cloning of the remaining full-length DNA resulted in an apparent 10-fold reduction in the error rate for PCR products. However, the existing approaches are not well suited for error removal in long synthetic DNA sequences where virtually all members in the population contain multiple errors.
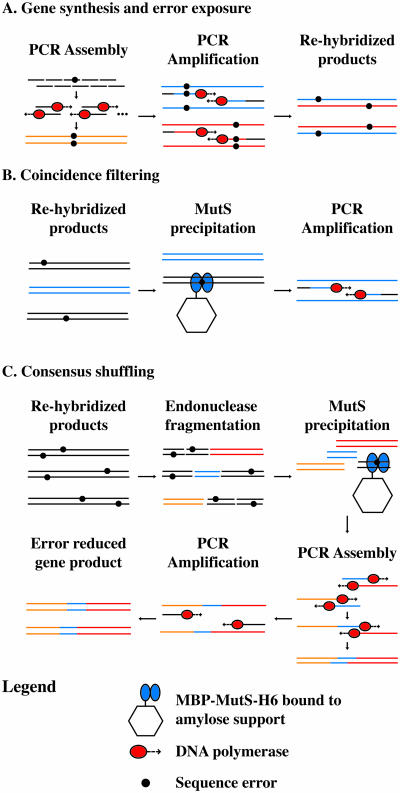
Error correction with MutS is outlined in Figure 1. The population of DNA molecules containing random errors is first re-hybridized to expose synthesis errors as mismatches (Figure 1A). Duplexes containing mismatches can then be removed from the population by affinity capture with immobilized MutS (Figure 1B), a process we term coincidence filtering, since both strands of the duplex must match to pass this filtering step. For long synthetic DNA sequences or for sequences with high error rates, coincidence filtering is ineffective, since the likelihood of both strands being perfectly matched after re-hybridization is very low. To generalize MutS error filtering for application on synthetic DNA, the synthetic DNA is cleaved into small overlapping fragments before MutS filtering. Fragments containing mismatches are selectively removed through absorption to an immobilized maltose-binding protein (MBP)–Thermus aquaticus (Taq) MutS–His6 fusion protein (MBP–MutS–H6) (18–20). The remaining mixture of fragments (enriched with fragments of the correct sequence) serves as a template for assembly PCR to produce the full-length product (Figure 1C). This process can be iterated until the consensus sequence emerges as the dominant species in the population. This approach is equivalent to DNA shuffling (21) with additional mismatch exposure and removal steps.
Figure 1.
Overview of gene synthesis, error exposure, coincidence filtering and consensus shuffling. (A) Gene synthesis from component oligonucleotides. PCR amplification of the PCR assembly reaction generates products that are re-hybridized to expose errors. Full-length genes: orange, blue and red lines. (B) Coincidence filtering on re-hybridized gene synthesis products containing few errors. Full-length genes containing errors are precipitated by MBP–MutS–H6 immobilized on amylose support. Error free gene: blue lines. (C) Consensus shuffling on re-hybridized gene synthesis products containing multiple errors. The re-hybridized gene synthesis products are fragmented, and error containing fragments are precipitated by MBP–MutS–H6 immobilized on amylose support. Error reduced fragments (orange, blue and red) are reassembled into the full-length gene followed by PCR amplification to generate error reduced products. Primers: black lines.
In this report, we assemble GFPuv from synthetic oligonucleotides and apply both coincidence filtering and consensus shuffling protocols to reduce errors in the resultant DNA populations. The error rates are characterized by gene function (fluorescence) and by DNA sequencing. We also provide a mathematical model describing the error reduction protocols to aid predictions about parameters influencing their effectiveness.
MATERIALS AND METHODS
Reagents
Chemicals were from Sigma. Restriction enzymes were from Promega and New England Biolabs. KOD Hot Start DNA Polymerase was from Novagen. Amylose resin was from NEB (catalog no. E8021S). Ni-NTA resin was from Novagen (catalog no. 70666). Ultrafiltration device from Millipore (catalog no. UFC900524). Slide-A-Lyzer dialysis membrane was from Pierce (catalog no. 66415).
Construction of a recombinant expression vector for MBP–MutS–H6
Full-length Taq MutS was amplified from template pETMutS (22) with primers 5′-AAA AAA CAT ATG GAA GGC ATG CTG AAG G-3′ and 5′-AAA AAT AAG CTT CCC CTT CAT GGT ATC CAA GG-3′ and cloned into the Nde1/HindIII sites of vector pIADL14 (23) to give plasmid pMBP–MutS–H6.
MBP–MutS–H6 purification
E.coli strain BL21(DE3) transformed with pMBP–MutS–H6 was grown to OD600 ∼1.0 and induced using 1 mM isopropyl-β-d-thiogalactopyranoside for 4 h at 37°C. Cells from 4 l of culture were pelleted and resuspended in 60 ml of buffer A (20 mM Tris–HCl, pH 7.4, 300 mM NaCl, 1 mM EDTA, 1 mM DTT and 1 mM phenylmethlysulfonyl fluoride). Cell suspension was sonicated on ice and insoluble material was removed by centrifugation at 50 000 g for 10 min at 4°C. Supernatant was applied to 5 ml amylose resin pre-equilibrated in buffer A. Bound MBP–MutS–H6 was washed three times using 20 ml buffer B (20 mM Tris–HCl, pH 7.4, 300 mM NaCl) and stored overnight at 4°C. MBP–MutS–H6 was eluted using 20 ml buffer B + 10 mM maltose. Eluate was applied to ∼4 ml of Ni-NTA resin pre-equilibrated in buffer B. Bound MBP–MutS–H6 was washed four times using 20 ml buffer B + 25 mM imidazole. Bound MBP–MutS–H6 was eluted using buffer B + 1 M imidazole. Eluate was concentrated via ultrafiltration using Amicon Ultra 5 kDa MWCO at 4°C. Concentrated sample was dialyzed extensively against 2× storage buffer (100 mM Tris–HCl, pH 7.5, 200 mM NaCl, 0.2 mM EDTA and 0.2 mM DTT) using a Slide-A-Lyzer 10 kDa MWCO cassette at 4°C. Protein concentration was determined using A280 and a calculated extinction coefficient of 119 070 M−1 cm−1. Dialyzed sample was diluted using an equal volume of glycerol and stored at −20°C. The final concentration of MBP–MutS–H6 (M.W. 135, 085) was ∼19.1 μM for a total yield of ∼1.5 mg of protein. Aliquots of sample were taken at each stage of the purification and resolved on an 8% SDS–PAGE gel (Supplementary Figure 1).
GFPuv assembly
Oligonucleotides were purchased from Qiagen with ‘salt-free’ purification. Sequence 261–1020 of pGFPuv (GenBank accession no. U62636 with T357C, T811A and C812G base substitutions) was assembled using 40mer (37) and 20mer (2) oligonucleotides with 20 bp overlap (Supplementary Table 1). Assembly reactions contained the following components: 64 nM each oligonucleotide, 200 μM dNTPs, 1 mM MgSO4, 1× buffer and 0.02 U/μl KOD Hot Start DNA Polymerase. Assembly was carried out using 25 cycles of 94°C for 30 s, 52°C for 30 s and 72°C for 2 min. PCR amplification of assembly products contained the following components: 10-fold dilution of assembly reaction, 25 μM of 20 bp outside primers, 200 μM dNTPs, 1 mM MgSO4, 1× buffer and 0.02 U/μl KOD Hot Start DNA Polymerase. PCR was carried out using 35 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 1 min followed by a final extension at 72°C for 10 min. PCR products were purified using the Qiagen QIAquick PCR purification kit with elution in dH2O followed by speed-vac concentration. Assuming an error rate of 1 × 10−6/bp/duplication for KOD DNA polymerase (24), 35 cycles of PCR would be expected to introduce ∼0.053 mutations per assembled GFPuv molecule.
Mismatch exposure and GFPuv fragmentation
Assembled GFPuv was diluted to 250 ng/μl in 10 mM Tris–HCl, pH 7.8, 50 mM NaCl and heated to 95°C for 5 min followed by cooling 0.1°C/s to 25°C. Heteroduplex for consensus filtering was split into three pools and digested to completion with NlaIII (NEB), TaqI (NEB) or NcoI plus XhoI (Promega) for 2 h following the manufacturer's protocols. Digests were purified using the Qiagen QIAquick PCR purification kit with elution in dH2O. Samples were pooled and the concentration was determined by measuring A260.
MBP–MutS–H6 binding
MBP–MutS–H6 binding reactions contained ∼11.5 ng/μl DNA and ∼950 nM MBP–MutS–H6 dimers in 1× binding buffer (20 mM Tris–HCl, pH 7.8, 10 mM NaCl, 5 mM MgCl2, 1 mM DTT and 5% glycerol). Reactions were allowed to incubate at room temperature for 10 min before incubation for 30 min with an equal volume of amylose resin pre-equilibrated in 1× binding buffer. Protein–DNA complexes were removed by low-speed centrifugation and aliquots of supernatant were removed for subsequent processing.
Reassembly, amplification and cloning
Supernatant (50 μl) from consensus filtering experiments was desalted using Centri-Sep spin columns (Princeton Separations) and concentrated. Purified and concentrated DNA fragments were reassembled as above with aliquots removed at varying cycles. Aliquots of assembly reactions were resolved on 2% agarose gels to monitor the reassembly process. Aliquots showing predominantly reassembled full-length GFPuv were PCR amplified as above. Aliquots of supernatant from coincidence filtering experiments were diluted 10-fold and PCR amplified as above. PCR products were digested with BamHI/EcoRI and ligated into the 2595 bp BamHI–EcoRI fragment of pGFPuv. Ligations were transformed into E.coli DH5 and fluorescent colonies were scored using a handheld 365 nm ultraviolet (UV) lamp.
Preparation of substrate for consensus shuffling from 10 non-fluorescent GFPuv clones
Ten non-fluorescent GFPuv clones were pooled in equal amounts. The nature and location of the mutations in these clones is shown in Figure 4. The GFP coding region was PCR amplified from the mixture and submitted to the consensus shuffling protocol with and without the application of the MBP–MutS–H6 error filter.
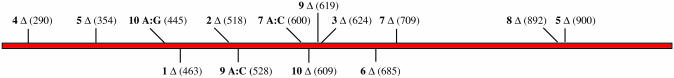
Figure 4.
Locations of mutations in the 10 non-fluorescent clones used as input for a consensus shuffling experiment. The number designation for each clone is followed by the type of mutation (Δ = 1 base deletion; X:X point mutations = GFPuv sense strand wt nucleotide:nucleotide substitution for wt nucleotide) and its position in assembled GFPuv. All 10 clones contain a single deletion mutation at distributed positions throughout the GFPuv open reading frame with 3/10 containing an additional point mutation. The generation of a GFPuv sequence encoding a fluorescent product is expected to coincide with the correction of all 10 deletion mutations. Therefore, percent fluorescent colonies are an indication of progress toward the consensus sequence of the population. The numbering system used is that of pGFPuv (GenBank accession no. U62636). Assembled GFPuv begins at position 261 and ends at position 1020.
RESULTS
To create an error filter, we constructed a fusion protein between MBP (19) and the mismatch binding protein from T.aquaticus (22) with a C-terminal His6 tag (MBP–MutS–H6). MBP–MutS–H6 was overexpressed and purified from E.coli to >95% purity (Supplementary Figure 1). MBP–MutS–H6 immobilized on amylose resin was shown to selectively retain a 40mer heteroduplex containing a deletion mutation over wild-type homoduplex (Supplementary Figure 2).
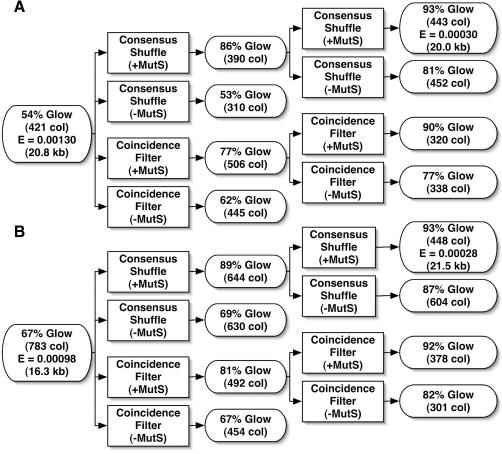
To demonstrate error correction, unpurified 40mer oligonucleotides were assembled by PCR (3) to produce a 760 bp gene encoding green fluorescent protein (25) (GFPuv). Two independent preparations of GFPuv containing typical gene synthesis errors (Figure 3 and Table 1) were re-hybridized and subjected to two iterations of coincidence filtering or consensus shuffling. For consensus shuffling, the GFPuv assembly product was split into three pools and digested into sets of overlapping fragments using distinct Type II restriction enzymes (Figure 2). The digests were pooled and subjected to error filtering with or without added MBP–MutS–H6. The unbound fragments were reassembled into full-length products and PCR amplified. For coincidence filtering, unbound full-length GFPuv was PCR amplified following treatment with the error filter. After cloning in E.coli, error rates were estimated by scoring colonies for fluorescence under a handheld UV lamp (Figure 3). The actual error rates of the input and consensus shuffled populations were determined by sequencing plasmid DNA from randomly selected colonies (Figure 3). The results show that two rounds of consensus shuffling increased the percentage of fluorescent colonies from ∼60 to >90% and reduced the error rate of the populations 4.3- and 3.5-fold from values of 1.3 to 0.3 and 0.98 to 0.28 errors/kb, respectively. MBP–MutS–H6 was required to increase the fraction of fluorescent colonies in each round of error filtering. The nature of the errors in the input and consensus shuffled populations are reported in Table 1.
Figure 3.
Consensus shuffling and coincidence filtering data for GFPuv. The percentages of fluorescent clones are reported as % glow with the total number of colonies counted in parentheses (# col). The experimentally determined error rates in errors/base, where determined, are reported as E with the total number of base pair sequenced in parentheses (# kb). (A) Process flow and data for gene assembly experiment 1. (B) Process flow and data for gene assembly experiment 2.
Table 1.
Sequence errors in input and consensus shuffled DNA
| Mutation | Deletion | Insertion | G/C to A/T | A/T to G/C | G/C to C/G | G/C to T/A | A/T to C/G | A/T to T/A |
|---|---|---|---|---|---|---|---|---|
| Mismatches | — | — | G:T/A:C | A:C/G:T | G:G/C:C | G:A/T:C | G:A/T:C | A:A/T:T |
| Input DNA | 18 | 1 | 15 | 1 | 3 | 4 | 1 | 0 |
| Consensus Shuffling | 3 | 0 | 2 | 0 | 2 | 5 | 0 | 0 |
Deletion, insertion, transition and transversion mutations were quantified after sequencing DNA from randomly selected colonies for the input populations and the populations after two rounds of consensus shuffling. The reported mutations are for the combined input populations (20.8 and 16.3 kb) and consensus shuffled populations (20.0 and 21.5 kb) reported in Figure 3. The two types of mismatched base pairs that arise after re-hybridization of the gene synthesis product are indicated below each transition or transversion mutation.
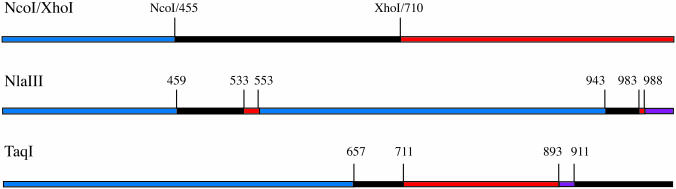
Figure 2.
Restriction enzyme cleavage sites used in consensus shuffling experiments. The numbering system used is that of pGFPuv (GenBank accession no. U62636). Assembled GFPuv begins at position 261 and ends at position 1020. After pooling the three digests, the average fragment size is 150 bp and the size range is 4–396 bp.
Although DNA shuffling has traditionally been used to create diversity through the combinatorial shuffling of mutations in a population, DNA shuffling also creates a sub-population of sequences with a reduction in diversity, as correct fragments can recombine to produce error-free sequences. Indeed, with consensus shuffling, it is possible to start with a population of DNA molecules wherein every individual in the population contains errors and create a new population where the dominant sequence is error free. To demonstrate this, 10 non-fluorescent GFPuv clones, each containing a deletion mutation (Figure 4), were pooled and subjected to either DNA shuffling alone or two iterations of consensus shuffling. Products were cloned in E.coli, and the percentage of fluorescent colonies was monitored as an indication of progress toward the consensus sequence. DNA shuffling alone (no MBP–MutS–H6) increased the percentage of fluorescent colonies to 30% (387 colonies total) similar to a previous report (26). Two rounds of consensus shuffling gave a new population that was 82% fluorescent (551 colonies total), indicating that the dominant species was likely the consensus sequence of the input population.
DISCUSSION
Both consensus shuffling and coincidence filtering protocols were effective in reducing errors in synthetic GFPuv populations (Figure 3). In both cases, two iterations of either consensus shuffling or coincidence filtering increased fluorescent colonies from average values of ∼60 to ≥90%. Sequencing data from two independent experiments showed a 4.3- and 3.5-fold reduction in the error rate for the consensus shuffled populations compared with the input populations giving final error rates of 0.3 and 0.28 errors/kb, respectively. These results demonstrate the usefulness of the MBP–MutS–H6 error filter in both consensus shuffling and coincidence filtering protocols. Taq MutS has previously been shown to bind to deletion mutations with high affinity (27), a mutation common in synthetic DNA. However, it is important to note that Taq MutS has lower affinity for specific point mutations and binds weakly to homoduplex DNA (27). These factors may limit the stepwise efficiency of the error filter. Moreover, specific point mutations may be refractory to removal even after multiple rounds of consensus shuffling. Two rounds of consensus shuffling using the MBP–MutS–H6 error filter proved most effective in reducing deletions and G/C to A/T transitions, consistent with previous reports for the selectivity of Taq MutS (27). However, it must be emphasized that each synthetic oligonucleotide point mutation would generate two heteroduplex DNA molecules containing unique mismatches after PCR amplification and re-hybridization (Figure 1A and Table 1). For example, a G to A transition mutation in a synthetic oligonucleotide would generate heteroduplexes with G–T or A–C mismatches after PCR amplification and re-hybridization. For consensus shuffling, either of these mismatch containing heteroduplexes could evade precipitation by the MBP–MutS–H6 error filter and participate in the reassembly of full-length GFPuv. Therefore, Table 1 lists the pair of mismatches that could give rise to the observed transition or transversion mutation. These results show that the MBP–MutS–H6 error filter was most effective at removing insertion/deletion loops and G–T/A–C mismatches from the population.
It should be possible to generalize the consensus shuffling protocol to a large number of synthetic DNA constructs. GFPuv was chosen as the synthetic construct in this study for its advantages as a fluorescent reporter gene. This allowed easy optimization of our protocol without the need to sequence thousands of base pairs of DNA. We expect the results reported here for consensus shuffling to readily translate to synthetic DNA constructs of varied sequence, greater overall length and/or higher initial errors/kb. Synthetic DNA constructs of varied sequence can be digested into a defined set of fragments using Type II restriction enzymes or fragmented into any desired size range using controlled DNase I digestion (26). Digestion and reassembly of a large number of different genes is expected to be as robust as the protocol of DNA shuffling (28), which has been broadly applied to a variety of gene sequences. Synthetic DNA constructs larger than GFPuv are expected to be amenable to error correction by consensus shuffling, as the error filtering is conducted on gene fragments before reassembly of the full-length gene. Thus, the errors/kb data presented in this study are expected to translate to larger genes with similar initial errors/kb (excepting mutations introduced by PCR amplification following the final application of the error filter). Synthetic DNA constructs of higher initial errors/kb are expected to be amenable for error correction by consensus shuffling. However, these constructs will require digestion into smaller sized gene fragments that may affect the efficiency of error correction. In contrast to consensus shuffling, an increase in the size of the synthetic DNA product or an increase in errors/kb would preclude the use of the coincidence filtering protocol, as every molecule in the population would contain one or more errors. As proof of the utility of the consensus shuffling protocol, 10 non-fluorescent GFPuv clones containing one or more errors (Figure 4) were converted into a population where 82% of the clones were fluorescent. It is important to note that DNA shuffling alone shows an improvement in percent fluorescent colonies in this example (from 0 to 30%). For synthetic DNA populations, DNA shuffling alone shows no improvement in percent fluorescent colonies (see Figure 3 ‘no MutS’ treatments). DNA shuffling alone improves the overall number of correct sequences only for small DNA populations with low error rates. For example, when shuffling 10 clones with a unique mutation in each clone, one would expect the fraction of correct products to be (9/10)10 = 35% (26), very close to the value of 30% that we observed. A mathematical model describing the error rates for shuffling and error filtering of synthetic DNA populations is presented below.
To estimate some parameters of consensus shuffling and coincidence filtering, a simple mathematical model (Equations 1–6) was constructed. An input population of dsDNA molecules of length N, containing E errors/base is re-hybridized, fragmented into shorter dsDNA fragments of average length S, error filtered and reassembled. P(F) is the probability a fragment of length S will have a correct sequence. We determine the probability that re-hybridized duplexes will have zero (C), one (H) or both (I) strands with errors. Equation 5 estimates the probability that a fragment will be correct after a cycle of MutS filtering, P(F′), by applying a MutS selectivity factor (M) to adjust the relative amounts of mismatch containing duplexes (I, H) while accounting for the total fraction of correct strands in the re-hybridized duplexes. The probability of obtaining an error free assembly product, P(A), is then given by Equation 6.
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
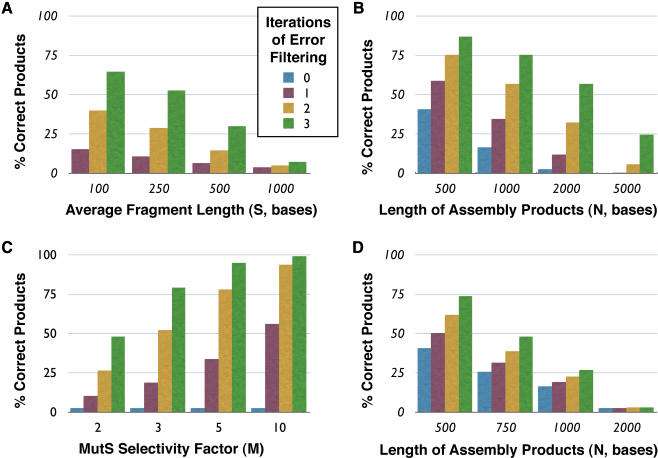
From our consensus shuffling error rate data (Figure 3), we estimate the MutS selectivity factor M to be ∼2.2. Figure 5 shows some predictions that emerge from this model assuming typical length (2 kb), fragment sizes (200 bp) and error rates (1.8 errors/kb). Consensus shuffling is predicted to be most effective with smaller fragment sizes (Figure 5A). As mentioned above, smaller fragment sizes could be obtained by controlled digestion with DNase I (21). In addition, multiple iterations of MutS filtering can have dramatic results on populations with few correct sequences (Figure 5B), although the model does not account for the differing specificity of MutS toward the various types of mismatches. The model also predicts that even modest improvements in the MutS selectivity factor through optimization of the MutS–DNA binding conditions and/or the use of a combination of MutS homologs with varying mismatch specificity (29) could dramatically improve the consensus shuffling protocol (Figure 5C). Coincidence filtering (N = S) is predicted to be effective for populations with low error rates per clone (Figure 5D) but becomes ineffective when the majority of re-hybridized duplexes contain mismatches.
Figure 5.
Mathematical modeling of consensus shuffling and coincidence filtering. Predictions from theoretical model of consensus shuffling calculated with the following parameters (unless otherwise specified): error rate of input population per base, E = 0.0018; length of product assembled, N = 2000; MutS selectivity factor, M = 2.2; average fragment size, S = 200. (A) Errors versus average digested fragment length for consensus shuffling. (B) Errors versus product length for consensus shuffling. (C) Errors versus MutS selectivity factor for consensus shuffling. (D) Errors versus product length for coincidence filtering (N = S).
We have demonstrated consensus shuffling and coincidence filtering as experimental methods to significantly reduce errors in synthetic DNA. Consensus shuffling should be generally applicable for error correction on synthetic genes of typical lengths and error rates. Two iterations of consensus shuffling (∼6 h/iteration) generated a population with ∼1 error/3500 bp. This reduction in error rate will allow the identification of a correct clone after sequencing DNA from a reduced number of colonies. Coincidence filtering is a simple and effective procedure to reduce errors in synthetic DNA populations with low error rates per clone. These methods should significantly increase the speed and decrease the cost of production of synthetic genes.
Note: While this manuscript was under review, Carr et al. (30) independently reported the application of Taq MutS in protocols for error reduction on synthetic DNA.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Dr P. Hsieh for the generous gift of clone pETMutS. This work was financially supported by DARPA, the NHGRI and the W.M. Keck Foundation. Funding to pay the Open Access publication charges for this article was provided by NIH R01 HG003275.
Conflict of interest statement. The authors declare a conflict of interest. A patent application has been filed based on the results presented in this manuscript. Peter Belshags is a co-founder of a company, Genetic Assemblies, Inc., that may commercialize this method.
REFERENCES
- 1.Matteucci M.D., Caruthers M.H. Nucleotide chemistry. 4. Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 1981;103:3185–3191. [Google Scholar]
- 2.Singh-Gasson S., Green R.D., Yue Y.J., Nelson C., Blattner F., Sussman M.R., Cerrina F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- 3.Stemmer W.P.C., Crameri A., Ha K.D., Brennan T.M., Heyneker H.L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 4.Richmond K.E., Li M., Rodesch M.J., Patel M., Lowe A.M., Kim C., Chu L.L., Venkataramaian N., Kaysen J., Belshaw P.J., et al. Amplification and assembly of chip eluted DNA (AACED): a method for high throughput gene synthesis. Nucleic Acids Res. 2004;32:5011–5018. doi: 10.1093/nar/gkh793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barany F. Genetic-disease detection and DNA amplification using cloned thermostable ligase. Proc. Natl Acad. Sci. USA. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cello J., Paul A.V., Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 7.Smith H.O., Hutchison C.A., Pfannkoch C., Venter J.C. Generating a synthetic genome by whole genome assembly: phi X174 bacteriophage from synthetic oligonucleotides. Proc. Natl Acad. Sci. USA. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferber D. Synthetic biology: microbes made to order. Science. 2004;303:158–161. doi: 10.1126/science.303.5655.158. [DOI] [PubMed] [Google Scholar]
- 9.Gustafsson C., Govindarajan S., Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Hest J.C.M., Tirrell D.A. Protein-based materials, toward a new level of structural control. Chem. Commun. 2001;19:1897–1904. doi: 10.1039/b105185g. [DOI] [PubMed] [Google Scholar]
- 12.Hoover D.M., Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong A.-S., Yao Q.-H., Peng R.-H., Li X., Fan H.-Q., Cheng Z.-M., Li Y. A simple, rapid, high-fidelity and cost-effective PCR-based two-step DNA synthesis method for long gene sequences. Nucleic Acids Res. 2004;32:e98. doi: 10.1093/nar/gnh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith J., Modrich P. Removal of polymerase-produced mutant sequences from PCR products. Proc. Natl Acad. Sci. USA. 1997;94:6847–6850. doi: 10.1073/pnas.94.13.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young L., Dong Q.H. Two-step total gene synthesis method. Nucleic Acids Res. 2004;32:e59. doi: 10.1093/nar/gnh058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy J.C., Jewell D.L., White K.I., Fox G.E., Willson R.C. Nucleic acid separations utilizing immobilized metal affinity chromatography. Biotechnol. Prog. 2003;19:982–986. doi: 10.1021/bp025563o. [DOI] [PubMed] [Google Scholar]
- 17.Kaur M., Makrigiorgos G.M. Novel amplification of DNA in a hairpin structure: towards a radical elimination of PCR errors from amplified DNA. Nucleic Acids Res. 2003;31:e26. doi: 10.1093/nar/gng026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schofield M.J., Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 19.Kellermann O.K., Ferenci T. Maltose-binding protein from Escherichia coli. Methods Enzymol. 1982;90:459–463. doi: 10.1016/s0076-6879(82)90171-9. [DOI] [PubMed] [Google Scholar]
- 20.Biswas I., Ban C., Fleming K.G., Qin J., Lary J.W., Yphantis D.A., Yang W., Hsieh P. Oligomerization of a MutS mismatch repair protein from Thermus aquaticus. J. Biol. Chem. 1999;274:23673–23678. doi: 10.1074/jbc.274.33.23673. [DOI] [PubMed] [Google Scholar]
- 21.Stemmer W.P.C. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 22.Biswas I., Hsieh P. Identification and characterization of a thermostable MutS homolog from Thermus aquaticus. J. Biol. Chem. 1996;271:5040–5048. doi: 10.1074/jbc.271.9.5040. [DOI] [PubMed] [Google Scholar]
- 23.McCafferty D.G., Lessard I.A.D., Walsh C.T. Mutational analysis of potential zinc-binding residues in the active site of the enterococcal D-Ala-D-Ala dipeptide VanX. Biochemistry. 1997;36:10498–10505. doi: 10.1021/bi970543u. [DOI] [PubMed] [Google Scholar]
- 24.Takagi M., Nishioka M., Kakihara H., Kitabayashi M., Inoue H., Kawakami B., Oka M., Imanaka T. Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl. Environ. Microbiol. 1997;63:4504–4510. doi: 10.1128/aem.63.11.4504-4510.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crameri A., Whitehorn E.A., Tate E., Stemmer W.P.C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 26.Stemmer W.P.C. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl Acad. Sci. USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schofield M.J., Brownewell F.E., Nayak S., Du C.W., Kool E.T., Hsieh P. The Phe-X-Glu DNA binding motif of MutS. The role of hydrogen bonding in mismatch recognition. J. Biol. Chem. 2001;276:45505–45508. doi: 10.1074/jbc.C100449200. [DOI] [PubMed] [Google Scholar]
- 28.Joern J.M. Methods in Molecular Biology. Vol. 231. Humana Press; 2003. DNA shuffling; pp. 85–89. [DOI] [PubMed] [Google Scholar]
- 29.Whitehouse A., Deeble J., Parmar R., Taylor G.R., Markham A.F., Meredith D.M. Analysis of the mismatch and insertion/deletion binding properties of Thermus thermophilus, HB8, MutS. Biochem. Biophys. Res. Commun. 1997;233:834–837. doi: 10.1006/bbrc.1997.6551. [DOI] [PubMed] [Google Scholar]
- 30.Carr P.A., Park J.S., Lee Y.J., Yu T., Zhang S., Jacobson J.M. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.