Abstract
The genes encoding several key fatty acid biosynthetic enzymes (called the fab cluster) are clustered in the order plsX-fabH-fabD-fabG-acpP-fabF at min 24 of the Escherichia coli chromosome. A difficulty in analysis of the fab cluster by the polar allele duplication approach (Y. Zhang and J. E. Cronan, Jr., J. Bacteriol. 178:3614–3620, 1996) is that several of these genes are essential for the growth of E. coli. We overcame this complication by use of the fab gene cluster of Salmonella typhimurium, a close relative of E. coli, to provide functions necessary for growth. The S. typhimurium fab cluster was isolated by complementation of an E. coli fabD mutant and was found to encode proteins with >94% homology to those of E. coli. However, the S. typhimurium sequences cannot recombine with the E. coli sequences required to direct polar allele duplication via homologous recombination. Using this approach, we found that although approximately 60% of the plsX transcripts initiate at promoters located far upstream and include the upstream rpmF ribosomal protein gene, a promoter located upstream of the plsX coding sequence (probably within the upstream gene, rpmF) is sufficient for normal growth. We have also found that the fabG gene is obligatorily cotranscribed with upstream genes. Insertion of a transcription terminator cassette (Ω-Cm cassette) between the fabD and fabG genes of the E. coli chromosome abolished fabG transcription and blocked cell growth, thus providing the first indication that fabG is an essential gene. Insertion of the Ω-Cm cassette between fabH and fabD caused greatly decreased transcription of the fabD and fabG genes and slower cellular growth, indicating that fabD has only a weak promoter(s).
The bacterial fatty acid biosynthetic pathway is a type II, or disassociated-enzyme, system, where each of the reactions of the pathway is catalyzed by a discrete cytoplasmic enzyme. Fatty acid biosynthesis in Escherichia coli is the paradigm type II system, and much has been learned about the pathway in recent years (12, 25). Recent work has shown that about half of the fatty acid biosynthesis (fab) genes are clustered as a set of contiguous genes at min 24 of the Escherichia coli chromosome in the order fabH-fabD-fabG-acpP-fabF (4, 19, 22, 33), whereas the rest of the fab genes are scattered around the chromosome as separately transcribed genes (12). The proteins encoded by the genes of the cluster are PlsX, β-ketoacyl-acyl carrier protein (ACP) synthase III, malonyl-coenzyme A (CoA):ACP transacylase, β-ketoacyl-ACP reductase, ACP, and β-ketoacyl-ACP synthase II, respectively. We consider the plsX gene (located immediately upstream of fabH) to be part of the E. coli cluster due to its role (albeit poorly understood) in phospholipid biosynthesis (10). The plsX phenotype is defined by a single mutant allele, plsX50, which confers sn-glycerol 3-phosphate auxotrophy on strains carrying mutations in plsB, the gene that encodes sn-glycerol 3-phosphate acyltransferase, the first enzyme of phospholipid synthesis. The cluster is delimited upstream by the rpmF gene, encoding the L32 ribosomal protein (19), and downstream by a gene (pabC) involved in p-aminobenzoic acid synthesis (7).
Similar fab gene clusters have recently been reported in other bacteria: Haemophilus influenzae Rd (fabH-fabD-fabG-acpP) (5), Vibrio harveyi (fabD-fabG-acpP-fabF) (27), and Rhodobacter capsulatus (plsX-fabH) (3). The recently completed genomic sequence of Helicobacter pylori also contains fab cluster homologs (31). However, the fab cluster homologs of H. pylori are split relative to the fab cluster genes of E. coli. The H. pylori genome contains adjacent plsX and fabH genes, with a ribosomal protein gene (rpmF) located upstream of plsX, as seen in R. capsulatus, while the remainder of the genes found in the E. coli fab cluster, fabD, fabG, acpP, and fabF, are clustered with accA (which encodes an acetyl-CoA carboxylase subunit) at a location 200 kb removed from the first cluster, with another ribosomal protein gene (rps21) located upstream of fabD. Among gram-positive bacteria, similar fab gene clusters have been reported in Bacillus subtilis (plsX-fabD-fabG-acpP) (17) and Streptomyces glaucescens (fabD-fabH-acpP-fabB; note that acpP was called fabC in this organism and that the last gene is as closely homologous to E. coli fabF as to E. coli fabB) (28).
Although all the proteins (except PlsX) encoded by the genes of the E. coli fab gene cluster have been extensively studied, the transcription and regulation of these genes have only recently been investigated (20, 21, 36). Podkovyrov and Larson (20) reported promoter probe studies suggesting that the rpmF-plsX genes are cotranscribed, that several promoters are present, and that some of these transcripts may continue into the fabHDG genes (20). However, these results were obtained with transcriptional fusions carried on multicopy plasmids and have not been confirmed by direct mapping of chromosomal transcription, nor has the physiological relevance of the various promoters been determined. These workers have also reported the presence of a promoter located within the plsX coding sequence that reads through downstream fab genes (21).
We began with the genes of the 3′ end of the cluster and reported transcriptional analyses of the fabD, fabG, acpP, and fabF genes (36). We also addressed the physiological relevance of the multiple acpP transcripts with a powerful genetic approach, polar allele duplication (Fig. 1). This method allows blockage of chromosomal transcription from sequences upstream of a given promoter without disruption of either coding sequences or downstream transcription. By use of this method, we showed that only one of the two major promoters that transcribe acpP is required for expression of physiological levels of this protein (36). In the present study, polar allele duplication was used to test the possibility that the upstream genes of the fab cluster (plsX-fabH-fabD-fabG) are transcribed as an operon. We were unable to isolate polar allele duplications of the fabD-fabG segment by the methods used for acpP, suggesting that transcription from an upstream promoter might be necessary for growth. In order to conduct a positive test of this hypothesis, we cloned the fab gene cluster from Salmonella typhimurium, since the cluster from this closely related bacterium should provide functional copies of the proteins needed for the growth of E. coli without providing a target for recombination with the E. coli gene segments needed to direct polar allele duplication (24). We report that in a plsX polar allele duplication strain, the S. typhimurium fab cluster plasmid, pYZ53, was not required for cell growth, indicating that the plsX gene has its own promoter(s) and that only that promoter(s) is required for expression of the PlsX protein and perhaps downstream Fab enzymes at physiological levels. In contrast, we found that the viability of a fabG polar allele duplication strain depended on expression of the S. typhimurium fabG gene and hence that distal promoters were required in order to obtain physiological levels of FabG. Polar allele duplication strains affecting fabD were found to grow very slowly, indicating that only a weak fabD promoter is present.
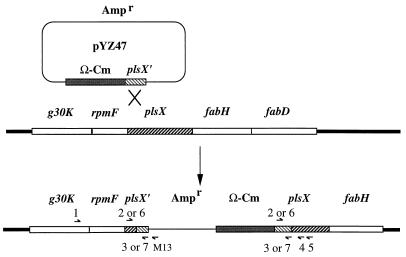
FIG. 1.
Polar allele duplication of plsX. Plasmid pYZ47 was transformed into strain YZ133 (strain UB1005 harboring plasmid pYZ53), followed by selection for transformants resistant to ampicillin, chloramphenicol, and kanamycin. Plasmid pYZ47 (which is unable to replicate in this strain) integrated into the E. coli chromosome in a single-crossover event via homologous recombination between the truncated plsX′ gene of pYZ47 and the intact plsX gene on the chromosome. All elements are indicated. The thick line represents the E. coli chromosome, and the thin line represents the plasmid. Half-arrows with numbers above or below represent the PCR primers used in the study (sequences are given in Materials and Methods). M13, M13 reverse-sequencing (−48) primer purchased from New England Biolabs.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage.
All bacterial strains are derivatives of E. coli K-12 or S. typhimurium LT2. The E. coli strains and plasmids used in this study are listed in Table 1. S. typhimurium MST2370 contains a locked-in Mud-P22 at min 21.5 (putA1019::MudQ) of the S. typhimurium linkage map which packages in the clockwise direction (2). The Mud-P22 phage DNA isolated from S. typhimurium MST2370 after induction with mitomycin C (35) was digested to completion with either EcoRV or NruI, and the fragments were ligated to pHSG575 cut with SmaI. The ligation products were transformed into an E. coli fabD mutant strain, LA2-89 (which is deficient in malonyl-CoA-ACP transacylase activity at 42°C), to select for complementing clones (13, 34). One each of the EcoRV- and NruI-derived plasmids (called pYZ48 and pYZ58, respectively), was retained and again transformed into strain LA2-89 to confirm complementation. Strain YZ133, which harbored plasmid pYZ53, containing S. typhimurium fab cluster DNA, was transformed with plasmid pYZ47 to produce the plsX polar duplication strain YZ137. Strain YZ152, which harbored plasmid pYZ60, containing S. typhimurium fab cluster DNA, was transformed with either plasmid pYZ37 or plasmid pYZ69 to produce the fabG (strain YZ157) or the fabD (strain YZ167) polar duplication strain, respectively. Plasmids pYZ47, pYZ37, and pYZ69 replicate from an R6Kγ replication origin and thus require the plasmid R6K-encoded Pir protein for replication. The wild-type E. coli recipient strains lack Pir, and thus transformants with plasmid-encoded antibiotic resistance (to ampicillin and chloramphenicol) result from integration of the plasmid DNA into the E. coli chromosome via homologous recombination between the 5′ portions of the fabG, plsX, or fabD genes. These recombinant strains were called YZ157, YZ137, and YZ167, respectively. The recA derivatives of strains YZ157, YZ137, and YZ167, called strains YZ158, YZ141 and YZ168, respectively, were constructed by transduction with a P1 phage lysate grown on strain JC10289 with selection for tetracycline resistance, followed by screening for UV sensitivity. Derivatives of strain YZ141 that were kanamycin sensitive (indicating loss of the S. typhimurium fab cluster plasmid pYZ53) were obtained by screening colonies that arose after cells were plated on rich broth (RB) agar plates lacking kanamycin. Strains YZ158 and YZ168 were cured of plasmid pYZ60 by transformation with the incompatible plasmids pYZ71 and pYZ72 and were then screened for colonies that were resistant to spectinomycin and sensitive to kanamycin to produce strains YZ166 and YZ170, respectively.
TABLE 1.
Plasmids and E. coli strains used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| UB1005 | F−metB1 relA1 spoT1 gyrA216 λr/λ− | Lab collection |
| LA2-89 | fabD(Am) supE1 [fabD(Ts) phenotype] | Lab collection (31) |
| WM95 | F′ 128::Tn10-11 lacIq ΔlacZM15/lDE3 ΔlacX74 uidA::pir recA1 rpsL | W. W. Metcalf (15) |
| JC10289 | thr-1 ara-14 leuB6 Δ(gpt-proA)62 thi-1 Δ(recA-srl)306 rpsL31 srlR301::Tn10-84 | CGSCa |
| YZ125 | LA2-89/pYZ48 | This work |
| YZ133 | UB1005/pYZ53 | This work |
| YZ137 | YZ133::pYZ47 (plsX polar duplication on chromosome) | This work |
| YZ141 | YZ137 Δ(recA-srl)306 srlR301::Tn10-84 | This work |
| YZ142 | LA2-89/pYZ58 | This work |
| YZ143 | Kans derivative of strain YZ141 (loss of pYZ53) | This work |
| YZ152 | UB1005/pYZ60 | This work |
| YZ157 | YZ152::pYZ37 (fabG polar duplication on chromosome) | This work |
| YZ158 | YZ157 Δ(recA-srl)306 srlR301::Tn10-84 | This work |
| YZ159 | UB1005 Δ(recA-srl)306 srlR301::Tn10-84 | This work |
| YZ166 | Transformation of pYZ71 into strain YZ158 to cure pYZ60 | This work |
| YZ167 | YZ152::pYZ69 (fabD polar duplication on chromosome) | This work |
| YZ168 | YZ167 Δ(recA-srl)306 srlR301::Tn10-84 | This work |
| YZ170 | Transformation of pYZ72 into strain YZ168 to cure pYZ60 | This work |
| Plasmids | ||
| pACYC177 | Apr Knr cloning vector; contains p15A replicon | Lab collection |
| pHSG575 | Cmr cloning vector; contains pSC101 replicon | 27 |
| pWM77 | Suicide vector derived from pJM703.1 | W. W. Metcalf (15) |
| pYZ37 | Insertion of the 700-bp PstI-EcoRV fragment (fabG′) of pKM22 into pWM77 with the 3.5-kb Ω-Cm fragment of pHP45Ω-Cm immediately upstream in the BamHI site | This work |
| pYZ46 | 500-bp plsX′ PCR product of the E. coli chromosome (amplified with primers 8 and 3) treated with T4 DNA Polymerase, then cut with SalI and inserted into pWM77 cut with SmaI and SalI | This work |
| pYZ47 | Insertion of the 3.5-kb Ω-Cm BamHI fragment of pHP45Ω-Cm into the BamHI site of pYZ46 | This work |
| pYZ48 | Insertion of the 3.8-kb EcoRV fab gene fragment of Mud-P22 phage DNA from S. typhimurium MST2370 into the SmaI site of pHSG575 | This work |
| pYZ53 | Insertion of the 3.8-kb BamHI-EcoRI fragment of pYZ48 into pACYC177 cut with BamHI and DraI | This work |
| pYZ58 | Insertion of the 2.2-kb NruI fab gene fragment of Mud-P22 phage DNA from S. typhimurium MST2370 into the SmaI site of pHSG575 | This work |
| pYZ59 | Insertion of the BamHI-PvuII fragment of pYZ48 and the PvuII-EcoRI fragment of pYZ58 together into pHSG575 cut with BamHI and EcoRI | This work |
| pYZ60 | Insertion of the 4.6-kb BamHI-EcoRI (filled-in) fragment of pYZ59 into pACYA177 cut with BamHI (filled in) and DraI | This work |
| pYZ63 | Insertion of the 652-bp PCR product of E. coli chromosomal DNA (amplified with primers 1 and 3) into pUC19 cut with BamHI and HindIII (filled in) | This work |
| pYZ64 | Plasmid pYZ63 was cut with HindIII and PstI and then blunt ended with T4 DNA polymerase and ligated | This work |
| pYZ65 | Insertion of the 459-bp PCR product of the E. coli chromosome (amplified with primers 2 and 5) into pUC19 cut with BamHI and HindIII (filled in) | This work |
| pYZ66 | Plasmid pYZ65 was cut with PstI and then ligated | This work |
| pYZ67 | Insertion of the 600-bp fabD′ PCR product of E. coli chromosomal DNA (amplified with primers 9 and 11) cut with EcoRI and HindIII into pBluescript II SK cut with EcoRI and HindIII | This work |
| pYZ68 | Insertion of the 3.5-kb Ω-Cm BamHI fragment of pHP45Ω-Cm into the BamHI site of pYZ67 | This work |
| pYZ69 | Insertion of the 4.2-kb SalI-NotI fragment of pYZ68 into pWM77 cut with SalI and NotI | This work |
| pYZ70 | Recircularization of the 4.5-kb XhoI fragment of pMPM-K6Ω (deletion of the Kanr gene) | This work and reference 14 |
| pYZ71 | Insertion of the 770-bp PCR product of pYZ58 (amplified with primers 16 and 17) into pYZ70 cut with NcoI and XmnI to construct a gene fusion with S. typhimurium fabG under the control of the arabinose promoter | This work |
| pYZ72 | Insertion of the 1,720-bp PCR product of pYZ58 (amplified with primers 18 and 17) into pYZ70 cut with NcoI and XmnI to construct a gene fusion with S. typhimurium fabD and fabG under the control of the arabinose promoter | This work |
CGSC, E. coli Genetic Stock Center, Yale University, New Haven, Conn.
Culture media and growth conditions.
Minimal E medium supplemented with 4,000 mg of glucose/liter, 100 mg of methionine/liter, and 10 mg of thiamine/liter or RB was used for growth of bacterial strains (16). Antibiotics were added at the following concentrations (in milligrams per liter): kanamycin, 25; ampicillin, 100; tetracycline, 30; and chloramphenicol, 34. Bacterial growth was monitored with a Klett-Summerson colorimeter with a green filter.
Plasmid isolation and recombinant DNA techniques.
Plasmid isolation was performed by either a modified alkaline lysis method (11) or Qiagen Spin minipreparations. Southern blot analyses were carried out according to the Genius System User’s Guide (Boehringer Mannheim Biochemicals). The probes were plasmid pYZ37, pYZ47, and pYZ69 labeled with digoxigenin (DIG)-dUTP via random-primed labeling with the Genius 2 DNA labeling kit, purchased from Boehringer Mannheim. S. typhimurium Mud-P22 phage lysate preparation and DNA isolation were performed according to the procedure of Youderian and coworkers (35). Low-stringency Southern blot analysis was performed with a DIG-dUTP-labeled PCR fragment (amplified with primers 10 and 13) that contained the E. coli fabD and fabG genes, plus a 5′ fragment of acpP. Other DNA manipulations were performed by standard procedures (26).
DNA sequencing of both strands of the S. typhimurium fab cluster genes on plasmid pYZ48 and pYZ58 was done by the Genetic Engineering Facility, University of Illinois at Urbana-Champaign, with Taq DNA polymerase cycle sequencing on an Applied Biosystems 373 DNA sequencer, with primers designed and synthesized by the facility staff.
RNA analyses.
Total RNA was isolated from exponentially growing cells by the rapid isolation method (1). Reverse transcriptase-coupled PCR (RT-PCR) was performed with the RETROscript kit, purchased from Ambion. The primer used for the first cDNA strand synthesis was the random decamer mixture provided in the kit. The primers used in PCR are listed below. Quantitative RT-PCR was carried out with the Ambion kit according to the protocol of Gilliland and coworkers (6). Briefly, plasmids pYZ64 and pYZ66 were used as templates with primers 1 and 7 and primers 6 and 5, respectively, to amplify the competitive DNA fragments. The concentrations of the competitive DNA fragments were then determined either by absorption at 260 nm or by comparing the fluorescence intensities with those of a DNA mass ladder (purchased from Gibco BRL) by densitometry of ethidium bromide-stained agarose gels. The reverse transcriptase (RT) reaction product (1 μl) and different concentrations of competitive DNA (as specified in the legend to Fig. 4) were added to a 25-μl PCR mixture. The same sets of primers used to amplify the competitive DNA were used in the RT-PCRs. The PCR products were separated on an agarose gel stained with ethidium bromide and quantitated by densitometry. The ratios of the fluorescence intensities of the PCR products of the competitive DNA to those of the RT-PCR products were plotted as a function of the concentration of the competitive DNA (6).
FIG. 4.
Analysis of plsX gene expression by quantitative RT-PCR. RT-PCRs were performed as described for Fig. 3, except that different masses of competitive DNA were added to a given mass of RT-PCR mixture. (A) Quantitative RT-PCR with primers 1 and 7. Different masses (15 × 10−5, 12.5 × 10−5, 10 × 10−5, 7.5 × 10−5, 5 × 10−5, 2.5 × 10−5, or 2.25 × 10−5 ng) of competitive DNA amplified from pYZ64 with primers 1 and 7 were added to the RT-PCR mixtures in lanes 1 through 8, respectively. (B) Quantitative RT-PCR with primers 6 and 5. Competitive DNA (7.5 × 10−5, 6.5 × 10−5, 4.5 × 10−5, 3.5 × 10−5, 2.5 × 10−5, 2 × 10−5, or 1 × 10−5 ng) was added to lanes 1 through 8, respectively. The fluorescence intensities of the bands on the agarose gel in each lane were quantified with densitometry following ethidium bromide staining. The ratio of the intensity of the PCR product of competitive DNA to that of the RT-PCR product was calculated for each reaction, and these ratios were plotted as a function of the competitive DNA concentration. (C and D) Plots of data from panels A and B, respectively.
For Northern blot analysis, whole-cell lysates were separated by electrophoresis on 0.8% formaldehyde agarose gels as described by Kornblum et al. (9). Northern transfer was performed by standard procedures (26). Hybridization, washing, and detection were carried out as described by the Genius System User’s Guide. Other experimental conditions are given in the legend to Fig. 6.
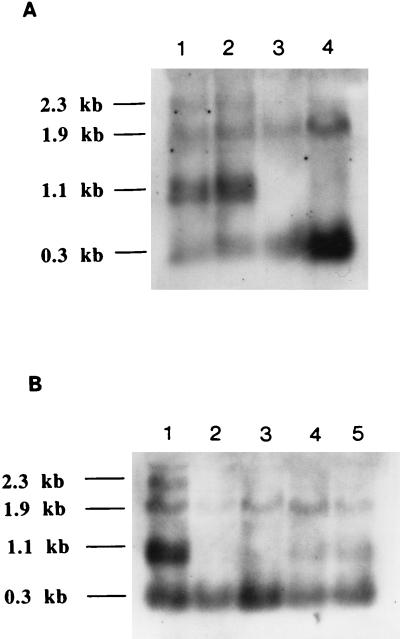
FIG. 6.
Northern analysis of fabG and fabD gene expression in the fabG and fabD polar allele duplication strains YZ166 and YZ170. The strains were grown to mid-log phase in RB that was either unsupplemented (strains UB1005 and YZ60), supplemented with 0.2% arabinose (strain YZ166), or supplemented either with 0.2% arabinose or with 0.4% glucose plus 0.002% fucose (strain YZ170). Cell lysates were prepared as described in Materials and Methods. The probe used was the DIG-labeled PCR product from the 3′ end of acpP obtained by amplification with primers 14 and 15. (A) Northern analysis of fabG expression. Lanes 1 and 2, strain UB1005; lane 3, strain YZ60; lane 4, strain YZ166. (B) Northern analysis of fabD expression. Lane 1, UB1005; lane 2, strain YZ60; lane 3, strain YZ166; lane 4, strain YZ170 grown in RB supplemented with glucose plus fucose; lane 5, strain YZ170 grown in RB supplemented with arabinose.
Primers used in RT-PCR and other manipulations.
In addition to the M13 16-mer reverse-sequencing primer from New England Biolabs, the primers used (sequences shown 5′ to 3′) were as follows: primer 1, GCAATGGTTGAAGATGAAATCATCC; primer 2, GTTAGATCATATGGGAGGG; primer 3, GACGTCGACGTTGTTCAAAGTCAG; primer 4, CTGACTGCGCAGGAATAATCTGC; primer 5, CACCAGCATTGTGCTGTCACAACT; primer 6, GTTAGATGTCATGGGAGGGGATTT; primer 7, GACGCGAACGTTGTTCAAAGTCAG; primer 8, TCGTTGGATCGGGGATAAACCG; primer 9, CTGGCGCGCACCTGCGATCCAA; primer 10, GGGAATTCTTGACCGTTCTCAACTGG; primer 11, CGCAACAGATGCAGTCAACAG; primer 12, GCGAATTCGAAACCAATGGTGATGC; primer 13, GGTCTTCAACCTAAGAAGCATTGTTGG; primer 14, GAAGTTACCAACAATGCTTC; primer 15, TCCTGATCAGACACGTTTGTCCTCCAGGGA; primer 16, GGAAAATCATGAGCTTTGAAGG; primer 17, CCCTAATAACGCAAATATTTTTC; and primer 18, GGATTTAATCATGACGCAA.
Genetic techniques.
Transduction was carried out according to the method of Miller (16). Allele duplication was done as described by Metcalf et al. (15). Plasmids pYZ37, pYZ47, and pYZ69 (Table 1), which contain the 5′ portions of the fabG, plsX, and fabD genes, respectively, were maintained in the Pir-containing strain, WM95, and were then transformed into the wild-type strains YZ152, YZ133, and YZ152, respectively (which lack Pir), followed by selection for transformants resistant to both ampicillin and chloramphenicol.
Nucleotide sequence accession number.
The nucleotide sequence of the S. typhimurium fab gene cluster has been submitted to GenBank under accession no. AF044668.
RESULTS
Cloning and sequencing of the S. typhimurium fab cluster.
As will be described below, we had failed to isolate various polar allele duplications within the E. coli fab cluster. This could be due to poor luck (successful transformations give only 10 to 20 colonies) or to disruption of essential transcription. In order to cope with the latter possibility, we cloned the fab cluster from S. typhimurium, a close relative of E. coli, and used plasmids carrying this DNA fragment to provide any essential proteins lost due to polar allele duplication. Our isolation of the S. typhimurium fab cluster was based on two assumptions: (i) that the overall organization of the S. typhimurium fab cluster would closely resemble that of E. coli (22), since the fab gene cluster is widely conserved among much more distantly related bacteria (5, 17, 27, 28) and (ii) that, given the similarities of the genetic maps of the two bacteria, the fab gene cluster of S. typhimurium would be located at about genome min 24. To test if these assumptions were correct, we used a “locked-in” Mud-P22 prophage (putA1019::MudQ) integrated at min 21.5 of the S. typhimurium genetic map (2, 35). Upon induction of this phage with mitomycin C, it cannot escape from the bacterial chromosome, and it packages successive phage headfuls of S. typhimurium chromosomal DNA in a clockwise direction (2). The phage particles in the lysate were isolated, and the encapsidated DNA was purified and digested with various restriction enzymes. Low-stringency Southern blot analysis was performed with a DIG-dUTP-labeled fragment (obtained by PCR with primers 10 and 13) which contains the complete E. coli fabD and fabG genes plus a 5′ fragment of the acpP gene. Positive bands were detected (data not shown), suggesting that the locked-in phage DNA did indeed contain the S. typhimurium fab cluster genes.
We cloned the S. typhimurium fab cluster genes from the phage particle DNA by complementation of an E. coli mutant deficient in malonyl-CoA-ACP transacylase activity at 42°C. Strain LA2-89 carries an amber mutation in the fabD gene together with a supE tRNA suppressor (34). The combination of these two characteristics results in both a temperature-sensitive malonyl-CoA-ACP transacylase and temperature-sensitive growth. The phage particle DNA was digested to completion with each of a variety of different restriction enzymes, and the fragments were then ligated to the low-copy-number vector pHSG575 (29). The resulting plasmids were transformed into a restriction-deficient E. coli strain, and plasmid preparations from pools of the resulting transformants were used to transform strain LA2-89, followed by selection for chloramphenicol-resistant clones that grew at 42°C. Only the plasmid pool constructed from EcoRV fragments gave transformants. One of these isolates, pYZ48, was sequenced and was found to contain homologs of E. coli rpmF, plsX, fabH, and fabD, plus two partial gene fragments, the 5′ end of fabG and the 3′ end of g30k, an open reading frame (ORF) of unknown function located upstream of rpmF. In order to obtain the remainder of the fabG gene, the phage particle DNA was digested with NruI, which cuts only once in the pYZ48 insert DNA (within the fabH gene), and was ligated to pHSG575 cut with SmaI, and plasmids that complemented E. coli LA2-89 were again selected. One such clone, pYZ58, was retained, sequenced, and used to construct a plasmid that carried a cluster with an intact fabG gene.
The deduced protein product of each S. typhimurium gene has the same number of residues as the E. coli homolog, except that the plsX ORF product is 3 residues longer than its E. coli homolog. Each of the deduced proteins has >90% amino acid identity to the analogous E. coli protein (Fig. 2), and thus the nomenclature of the S. typhimurium fab genes is the same as that of the E. coli genes. The only noteworthy difference between the fab gene clusters of the two organisms was a 55-bp deletion within the S. typhimurium fabG-acpP intergenic region compared to that of E. coli (the intergenic regions between other S. typhimurium fab cluster genes were very similar to those of E. coli).
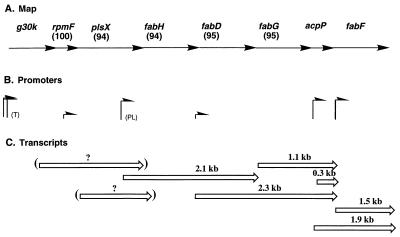
FIG. 2.
Organization of the fab cluster genes, their promoters, and transcripts. (A) The order of the genes in E. coli and S. typhimurium is identical. The percentages of amino acid residues identical in the S. typhimurium and E. coli fab cluster proteins are given above the arrows representing the individual genes. The S. typhimurium nucleotide sequence is about 85% identical to the E. coli fab gene cluster sequences. (B) The known promoters of the region are depicted (the heights of the arrows are intended to give a crude idea of relative promoter strengths). The promoters upstream of g30K (marked T) are those identified by S1 nuclease mapping by Tanaka and coworkers (30). The promoter within plsX (PL) is that identified by primer extension studies by Podkovyrov and Larson (21). (C) The transcripts identified in this study and our prior studies (36) are shown. The plsX transcripts were detected only by RT-PCR, and thus the lengths of these transcripts are unknown. The scarcity of the longer plsX transcript could be due to termination of most of the transcripts initiated at the promoters upstream of g30K at the terminator mapped downstream of rpmF (30).
Construction of a plsX gene polar allele duplication.
We used polar allele duplication (Fig. 1) to demonstrate that the promoter located immediately upstream of the acpP gene is sufficient for expression of ACP at physiological levels (36). In the present study, we extended this approach to the upstream genes of the E. coli fab cluster. A PCR product beginning 40 bp upstream of the small rpmF coding sequence and ending 150 bp within the plsX coding sequence was inserted into the oriR6Kγ plasmid, pWM77 (15), immediately downstream of an Ω-Cm cassette that blocks transcription from upstream genes. We transformed the resulting plasmid, pYZ47, into the wild-type E. coli strain UB1005, which lacks Pir (and is therefore unable to replicate pYZ47), and transformants resistant to both chloramphenicol and ampicillin were selected. Such transformants can be formed only by single crossover of the plasmid into the chromosome (Fig. 1). Several failed attempts to construct this strain, together with the data of Podkovyrov and Larson (20), suggested that cotranscription of the plsX gene and downstream fab genes with the upstream ribosomal protein gene, rpmF, might be required for growth.
To test this possibility, plasmid pYZ53, containing the complete S. typhimurium rpmF, plsX, fabH, and fabD genes, plus the 5′ end of the S. typhimurium fabG gene (the insert DNA is the same as that of plasmid pYZ48), was transformed into the wild-type strain UB1005 to produce strain YZ133. This strain was then transformed with pYZ47 to obtain the polar allele duplication. The rationale was that the S. typhimurium fab genes would provide any E. coli chromosomal functions lost as a result of the formation of the polar allele duplication but would not be a substrate for recombination with the E. coli sequences of plasmid pYZ47. Colonies resistant to kanamycin (indicating the presence of the S. typhimurium fab cluster plasmid pYZ53), chloramphenicol, and ampicillin (Fig. 1) were obtained. PCR analyses using primer 1 and the M13 reverse-sequencing primer (Fig. 1), plus Southern analysis of one of such recombinant strain, YZ137, verified the expected integration of plasmid pYZ47 into the E. coli chromosome (data not shown). A recA mutation was then transduced into the strain to produce the stabilized strain YZ141.
Strain YZ141 was grown on RB agar lacking kanamycin (in agreement with the observations of prior workers [23], it was found that p15A origin plasmids were not stably maintained without selection) to test if pYZ53 (which contains the S. typhimurium fab genes) is essential for cell viability. About 300 colonies were screened for kanamycin resistance, and about 20% of the colonies were kanamycin sensitive. One of these kanamycin-sensitive derivatives (called YZ143) was further tested by plasmid isolation and Southern analysis to confirm both the loss of S. typhimurium fab gene-containing plasmid pYZ53 and the presence of the expected plsX polar allele duplication on the E. coli chromosome (data not shown). This strain had the expected chromosomal map, indicating that a promoter(s) located upstream of the plsX gene provides sufficient transcription to support cell viability. This promoter probably lies within the rpmF gene, based on the studies of Podkovyrov and Larson (20), whereas the longer plsX transcript probably originates at the two promoters mapped upstream of g30k by Tanaka and coworkers (30).
Transcription of the plsX gene.
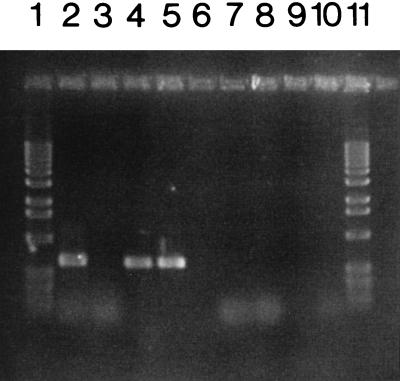
RT-PCR was used to detect and quantitate plsX transcription in strains UB1005 and YZ143, since several attempts to perform Northern analysis of plsX transcription failed due to the scarcity of the transcripts. When the primer pair 1 and 3 (see Fig. 1) was used, both strain UB1005 and strain YZ143 gave RT-PCR products of 652 bp (Fig. 3, lanes 4 and 5), consistent with cotranscription of the plsX gene with the upstream ribosomal protein gene, as suggested by Podkovyrov and Larson (20). When primers 1 and 4 (Fig. 1) were used to prime RT-PCR, a product of the expected length was detected in strain UB1005 (Fig. 3, lane 2) but not in strain YZ143 (Fig. 3, lane 3), demonstrating that the polar allele duplication indeed blocked transcription from upstream. These total-RNA preparations were also tested in direct PCRs (in the absence of RT) with the same sets of primers (Fig. 3, lanes 7 to 10) to rule out the possibility of DNA contamination of the RNA preparations. Primers 2 and 5 (Fig. 1) were also used in RT-PCR analysis, and products of the expected length were detected both in strain UB1005 and in strain YZ143 (Fig. 4 and data not shown), a result consistent with the viability of strain YZ143.
FIG. 3.
RT-PCR analysis of plsX gene expression in the plsX polar allele duplication strain. The E. coli plsX polar allele duplication strain YZ143 and the wild-type strain UB1005 were grown to mid-log phase in RB, and total RNA was prepared as described in Materials and Methods. RT-PCRs were performed by using the Ambion RETROscript kit as directed. Lanes 2 through 5, RT-PCRs; lanes 7 through 10, PCRs with approximately 0.5 μg of total RNA as the template, but lacking RT. One microliter of RT reaction product, which corresponded to approximately 0.5 μg of total RNA, was used for each RT-PCR. The primer pairs used were 1 and 4 (lanes 2, 3, 7, and 8) or 1 and 3 (lanes 4, 5, 9, and 10). In lanes 2, 4, 7, and 9, RNA from strain UB1005 was analyzed. In lanes 3, 5, 8, and 10, RNA from strain YZ143 was analyzed. Lanes 1 and 11 are the 1-kb DNA ladder from BRL, and lane 6 was left vacant.
Quantitative RT-PCR (6) was used to assess the ratio of the level of the transcript containing both plsX and rpmF to that of the transcript containing only plsX. The principle of this method is to utilize a known concentration of a DNA fragment (obtained by amplification with the same primers used in the RT-PCRs) to compete with the RT-PCR product. To obtain the needed competitive DNA fragments, 652- and 459-bp PCR fragments were amplified from E. coli chromosomal DNA with primer pair 1 and 3 and primer pair 2 and 5, respectively, and the amplified fragments were cloned into vector pUC19. A HindIII-PstI fragment and a PstI fragment, respectively, were deleted from the plasmid inserts to produce plasmids pYZ64 and pYZ66, respectively. These plasmids generate amplification products of 522 and 346 bp, respectively, as competitive DNA fragments. Two primers, 6 and 7, were paired with primers 5 and 1, respectively; primer pair 6 and 5 and primer pair 1 and 7 were used to amplify competitive DNA fragments from plasmids pYZ66 and pYZ64, respectively, and the concentrations of the competitive DNA fragment solutions were determined as described in Materials and Methods. RT-PCR was carried out with known concentrations of competitive DNA added to the reaction mixtures. A total-RNA preparation from strain UB1005 was used as the RT template, and the concentration of cDNA formed was taken to be proportional to the mRNA concentration. Primer 1 anneals to a sequence in g30k upstream of rmpF, and primer 6 anneals to a sequence at the 5′ end of the plsX gene, whereas both primer 5 and primer 7 anneal to sequences in the center of the plsX gene (Fig. 1). Therefore, the RT-PCR product from primer pair 1 and 7 represents only the products of cotranscription of rmpF and plsX, whereas the product from primer pair 6 and 5 represents the total of the plsX transcripts. When decreasing concentrations of the 522-bp competitive DNA fragment synthesized with primers 1 and 7 were added to the reaction mixtures for RT-PCR of strain UB1005 with the same primer pair, it was shown that the concentration of the 652-bp RT-PCR product increased while the concentration of the 522-bp PCR product from the competitive DNA template decreased (Fig. 4A, lanes 1 to 8). The ratios of the fluorescence intensities of the PCR products to those of the RT-PCR products were plotted as a function of the concentration of the competitive DNA (Fig. 4C). (When the ratio is 1, the molar concentration of the competitive DNA added to the reaction mixture is identical to the molar concentration of the plsX cDNA, which is proportional to the plsX mRNA concentration). The level of cDNA synthesized from the rpmF-plsX cotranscripts was 6.40 × 10−5 ng/mg of UB1005 total RNA (Fig. 4C). Likewise, when the 346-bp competitive DNA fragment obtained with primers 6 and 5 from pYZ66 was added to the RT-PCR mixture (Fig. 4B and D), the concentration of cDNA synthesized by using the plsX total transcripts as the original template was 7.23 × 10−5 ng/mg of UB1005 total RNA. Therefore, when converted to molar quantities, these RT-PCR data indicate that about 60% of plsX transcription initiated at the promoters mapped upstream of the g30K gene by Tanaka and coworkers (30), whereas only 40% originated from the plsX-specific promoter (mean of three experiments).
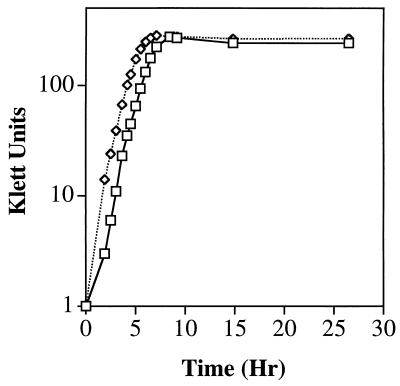
The normal growth rate of the plsX polar allele duplication strain YZ143 (Fig. 5) indicated that transcription from the promoter(s) upstream of rpmF was not required for growth.
FIG. 5.
Growth curve of the plsX polar allele duplication strain YZ143. Strain YZ143 (□) and strain YZ159 (a recA derivative of strain UB1005) (◊) were grown in the supplemented minimal E medium described in Materials and Methods. Growth was monitored with a Klett-Summerson colorimeter with a green filter.
Construction of fabG and fabD polar allele duplications.
Initial attempts to construct polar allele duplications upstream of fabG or fabD were unsuccessful, and to avoid the possibility of disrupting essential transcription, we constructed a plasmid that contained intact copies of the S. typhimurium rpmF, plsX, fabH, fabD, and fabG genes. This construct was assembled in vector pHSG575 by a tripartite ligation using the inserts of plasmid pYZ48 and pYZ58 and then was subcloned into the kanamycin-resistant p15A vector, pACYC177. The resulting plasmid, pYZ60, was transformed into the wild-type strain UB1005 to give strain YZ152. Strain YZ152 was then transformed with plasmid pYZ37, which contains a 240-bp segment of the fabG coding sequence plus 300 bp of upstream sequence (a chromosomal PstI-EcoRV fragment) with an Ω-Cm cassette upstream of fabG′, or with plasmid pYZ69, which contains a 90-bp segment of the fabG coding sequence plus 470 bp of upstream sequence (a chromosomal EcoRI-HindIII fragment) with an Ω-Cm cassette inserted upstream of fabD′, and transformants resistant to both chloramphenicol and ampicillin were selected. The expected integration events were confirmed by PCR with the M13 reverse-sequencing primer plus either primer 10 (for fabG) or primer 6 (for fabD) and by Southern analysis as described above (data not shown). Strains YZ157 (fabG duplication) and YZ167 (fabD duplication) were then stabilized by introduction of a recA mutation to produce strains YZ158 and YZ168, respectively.
Transcription of the fabD and fabG genes in the polar allele duplication strains.
Strain YZ158, containing a fabG polar allele duplication, and strain YZ168, containing a fabD polar allele duplication, were first tested to see if plasmid pYZ60, which carries the S. typhimurium fab gene cluster, was required for growth. We first grew the strains without kanamycin selection in liquid medium and then screened for kanamycin-sensitive colonies without success (300 colonies of each strain were screened). We then cloned the S. typhimurium fabG gene into pYZ70, a kanamycin-sensitive derivative of vector pMPM-K6Ω (14) in which the intact gene was positioned such that it was transcribed exclusively from the vector arabinose-regulated araBAD promoter and was translated by using the vector ribosome binding site. The resulting spectinomycin-resistant plasmid, pYZ71, was used to transform strain YZ158, with selection for transformants resistant to ampicillin, chloramphenicol, tetracycline, and spectinomycin, followed by screening for kanamycin sensitivity. All these steps were carried out in RB medium supplemented with 0.2% arabinose to induce expression of S. typhimurium fabG. Since plasmids pYZ60 and pYZ71 share the p15A replication origin, plasmid incompatibility due to the presence of pYZ71 was expected to cure this strain of pYZ60. One such cured strain (YZ166) resistant to ampicillin, chloramphenicol, tetracycline, and spectinomycin but sensitive to kanamycin was retained, and the presence of a single plasmid, pYZ71, was confirmed by plasmid isolation and restriction analysis (data not shown).
Strain YZ166 was first assayed for dependence on arabinose-induced S. typhimurium fabG gene expression by streaking the strain on media supplemented with either 0.2% arabinose to induce the arabinose promoter or 0.4% glucose plus 0.002% fucose to decrease basal expression of the arabinose promoter (18). We found that strain YZ166 gave no detectable growth on plates supplemented with glucose-fucose, whereas it grew as well as the wild-type strain, UB1005, on plates supplemented with arabinose. Moreover, strain YZ166 failed to grow in the absence of arabinose, indicating that basal expression from the araBAD promoter was unable to support growth. Therefore, blocking of the transcription of fabG from upstream promoters was lethal to the cell unless another source of FabG was supplied.
Northern analysis was also performed to examine fabG transcription in strain YZ166. We did not use a fabG-specific probe because it could be difficult to distinguish transcription of the E. coli chromosomal fabG gene from that of the plasmid-borne S. typhimurium fabG gene. Instead, we used a probe that contained only the 3′ end of the acpP gene. We previously found (36) that fabG and acpP are cotranscribed to give two products of 1.1 and 2.3 kb (see Fig. 6A, lanes 1 and 2). Both transcripts disappeared in the acpP polar duplication strain, although the other transcripts, of 0.3 and 1.9 kb, which initiate at the acpP promoter (36), were unaffected (Fig. 6A, lane 3). Essentially the same result was given upon insertion of the Ω-Cm cassette between the fabD and fabG genes (Fig. 6A, lane 4). Therefore, in contrast to our findings with acpP and plsX, the fabG gene lacks a promoter immediately upstream of its coding sequence that is sufficiently strong to provide sufficient levels of gene product to support growth. These results also indicate that the 1.1-kb mRNA is produced by processing of a longer transcript(s). This also is the first evidence demonstrating fabG to be an essential gene.
Similar constructions gave pYZ72, in which the S. typhimurium fabD and fabG genes were placed under the control of the araBAD promoter. This plasmid was used to transform strain YZ168, and colonies resistant to ampicillin, chloramphenicol, tetracycline, and spectinomycin were selected and then screened for kanamycin sensitivity in the presence of arabinose as described above. The resulting strain, YZ170, was tested for the presence of plasmids, and only plasmid pYZ72 was detected. Strain YZ170 was assayed for dependence on the S. typhimurium fabD fabG genes carried by plasmid pYZ72 as described above. To our surprise, we found that there was detectable growth of strain YZ170 on the glucose-fucose medium, although the colonies were much smaller and grew less densely than those formed on medium supplemented with arabinose (data not shown). This slow growth seemed to be due to low-level expression of fabD (rather than of fabD plus fabG or of fabG alone), since substitution of pYZ71, the arabinose-regulated fabG plasmid, for the fabD fabG plasmid, pYZ72, produced only a very modest increase in growth upon the addition of arabinose, indicating that FabG levels did not limit growth under these conditions.
The slow growth of the fabD polar duplication strain indicated that a weak promoter(s) located immediately upstream of the fabD coding sequence transcribes fabD and fabG. The weak nature of this promoter(s) was evident in Northern analyses. The same acpP-specific probe used in the Northern analysis of fabG transcription (see above) was used to assay expression of the fabD gene in the fabD polar duplication strain, YZ170. In contrast to results with the acpP and fabG duplication strains, insertion of the Ω-Cm cassette between the fabH and fabD genes resulted in greatly decreased levels of the 1.1- and 2.3-kb transcripts (the 2.3-kb transcript was virtually undetectable) (Fig. 6B, lanes 4 and 5), a result consistent with the slow growth of strain YZ170. These results indicated that the promoter located immediately upstream of fabD is only weakly functional and that most of the 1.1-kb transcript was the processed product of a longer transcript(s) initiated at promoters well upstream of fabD.
DISCUSSION
We developed the polar duplication approach to overcome a deficiency of standard transcriptional mapping that arises when a gene is found to have multiple transcripts. The usual assumption is that protein production is a direct function of transcript abundance, and hence, major transcripts are considered more important than minor transcripts. However, this is not necessarily valid, since a minor transcript may be more efficiently translated than a major transcript, or a major transcript might be the processed product of a longer transcript, which consequently becomes scarce. On the other hand, a minor transcript may reflect only the incomplete nature of most transcription terminators and hence may have no physiological importance. Another complication of standard transcriptional mapping is the processing of the primary products of transcription, which can be detected by the polar duplication method (see below). Use of the S. typhimurium homologs to provide possible essential functions during the construction of polar duplication strains increases the applicability of the approach. We chose S. typhimurium based on its close relatedness with E. coli (and hence their interchangeable gene expression signals) and the known lack of recombination between homologous genes in these organisms, due largely to mismatches at the third positions of codons and the resulting inhibition of recombination by mismatch repair (24). However, genes from other organisms, such as H. influenzae, could also be useful.
Our application of the polar duplication approach to the fab gene cluster shows that, although this gene cluster shows obligatory cotranscription of some pairs of genes, some genes have a promoter located immediately upstream of the coding sequence that provides sufficient transcription for normal growth. Examples of such genes are acpP and plsX. In the case of plsX, our data bear on the argument of Podkovyrov and Larson (20) that cotranscription of rpmF and plsX could play an important role in coordinating ribosome synthesis with cell membrane synthesis. If rpmF-plsX cotranscription is important, its lack might be expected to slow or block growth. However, this conclusion is tempered by the lack of information on plsX function. This gene was discovered by the ability of a mutant allele, plsX50, to allow effective supplementation of plsB mutants of E. coli with sn-glycerol 3-phosphate on certain carbon sources (10). Only one plsX allele has been characterized, and the interactions studies involved a single plsB allele. The plsX50 mutation has been reported to be a single-base pair deletion upstream of the coding sequence (GenBank accession no. M96793), and it is unclear whether this mutation causes a gain of function or a loss of function relative to the wild-type gene, since no complementation studies have been reported. On the other hand, most bacterial genomes sequenced to date encode a PlsX homolog, and thus, this protein seems likely to play an important role in cellular physiology. The prevalence of plsX-like genes in bacterial genomes indicates that further study of this enigmatic E. coli gene is required.
In contrast to plsX and acpP, fabG lacks a proximal promoter. Insertion of the Ω-Cm cassette between fabD and fabG abolished the synthesis of both the 1.1-kb mRNA, a cotranscript of fabG and acpP, and the 2.3-kb mRNA, a transcript of fabD, fabG, and acpP (Fig. 6). These results indicate that the abundant 1.1-kb mRNA is not initiated from a promoter located immediately upstream of the fabG gene but is produced by processing of longer transcripts. This is also consistent with the fact that although the 1.1-kb mRNA is very abundant, a strong promoter could not be detected immediately upstream of the fabG coding sequence. Several DNA fragments containing the overlapping regions immediately upstream of the fabG coding sequence were cloned into a promoter detection vector in which the inserts can drive lacZ expression. None of the fabG fragments resulted in β-galactosidase levels significantly higher than background (data not shown). We conclude that cotranscription of fabG with upstream genes is required for growth.
Transcription of fabD provides a middle ground between the extremes of acpP-plsX and fabG. Our previous Northern analyses of fabD transcription were inconclusive. In repeated attempts, only faint and diffuse bands were detected with a fabD probe (36). Our present data show that although fabD retains a proximal promoter within 370 bp of its coding sequence, this promoter is not sufficiently strong to support normal growth, and therefore cotranscription of fabD with upstream genes is needed. These transcripts could initiate at the promoters located upstream of g30k (29) and/or at the promoter mapped within plsX in the primer extension studies of Podkovyrov and Larson (21). The presence of a weak fabD promoter is consistent with the data of Podkovyrov and Larson (20).
The fabG polar allele duplication strain carrying a plasmid with the S. typhimurium fabG gene under the control of the araBAD promoter also showed no detectable growth unless the S. typhimurium fabG was induced. These data indicate that the fabG gene is essential for growth, a conclusion that is of interest, since several ORFs in addition to fabG have been classified as β-ketoacyl-ACP reductases by various annotators of the E. coli genomic sequence. These classifications could be explained if β-ketoacyl-ACP reductases exist that were specific either for different acyl chain lengths or for synthesis of saturated versus unsaturated fatty acids existed. However, fractionation of E. coli cell extracts gave only a single enzymatic activity that functioned with all acyl chains tested (32), and purified FabG catalyzes all the β-ketoacyl-ACP reductions required in the de novo synthesis of the long-chain fatty acids of E. coli in a reconstituted in vitro system (8). For these reasons we doubt that these other ORFs play a role in membrane lipid synthesis; instead, we suggest that they function in reductions of β-ketoacyl-CoA intermediates in other pathways (e.g., poly-β-hydroxybutyrate synthesis). The fact that the fabG polar allele duplication strain YZ166 is a conditionally lethal mutant (the strain cannot grow in medium lacking arabinose) should allow the determination of the stage at which the fatty acid biosynthetic pathway is arrested upon depletion of FabG protein in strain YZ166.
It is not surprising that the S. typhimurium fatty acid biosynthetic gene cluster has very high sequence identity to E. coli homologs at both the nucleic acid and amino acid levels (Fig. 1). However, the 55-bp deletion within the intergenic region between the fabG and acpP genes of S. typhimurium relative to that of E. coli was unexpected, especially given that the other fab cluster intergenic regions are very similar in the two bacteria. The deletion removes 55 bp located upstream of a sequence which is identical to that of the E. coli acpP promoter we identified previously (36). Therefore, S. typhimurium acpP transcription may differ somewhat from that of E. coli.
Why are these fab genes clustered when some of the genes retain their own promoters? Since E. coli fatty acid synthesis is a very tightly coupled pathway in which only traces of intermediates are seen (12), it seems unlikely that there would be a need to alter the ratios of the proteins encoded by these genes. Internal promoters could provide the means to combat the natural polarity seen in operons and also to increase the expression of a noncatalytic protein like ACP, which is needed in large quantities. However, the effects of natural polarity can also be canceled by increasing the relative efficiencies of translation of downstream ORFs. It will be interesting to see if the fab clusters of other bacteria utilize the E. coli mix of multigenic and monogenic transcription.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI15650.
We thank Charles Miller and Stanley Maloy for useful suggestions regarding this work.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Sudman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1987. pp. 4.8.1–4.8.3. [Google Scholar]
- 2.Benson N R, Goldman B S. Rapid mapping in Salmonella typhimurium with Mud-P22 prophages. J Bacteriol. 1992;174:1673–1681. doi: 10.1128/jb.174.5.1673-1681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carty S M, Colbeau A, Vignais P M, Larson T J. Identification of the rpmF-plsX-fabH genes of Rhodobacter capsulatus. FEMS Microbiol Lett. 1994;118:227–231. doi: 10.1111/j.1574-6968.1994.tb06832.x. [DOI] [PubMed] [Google Scholar]
- 4.Cronan J E, Jr, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 612–636. [Google Scholar]
- 5.Fleischmann R D, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 6.Gilliland G, Perrin S, Bunn H F. Competitive PCR for quantitation of mRNA. In: Innis M, et al., editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 60–80. [Google Scholar]
- 7.Green J M, Merkel W K, Nichols B P. Characterization and sequence of Escherichia coli pabC, the gene encoding aminodeoxychorismate lyase, a pyridoxal phosphate-containing enzyme. J Bacteriol. 1992;174:5317–5323. doi: 10.1128/jb.174.16.5317-5323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heath R J, Rock C O. Roles of the FabA and FabZ beta-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J Biol Chem. 1996;271:27795–27801. doi: 10.1074/jbc.271.44.27795. [DOI] [PubMed] [Google Scholar]
- 9.Kornblum J S, Projam S J, Moghazek S L, Novik R P. A rapid method to quantitate non-labeled RNA species in bacterial cells. Gene. 1988;63:75–85. doi: 10.1016/0378-1119(88)90547-1. [DOI] [PubMed] [Google Scholar]
- 10.Larson T J, Ludtke D N, Bell R M. sn-Glycerol-3-phosphate auxotrophy of plsB strains of Escherichia coli: evidence that a second mutation, plsX, is required. J Bacteriol. 1984;160:711–717. doi: 10.1128/jb.160.2.711-717.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S-Y, Rasheed S. A simple procedure for maximum yield of high quality plasmid DNA. BioTechniques. 1990;9:676–679. [PubMed] [Google Scholar]
- 12.Magnuson K, Jackowski S, Rock C O, Cronan J E., Jr Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magnuson K S, Oh W, Larson T J, Cronan J E., Jr Cloning and nucleotide sequence of the fabD gene encoding malonyl-coenzyme A-acyl-carrier protein transacylase of Escherichia coli. FEBS Lett. 1992;299:262–266. doi: 10.1016/0014-5793(92)80128-4. [DOI] [PubMed] [Google Scholar]
- 14.Mayer M P. A new set of useful cloning and expression vectors derived from pBlueScript. Gene. 1995;163:41–46. doi: 10.1016/0378-1119(95)00389-n. [DOI] [PubMed] [Google Scholar]
- 15.Metcalf W W, Jiang W, Wanner B L. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6Kγ origin plasmids at different copy numbers. Gene. 1994;138:1–7. doi: 10.1016/0378-1119(94)90776-5. [DOI] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 17.Morbidoni H R, de Mendoza D, Cronan J E., Jr Bacillus subtilis acyl carrier protein is encoded in a cluster of lipid biosynthesis genes. J Bacteriol. 1996;178:4794–4800. doi: 10.1128/jb.178.16.4794-4800.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogden S, Haggerty D, Stoner C M, Kolodrubetz D, Schleif R. The Escherichia colil-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci USA. 1980;77:3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh W, Larson T J. Physical locations of genes in the rne (ams)-rpmF-plsX-fab region of the Escherichia coli K-12 chromosome. J Bacteriol. 1992;174:7873–7874. doi: 10.1128/jb.174.23.7873-7874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Podkovyrov S, Larson T J. Lipid biosynthetic genes and a ribosomal protein gene are cotranscribed. FEBS Lett. 1995;368:429–431. doi: 10.1016/0014-5793(95)00702-b. [DOI] [PubMed] [Google Scholar]
- 21.Podkovyrov S, Larson T J. Identification of promoter and stringent regulation of transcription of the fabH, fabD and fabG genes encoding fatty acid biosynthetic enzymes of Escherichia coli. Nucleic Acids Res. 1996;24:1747–1752. doi: 10.1093/nar/24.9.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawlings M, Cronan J E., Jr The gene encoding Escherichia coli acyl carrier protein lies within a cluster of fatty acid biosynthetic genes. J Biol Chem. 1992;267:5751–5754. [PubMed] [Google Scholar]
- 23.Ray A, Skurray R. Stabilization of the cloning vector pACYC184 by insertion of F plasmid leading region sequences. Plasmid. 1984;11:272–275. doi: 10.1016/0147-619x(84)90036-2. [DOI] [PubMed] [Google Scholar]
- 24.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 25.Rock C O, Cronan J E., Jr Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Shen Z, Byers D M. Isolation of Vibrio harveyi acyl carrier protein and the fabG, acpP, and fabF genes involved in fatty acid biosynthesis. J Bacteriol. 1996;178:571–573. doi: 10.1128/jb.178.2.571-573.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summers R G, Ali A, Shen B, Wessel W A, Hutchinson C R. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: a possible link between fatty acid and polyketide biosynthesis. Biochemistry. 1995;34:9389–9402. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]
- 29.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka Y, Tsujimura A, Fujita N, Isono S, Isono K. Cloning and analysis of an Escherichia coli operon containing the rpmF gene for ribosomal protein L32 and the gene for a 30-kilodalton protein. J Bacteriol. 1989;171:5707–5712. doi: 10.1128/jb.171.10.5707-5712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomb J F, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 32.Toomey R E, Wakil S J. Studies on the mechanism of fatty acid synthesis XV: preparation and general properties of β-ketoacyl acyl carrier protein reductase from Escherichia coli. Biochim Biophys Acta. 1966;116:189–197. doi: 10.1016/0005-2760(66)90001-4. [DOI] [PubMed] [Google Scholar]
- 33.Tsay J-T, Oh W, Larson T J, Jackowski S, Rock C O. Isolation and characterization of the β-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J Biol Chem. 1992;267:6807–6814. [PubMed] [Google Scholar]
- 34.Verwoert I I, Verhagen E F, van der Linden K H, Verbree E C, Nijkamp H J, Stuitje A R. Molecular characterization of an Escherichia coli mutant with a temperature-sensitive malonyl coenzyme A-acyl carrier protein transacylase. FEBS Lett. 1994;348:311–316. doi: 10.1016/0014-5793(94)00630-x. [DOI] [PubMed] [Google Scholar]
- 35.Youderian P, Sugiono P, Brewer K L, Higgins N P, Elliott T. Packaging specific segments of the Salmonella chromosome with locked-in Mud-P22 prophages. Genetics. 1988;118:581–592. doi: 10.1093/genetics/118.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Cronan J E., Jr Polar allele duplication for transcriptional analysis of consecutive essential genes: application to a cluster of Escherichia coli fatty acid biosynthetic genes. J Bacteriol. 1996;178:3614–3620. doi: 10.1128/jb.178.12.3614-3620.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]