Abstract
The ATPase of Ilyobacter tartaricus was solubilized from the bacterial membranes and purified. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the purified enzyme revealed the usual subunit pattern of a bacterial F1Fo ATPase. The polypeptides with apparent molecular masses of 56, 52, 35, 16.5, and 6.5 kDa were identified as the α, β, γ, ɛ, and c subunits, respectively, by N-terminal protein sequencing and comparison with the sequences of the corresponding subunits from the Na+-translocating ATPase of Propionigenium modestum. Two overlapping sequences were obtained for the polypeptides moving with an apparent molecular mass of 22 kDa (tentatively assigned as b and δ subunits). No sequence could be determined for the putative a subunit (apparent molecular mass, 25 kDa). The c subunits formed a strong aggregate with the apparent molecular mass of 50 kDa which required treatment with trichloroacetic acid for dissociation. The ATPase was inhibited by dicyclohexyl carbodiimide, and Na+ ions protected the enzyme from this inhibition. The ATPase was specifically activated by Na+ or Li+ ions, markedly at high pH. After reconstitution into proteoliposomes, the enzyme catalyzed the ATP-dependent transport of Na+, Li+, or H+. Proton transport was specifically inhibited by Na+ or Li+ ions, indicating a competition between these alkali ions and protons for binding and translocation across the membrane. These experiments characterize the I. tartaricus ATPase as a new member of the family of FS-ATPases, which use Na+ as the physiological coupling ion for ATP synthesis.
F1Fo ATPases (ATP synthases) catalyze ATP synthesis in bacteria, mitochondria, or chloroplasts under consumption of the free energy of a transmembrane electrochemical gradient of protons or, in some cases, Na+ ions (1–3, 19). Accordingly, the proton- or sodium ion-translocating ATPases have been classified as FP-ATPases and FS-ATPases, respectively (8). The water-soluble F1 part of these enzymes catalyzes the hydrolysis of ATP and has the subunit composition α3β3γδɛ. The membrane-embedded Fo component is responsible for the translocation of the coupling ions across the membrane and in bacteria has the subunit composition ab2c9–12. Except for the extended coupling ion specificity, FS- and FP-ATPases are closely related with respect to structure and function. This has been most impressively demonstrated by the construction of functional hybrids between the FP-ATPase of Escherichia coli and the FS-ATPase of Propionigenium modestum (5, 15).
FS-ATPases are rare in nature, and until now, the only well-characterized example is the enzyme from P. modestum (12–14). Another organism harboring a Na+-translocating ATPase is Acetobacterium woodii (17). As this enzyme was reported to consist of only six instead of the usual eight subunits, its belonging to the family of FS-ATPases remains to be proven.
Nevertheless, there is good reason to believe that P. modestum is not the only organism synthesizing an ATPase of the FS type. A Na+-translocating F1Fo ATPase is probably advantageous for P. modestum, which thrives on ATP synthesis by the ΔμNa+ that is generated during succinate fermentation by the methylmalonyl coenzyme A decarboxylase Na+ pump (4). A related ATP synthesis mechanism may be operating in Ilyobacter tartaricus, which is a close relative to P. modestum (15a). The bacterium grows from the fermentation of l-tartrate or citrate to acetate, formate, and CO2 (18). Oxaloacetate, the first degradation product of both substrates, is probably further degraded to pyruvate by an oxaloacetate decarboxylase Na+ pump. This enzyme has been well characterized for Klebsiella pneumoniae or Salmonella typhimurium, where it participates in the fermentation of citrate or l-tartrate (3). To investigate this possibility, the F1Fo ATPase of I. tartaricus has been purified. The enzyme was characterized as an FS-ATPase with clear relationships to that of P. modestum.
MATERIALS AND METHODS
Cell growth.
I. tartaricus (DSM 2382) was grown anaerobically in a medium containing, per liter of H2O, the following: 20 g of NaCl, 3 g of MgCl2 · 6H2O, 0.5 g of KCl, 0.25 g of NH4Cl, 0.2 g of KH2PO4, 0.15 g of CaCl2 · 2H2O, 2 g of l-tartrate, 2.5 g of NaHCO3, and 360 mg of Na2S · 9H2O. The medium further contained trace element solution, seven-vitamin solution, and selenite-tungsten solution (20). Cells were usually grown at 30°C in gastight bottles containing 1 liter of medium. For mass production of cells, 200 liters of medium in a fermentor was inoculated with 5 liters of a well-grown culture. After 18 h of growth, the cells (∼80 g [wet mass]) were collected by continuous centrifugation and frozen for storage in liquid nitrogen.
ATPase purification.
The I. tartaricus cells (5 g [wet mass]) were suspended in 15 ml of extraction buffer (50 mM potassium phosphate [pH 8.0], 1 mM dithiothreitol; 0.1 mM diisopropyl fluorophosphate, 0.1 mg of DNase I per ml). After homogenization, the cells were disrupted by two passages through a French pressure cell at 400 kPa (6,000 lb/in2). Unbroken cells and large debris were removed by centrifugation (15 min, 7,700 × g). The supernatant was diluted to 25 ml with extraction buffer, and the membranes were sedimented by ultracentrifugation (30 min, 145,000 × g). After the membranes were washed with 25 ml of extraction buffer, they were suspended in 10 ml of 50 mM MOPS (morpholinepropanesulfonic acid)-KOH buffer, pH 7.0, containing 1% Triton X-100. After 15 min with occasional mixing at 0°C, the solubilized membrane proteins were isolated by ultracentrifugation (1 h, 200,000 × g). The solubilized ATPase was supplied with 0.5 ml of 1 M MgCl2. Contaminating proteins were precipitated with polyethylene glycol 6000 (approximately 0.4 ml of 50% [wt/wt] polyethylene glycol 6000 was added to 10 ml of enzyme solution). The precipitate was removed by centrifugation (15 min, 39,000 × g) when 90% of the activity was still present in the supernatant. To 10.5 ml of the supernatant, 1.5 ml of 50% polyethylene glycol 6000 was added to precipitate the ATPase. This was collected by centrifugation and dissolved in 1 ml of 5 mM potassium phosphate (pH 7.0) containing 1 mM dithiothreitol, 0.1 mM diisopropyl fluorophosphate, and 0.05% Triton X-100. Insoluble material was removed by centrifugation, and each 200 μl of the ATPase (0.4 to 0.5 mg) was subjected to gel chromatography (0.2 ml/min) with a Superose-6 HR10/30 column preequilibrated with 50 mM potassium phosphate–150 mM KCl–5 mM MgCl2–0.05% Triton X-100, pH 7.0.
Preparation of reconstituted proteoliposomes.
A suspension of 60 mg of phosphatidylcholine (Sigma; Type II S) in 1.9 ml of 50 mM potassium phosphate–1 mM MgCl2–1 mM dithiothreitol, pH 7.0, was sonicated twice for 1 min each in a water bath sonicator. Purified ATPase (0.1 ml; 0.2 to 0.3 mg of protein) was added to the suspension, and the mixture was incubated for 10 min at 25°C with occasional shaking, frozen in liquid nitrogen, and thawed within 1 h at 0°C. The proteoliposomes were sonicated twice for 5 s each, collected by ultracentrifugation (50 min, 200,000 × g), and resuspended in 0.3 ml of 5 mM potassium phosphate buffer, pH 7.0, containing 1 mM MgCl2.
Determination of ATPase activity.
ATPase activity was determined by a coupled spectrophotometric assay. The reaction mixture contained 50 mM potassium phosphate (pH 8.0) or 50 mM potassium borate (pH 9.2), 100 mM K2SO4, 0.25 mM dipotassium NADH, 3 mM potassium phosphoenolpyruvate, 15 U of lactate dehydrogenase per ml, 10 U of pyruvate kinase per ml, 2.5 mM Mg-ATP, and F1Fo-ATPase (10 to 50 mU/ml). In the case of the solubilized protein, 0.01% Triton X-100 was added to the assay mixture. The reaction was started by addition of the ATPase. NADH oxidation was continuously monitored at 340 nm.
Determination of proton transport.
Proton transport into reconstituted proteoliposomes was measured by the quenching of 9-amino-6-chloro-2-methoxyacridine (ACMA). The reaction mixture contained 5 mM potassium phosphate (pH 7.0), 5 mM MgCl2, 100 mM K2SO4, 1 μM ACMA, and 50 μl of reconstituted proteoliposomes (7.5 mg of phospholipid) per ml. After the fluorescence signal had stabilized, the reaction was started by the addition of 1.7 mM ATP. Fluorescence was measured with an excitation wavelength of 410 nm and an emission wavelength of 480 nm.
Determination of Na+ or Li+ transport.
Sodium ion transport into reconstituted proteoliposomes was measured as described elsewhere (5). The reaction mixture (0.75 ml) contained 5 mM potassium phosphate, 100 mM K2SO4, 5 mM MgCl2, 6 mM potassium phosphoenolpyruvate, 20 U of pyruvate kinase, 2 mM 22NaCl (380 to 420 cpm/nmol), and 100 μl of proteoliposomes (10 mg of lipid), pH 7.0. After 5 min, the transport was started by the addition of 2.5 mM Mg-ATP. Samples of 90 μl were passed over Dowex 50 K+ columns at various times. The proteoliposomes were eluted twice with 0.6 ml each of 2 mM Tricine-KOH (pH 7.0)–200 mM sucrose–5 mM MgCl2. The amount of 22Na+ entrapped within the proteoliposomes was subsequently determined by measuring the γ radiation. For lithium ion transport measurements, the reaction mixtures contained 2 mM LiCl instead of 22NaCl. External Li+ ions were removed by passing the reaction mixtures over Dowex 50 K+ columns, and the amount of Li+ entrapped inside the proteoliposomes was subsequently determined by flame emission spectroscopy.
RESULTS AND DISCUSSION
Purification of the F1Fo ATPase of I. tartaricus.
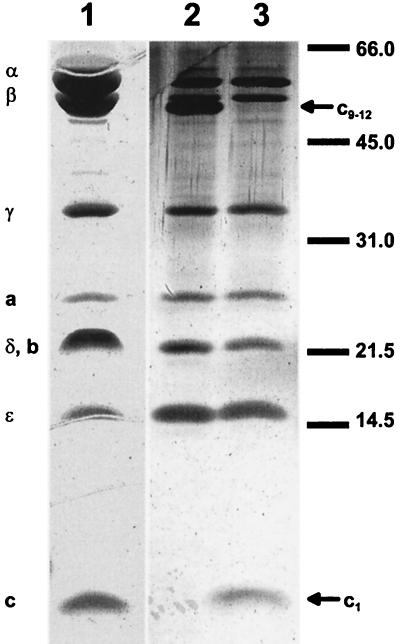
The ATPase of I. tartaricus was isolated as described in Materials and Methods and in Table 1. Figure 1 shows the subunit composition of the purified enzyme as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The pattern is typical for an F1Fo ATPase. According to the molecular masses, the individual polypeptide bands could tentatively be identified as ATPase subunits as follows: α (56 kDa), β (52 kDa), γ (35 kDa), δ (22 kDa), ɛ (16.5 kDa), a (25 kDa), b (22 kDa), c (monomer) (6.5 kDa), and c (multimer) (50 kDa). Individual subunits are well separated on the gel except for the δ and b subunits, which probably move together in the 22-kDa band (broadened on the Coomassie blue-stained gel). A polypeptide with the size of monomeric subunit c was clearly seen, if the ATPase was precipitated with trichloroacetic acid prior to SDS-PAGE (16). Without this treatment, however, the 6.5-kDa band was lacking, and instead, one moving with an apparent molecular mass of 50 kDa was observed. The c subunits of the I. tartaricus ATPase therefore form a very strong complex that resists boiling with SDS for 5 min. This property of the I. tartaricus c subunits is similar to the properties of those from the P. modestum (13) or A. woodii (17) ATPases.
TABLE 1.
Purification of the ATPase of I. tartaricus (starting material, 5 g of wet packed cells)
| Step | Protein (mg) | Activity (U) | Sp act (U/mg) | Yield (%) |
|---|---|---|---|---|
| Triton extraction | 53.5 | 68 | 1.28 | 100 |
| Polyethylene glycol fractionation | 2.8 | 47 | 17 | 69 |
| Gel filtration | 1.3 | 26 | 20.7 | 38 |
FIG. 1.
SDS-PAGE of the purified ATPase of I. tartaricus. Lane 1, 30 μg of ATPase, stained with Coomassie brilliant blue; lanes 2 and 3, 1 μg of ATPase, stained with silver; samples applied to lanes 1 and 3 were precipitated with trichloroacetic acid. The mobilities of molecular mass markers (in kilodaltons) and the assignments of the polypeptide bands to individual ATPase subunits are indicated.
The polypeptide bands seen in Fig. 1 were further identified as subunits of the I. tartaricus ATPase by N-terminal protein sequencing (Table 2). The N-terminal sequences of the α (24 residues), β (24 residues), γ (26 residues), and ɛ (35 residues) subunits were 88, 79, 77, and 63% identical, respectively, to the N termini of the corresponding subunits from the P. modestum ATPase. Two overlapping sequences were obtained for the polypeptides moving with an apparent molecular mass of 22 kDa, in accord with the supposition that this band represents the unresolved b and δ subunits. No sequence was obtained for the putative a subunit. About 70% of the entire c subunit sequence was identified by N-terminal protein sequencing. An alignment of this sequence with the corresponding ones from the ATPase of P. modestum, A. woodii, and E. coli is shown in Fig. 2. This part of the I. tartaricus c subunit was 93, 56, and 29% identical to that from P. modestum, A. woodii, and E. coli, respectively. Q32, which has previously been identified as one of the Na+-binding ligands (8), is conserved in the FS-ATPases, whereas the E. coli ATPase has I at this position. The polypeptide with the apparent molecular mass of 6.5 kDa is therefore clearly identified as the c subunit. The data also indicate a close phylogenetic relationship between I. tartaricus and P. modestum, whereas A. woodii and E. coli are more distantly related organisms.
TABLE 2.
N-terminal sequences of the subunits of the I. tartaricus ATPase and identity with the corresponding subunit of the P. modestum ATPase
| Subunit | N-terminal sequence | % Identity to corresponding P. modestum subunit |
|---|---|---|
| α | MKIRPEEIXXIIKTEIENYKLGLD | 88 |
| β | MRNKGILTQIIGPVVDVSFDNELP | 83 |
| γ | AGGKELKGRIKSVQXTHQITKAMEIV | 76 |
| ɛ | ATFKLEVITPLKKIIEKXVVMIVLRTTEGDMGVLA | 63 |
| c | MDMLFAKTVVLAASAVGAGTAMIAGIGPGVGQGYAAGKAVESVARQPEAKGDIIQ | 93 |
FIG. 2.
Sequence alignment of the N-terminal portions of the c subunits from the ATPases of I. tartaricus (ita-c), P. modestum (pmo-c), A. woodii (awo-c), and E. coli (eco-c).
Catalytic properties of the ATPase.
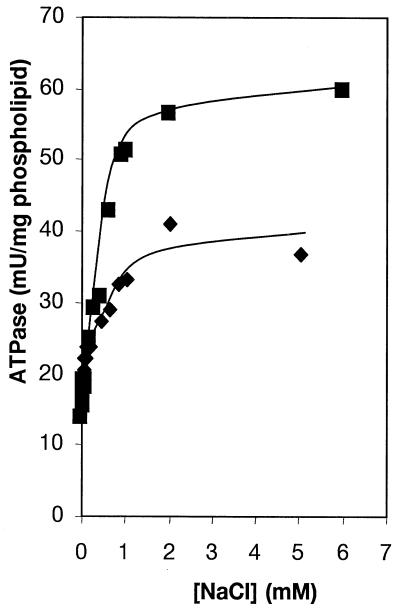
A characteristic of FS-ATPases is the specific activation by Na+ ions. Figure 3 shows the activation profile of the reconstituted I. tartaricus ATPase by Na+ ions at pH 8.0 and 9.2. Half-maximal activation of the ATPase occurred at 270 or 230 μM Na+ at pH 8.0 or 9.2, respectively. Hill plot analyses indicated positive cooperativity with nH being 1.8 at pH 8.0 and being 2.6 at pH 9.2. These data indicate that the ATPase acquires maximal activation after at least two or three binding sites have been simultaneously occupied with Na+ ions. The residual ATPase activity observed without NaCl addition was completely inhibited after preincubation of the enzyme with N, N′-dicyclohexylcarbodiimide (DCCD). It reflects either partial activation by residual Na+ ions or the Na+-independent activity of this enzyme. Low Na+ concentrations affect the I. tartaricus ATPase more significantly than the P. modestum enzyme, because the latter requires five-times-higher Na+ concentrations for half-maximal saturation (10, 11). The I. tartaricus ATPase was activated about twofold by 10 to 20 mM LiCl (data not shown). Half-maximal activation was observed at 830 μM LiCl. The affinity of the I. tartaricus ATPase for Li+ was therefore about three times lower than that for Na+. With a 10-times-lower affinity for Li+ than for Na+, the P. modestum ATPase discriminates more significantly between these alkali ions (10). In the absence of Na+ addition, the reconstituted ATPase had its optimum at pH 6.7. About half of this activity was found at pH 6 or 8, respectively. At pH 6.0, ATPase was not activated by 5 mM NaCl, but at pH 7 to 8, 5 mM NaCl activated the ATPase twofold. These results are compatible with a competition among Na+, Li+, and H+ for binding at the same site of the enzyme, thereby eliciting its activation, as has already been described in detail for the P. modestum ATPase (6, 11, 14).
FIG. 3.
Effect of Na+ concentration on the activity of reconstituted I. tartaricus ATPase at pH 8.0 (⧫) and 9.2 (▪).
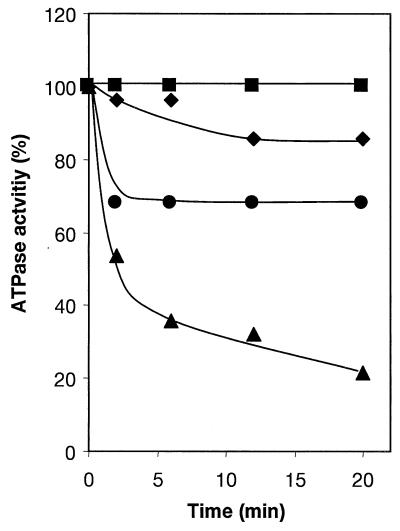
Incubation of the I. tartaricus ATPase with 200 μM DCCD in phosphate buffer, pH 7.0, containing 20 mM KCl led to 80% inhibition of the ATPase after 20 min (Fig. 4). The ATPase was specifically protected from this inhibition by 2 mM NaCl or 20 mM LiCl, similar to the P. modestum enzyme (10, 11). This implies that the DCCD-reactive amino acid (cE65 in the case of the P. modestum ATPase) is part of a Na+-specific binding site. Occupation of this site by Na+ or Li+ interferes with the modification by DCCD and therefore protects the ATPase from inhibition.
FIG. 4.
Inhibition of the I. tartaricus ATPase by DCCD and protection from this inhibition by Na+ or Li+ ions. ATPase was incubated at 25°C in 50 mM potassium phosphate buffer, pH 7.0, with 200 μM DCCD and 2 mM NaCl (⧫), 20 mM LiCl (•), or 20 mM KCl (▴). The residual ATPase activity was determined with samples taken at the indicated times. DCCD was absent in the control (▪).
ATP-driven transport of H+, Na+, or Li+.
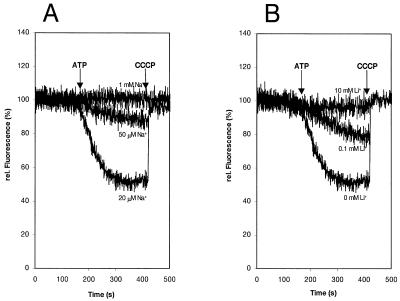
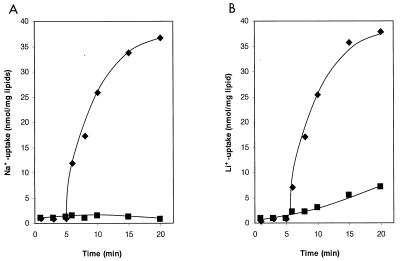
The coupling ion specificity of the I. tartaricus ATPase was further investigated by transport experiments with proteoliposomes reconstituted with the purified enzyme. Proton translocation was determined with the ACMA fluorescence quenching technique. The results in Fig. 5 indicate that the quenching response is elicited by ATP addition and is completely reversed with the uncoupler carbonyl cyanide m-chlorophenylhydrazone. The accumulation of protons inside the proteoliposomes was severely inhibited by 50 μM NaCl and completely abolished at 1 mM NaCl. Inhibition of proton transport was also observed in the presence of 0.1 mM LiCl, and 10 mM (this) alkali salt completely prevented proton uptake. These results thus indicate that the I. tartaricus ATPase pumps protons and that Na+ or Li+ ions specifically interfere with this transport. H+, Na+, or Li+ therefore probably competes for binding and translocation by the ATPase. Direct measurements of Na+ and Li+ transport by the ATPase are shown in Fig. 6. After ATP addition, the internal Na+ concentration increased within 20 min from 1 to 37 nmol/mg of lipid, while no Na+ accumulation occurred in the control without ATP. Li+ ions were accumulated within 20 min to 38 and 7 nmol/mg of lipid in the presence or absence of ATP, respectively.
FIG. 5.
Proton transport into proteoliposomes containing the purified I. tartaricus ATPase and inhibition of H+ transport by Na+ (A) or Li+ (B) ions. Proton transport into the proteoliposomes was determined by the ACMA fluorescence quenching technique. Quenching was elicited by ATP addition; the signal was completely reversed with the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP).
FIG. 6.
Kinetics of Na+ (A) or Li+ (B) transport into proteoliposomes containing the purified I. tartaricus ATPase. The transport reactions were initiated by ATP addition, and Na+ or Li+ ions accumulated within the proteoliposomes were determined as described in Materials and Methods. Controls in the absence of ATP are indicated (▪).
We have recently shown that, besides subunit c, subunit a of the P. modestum ATPase contributes to the extended coupling ion specificity of this enzyme. The ATPase with a triple mutation in its a subunit loses the capacity to translocate Na+ with retention of its H+- or Li+-translocating properties (7, 9). It is not known, however, which residues determine the extended Na+-translocating properties of the a subunit of the P. modestum ATPase. The identification of the I. tartaricus ATPase as another FS-ATPase now offers the possibility of determining, by sequencing of its a subunit, those residues that have been uniquely conserved within the a subunits of I. tartaricus and P. modestum. Such residues are obvious candidates for determining the enzyme’s function as a Na+-translocating ATPase.
REFERENCES
- 1.Boyer P D. The ATP synthase—a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 2.Deckers-Hebestreit G, Altendorf K. The F0F1-type ATP synthases of bacteria. Structure and function of the F0 complex. Annu Rev Microbiol. 1996;50:791–824. doi: 10.1146/annurev.micro.50.1.791. [DOI] [PubMed] [Google Scholar]
- 3.Dimroth P. Primary sodium ion translocating enzymes. Biochim Biophys Acta. 1997;1318:11–51. doi: 10.1016/s0005-2728(96)00127-2. [DOI] [PubMed] [Google Scholar]
- 4.Hilpert W, Schink B, Dimroth P. Life by a new decarboxylation-dependent energy conservation mechanism with Na+ as coupling ion. EMBO J. 1984;3:1665–1670. doi: 10.1002/j.1460-2075.1984.tb02030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaim G, Dimroth P. Formation of a functionally active sodium-translocating hybrid F1F0 ATPase in Escherichia coli by homologous recombination. Eur J Biochem. 1993;218:937–944. doi: 10.1111/j.1432-1033.1993.tb18450.x. [DOI] [PubMed] [Google Scholar]
- 6.Kaim G, Dimroth P. A double mutation in subunit c of the Na+-specific F1F0 ATPase of Propionigenium modestum results in a switch from Na+ to H+-coupled ATP synthesis in the Escherichia coli host cells. J Mol Biol. 1995;253:726–738. doi: 10.1006/jmbi.1995.0586. [DOI] [PubMed] [Google Scholar]
- 7.Kaim G, Dimroth P. A triple mutation in the a subunit of the Escherichia coli/Propionigenium modestum F1F0 ATPase hybrid causes a switch from Na+ stimulation to Na+ inhibition. Biochemistry. 1998;37:4626–4634. doi: 10.1021/bi973022f. [DOI] [PubMed] [Google Scholar]
- 8.Kaim G, Wehrle F, Gerike U, Dimroth P. Molecular basis for the coupling ion selectivity of F1F0 ATP synthases: probing the liganding groups for Na+ and Li+ in the c subunit of the ATP synthase from Propionigenium modestum. Biochemistry. 1997;36:9185–9194. doi: 10.1021/bi970831q. [DOI] [PubMed] [Google Scholar]
- 9.Kaim G, Matthey U, Dimroth P. Mode of interaction of the single a subunit with the multimeric c subunits during the translocation of the coupling ions by F1F0 ATPases. EMBO J. 1998;17:688–695. doi: 10.1093/emboj/17.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kluge C, Dimroth P. Specific protection by Na+ or Li+ of the F1F0-ATPase of Propionigenium modestum from the reaction with dicyclohexylcarbodiimide. J Biol Chem. 1993;268:14557–14560. [PubMed] [Google Scholar]
- 11.Kluge C, Dimroth P. Kinetics of inactivation of the F1F0 ATPase of Propionigenium modestum by dicyclohexylcarbodiimide in relationship to H+ and Na+ concentration: probing the binding site for the coupling ions. Biochemistry. 1993;268:10378–10386. doi: 10.1021/bi00090a013. [DOI] [PubMed] [Google Scholar]
- 12.Laubinger W, Dimroth P. Characterization of the Na+-stimulated ATPase of Propionigenium modestum as an enzyme of the F1F0-type. Eur J Biochem. 1987;168:475–480. doi: 10.1111/j.1432-1033.1987.tb13441.x. [DOI] [PubMed] [Google Scholar]
- 13.Laubinger W, Dimroth P. Characterization of the ATP synthase of Propionigenium modestum as a primary sodium pump. Biochemistry. 1988;27:7531–7537. doi: 10.1021/bi00419a053. [DOI] [PubMed] [Google Scholar]
- 14.Laubinger W, Dimroth P. The sodium ion translocating adenosinetriphosphatase of Propionigenium modestum pumps protons at low sodium ion concentrations. Biochemistry. 1989;28:7194–7198. doi: 10.1021/bi00444a010. [DOI] [PubMed] [Google Scholar]
- 15.Laubinger W, Deckers-Hebestreit G, Altendorf K, Dimroth P. A hybrid adenosinetriphosphatase composed of F1 of Escherichia coli and F0 of Propionigenium modestum is a functional sodium ion pump. Biochemistry. 1990;29:5458–5463. doi: 10.1021/bi00475a008. [DOI] [PubMed] [Google Scholar]
- 15a.Ludwig, W. Personal communication.
- 16.Matthey U, Kaim G, Dimroth P. Subunit c from the sodium-ion-translocating F1F0-ATPase of Propionigenium modestum. Production, purification and properties of the protein in dodecylsulfate solution. Eur J Biochem. 1997;247:820–825. doi: 10.1111/j.1432-1033.1997.t01-1-00820.x. [DOI] [PubMed] [Google Scholar]
- 17.Reidlinger J, Müller V. Purification of ATP synthase from Acetobacterium woodii and identification as a Na+-translocating F1F0-type enzyme. Eur J Biochem. 1994;223:275–283. doi: 10.1111/j.1432-1033.1994.tb18992.x. [DOI] [PubMed] [Google Scholar]
- 18.Schink B. Fermentation of tartrate enantiomers by anaerobic bacteria, and description of two new species of strict anaerobes, Ruminococcus pasteurii and Ilyobacter tartaricus. Arch Microbiol. 1984;139:409–414. [Google Scholar]
- 19.Weber J, Senior A E. Catalytic mechanism of F1-ATPase. Biochim Biophys Acta. 1997;1319:19–58. doi: 10.1016/s0005-2728(96)00121-1. [DOI] [PubMed] [Google Scholar]
- 20.Widdel F, Pfennig N. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov. sp. nov. Arch Microbiol. 1981;129:395–400. doi: 10.1007/BF00406470. [DOI] [PubMed] [Google Scholar]