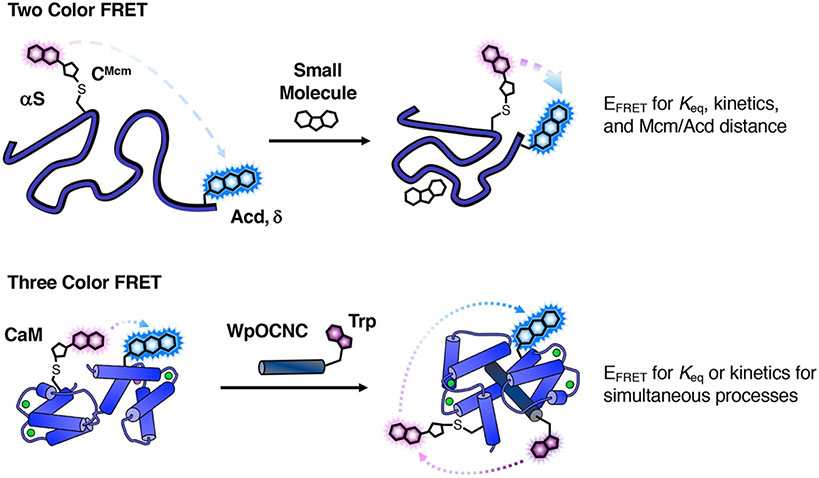
Abstract
Site-specific protein labeling can be used to monitor protein motions and interactions in real time using Förster resonance energy transfer (FRET). While there are many fluorophores available for protein labeling, few FRET pairs are suitable for monitoring intramolecular protein motions without being disruptive to protein folding and function. Here, we describe the synthesis and use of a minimally perturbing FRET pair comprised of methoxycoumarin maleimide (Mcm-Mal) and acridonylalanine (Acd). Acd can be incorporated into a protein through unnatural amino acid mutagenesis. Mcm-Mal is fluorogenic when reacted with cysteine and can label cysteine/Acd double mutant proteins. This labeling strategy provides an easy to install FRET pair with a working range or 15-40 Å, making it ideal for monitoring most intramolecular motions. Additionally, Mcm/Acd FRET can be combined with tryptophan fluorescence for monitoring multiple protein motions via three color FRET.
1. Introduction
Fluorescence spectroscopy methods provide the chance to observe protein motions in real time under physiological conditions.[1] If a protein is labelled with two fluorescent probes, dynamic structural information can be obtained using distance-dependent interactions such as Förster resonance energy transfer (FRET) and quenching by photo-induced electron transfer.[2] An important area of research in bioorganic chemistry is the development of small fluorescent probes that can be easily installed for multiple applications. Herein, we have outlined the synthesis of methoxycoumarin maleimide (Mcm-Mal, Scheme 1) and acridonylalanine (Acd, Scheme 2) and described their installation and use as a site-specific FRET pair. Mcm/Acd labelling can also be combined with a single Trp mutant for three color FRET experiments. This allows for the simultaneous monitoring of two interactions such as a protein/protein interaction and a conformational change within one of the proteins.
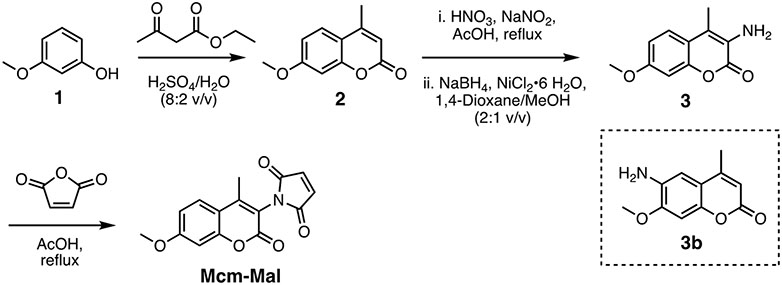
Scheme 1.
Synthesis of Methoxycoumarin maleimide (Mcm-Mal).
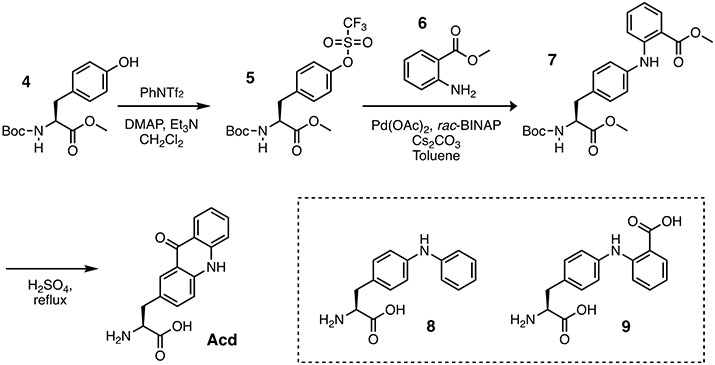
Scheme 2.
Synthesis of Acridonylalanine (Acd)
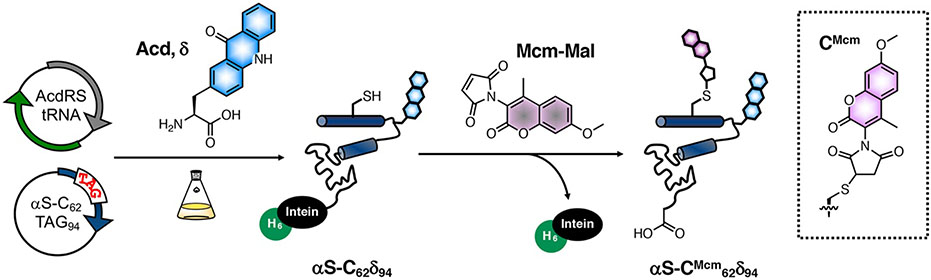
The Mcm/Acd FRET pair is installed by using site-directed mutagenesis and amber codon suppression to generate constructs with single Cys and Acd mutations, followed by reaction of Cys with Mcm-Mal. (Fig. 1) This labelling strategy provides advantages over other methods, such as a fluorescent protein fusion, which can disrupt protein function due to the size of the label, or doubly labelling using click chemistry, which can result in low protein yields.[3] Both Acd and Mcm are small probes, roughly the same size as endogenous amino acids, and therefore are minimally disruptive to protein folding, motion, and function.[4] Importantly, Mcm-Mal can quickly and efficiently label a protein at a mutant Cys site without other reagents or resorting to ligation methods used previously to introduce Mcm derivatives.[5, 6] Acd is an unnatural amino acid (Uaa) that can be incorporated by using site-directed mutagenesis and amber codon suppression, and therefore can be inserted on the interior of a protein without the need to unfold and refold the protein.[7, 8] To further facilitate labelling, we commonly use C-terminal intein-His6 tags to isolate full-length proteins containing Uaas from truncated versions that terminate at the amber codon.[9] This intein-His6 tag modification can be cleaved with β-mercaptoethanol (BME) either before or after Mcm-Mal labelling.
Figure 1.
Mcm/Acd labelling for FRET experiments. Protein expressed with Cys and acridonylalanine (Acd, δ) mutants is harvested as a His6-tagged intein fusion and reacted with methoxycoumarin maleimide (Mcm-Mal) with cleavage of the intein purification tag. α-synuclein (αS) with Mcm labeling at Cys62 and Acd labeling at position 94 is shown as an example.
Mcm/Acd labelling can also improve FRET measurements over commonly used fluorophores such as fluorescein and rhodamine. Both fluorescein and rhodamine are larger in size, which can be disruptive to protein folding and both probes are normally installed using maleimide labelling and/or biorthogonal reactions such as copper-catalyzed click chemistry. As a result, each fluorophore has a relatively long linker separating the fluorophore from the protein backbone. This adds uncertainty to each FRET measurement. Additionally, the fluorescein/rhodamine FRET has a larger Förster radius (, the distance of half-maximal energy transfer) of ~50 Å. With FRET working distances in the 30-90 Å range, it is difficult to use fluorescein and rhodamine as a FRET pair to study and intraprotein distances, which are often shorter than 40 Å.
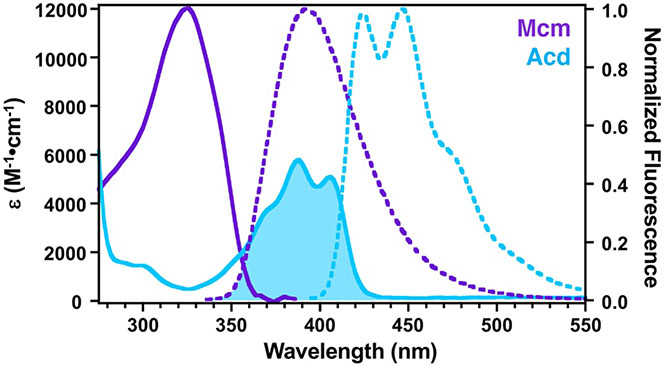
Our laboratory has worked to develop non-perturbing, short/medium-range probe pairs that can be selectively excited in proteins and are better suited to studying these protein regions. Acd is excited at 380-400 nm with emission at 420-450 nm. We have shown that Acd can be a valuable FRET acceptor from Mcm (excited at 325 nm) with a working range of 15-40 Å.[7] Mcm is an excellent FRET donor to Acd since it has a high extinction coefficient with a peak in the absorption spectrum that coincides with a minimum in the Acd absorption spectrum to avoid undesired direct excitation of the acceptor Acd (Fig. 5). Both Mcm and Acd have longer excitation wavelengths exceeding 300 nm and overlap with tryptophan emission, and therefore can be used to perform three color FRET experiments, monitoring binding and conformational change events in the same experiment. Although deconvolution to extract distance information is not reliable, three color FRET can still be used to measure binding affinity or kinetics.
Figure 5.
Spectral Overlap of Mcm/Acd FRET Pair. Ac-CysMcm (purple) and Acd (blue) absorption spectra (solid lines) were measured in 20 mM Tris 100 mM NaCl, pH. 7.5. Ac-CysMcm (purple) and Acd (blue) fluorescence emission spectra (dashed lines) were measured with excitation at 325 nm for Ac-CysMcm and 386 nm for Acd. Spectral overlap between Ac-CysMcm emission and Acd absorption is indicated by the shaded area.
Within we describe the synthesis and purification of Mcm and Acd singly and doubly labeled proteins and provide general considerations for FRET experiments using the proteins calmodulin (CaM) and α-synuclein (αS) as examples. CaM is a calcium sensor protein that undergoes a dramatic conformational change in the presence of calcium and helical peptide binding partners.[10, 11] αS is a disordered protein that contributes to the pathogenesis of Parkinson’s disease.[12, 13] These provide examples of the value of FRET to monitor conformational change and protein/protein interactions.
2. Experimental Design
2.1. Choice of fluorescence experiment.
The ability to multiply label proteins for FRET permits applications that can be used to study a variety of protein processes, most significantly conformational changes and binding interactions. For conformational changes, one is typically interested in either determining distances between probe pairs on the protein or measuring the rate of the change. For binding interactions, one is typically interested in determining the affinity of the interaction through titration of one of the partners or measuring the rate of association and/or dissociation. The excitation and emission spectra of Mcm and Acd allow one to perform fluorescence experiments in a variety of media and formats, including fluorescence emission or fluorescence lifetime measurements in a cuvette-based instrument, or emission measurements in a multi-well plate reader format. Both of these types of instruments can typically acquire “steady state” fluorescence data on the seconds timescale, which may not be fast enough to usefully measure kinetic phenomena. Therefore, rapid mixing or stopped flow instruments are used to acquire data on the millisecond or faster timescale. We will briefly describe two sets of experiments with Mcm/Acd labeled proteins to illustrate the considerations that one would make in such experiments.
2.2. Design of intramolecular FRET experiments to monitor conformational change.
As noted above, Mcm/Acd FRET pairs have a useful working range of 15-40 Å. Therefore, the pair is not suited to quantitively measure interprobe distance for every conformational change in a folded protein, but most significant domain-level conformational changes or loop refolding events will fall within this range. Disordered proteins, on the other hand, can occupy much more extended conformations, and for disordered proteins greater than 50 amino acids in length, Mcm/Acd labeling is suited to measuring dynamics within regions of the protein rather than the protein’s full span.
FRET measurements of interlobe dynamics of CaM are illustrative of the type of experiment that one might typically perform with a folded protein, in which Ca2+ binding to EF-hand motifs within the lobes and/or peptide binding to the linker region between the lobes can induce a compact conformation of the protein.[10, 11] The movement can be monitored using an Mcm/Acd pair within CaM, and this can be combined with a probe in the peptide (below). For a folded protein such as CaM, one anticipates that one or more X-ray crystal, NMR, or cryo-electron microscopy (cryo-EM) structures would be available, allowing one to choose locations for Mcm/Acd so that a significant change in FRET efficiency () will occur in response to a stimulus, in this case Ca2+ or peptide. For CaM, we used existing crystal and NMR structures to determine that a significant change in would occur due to a decrease in distance between positions 12 and 112 upon peptide binding.[14, 15] Our choice of labelling position was also informed by prior experiments showing that these positions were tolerant to modification (labelled proteins had similar circular dichroism spectra and Ca2+/peptide binding affinity to wild-type protein). While tolerance for Mcm or Acd modification is potentially predictable based upon molecular modelling or bioinformatic considerations, our recent evaluation of more than 50 positions in the proteins LexA and RecA showed that existing methods do not necessarily provide reliable predictors of tolerance for Acd.[8] Thus, while we continue to investigate computational approaches for predicting Uaa tolerance, we must currently recommend an empirical approach to choosing non-perturbing Mcm and Acd sites where labelled proteins should be tested in a functional assay that requires proper folding. For a disordered protein like αS lacking a clear functional assay, taking a structure-guided approach to designing probe locations may not seem feasible, but available computational models as well as previous data on mutational tolerance can be used to choose locations, and we have used assays such as vesicle binding or aggregation to confirm that labelled proteins have similar properties to the wild-type protein.[16, 17]
Binding of WpOCNC, a peptide derived from a CaM-regulated cyclic nucleotide-gated ion channel, induces a conformational change in CaM.[14, 18] Monitoring Mcm/Acd FRET as a function of WpOCNC concentration allowed us to determine a half maximal effective concentration (EC50) or dissociation constant (Kd) for the peptide, a procedure that would be generalizable to any allosteric modulator. Fluorescence spectra for all of the single (Mcm only and Acd only) and double-labelled constructs were collected under conditions where the peptide was fully bound as well as in its absence. These singly-labelled protein spectra were used to analyze the double-labelled spectra to determine . This allows one to correct for changes in quantum yield or spectral overlap when the chromophore changes environment, which can be an important and often overlooked factor when interpreting FRET data. Interprobe distances were then extracted from these data using procedures that can easily be applied to Mcm/Acd FRET experiments for most proteins.
In our αS experiments, we have studied the effects of small molecules on the conformation of αS monomer using FRET, in order to identify aggregation resistant protein conformations.[19, 20] A full set of FRET-based distance constraints requires ~25 experiments, and is best performed with multiple probe pairs that have different working distance ranges.[19] A description of these methods is beyond the scope of this chapter, but αS experiments illustrate the use of fluorescence lifetimes to measure αS Mcm/Acd FRET, which can be useful in aggregating systems where matching the concentration of singly labelled control proteins to the double labelled protein may not be feasible.
2.3. Design of intra- and intermolecular FRET experiments to monitor binding and conformational change kinetics.
The Mcm/Acd CaM construct can also be used to illustrate other types of experiments. While Mcm/Acd FRET changes as a function of WpOCNC concentration could be fit to determine a Kd or EC50, one can also take advantage of the spectral overlap of Trp emission with Mcm absorption to monitor peptide binding through FRET. Mcm energy transfer to Acd allows one to perform three color FRET experiments, monitoring two processes, such as binding and conformational change, in the same experiment. Extraction of two sets of distance information is not practical, because there is some direct Trp/Acd FRET, so the number of control experiments necessary to deconvolute the three FRET processes exceeds the complexity of simply measuring the two FRET interactions in separate experiments. However, two step FRET can be useful to simultaneously measure binding and conformational change kinetics. Using a stopped flow fluorometer, we observed rapid binding of WpOCNC through 295 nm excitation, monitoring Acd emission at 420 nm. In the same experimental setup, we excited at 325 nm to monitor the conformational change in CaM alone. Our EFRET and kinetic data supported the idea that WpOCNC stabilizes existing compact CaM conformations rather than binding and then inducing a large conformational change, consistent with previous observations.[14] We describe the process of fitting these data in section 6.
3. Chemical Synthesis
3.1. Reagents, equipment, and general procedures.
The following supplies and instruments were used in chemical synthesis in our laboratory. In many cases, such as instruments, appropriate substitutions will be necessary. Milli-Q filtered (18 MΩ) water was used for all solutions (Millipore). Low resolution electrospray ionization mass spectra (LRMS) were obtained on a Waters Acquity Ultra Performance LC connected to a single quadrupole detector (SQD) mass spectrometer. High resolution electrospray ionization mass spectra (HRMS) were collected with a Waters LCT Premier XE liquid chromatograph/mass spectrometer. Nuclear magnetic resonance (NMR) spectra were obtained on a Bruker 400 MHz instrument.
Synthetic Reagents
Sulfuric acid (H2SO4)
Ethyl acetoacetate
3-Methoxyphenol
Sodium nitrite (NaNO2)
Sodium borohydride (NaBH4)
Nickel (II) chloride hexahydrate (NiCl2•6 H2O)
Nitric acid (70%, HNO3)
Acetic acid
Maleic anhydride
Sodium carbonate (Na2CO3)
Acetic anhydride
Sodium acetate (NaOAc)
Boc-L-Tyrosine methyl ester (Chem-Impex #04026)
Triethylamine (Et3N)
4-Dimethylaminopyridine (DMAP)
N-Phenyl-bis(trifluoromethanesulfonimide) (Oakwood Chemical #024773)
Methyl 2-aminobenzoate (Sigma #236454)
2,2′-Bis(diphenylphosphino)-1,1′-binaphthalene (rac-BINAP) (Sigma #481084)
Palladium (II) acetate (Pd(OAc)2) (Sigma #683124)
Cesium carbonate (Cs2CO3) (Sigma #441902)
Solvents and Purification Materials
3-Methoxyphenol
Methanol
HPLC-grade water
1,4-Dioxane
Acetone
Dichloromethane (CH2Cl2)
Sodium sulfate (Na2SO4)
Ethyl acetate (EtOAc)
Acetic anhydride
Chloroform (CHCl3)
Ammonium hydroxide (NH4OH)
Dowex® 50WX8 hydrogen form, strongly acidic cation exchange resin (Sigma #217514)
Silica gel 60 (230–400 mesh)
3.2. Synthesis of Methoxycoumarin maleimide (Mcm-Mal).
The Mcm-Mal was synthesized from 3-methoxyphenol in three steps. In the first step, we performed the Pechmann condensation between 3-methoxyphenol and ethyl acetoacetate to give compound 2. Then compound 3 was prepared by nitration of 2 with 70% HNO3 in AcOH, followed by reduction using NaBH4 in the presence of NiCl2•6 H2O. Finally, Mcm-Mal was synthesized by the condensation of compound 3 and maleic anhydride in AcOH. The reported yields are representative.
3.2.1. Synthesis of 7-methoxy-4-methyl-2H-chromen-2-one (2).
Dissolve 3-methoxyphenol (1 g, 8.06 mmol) and ethyl acetoacetate (1.26 g, 9.67 mmol, 1.2 equiv.) in 80% H2SO4 (10 mL) slowly. Stir the reaction mixture at room temperature for 6 hours. Pour the reaction mixture onto crushed ice (250 g) and stir for 1 hour. Collect the precipitate on a Buchner funnel and wash it with cold water until it reaches neutral pH. Dry the white solid with a high vacuum pump, redissolve it in dichloromethane (CH2Cl2), and purify it by silica gel flash chromatography (n-hexane/EtOAc–7:3 v/v) to give 2 as a white solid (1.103 g, 72%). 1H NMR (400 MHz, CDCl3) δ 7.49 (d, J = 8.8 Hz, 1H), 6.85 (d, J = 10.7 Hz, 1H), 6.81 (s, 1H), 6.12 (s, 1H), 3.86 (s, 3H), 2.39 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 162.8, 161.5, 155.5, 152.7, 125.7 (+), 113.8, 112.5 (+), 112.1 (+), 101.0 (+), 55.9 (+), 18.9 (+). Rf value 0.79 (3:2 n-hexane/ EtOAc). HRMS (ESI)+ m/z calc’d for C11H10O3, 190.0630, found 190.0624.
3.2.2. Synthesis of 3-amino-7-methoxy-4-methyl-2H-chromen-2-one (3).
To a slurry of 2 (1 g, 5.25 mmol) in AcOH (25 mL) slowly add 70% HNO3 (6 mL) and NaNO2 (0.508 g, 7.36 mmol) on an ice water bath. Then fit the reaction mixture with a condenser and heat it to 80 °C for one hour to give a mixture of nitro regioisomers. Cool the reaction mixture to 0 °C, and filter the resulting needle-like white crystals using a Buchner funnel. Dilute the resulting filtrate with CH2Cl2 and add saturated Na2CO3 in Milli-Q H2O (100 mL) carefully. Wash the organic layer with the saturated Na2CO3 solution three times (100 mL × 3). Wash the organic layer with brine and dry it over Na2SO4. Filter the solution and evaporate the solvent to give a mixture of nitro isomers as yellow solid. Use this mixture for the next reaction without further purification.
Place the mixture in a 250 mL two-necked round bottom flask and dissolve it in 1,4 dioxane/MeOH/ (25 mL, 2:1 v/v). To the resulting solution add NiCl2•6 H2O (164 mg, 0.3 equivalents), then NaBH4 (174 mg, 2 equivalents) in portions over 5 minutes at 0 °C. Onset of the reaction is manifested by the formation of a black precipitate typical of nickel boride. Stir the reaction mixture at 25 °C for 10 minutes and pour it into ice-cold water. Extract the resulting mixture with EtOAc (200 mL) and water (200 mL). Dry the organic layer over Na2SO4 and concentrate it under reduced pressure. Purify the crude mixture by column chromatography using n-hexane/EtOAc (7:3 v/v) to yield 3 (0.336 g, 36%). %). 1H NMR (400 MHz, CDCl3) δ 7.37 (d, J = 8.7 Hz, 1H), 6.86 (d, J = 8.8 Hz, 1H), 6.83 (s, 1H), 3.97 (s, 2H), 3.84 (s, 3H), 2.21 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 159.6, 159.4, 150.0, 127.1, 123.6 (+), 120.6, 115.3, 112.5 (+), 101.1 (+), 55.9 (+), 12.1 (+). Rf value for compound 3 is 0.43 and for 3b is 0.24 (3:2 n-hexane/ EtOAc). HRMS (ESI)+ m/z calc’d for C11H11NO3, 205.0739, found 205.0732.
3.2.3. Synthesis of 1-(7-methoxy-4-methyl-2-oxo-2H-chromen-3-yl)-1H-pyrrole-2,5-dione (Methoxycoumarin maleimide, Mcm–Mal).
To a solution of 3 (230 mg, 1.121 mmol) in CHCl3 (8 mL) add maleic anhydride (126 mg, 1.233 mmol) and stir the reaction mixture overnight at room temperature. Concentrate the reaction mixture using a rotary evaporator. Suspend the residue in Ac2O (12 mL) and add NaOAc (28 mg, 0.341 mmol, 0.3 equiv.). Stir the reaction mixture at 100 °C for 15 minutes. After cooling, dilute the reaction mixture with EtOAc (>100 mL) and wash it with saturated Na2CO3 in Milli-Q water (50 mL × 4). Wash the organic layer with brine and dry it over Na2SO4. Filter and evaporate this solution, the redissolve it in CH2Cl2 and purify it by silica gel flash chromatography (n-hexane/ EtOAc–6:4 v/v) to give Mcm-Mal (0.268 g, 84%) as yellow crystalline solid. 1H NMR (400 MHz, CDCl3) 7.57 (d, J = 8.9 Hz, 1H), 6.93 (dd, J = 2.6 Hz, 8.9 Hz, 1H), 6.91 (s, 2H), 6.86 (d, J = 2.6 Hz, 1H), 3.90 (s, 3H), 2.32 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 169.0, 163.5, 157.8, 154.5, 152.8, 135.0 (+), 126.6 (+), 113.4, 113.1 (+), 112.9, 100.9 (+), 55.9 (+), 14.5 (+). Rf value 0.61 (n-hexane/ EtOAc = 1/1). HRMS (ESI)+ calc’d for C15H11NO5 [M+H]+ 286.0710, found 286.0704.
3.3. Synthesis of Acridonylalanine (Acd).
The fluorescent unnatural amino acid Acd can prepared in three steps from protected L-tyrosine methyl ester (4) following the procedure reported by Sungwienwong et al.[21] The synthetic route is shown below.
3.3.1. Synthesis of Methyl (S)-2-((tert-butoxycarbonyl)amino)-3-(4-(((trifluoromethyl)sulfonyl) oxy)phenyl) propanoate (5).
Dissolve N-Boc-L-tyrosine methyl ester 4 (4.50 g, 15.24 mmol) in CH2Cl2 (30 mL). Add triethylamine (6.38 mL, 45.72 mmol), 4-(dimethylamino) pyridine (0.186 g, 1.524 mmol), and N-phenyl bis(trifluoro-methanesulfonimide) (7.62g, 21.34mmol). Purge the reaction flask with argon. Stir the reaction at room temperature under an argon balloon for 16 hours. Concentrate the reaction mixture under reduced pressure. Add 30 mL saturated NH4Cl solution and extract with 3x30 mL EtOAc. Concentrate the EtOAc solution by rotary evaporation and purify it by silica flash chromatography, eluting with 8:2 hexanes/EtOAc. (Rf = 0.26) to give the purified product 5 a colorless oil (5.77 g, 88.62% yield). 1H NMR (400 MHz, CDCl3) δ 7.27 – 7.21 (m, 4H), 5.04 (d, J = 7.0 Hz, 1H), 4.62 (q, J = 6.6, 6.2 Hz, 1H), 3.74 (s, 3H), 3.19 (dd, J = 13.8, 5.6 Hz, 1H), 3.06 (dd, J = 13.7, 6.4 Hz, 1H), 1.43 (s, 9H). 13C NMR (101 MHz, CDCl3) δ 172.1, 155.2, 148.8, 137.2, 131.3 (+), 121.5 (+), 80.3, 54.4 (+), 52.5 (+), 38.0 (−), 28.4 (+). HRMS (ESI)+ calc’d for C16H20F3NO7S [M+Na]+ 450.0805, found 450.0827.
3.3.2. Synthesis of (S)-Methyl 2-((4-(2-((tert-butoxycarbonyl)amino)-3-methoxy-3-oxopropyl) phenyl)amino) benzoate (6).
Add triflate 5 (3.000 g, 7.02 mmol) to 75 mL degassed toluene in an oven-dried round-bottom flask, followed by methyl 2-aminobenzoate (1200 μL, 9.27 mmol). Degas the solution with argon for 5 minutes. Add palladium(II) acetate (0.082 g, 0.365 mmol) and racemic 2,2’- bis(diphenyl-phosphino)-1,1’-binaphthyl (0.054 g, 0.087 mmol) to the flask and stir for 3 minutes while under argon. Add cesium carbonate (6.88 g, 21.1 mmol) to the flask and degas the solution again with argon. Fit the flask with a reflux condenser and heat it to 135 °C for 23 hours. Allow the solution to cool to ambient temperature, then filter the contents through a short plug of silica gel using CH2Cl2 to transfer the material to the silica (200 mL), and then elute the product with EtOAc (300 mL). Concentrate the clarified solution under reduced pressure purify it by flash column chromatography (9:1 Hexane/EtOAc) to afford compound 6 (2.759 g, 6.44 mmol, 91.7%) as a yellow oil. (Rf = 0.2 in 85:15 Hexane/EtOAc) 1H NMR (500 MHz, CDCl3) δ 9.44 (s, 1H), 7.96 (d, J = 8.0 Hz, 1H), 7.31 (t, J = 7.9 Hz, 1H), 7.23 (d, J = 8.5 Hz 1H), 7.18 (d, J = 8.2 Hz, 2H), 7.10 (d, J = 8.1 Hz, 1 H), 6.73 (t, J = 7.5 Hz, 1 H), 5.05 (d, J = 7.9 Hz, 1 H), 4.61 – 4.57 (m, 1 H), 3.90 (s, 3H), 3.74 (s, 3H), 3.07 (dd, J = 26.5, 5.7 Hz, 2 H), 1.43 (s, 9H); 13C NMR (125 MHz, CDCl3): δ 172.5 (np), 169.0 (np), 155.3 (np), 148.0 (np), 139.8 (np), 134.2 (+), 131.8 (+), 131.3 (np), 130.4 (+), 122.6 (+), 117.3 (+), 114.2 (+), 112.1 (np), 80.0 (np), 54.6 (+), 52.4 (+), 51.9 (+), 38.0 (−), 28.5 (+). HRMS (ESI)+ calc’d for C23H28N2O6 [M + H]+ 429.2000, found 429.2026.
3.3.3. Synthesis of (S)-2-amino-3-(9-oxo-9,10-dihydroacridin-2-yl)propanoic acid (Acridonylalanine, Acd).
Add a solution of 13.5 M sulfuric acid (12 mL) to a flask containing compound 6 (1.02 g, 2.38 mmol). Fit the flask with a reflux condenser and heat it to 115 °C for 16 hours in an oil bath. Add 80 mL water to the flask and stir for 15 minutes. Remove the reaction from the hot oil bath and allow it to cool. After the solution reaches ambient temperature, cool it to 4 °C and allowed it to stand for 2 hours. Make 100 g of ion-exchange resin (Dowex® 50WX8 hydrogen form, strongly acidic cation exchange resin) into a slurry with 1.8 M aqueous H2SO4 and apply it to a flash chromatography column. Wash the resin with 350 mL 1.8 M aqueous H2SO4, 2 L of water, 1 L of 1.5 M aqueous NH4OH, and 4 L of water. Following each wash, dry the resin by blowing air through the column. Vacuum filter the cooled Acd solution on a Büchner funnel to remove precipitated material, and apply the clarified solution to the washed and dried ion-exchange resin. Shake the resulting resin slurry in the chromatography column for 5 minutes before draining the solution. Reapply this solution to the dried resin and shake it for an additional 5 minutes. Set aside the twice-passed solution. Wash the loaded resin with 4 L water, then elute the compound of interest with 1.45 L of 1.5 M NH4OH. Concentrate the solution to 50 mL by rotary evaporation, and then lyophilize it to dryness, to yield a crop of Acd as a yellow powder (0.6375 g, 2.26 mmol, 94.9%). The ion-exchange resin can be recycled by washing it with 4 L of water and drying it until further use. Both byproducts 8 and 9 can be separated from the product, as they do not bind to the ion exchange resin. 1H NMR (500 MHz, CD3OD) δ 8.33 (dd, J = 1.8, 1.1 Hz, 1H), 8.25 (d, J = 1.8 Hz, 1H), 7.75 (m, 1 H), 7.68 (dd, J = 8.6, 2.1 Hz, 1H), 7.53 (dd, J = 8.5, 3.8 Hz, 2H), 7.3 (m, 1H), 4.28 (dd, J = 7.7, 5.4 Hz, 1H), 3.54 (m, 2H); 13C NMR (125 MHz, CD3OD): δ 180.1, 171.5, 142.74, 142.28, 136.1 (+), 135.3 (+), 129.0, 128.3 (+), 127.5 (+), 123.0 (+), 119.6 (+), 119.3, 118.6 (+), 117.0, 55.3 (+), 37.2 (−). HRMS (ESI)+ calcd for C16H15N2O3 [M+H]+ 283.1083, found 283.1093.
4. Preparation of α-Synuclein (αS) Constructs for Intramolecular FRET
4.1. Reagents, equipment, and general procedures.
The following supplies and instruments were used in protein production in our laboratory. In many cases, such as instruments, appropriate substitutions will be necessary.
PCR Reagents and Equipment
HiFi DNA Assembly Master Mix (New England Biolabs #E2621S)
QuickChange® site-directed mutagenesis kit (Stratagene #200521)
PCR nucleotide mix (Promega #280175)
Pfu turbo polymerase (Stratagene #600250)
Biorad T100 Thermal Cycler
pTXB1Vector (New England Biolabs #N6707S)
NEB10b cells (New England Biolabs #C3019H)
Protein Expression Equipment
3 kDa MWCO Amicon Ultra centrifugal filter units (EMD Millipore #UFC900324)
Sorvall RC-5 centrifuge with SS-34 and GS3 rotors
QSonica Q700 sonicator for cell lysis
Bruker Ultraflex III Matrix assisted laser desorption/ionization instrument (MALDI)
Thermo Scientific Genesis 150 UV-Vis
ÄKTA FPLC for protein purification
Labconco Lyophilizer for drying proteins
Protein Expression Reagents
E. coli BL21(DE3) cells (Stratagene)
Milli-Q filtered (18 MΩ) water (Millipore)
Ampicillin Sodium salt (Fisher Scientific #BP1760-25)
Dialysis Tubing, Nominal MWCO 3,500 (Fisher Scientific #21-152-10)
Reagents for M9 salts (See recipie below for additional reagents)
Protease inhibitor cocktail pill cOmplete mini tablets, EDTA-free, Easy Pack, (Roche #04693159001)
Nickel resin (Gold bio, # H-320-100)
β-mercaptoethanol (BME) (Biorad #1610710)
5 mL HiTrap Q HP column (GE Healthcare #17-1154-01)
DMSO for molecular biology (Sigma Aldrich #D8418)
Bond-Breaker TCEP Solution, Neutral pH (ThermoFisher Scientific #77720)
Isopropyl-β-D-1-thiogalactopyranoside Lab Scientific (BioKEMIX, Inc. #I-555-10)
Buffers: Sterile filtered, and if for FPLC Degassed
50 mM HEPES, pH 7.5
50 mM HEPES, 10 mM imidazole, pH 7.5
50 mM HEPES, 300 mM imidazole, pH 7.5
20 mM Tris, pH 8.0
20 mM Tris, pH 8.0 100 mM NaCl
4.1.2. Recipies
SOC Media
Tryptone, 5 g
Sodium chloride, 1.25 g
Yeast extract, 1.25 g
250 mM potassium chloride, 2.5 mL
Milli-Q filtered water, fill to 250 mL final volume
Autoclave the solution with the components above. Once the solution is cool, add the components below:
1 M MgCl2, 2.5 mL
40% w/v Glucose, 2.5 mL
M9 Media
Dilute 50 mL 10x M9 salts (recipe below), dilute to 500 mL total volume with MilliQ water and autoclave. Once cooled to room temperature add M9 additional components (list below).
M9 Salts (10X):
Sodium phosphate dibasic: 60 g/L (0.42 mol/L)
Potassium phosphate monobasic: 30 g/L (0.22 mol/L)
Sodium chloride: 5 g/L (0.086 mol/L)
Ammonium chloride: 10 g/L (0.19 mol/L)
Additional M9 Components (diluted in Milli-Q water and sterilized with 0.22 μM bottle-top filter unless otherwise noted):
1 M Magnesium sulfate, 1 mL
15 mg/mL Iron (II) chloride, 500 μL
15 mg/mL Zinc (II) chloride, 500 μL
0.01 M Calcium chloride, 100 μL
1 mL Yeast extract, 10% w/v (sterilize by autoclave)
40% w/v Glucose solution, 6.25 mL
4.2. Cloning of αS Constructs.
4.2.1. Gene insertion:
Insert the target protein gene block into the pTXB1 target vector using Gibson assembly.
Mix together 100 ng of pTXB1 vector and 2 equivalents of insert (5 equivalents for inserts more than 200 base pairs long) in a PCR tube. Add Sterile water up to 10 μL.
Add 10 μL of 2x Hifi Assembly Master Mix.
Set incubate reaction mixture in thermocycler at 50 °C for 45-60 minutes.
Incubate reaction mixture on ice for 5 minutes.
Add 2 μL of your assembled plasmid to 50 μL of NEB10b cells and incubate on ice for 30 minutes.
Heat shock cells by incubating in a 42 °C water bath for 45 seconds, and return to ice for 5 minutes.
Add 950 μL of SOC outgrow medium and incubate 37°C, shaking at 250 rpm for 60 minutes.
Plate onto a LB/ampicillin/streptomycin plate and incubate at 37 °C overnight.
4.2.2. PCR:
Use the following primers to introduce cysteine and amber stop (TAG) mutations into αS.
-
4A.
Mutation Q62C
Forward: 5' – TGCTCCTCCAACATTTGTCACCCACTCTTTGGTCTTCTCAGCCAC – 3'
Reverse: 5' – GTGGCTGAGAAGACCAAAGAGTGGGTGACAAATGTTGGAGGAGCA – 3'
-
4B.
Mutation E114C
Forward: 5' – CCCCACAGGAAGGAATTCTGTGCGATATGCCTGTGGATCCTGA – 3'
Reverse: 5' – TCAGGATCCACAGGCATATCGCACAGAATTCCTTCCTGTGGGG – 3'
-
4C.
Mutation F94TAG
Forward: 5' – GCATTGCAGCAGCCACTGGCTAGGTCAAAAAGGACCAGTTGGG – 3'
Reverse: 5' – CCCAACTGGTCCTTTTTGACCTAGCCAGTGGCTGCTGCAATGC – 3'
Combine 1 μL of wild-type plasmid, 0.5 μL of forward primer (50 μM), 0.5 μL of reverse primer (50 μM), 0.5 μL Phusion Polymerase, 1 μL dNTP stock, 10 μL of polymerase reaction buffer, and 36.6 μL of water in a PCR tube on ice. The final volume should be 50 μL.
- Heat the PCR tube in a thermocycler using the method below:
Thermocycler program Step # Temperature Duration 1 98 °C 00:30 min. 2 98 °C 00:10 min. 3 Annealing Temp. 00:30 min. 4 72 °C 00:30 min./kb 5 Go to step 2 29 times 6 72 °C 10:00 min. 7 4 °C ∞ hold Add 1 μL of DPNI to the reaction buffer and incubate at 37 °C for 1 hour.
Incubate 5 μL of the PCR reaction mixture and 50 μL of brewed NEB10b cells on ice for 30 minutes
-
b.
Heat shock cells by incubating in a 42 °C water bath for 45 seconds, and return to ice for 5 minutes.
-
c.
Add 450 μL of SOC media and place in a 37 °C incubator for 1 hour, shaking at 250 rpm.
-
d.
Plate 300 μL onto an LB plate supplemented with 100 μg/mL ampicillin. Place in a 37 °C incubator overnight.
-
e.
Repeat for every desired mutation.
4.3. Expression of Cys Mutant αS for Mcm Labeling (αS-C62 and αS-C114).
4.3.1. Transformation of αS mutant plasmids (pTXB1_αS-C62_Mxe-H6 or pTXB1_αS-C114_Mxe-H6)
Add 2 μL of desired plasmid DNA to 50 μL of BL21-DE3 E. coli cells that were previously allowed to thaw on ice. Incubate on ice for 30 minutes.
Heat shock cells by incubating in a 42 °C water bath for 45 seconds, and return to ice for 5 minutes.
Add 450 μL of SOC media and place in a 37 °C incubator for 1 hour, shaking at 250 rpm.
Plate 300 μL onto an LB plate supplemented with 100 μg/mL ampicillin. Place in a 37 °C incubator overnight.
4.3.2. Primary culture
Pick a single colony from the LB/ampicillin plate using a sterilized toothpick and inoculate 5 mL of LB media supplemented with 5 μL of Ampicillin stock (100 μg/mL).
Incubate primary cultures at 37 °C, shaking at 250 rpm, for 4-5 hours. The cultures should look visibly cloudy, with an OD600 ≈ 0.6-0.7, measured on UV-Vis instrument.
Prepare 1 L of LB media, supplemented with 100 μg/mL ampicillin for secondary culture.
4.3.3. Inoculation and induction of secondary culture
Inoculate 1 L of LB media with the 5 mL primary culture. Place in a 37 °C incubator with shaking at 250 rpm.
Grow until OD600 = 0.6-1.0 as measured on UV-Vis instrument.
Add isopropyl-β-D-1-thiogalactopyranoside (IPTG) to final concentration of 1 mM (238.30 g/mol) and place in incubator at 18 °C overnight (16-18 hours).
4.4. Expression of Acd Mutant αS and Cys/Acd Mutant αS for Double Labeling (αS-δ94, αS-C62δ94, and αS-δ94C144).
αS mutant plasmids (pTXB1_αS-TAG94_Mxe-H6, pTXB1_αS-C62TAG94_Mxe-H6, pTXB1_αS-TAG94C114_Mxe-H6) are expressed using Uaa mutagenesis methods.[22] When incorporating a Uaa into a protein, it is required to transform in the synthetase machinery. For this study, a pDule2 plasmid containing the A9 Acd synthetase which has streptomycin resistance was simultaneously transformed into BL21 DE3 Gold cells in addition to the αS pTXB1 plasmid. The secondary culture must be inoculated into M9 minimal media to ensure the high Acd incorporation. We have found that smaller growths tend to yield more protein, i.e. a 1 L growth will yield roughly half the material of 2 x 500 mL growths.
4.4.1. Transformation
Follow the general procedure above, but add 2 μL each pTXB1 αS plasmid and pDule2 AcdRS plasmid to 50 μL BL21-DE3 E. coli cells. Incubate on ice for 30 minutes, then heat shock.
Recover cells and incubate with SOC media at 37 °C for 1 hour.
Plate onto a LB/ampicillin/streptomycin plate and incubate at 37 °C overnight.
4.4.2. Primary culture
Follow the previous procedure (4.3.2), but grow the cells in LB supplemented with 100 μg/mL of ampicillin and 50 μg/mL of streptomycin until OD600 ≈ 0.6-0.7 or until visibly cloudy.
Prepare M9 media for inoculation (see recipie in section 4.1.2)
4.4.3. Inoculation and induction of secondary culture
Add 2 mL primary culture to 500 mL M9 media, supplemented with 100 μg/mL ampicillin and 50 μg/mL streptomycin.
Incubate at 37 °C with shaking at 250 rpm until OD600 = 0.7-1.0, measured by UV-Vis.
Weigh Acd (282.30 g/mol; for 500 mL culture add 141 mg) for a final concentration of 1 mM in M9 media. Dissolve in 1-2 mL sterile MilliQ water with a few drops of 5 M NaOH to solubilize Acd. Add this solution to the secondary culture and incubate for 5 minutes.
-
4.
Add IPTG to a final concentration of 1 mM (238.30 g/mol; for 500 mL culture add 119 mg, or 500 μL from 1 M stock solution)
-
5.
Incubate at 18 °C overnight (16-18 hours).
4.5. Purification of αS Constructs.
After overnight culture growth, the following procedures are used to purify αS proteins, with or without Uaa incorporation.
4.5.1. Cell pelleting and lysis
Divide culture into 500 mL centrifuge bottles. Ensure that they are balanced. Centrifuge using Sorvall GS-3 rotor at 4,000 rpm for 20 minutes at 4 °C.
Resuspend cell pellets in 15 mL lysis buffer (40 mM Tris, 5 mM EDTA, pH 8.0) containing one Roche protease inhibitor cocktail pill.
Lyse resuspended cells by sonication (amplitude 32, 5 minutes total, 1 second on, 2 second off). Place vessel in ice/water bath to avoid overheating.
Pellet the cell debris by centrifugation (14,000 rpm, 20 minutes, 4 °C) in balanced ultracentrifuge tubes using Sorvall SS-34 rotor.
4.5.2. Ni-NTA purification and intein cleavage
Add nickel resin 50% slurry to a fritted column for a total bed volume of 3 mL.
Rinse nickel resin with 15 mL 50 mM HEPES buffer, pH 7.5.
Add cell supernatant to nickel resin and incubate at 4 °C for 1 hour while shaking.
Allow Ni-NTA supernatant to flow through.
Wash resin with 3 x 5 mL of buffer (50 mM HEPES, pH 7.5).
Wash resin with 2 x 10 mL of wash buffer (50 mM HEPES, 10 mM imidazole, pH 7.5)
Elute protein with 3 x 4 mL of elution buffer (50 mM HEPES, 300 mM imidazole, pH 7.5).
Add β-mercaptoethanol (BME) to 200 mM final concentration (14.2 M stock solution; 169 μL per 12 mL solution). Let BME react at room temperature for 20 hours on rotisserie.
Dialyze at 4 °C in 20 mM Tris, pH 8.0 for 7-10 hours.
Incubate with Ni-NTA resin on ice for 1 hour. Collect flowthrough, then dialyze overnight at 4 °C in 20 mM Tris pH 8.0 (Buffer A for FPLC).
4.5.3. FPLC purification for all αS constructs via ion exchange chromatography
If the construct has a cysteine, prior to FPLC purification, 10 μL of 0.5 M TCEP Bond Breaker™ should be added to reduce any aberrant disulfides formed between αS proteins and/or BME.
Prepare, sterile filter and degas FPLC buffers A (20 mM Tris, pH 8.0) and B (20 mM Tris, 1 M NaCl, pH 8.0)
Load 5 mL of the protein sample onto a HiTrap Q HP column (5 mL) on an ÄKTA FPLC using a 100 minute NaCl gradient (0 to 100 mM NaCl in 20 mM Tris, pH 8.0). The protein elutes at roughly 30% buffer B.
- Identify the fractions containing the product by MALDI MS
αS Construct: Calculated
MWWT 14,460 Da C62 14,435 Da C114 14,434 Da C62δ94 15,572 Da δ94CMcm114 14,571 Da Any proteins that do not need to be further labeled (such as αS-δ94) can be dialyzed at 4 °C against αS buffer (20 mM Tris, 100 mM NaCl, pH 7.5) overnight and stored at −80 °C in 1.5 mL aliquots and thawed for fluorescence experiments. Concentrate the dialyzed solution by centrifugation with a 3 kDa cutoff filter. Monitor the concentration of Acd labeled αS by UV-Vis until the stock solution is greater than 20 μM. Concentrations can be determined by UV-Vis using the extinction coefficient of Acd (Ɛ386 = 5,700 M−1 cm−1).
Cys containing mutants should be labeled directly after FPLC purification.
4.6. Cys αS labelling with Mcm-Mal
After FPLC purification, add 20 μL Bond Breaker solution to the protein solution (10-20 mL) and stir at room temperature on a rotisserie for 10-20 minutes.
Add 200–300 μL of 25 mM Mcm-Mal in DMSO to the protein solution and incubate at room temperature on the rotisserie.
Monitor labeling progress by MALDI MS. The labeling should be complete in 1-3 hours.
After labelling with Mcm-Mal, dialyze the solution at 4 °C against Tris buffer (20 mM, pH 8.0) overnight.
Add 20 uL Bond Breaker solution to the protein solution and mix on rotisserie for 10-20 minutes.
Purify the protein on the FPLC using the gradient from 4.5.3. This will remove excess dye still be present after dialysis.
Identify protein fractions using MALDI MS (see table in 4.5.3 for desired masses)
Dialyze the protein solution against 20 mM Tris, 100 mM NaCl, pH 7.5 buffer overnight.
Concentrate the dialyzed solution by centrifugation with a 3 kDa cutoff filter. Monitor the concentration of Mcm labeled αS by UV-Vis until it is greater than 20 μM (Mcm extinction coefficient: Ɛ325 = 12,000 M−1 cm−1).
Store concentrated samples at −80 °C in 1.5 mL aliquots and thaw once for fluorescence experiments.
5. Preparation of Calmodulin (CaM) Constructs for Intramolecular and Intermolecular FRET.
In general, CaM constructs can be prepared using the same methods and equipment described for the αS constructs above. Differences are highlighted below.
5.1. Reagents, equipment, and general procedures.
Use the reagents listed in 4.1, for the expression of CaM constructs. CaM expression also requires a lyophilizer.
5.2. Cloning of CaM Constructs.
Follow the procedure outlined in section 4.2 for insertion and cloning of CaM mutants into the pTXB1 vector. For Cys and Acd mutations, use the primer sequences below.
-
5A.
Phe12 to Cys
Forward: 5’-GAGCAGATTGCAGAATGCAAAGAAGCTTTTTCACTA-3’
Reverse: 5’-TAGTGAAAAAGVTTVTTTGCATTCTGCAATCTGCTC-3’
-
5B.
Leu112 to TAG
Forward: 5’-GCAGAACTTCGTCATGAGATGACAAATTAGGGGGAGAAGCTAACA-3’
Reverse: 3’-TGTTAGCTTCTCCCCCTAATTTGTCATCACATGACGAAGTTCTGC-3’
5.3. Expression of Cys Mutant CaM for Mcm Labeling (CaM-C12-GyrA-H6).
-
5.3.1
Using the same procedure as the 4.3.1, transform the plasmid encoding CaM F12C-GyrA into BL21-Gold (DE3) E. Coli cells and grow against ampicillin (Amp, 100 μg/mL) on an LB-agar plate.
-
5.3.2
Using the same protocol at 4.3.2 for primary culture
-
5.3.3
Using the same protocol as 4.4.3 for inoculation of secondary culture and IPTG induction
5.4. Expression of Acd Mutant CaM and Cys/Acd Mutant CaM for Double Labeling (CaM-δ112 and CaM-C12δ112)
-
5.4.1
Use the same transformation procedure as 4.4.1 for plasmids encoding CaM-δ112-GyrA-H6,CaM-C12δ112-GyrA-H6. Each transformation should also contain a plasmid encoding for an orthogonal Acd synthetase 2b (AcdRS2b, also referred to as clone A9) and tRNACUA on (Strep, 100 μg/mL) and Amp (100 μg/mL) LB-agar plates.
-
5.4.2
Follow the procedure 4.4.2 for primary cultures
-
5.4.3
follow procedure 4.4.3 for inoculation and induction.
5.5. Purification of CaM Constructs.
Cells should be harvested using the same centrifugation, nickel column purification, and BME cleavage procedures outlined in section 4.5.
After the resulting cleavage solution is dialyzed against 20 mM Tris pH 8.0 (2L) overnight, add 10 μL of 0.5 M TCEP Bond Breaker™ was added to the CaM-C12 and CaM-C12δ112 constructs to reduce any aberrant disulfides formed between CaM proteins and/or βME.
Purify each CaM construct over a HiTrap Q column using a 120 min NaCl gradient (0.1 M to 0.8 M NaCl in 20 mM Tris, pH 8.0).
- Confirm fractions containing the product peak by MALDI and dialyze twice against water at 4 °C (2 L, 2 hours each).
CaM Contruct: Calculated
MWWT 16,838 Da C12 16,665 Da δ112 16,881 Da CMcm12δ112 16,838 Da Flash freeze samples containing the protein in liquid N2 and lyophilize to a powder.
5.6. Cys CaM Labelling with Mcm-Mal
Make a stock solution of 25 mM Mcm-Mal by dissolving 4.6 mg Mcm-Mal in 645 μL DMSO.
Dissolve CaM-C12 and/or CaM-C12δ112 in 1 mL of 20 mM Tris, pH 7.5 buffer.
To each sample add 10 equivalents of Mcm-Mal solution (based off UV-Vis quantification of protein solutions)
Incubate samples at 37 °C with shaking at 200 rpm for 3 hours.
Monitor labeling progress with MALDI MS. Efficient labeling will show a +285 Da increase in mass.
Dilute each sample in 2 mL of 20 mM Tris, pH 8.0 buffer and perform a second round of FPLC purification using the gradient described in section 5.5.
Analyze fractions by MALDI MS. Pool and dialyze fractions containing the desired mass for Cys-Mcm conjugated product against 2 x 2 L water before flash freezing in liquid nitrogen and lyophilizing to a powder for storage at 4 °C.
6. Fluorescence Experiments and FRET Calculations
6.1. Reagents, equipment, and general procedures.
In addition to proteins expressed, labeled, and purified as described above, the following experiments need several spectrometers. UV-Vis absorption spectra were acquired on a Hewlett-Packard (HP) 8452A diode array spectrophotometer. Fluorescence spectra were collected with a Tecan M1000 plate reader or a Photon Technologies International (PTI) QuantaMaster40 fluorometer. Fluorescence lifetime measurements were made on the PTI Quantamaster40 using pulsed LED light sources. Stopped flow fluorescence measurements were made on a KinTek AutoSF-120 stopped flow instrument. Since most laboratories will not have exactly these instruments, we provide general procedures for analyzing data that can be adapted for data collected with a given instrument.
FRET Reagents and Equipment
7-Methoxycoumarin-4-yl acetic acid (Mcm-Ac) (MilliporeSigma #235199)
HP 8452A diode array spectrophotometer (currently Agilent Technologies)
Tecan M1000 plate reader
PTI QuantaMaster40 fluorometer (currently Horiba Scientific)
KinTek AutoSF-120 stopped flow instrument
6.1. Absorption and emission spectra of FRET constructs and singly-labeled control constructs.
Some desired equilibrium information such as a Kd or EC50 can be determined simply based on a concentration-dependent change in FRET (measured by changes in donor or acceptor emission) without rigorous determination of . For determining interprobe distances, and calculations are required. These calculations are often performed using generic measurements of the spectral properties of the chromophores, for example from a solution of an Mcm-Cys small molecule and a solution of free Acd amino acid. However, such an approach misses important photophysical effects that can dramatically impact the value, like changes in spectral shape or changes in donor quantum yield that depend on the chromophore’s local environment. Therefore, we acquire not only the emission spectrum of the Mcm/Acd FRET construct, , but also the following spectra:
-
6A.
Absorption spectrum of acceptor only control construct,
-
6B.
Emission spectrum of donor-only control construct (excited at donor ),
-
6C.
Emission spectrum of acceptor-only control construct (excited at donor ),
-
6D.
Emission spectrum of a solution of Mcm-Ac (excited at ),
where is the excitation wavelength of the donor (Mcm). Note that should not necessarily be the maximum in the donor absorption spectrum, but the wavelength that provides the highest ratio of donor excitation to direct acceptor excitation (measured in the emission spectrum of the acceptor-only control). The Mcm excitation maximum coincides with a minimum in the Acd excitation spectrum, which is an ideal scenario. It is also beneficial that Mcm and Acd have comparable extinction coefficients and that their spectral shapes are relatively insensitive to environment. Thus, does not vary dramatically during the course of a conformational change. One aspect of the Mcm/Acd pair that is not ideal is that the Acd extinction coefficient decreases by roughly 3-fold on going from an aqueous to hydrophobic environment, which will impact distance calculations.
To determine , the molar extinction coefficient of the acceptor at each wavelength , one must normalize the absorption spectrum of the acceptor-only construct, , to the value at ~386 nm (slight shifts in the absorption maximum may occur), and multiply by the molar extinction coefficient of Acd (5,700 M−1cm−1 at 386 nm). To further correct for any change to the Acd extinction coefficient with environment, one must measure the concentration of the acceptor-only construct in an independent assay such as a BCA assay and compare the absorption to that of an equimolar concentration of free Acd.
To determine the donor (Mcm) quantum yield (ΦD), one must integrate the emission spectrum of the donor-only construct, , and compare this to the integrated emission spectrum of an equimolar solution of an Mcm-Ac in the same buffer, . The ratio of the integrated and spectra is then multiplied by the quantum yield of the standard (0.18 for Mcm-Ac) to obtain the quantum yield of Mcm in the protein (ΦD).[23]
6.2. Calculation of .
Converting an measurement into an interprobe distance requires calculation of . depends on the properties of the chromophores and their locations within the protein and is given by Equation (1)
| (1) |
where is a geometrical factor that relates the orientation of the donor (Mcm) and acceptor (Acd) electronic transition moments, ΦD is the quantum yield of the donor, n is the index of refraction of the solvent, NA is Avogadro’s number, and J is the spectral overlap integral defined in units of M−1•cm−1•nm4. J is formally defined as
| (2) |
where is the molar extinction coefficient of the acceptor at each wavelength (determined in section 6.1) and is the normalized donor-only emission spectrum given by
| (3) |
where is the fluorescence intensity of the donor at each wavelength (determined in section 6.1). Substituting these results into Equation (1), as well as the donor (Mcm) quantum yield (determined in section 6.1), 1.33 for the index of refraction of water, and 2/3 for , gives the Förster distance, . These values can then be used to convert values to interprobe distances (R) using Equation (4).
| (4) |
can be determined either from steady state fluorescence spectra or from fluorescence lifetime measurements.
6.3. Steady state EFRET measurements.
Our approach for determining uses spectral fitting to minimize error due to small variations in emission maxima and corrects for artifacts due to direct excitation of the Acd acceptor. Even for the nearly ideal Mcm/Acd FRET pair, some small amount of direct Acd excitation will still occur. This can be corrected for by measuring the emission of an Acd-only construct with excitation at 325 nm. Fitting of spectra containing Mcm/Acd double-labelled protein can be performed by minimizing the square difference between the spectra from the double-labelled protein and a weighted sum of single-labelled spectra:
| (5) |
Here, , , and represent the fluorescence intensity at a given wavelength from protein which was double-labelled, single-labelled with only the FRET donor (Mcm), and single-labelled with only the FRET acceptor (Acd), respectively. A and B are wavelength invariant weights. This can be done simply using the Solver function in Microsoft Excel to vary the values of A and B to minimize the sum of this square difference across all emission wavelengths. The resulting, fitted equation would be
| (6) |
can then be directly obtained from the linear weight of donor spectrum, A, where
| (7) |
Shown below are example spectra for Mcm-only, Acd-only, and Mcm/Acd labeled αS. As one can see, simply measuring Mcm emission in the doubly labeled construct and comparing it to the Mcm-only construct can be subject to artifacts
6.4. Fluorescence lifetime EFRET measurements.
Fluorescence lifetime measurements allow one to determine under conditions where it is difficult to match the concentrations of doubly labelled and donor-only constructs, such as during protein aggregation. For lifetime measurements simply requires measurements of donor lifetimes for the doubly labeled and donor-only constructs. In our studies, lifetime measurements were made via TCSPC using a 340 nm LED light source. TCSPC data were fit to single or bi-exponential decays using PowerFit-10. was calculated as follows
| (8) |
where is the lifetime of the Mcm/Acd construct and is the lifetime of the Mcm construct. If the constructs occupy multiple conformational states, one can use the ability to fit lifetime data to a bi-exponential decay to analyze for one of the states. For example, if the Mcm/Acd bi-exponential fit indicates lifetimes and , where is identical to , one can determine for alone, and convert that to a distance R for that conformational subpopulation using Equation (4).
6.5. Stopped flow experiments.
Stopped-flow experiments can be performed to determine rapid association or dissociation kinetics, monitoring at two emission wavelengths to explicitly monitor both donor and acceptor emission for two color FRET or to measure the change in two different donor emissions for three color FRET. Choosing appropriate protein concentrations for these experiments must balance considerations of chromophore brightness with some prior knowledge of the equilibrium association constants. For example, the CaM experiments in Ferrie et al. were performed with 1 μM Mcm/Acd labelled CaM and 2 μM pOCNC peptide, concentrations expected to lead to complete binding with rates measurable on the millisecond timescale.[5] Typically, each data set is measured in triplicate or greater and data are averaged to achieve reasonable signal-to-noise ratios. For each mixing event, the fluorescence emission of Mcm (390 ± 25 nm) and Acd (440 ± 40 nm) are measured with excitation of Trp at 295 nm or Mcm at 325 nm, collecting data over a sufficient time range for the reaction to achieve equilibrium. Nonlinear fits to the appropriate kinetic expressions can then be performed using a program such as Prism (Graphpad Software).
7. Conclusions
In this chapter, we have provided detailed experimental procedures for the synthesis of Mcm-Mal and Acd, which enable the use of a combination of Cys labeling by Mcm-Mal and Acd genetic incorporation to easily introduce a FRET pair that is selectively excitable and minimally perturbing in proteins. Mcm/Acd FRET pairs can be used to monitor conformational changes in the 15-40 Å range, a useful scale for tracking motions within protein domains or among domains of moderately sized proteins. The given step-wise protocols for the preparation of CaM and αS constructs to study intramolecular and/or intermolecular FRET can be readily adapted to the preparation and labelling of other proteins with a broad range of applications. Likewise, we have provided descriptions of fluorescence data analysis that should allow the reader to study binding and conformational changes to either extract equilibrium constants and kinetic parameters or to rigorously extract interchromophore distance information. To do so, the spectra of Mcm- or Acd-only proteins must be used to correct for changes in quantum yield and/or spectral shape to accurately determine and . Additionally, Trp FRET with both Mcm and Acd can be used in combination with Mcm/Acd FRET to monitor the thermodynamics or kinetics of two processes simultaneously. As we note, attempts to extract distance information in the three chromophore system are not advised since the number of control experiments would be impractical compared to simply performing two standard FRET experiments. Importantly, although we use a simple example with a peptide that has a single Trp residue, the three-way FRET protocol can be implemented with any tryptophan containing protein, including those with multiple Trp residues. The Trp/Mcm/Acd trio balances small probe size with selective excitation, and we are currently working on the development of complementary red-shifted minimalist fluorescence probes to further expand the scope of experiments that are possible using simple labeling schemes similar to those presented here.[24]
Figure 2.
FRET Experiment Schemes. Two Color FRET: αS is treated with osmolyte TMAO, causing the protein to compact and increasing EFRET between Mcm and Acd. EFRET can be rigorously converted to an interchromophore distance. Three Color FRET: Mcm/Acd-labeled CaM binds the WpOCNC peptide, resulting in FRET from Trp in WpOCNC to Mcm in CaM. Trp will also undergo FRET with Acd, so FRET analysis should be restricted to binding affinity and kinetic parameter determination.
Figure 3:
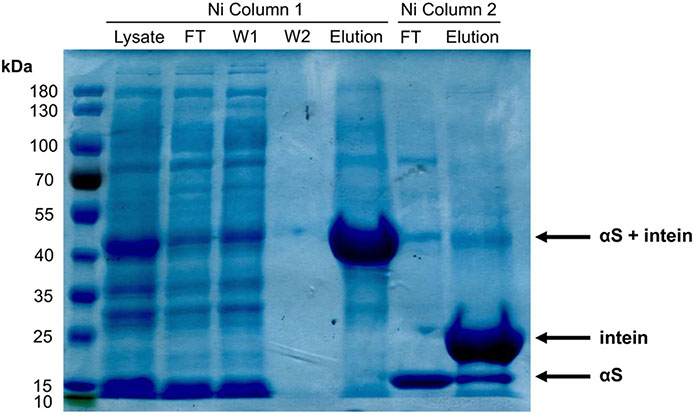
Protein Purification and Cleavage of Intein Handle. αS-δ94C114 nickel purification visualized by a Coomassie stained 10% SDS-Page gel. Lane 6 contains14 kDa synuclein, showing successful cleavage from the C-terminal MxeGyrA intein using β-mercaptoethanol.
Figure 4:
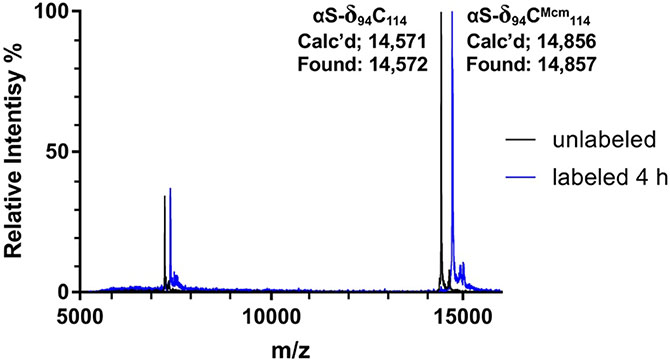
Monitoring Cys Modification by Mcm-Mal for Protein Double Labeling. MALDI-TOF spectra, monitoring of Mcm-Mal labeling of αS-δ94C114. The calculated protein mass for the unlabeled protein (αS-δ94C114) was 14,571 Da and was measured to be 14,572 Da. After 4 hours, more than 95% of the protein was labeled, signified by a +285 Da mass shift (αS-δ94CMcm114).
Figure 6.
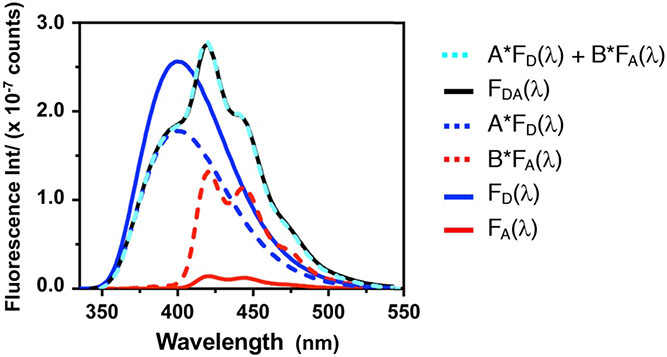
Fitting Double Labeled Protein Emission Spectra to Determine . Fluorescence emission spectra (325 nm excitation) of 1 μM concentrations of αS-CMcm62δ94 (double labeled, FDA) and the corresponding singly-labelled αS-CMcm62 (donor only, FD) and αS-δ94 (acceptor only, FA) constructs. Deconvolution of the doubly labelled spectrum by fitting to a weighted sum of the singly-labelled spectrum performed using Equation (5).
References
- [1].Giepmans BNG, Adams SR, Ellisman MH, Tsien RY, The Fluorescent Toolbox for Assessing Protein Location and Function, Science 312(5771) (2006) 217–224. [DOI] [PubMed] [Google Scholar]
- [2].Lakowicz JR, Principles of Fluorescence Spectroscopy, 3 ed., Springer US; 2006. [Google Scholar]
- [3].Haney CM, Wissner RF, Petersson EJ, Multiply Labeling Proteins for Studies of Folding and Stability, Current Opinion in Chemical Biology 28 (2015) 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Speight LC, Samanta M, Petersson EJ, Minimalist Approaches to Protein Labelling: Getting the Most Fluorescent Bang for Your Steric Buck, Australian Journal of Chemistry 67(5) (2014) 686–700. [Google Scholar]
- [5].Ferrie JJ, Ieda N, Haney CM, Walters CR, Sungwienwong I, Yoon J, Petersson EJ, Multicolor Protein Fret with Tryptophan, Selective Coumarin-Cysteine Labeling, and Genetic Acridonylalanine Encoding, Chemical Communications 53(80) (2017) 11072–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tanaka T, Wagner AM, Warner JB, Wang YJ, Petersson EJ, Expressed Protein Ligation at Methionine: N-Terminal Attachment of Homocysteine, Ligation, and Masking, Angewandte Chemie International Edition 52(24) (2013) 6210–6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Speight LC, Muthusamy AK, Goldberg JM, Warner JB, Wissner RF, Willi TS, Woodman BF, Mehl RA, Petersson EJ, Efficient Synthesis and in Vivo Incorporation of Acridon-2-Ylalanine, a Fluorescent Amino Acid for Lifetime and Förster Resonance Energy Transfer/Luminescence Resonance Energy Transfer Studies, Journal of the American Chemical Society 135(50) (2013) 18806–18814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hostetler ZM, Ferrie JJ, Bornstein MR, Sungwienwong I, Petersson EJ, Kohli RM, Systematic Evaluation of Soluble Protein Expression Using a Fluorescent Unnatural Amino Acid Reveals No Reliable Predictors of Tolerability, ACS Chemical Biology 13(10) (2018) 2855–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Batjargal S, Walters CR, Petersson EJ, Inteins as Traceless Purification Tags for Unnatural Amino Acid Proteins, Journal of the American Chemical Society 137(5) (2015) 1734–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chin D, Means AR, Calmodulin: A Prototypical Calcium Sensor, Trends Cell Biol 10(8) (2000) 322–328. [DOI] [PubMed] [Google Scholar]
- [11].Vetter SW, Leclerc E, Novel Aspects of Calmodulin Target Recognition and Activation, Eur J Biochem 270(3) (2003) 404–414. [DOI] [PubMed] [Google Scholar]
- [12].Lashuel HA, Overk CR, Oueslati A, Masliah E, The Many Faces of Alpha-Synuclein: From Structure and Toxicity to Therapeutic Target, Nature Reviews Neuroscience 14(1) (2013) 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Jakes R, Goedert M, Alpha-Synuclein in Lewy Bodies, Nature 388(6645) (1997) 839–840. [DOI] [PubMed] [Google Scholar]
- [14].Contessa GM, Orsale M, Melino S, Torre V, Paci M, Desideri A, Cicero DO, Structure of Calmodulin Complexed with an Olfactory Cng Channelfragment and Role of the Central Linker: Residual Dipolar Couplingsto Evaluate Calmodulin Binding Modes Outside the Kinase Family, Journal of Biomolecular NMR 31(3) (2005) 185–199. [DOI] [PubMed] [Google Scholar]
- [15].Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Mei Ono A, Güntert P, Optimal Isotope Labelling for Nmr Protein Structure Determinations, Nature 440(7080) (2006) 52–57. [DOI] [PubMed] [Google Scholar]
- [16].Haney CM, Wissner RF, Warner JB, Wang YXJ, Ferrie JJ, Covell DJ, Karpowicz RJ, Lee VMY, Petersson EJ, Comparison of Strategies for Non-Perturbing Labeling of Alpha-Synuclein to Study Amyloidogenesis, Organic & Biomolecular Chemistry 14(5) (2016) 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Haney CM, Cleveland CL, Wissner RF, Owei L, Robustelli J, Daniels MJ, Canyurt M, Rodriguez P, Ischiropoulos H, Baumgart T, Petersson EJ, Site-Specific Fluorescence Polarization for Studying the Disaggregation of A-Synuclein Fibrils by Small Molecules, Biochemistry 56(5) (2017) 683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu MY, Chen TY, Ahamed B, Li J, Yau KW, Calcium-Calmodulin Modulation of the Olfactory Cyclic Nucleotide-Gated Cation Channel, Science 266(5189) (1994) 1348–1354. [DOI] [PubMed] [Google Scholar]
- [19].Ferrie JJ, Haney CM, Yoon J, Pan B, Lin Y-C, Fakhraai Z, Rhoades E, Nath A, Petersson EJ, Using a Fret Library with Multiple Probe Pairs To drive Monte Carlo Simulations of A-Synuclein, Biophysical Journal 114(1) (2018) 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Daniels MJ, Nourse JB, Kim H, Sainati V, Schiavina M, Murrali MG, Pan B, Ferrie JJ, Haney CM, Moons R, Gould NS, Natalello A, Grandori R, Sobott F, Petersson EJ, Rhoades E, Pierattelli R, Felli I, Uversky VN, Caldwell KA, Caldwell GA, Krol ES, Ischiropoulos H, Cyclized Ndga Modifies Dynamic A-Synuclein Monomers Preventing Aggregation and Toxicity, Scientific Reports 9(1) (2019) 2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sungwienwong I, Hostetler ZM, Blizzard RJ, Porter JJ, Driggers CM, Mbengi LZ, Villegas JA, Speight LC, Saven JG, Perona JJ, Kohli RM, Mehl RA, Petersson EJ, Improving Target Amino Acid Selectivity in a Permissive Aminoacyl Trna Synthetase through Counter-Selection, Organic & Biomolecular Chemistry 15(17) (2017) 3603–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dumas A, Lercher L, Spicer CD, Davis BG, Designing Logical Codon Reassignment - Expanding the Chemistry in Biology, Chemical Science 6(1) (2015) 50–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brun MP, Bischoff L, Garbay C, A Very Short Route to Enantiomerically Pure Coumarin-Bearing Fluorescent Amino Acids, Angewandte Chemie-International Edition 43(26) (2004) 3432–3436. [DOI] [PubMed] [Google Scholar]
- [24].Sungwienwong I, Ferrie JJ, Jun JV, Liu C, Barrett TM, Hostetler ZM, Ieda N, Hendricks A, Muthusamy AK, Kohli RM, Chenoweth DM, Petersson GA, Petersson EJ, Improving the Fluorescent Probe Acridonylalanine through a Combination of Theory and Experiment, Journal of Physical Organic Chemistry 31(8) (2018) e3813. [DOI] [PMC free article] [PubMed] [Google Scholar]