Abstract
Complementation of an Escherichia coli cya mutant with a genomic library from Aeromonas hydrophila allowed isolation of clones containing two different cya genes. Whereas one of these genes (cyaA) coded for an adenylyl cyclase (AC1) belonging to the previously described class I adenylyl cyclases (ACs), the second one (cyaB) coded for a protein (AC2) that did not match any previously characterized protein when compared to protein sequence databases. In particular, it did not align with any of members of the three known classes of ACs. The purified AC2 enzyme exhibited remarkable biochemical characteristics, namely, an optimum activity at a high temperature (65°C) and at an alkalinic pH (9.5). In order to investigate the functions of both cyclases in A. hydrophila, each gene was inactivated in the chromosome and the resulting mutant strains were examined for physiological alterations. It was shown that, in contrast to cyaA, the cyaB gene was not expressed under usual laboratory growth conditions. However, introduction of a plasmid harboring the cyaB gene in a cyaA mutant, as well as in a cyaA cyaB mutant, allowed cyclic AMP production. AC2 is the first member of a new class of previously unrecognized ACs, and to date, no functional counterpart has been demonstrated in other organisms. However, scanning databases revealed a significant similarity between AC2 and the gene product of three hyperthermophilic archaebacteria: Methanobacterium thermoautotrophicum, Archaeglobus fulgidus, and Methanococcus jannaschii. The possibility of a gene transfer between such phylogenetically divergent bacteria is discussed.
Adenylyl cyclase (AC) activity exists in a broad range of procaryotic to eucaryotic species in which cyclic AMP (cAMP) controls many major cellular functions. Comparative analysis of cya gene sequences from a variety of organisms led us to classify AC enzymes according to the features they commonly shared. Three well-defined classes have thus emerged, raising an interesting question about evolutionary convergence or divergence of cAMP-synthesizing enzymes (10). The three types of ACs are: (i) the enterobacterial class, or class I; (ii) the toxic class, or class II, including the Bacillus anthracis and Bordetella pertussis calmodulin-activated toxins; and (iii) the universal class, or class III, comprising ACs and guanylyl cyclases from a variety of eucaryotic and procaryotic organisms.
The characterization of class I ACs resulted from Escherichia coli AC description and a comparative study of the cya locus in six other enterobacteria and three bacteria from families related to enterobacteria. Among the latter is the bacterium Aeromonas hydrophila. This facultatively anaerobic, gram-negative rod lives in fresh and estuarine water and belongs to the enteric phylogenetic subgroup. It is a primary and/or opportunistic pathogen of a variety of aquatic and terrestrial animals, including humans (the clinical manifestations of infection range from gastroenteritis and wound infections to septicemia and meningitis) (11). Presence of cAMP and its intracellular receptor, the catabolic activator protein (CAP), has been demonstrated in A. hydrophila, suggesting the presence of AC activity (4). Indeed, an AC gene (cyaA) coding for an 843-amino-acid AC (AC1), 73% similar to that of E. coli and belonging to the enterobacterial class I, was cloned from A. hydrophila (26). The surprise, reported here, came from the identification of another AC, AC2, coded by a second gene that we called accordingly cyaB.
We present here the characterization of this small and unexpected enzyme, which exhibits an unusual alkalinophilic and thermophilic activity. As a most prominent feature, this AC cannot enter the previous AC classification. Thus, this protein can be considered the first member of a fourth class of ACs. Its predicted polypeptide sequence is not similar to any protein exhibiting a currently known function. Publication of the genome sequence of the archaeon Methanococcus jannaschii (8) and more recently of Methanobacterium thermoautotrophicum (24) and Archaeoglobus fulgidus (12) revealed the presence of genes of unknown function that code for homologous proteins displaying significant similarity with AC2. However, expression of the M. jannaschii protein in E. coli did not restore cAMP synthesis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli S17.1 | recA thi pro hsdR chromosomal RP4-2Tn1::ISR1, Tc::Mu, Km::Tn7 | 23 |
| E. coli TG1 | lac proAB thi supE hsdD5 F′ traD36 proAB+ lacIqlacZΔM15 | Stratagene (200123) |
| E. coli TP610 | F−thi-1 thr-1 leuB6 pro lacY1 tonA21 supE44 hsdR hsdM recBC lop11 lig+ cya-610 | L. Hedegaard |
| A. hydrophila 218 | M. Y. Popoff | |
| A. hydrophila 543 | M. Y. Popoff | |
| Plasmids | ||
| pLA2917 | IncP, Tcr Kmr | L. N. Allen |
| pSUP202 | Tcr Cmr Apr | 23 |
| pTrc99A | Apr | Pharmacia (27-0971-01) |
| pUC4K | With Km cartridge (Kmr) | Pharmacia (27-4987-01) |
| pHP45Ω | With streptomycin cartridge (Smr) | R. Fellay |
| pDIA6589 | 2.9-kb Sau3A fragment of A. hydrophila DNA containing cyaA into BamHI site of pBR322 | This study |
| pDIA6526 | 1.2-kb Sau3A fragment of A. hydrophila DNA containing cyaB into BamHI site of pBR322 | This study |
| pDIA6535 | AflIII-BamHI-amplified PCR fragment of the cyaB CDSa into pTrc99A cut with NcoI-BamHI | This study |
| pDIA8516 | cyaA disrupted by Smr cartridge subcloned into pSUP202 | This study |
| pDIA8507 | cyaB disrupted by Kmr cartridge subcloned into pSUP202 | This study |
| pDIA6539 | cyaB (PCR product) into BglII site of pLA2917 | This study |
| pDIA6540 | cyaB gene disrupted by Kmr cartridge into BglII site of pLA2917 | This study |
| AMJET17 | BamHI-EcoRI fragment of M. jannaschii chromosomal DNA with the MJ0240 CDS in pUC18 | 8 |
| pDIA8514 | PstI-amplified PCR fragment of AMJET17 cloned into PstI site of pBR322 | This study |
| pDIA8515 | BspHI-PstI-amplified PCR fragment of AMJET17 into pTrc99A digested with NcoI-PstI | This study |
CDS, coding sequence.
Media and growth conditions.
Cultures were grown aerobically at 30 or 37°C in Luria-Bertani (LB) medium, 2YT medium, or minimal medium M63 (15) supplemented either with glucose (0.4%), glycerol (0.4%), or succinate (0.2%) as a carbon source. When required, cAMP and/or isopropyl-β-d-thiogalactopyranoside (IPTG) were added to a final concentration of 0.5 and 1 mM, respectively. Antibiotics were added at final concentrations (in micrograms per milliliter) as follows: ampicillin (Ap), 50; chloramphenicol (Cm), 40; kanamycin (Km), 200 for A. hydrophila and 25 for E. coli; tetracycline (Tc), 10; and streptomycin (Sm), 100. Solid media contained 1.5% Difco agar. Screening for the ability to ferment maltose was performed on MacConkey agar indicator plates containing 1% maltose, allowing visual observation of sugar fermentation.
Carbon source utilization tests were performed on Biotype 100 strips from BioMérieux as described by the supplier (reference no. 88818).
Swarm plates containing 0.5% Casamino Acids in M63 medium and 0.3% agar were used to assess the motility of A. hydrophila strains. Approximately 105 exponentially growing cells were inoculated onto the plates and incubated at 30°C for 17 h; the diameter of the swarming colony was then measured in millimeters.
Genetic techniques.
Transformation of E. coli was performed by electroporation or by the CaCl2 method (14).
Bacterial matings were performed as follows. The donor was an overnight culture of E. coli S17.1 strain containing derivatives of plasmid pLA2917 or pSUP202. The recipient was an overnight culture of A. hydrophila 218 grown in LB medium. Both cultures were centrifuged, washed three times, and resuspended in LB medium. Donor (0.1 ml) and recipient (0.1 ml) suspensions were spread onto LB plates and incubated at 37°C for 18 to 20 h. The mating mixture was harvested, washed with minimal medium, and then plated onto minimal-medium plates supplemented with glucose as the carbon source. Antibiotics (Km, 200 μg/ml; Sm, 100 μg/ml) were used to allow the positive selection of A. hydrophila exconjugants. After each cross, eight colonies of exconjugants were isolated and checked by DNA hybridization for the expected location of the cartridge insertions in cyaA and cyaB chromosomal genes.
DNA manipulations and sequencing.
Large-scale DNA isolation was carried out using the plasmid Maxi-Kit (Qiagen, Inc.) as recommended by the manufacturer. Small-scale DNA preparation was performed according to the method of Birnboim and Doly (3). Endonuclease digestions, alkaline phosphatase treatments, ligations, Southern blotting, and hybridization methods were performed as described by Maniatis et al. (14).
An M. jannaschii DNA fragment coding for a protein homologous to AC2 (MJ0240), AMJET17, was isolated from a genome library provided by the TIGR/ATCC Microbial genome special collection (8).
DNA sequencing was performed by the dideoxy chain method of Sanger et al. (19) with the Pharmacia sequencing kit and [α-35S]thio-dATP. Universal primers or specific synthetic oligonucleotides were employed in this procedure.
Sequence comparisons were performed by using BLAST programs (1).
Cloning of the cyaA and cyaB genes.
A Sau3A partial digestion was performed on A. hydrophila genomic DNA. The fragments ranging from 1 to 6 kb were ligated into plasmid pBR322 cut with the enzyme BamHI. The E. coli TP610, defective in AC, was transformed and screened for its ability to metabolize maltose on MacConkey agar plates. Wild-type E. coli is able to ferment maltose by expressing the maltose regulon, which requires cAMP for its expression (27). The cya mutant TP610 cannot activate expression of the maltose regulon and thus is not able to metabolize maltose. Mal+ and Mal− phenotypes are easily distinguishable on MacConkey maltose agar plates that include a pH indicator. Mal+ cells form red colonies because of acid production during fermentation, whereas Mal− cells remain white. Upon introduction of an A. hydrophila library into TP610, most transformants formed white colonies on MacConkey maltose agar plates containing ampicillin. However, a few red colonies were found. Two plasmids, pDIA6589 and pDIA6526, were isolated from red transformants and analyzed.
Production and purification of the AC2 protein.
In order to overproduce and purify the AC2 enzyme (which was shown to be cytoplasmic), the cyaB coding sequence was amplified by PCR with two oligonucleotides, 5′ GCCTGCACATGTCATCACAACAC 3′ and 5′ AGAGCAGTTGCGGATCCTGGA 3′. The PCR product, digested with AflIII and BamHI, was then cloned under the control of the inductible trc promoter in the pTrc99A expression vector and digested by NcoI-BamHI, yielding plasmid pDIA6535.
A 1-liter culture of E. coli TP610 containing pDIA6535 was grown in 2YT medium supplemented with Ap (100 μg/ml) and Cm (30 μg/ml). When the optical density at 600 nm reached 0.5, IPTG (1 mM) was added to the medium in order to induce AC2 expression. At 3 h after induction, bacteria were harvested by centrifugation. The bacterial pellet, resuspended in 50 ml of Tris-HCl buffer (50 mM, pH 7.4) supplemented with 1 mM dithiothreitol (DTT), was sonicated and centrifuged for 15 min at 12,000 × g. The supernatant was then kept frozen for further purification and AC assays.
The three-step purification procedure consists of chromatography on Blue-Sepharose, DEAE-Sephacel, and Ultrogel ACA54. Then 250-mg portions of crude bacterial extracts were first applied on a Blue-Sepharose column equilibrated with 50 mM Tris-acetate buffer (pH 6). Most of the AC activity was found in the column flowthrough, which contained 10% of the total applied proteins. This fraction, which contained AC2, was applied to a DEAE-Sephacel column equilibrated and washed with the Tris-acetate buffer. AC2 was recovered after elution with 0.2 M NaCl added to the buffer. The AC2-containing fraction was then applied on a ACA54 gel filtration column and equilibrated with Tris-acetate buffer for further purification. All the washing, equilibration, and elution steps were performed in the presence of 1 mM DTT at 4°C. The partially purified protein was aliquoted and kept in 50 mM Tris-acetate buffer supplemented with DTT at −20°C. The protein concentration was determined according to the method of Bradford (7) with a Bio-Rad protein assay kit (no. 500-0006). This three-step procedure allowed a 10-fold purification of AC2. The protein was shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to be more than 95% pure (data not shown). The identity of the purified protein was proven by microsequencing its N-terminal peptide, SSQHF, which was found to correspond to the cyaB nucleotide sequence.
In vitro assays of AC and cAMP.
AC was assayed as described by Ladant et al. (13). After tests under different conditions were performed, the assays were done at 30°C in a volume of 100 μl containing 25 mM glycine (pH 9.5), 20 mM MgCl2, and 10 mM [α-32P]ATP (about 105 cpm/assay) and 0.1 mM cAMP (104 cpm/assay). One unit of AC corresponds to 1 nmol of cAMP formed in 1 min at 30°C.
Total cAMP production was quantified by a radioimmunological assay (25) with overnight culture supernatants heated to 100°C for 5 min. The amount of cAMP was expressed as picomoles per milligram (dry weight) of bacteria.
Nucleotide sequence accession numbers.
The DNA sequences reported in this manuscript are deposited in the GenBank/EMBL/DNA Data Bank of Japan. The AC1 accession number is X95881, and the AC2 accession number is AJ223730.
RESULTS
A. hydrophila cya genes.
Complementing the cya defect of an E. coli mutant with an A. hydrophila (strain 218) DNA genomic library (constructed and screened as described in Materials and Methods) led to the isolation of two distinct fragments (cloned into plasmids pDIA6589 and pDIA6526; see Table 1). Their nucleotide sequences were determined, and the corresponding polypeptides were translated by using the universal genetic code.
The first 2,896-bp fragment contained a gene, cyaA, coding for a polypeptide of 843 amino acids, AC1, which displayed a 73% amino acid similarity (52% of which were identical) to the E. coli AC. The second fragment was 1,451 bp. It contained two open reading frames that comprised the coding sequences of 162- and 191-amino-acid polypeptides, respectively. We found a significant bias in RNY codons for the longer one (34.6% instead of 25%) (21). The sequence of the 191-amino-acid polypeptide was shown to be significantly homologous to the gene products of three hyperthermophilic archaebacteria. Moreover, disruption of this coding sequence abolished the cya complementation ability of plasmid pDIA6535 (data not shown). Thus, the longer coding sequence, cyaB, was considered to code for a genuine AC (see also below). The calculated molecular size of the corresponding polypeptide, AC2, was 21.5 kDa, and its isoelectric point was 4.85. No sequence similarity was found between this protein and AC1. The E. coli cya mutant expressing the cyaB gene cloned into pBR322 [TP610(pDIA6526)] synthesized 54,000 pmol of cAMP per mg (dry weight) of bacteria (i.e., 15 times more cAMP than was synthesized by the E. coli cya mutant expressing the cyaA gene cloned into the same vector [TP610 carrying pDIA6589]; Table 2). The complementation ability of AC2 in terms of cAMP synthesis strongly suggests that AC2 was an AC.
TABLE 2.
cAMP production measured in different strains and constructions
| Strain | Culture conditions | cAMP production (pmol/mg [dry wt] of bacteria) |
|---|---|---|
| TP610a | LB | 33 |
| TP610(pDIA6589) | LB | 600 |
| TP610(pDIA6589) | Glucose | 3,700 |
| TP610(pDIA6526) | LB | 10,500 |
| TP610(pDIA6526) | Glucose | 54,000 |
| TP610(pDIA6535) | LB | 5,500–9,000 |
| TP610(pDIA6535) | LB + IPTG | 91,000 |
| A. hydrophila wild type | Glucose | 226–240 |
| A. hydrophila wild type | Gluconate | 1,070 |
| A. hydrophila cyaA | Glucose | 70 |
| A. hydrophila cyaA | Gluconate | No growth |
| A. hydrophila cyaB | Glucose | 160 |
| A. hydrophila cyaB | Gluconate | 730 |
| A. hydrophila cyaA cyaB | Glucose | 30 |
| A. hydrophila cyaA cyaB | Gluconate | No growth |
| A. hydrophila cyaA cyaB pLA2917 | Glucose | 60 |
| A. hydrophila cyaA cyaB pDIA6539 | Glucose | 1,800 |
| A. hydrophila cyaA cyaB pDIA6539 | Gluconate | 2,300 |
TP610 is the E. coli cya mutant strain.
The presence of a cyaA gene coding for a class I AC in A. hydrophila was not unexpected because of the closeness of this strain to enterobacteria. In contrast, the presence of cyaB was completely unexpected. We therefore investigated whether this gene was present in other Aeromonas strains. An amplified labeled probe (260 bp) internal to the cyaB gene was used to screen Southern blots obtained from Aeromonas species. Several Aeromonas species were found to be positive (A. hydrophila 218 and 543, A. caviae, and A. trota), whereas other species were negative (A. eucrenophila, A. schubertii, A. sobria, and A. veronii). In other species related to enterobacteria, such as Vibrio cholerae and E. coli, no hybridization could be detected (data not shown).
In vitro AC activity of AC2.
In order to substantiate the identity of AC2 as an authentic AC, the AC2 protein was purified from E. coli cells containing plasmid pDIA6535 (as described in Materials and Methods). Microsequencing of its N-terminal end produced the sequence SSQHF, which is identical to that of the N terminus predicted from the cyaB gene sequence (except for removal of the N-terminal methionine residue). Different substrates and assay media were then tested for their effects on AC activity.
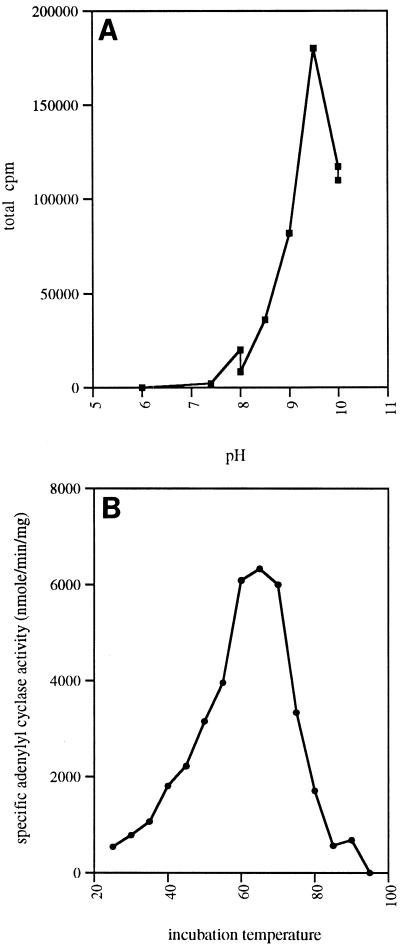
The optimal concentration of ATP was 10 mM. AC2 did not exhibit any guanylyl cyclase activity, and the addition of 5 mM GTP was found to abolish the enzyme activity completely. The influences of ions, salt concentration, pH, and temperature on the activity of purified AC2 were tested further. It was found that CaCl2 and MgCl2 increased the specific activities 1.3 and 4 times, respectively. The maximum activity occurred when the MgCl2 concentration was 20 mM. Remarkably, the optimal pH range for activity was 8.5 to 10. The maximum activity of this enzyme was obtained in a 25 mM glycine buffer at pH 9.5 (Fig. 1A). At this pH, the specific activity reached 800 nmol/min/mg, which is 25 times greater than the value obtained under the same conditions (2 mM ATP, 20 mM MgCl2) at pH 7.4. Moreover, when a range of temperatures from 25 to 90°C was tested, it was shown that a maximum activity of 6,300 nmol/min/mg (about 8 times the activity observed at 30°C) was reached at 65°C (Fig. 1B).
FIG. 1.
AC2 specific activity. (A) Influence of pH. (B) Influence of temperature.
Biological functions of AC1 and AC2 in A. hydrophila.
In order to explore the respective contributions of the two ACs in cAMP-dependent functions, we disrupted both cya genes in the chromosome. Knockout mutations of cyaA, cyaB, and cyaA cyaB were constructed. The phenotype of each strain was subsequently investigated.
(i) Disruption of the cyaA and cyaB genes.
A. hydrophila strains harboring a disrupted cyaA and/or cyaB gene were constructed by chromosome in situ recombination with a suicide vector. The conjugative cosmid pSUP202 was chosen for these experiments because this pBR325 derivative replicates only in E. coli (23). The cyaA and cyaB genes were first cloned into pSUP202 and subsequently disrupted by inserting a Smr or Kmr cartridge into each gene, yielding plasmids pDIA8516 and pDIA8507, respectively. These plasmids were then transferred by conjugation from the E. coli donor strain (S17.1) into the A. hydrophila recipient strain as described in Materials and Methods. Selection of the recombinants was performed with Sm and Km as selective agents and cyaA, cyaB, and cyaA cyaB recombinant strains were isolated. The presence of the inserts at the expected position was controlled by Southern hybridization as described in Materials and Methods.
(ii) Phenotype of the different A. hydrophila cya mutant strains.
In the first set of experiments, the growth of the A. hydrophila exconjugants was tested on 100 different carbon sources by using Biotype 100 strips. Compared to the A. hydrophila wild type, the cyaA and cyaA cyaB mutants failed to grow on about 20 substrates (data not shown). Inability to grow on maltose, galactose, glycerol, and succinate is a typical manifestation of cAMP defect as previously demonstrated in E. coli (5, 27). Further experiments showed that addition of cAMP to the media restored growth of the cyaA and cyaA cyaB mutants on all of these carbon sources. In contrast, the A. hydrophila wild type and its cyaB derivative did not show any growth differences on the different carbon sources.
In E. coli, bacterial motility depends on the presence of flagella, and the master operon which controls the expression of flagellum synthesis has been shown to be positively regulated by the cAMP-CAP complex (2). This highly sensitive cAMP-dependent function prompted us to test the swarming ability of the A. hydrophila exconjugants. Our results showed that the motility of the cyaA and cyaA cyaB mutants was abolished, indicating that it is a cAMP-dependent process as in E. coli. In contrast, motility of the cyaB mutant was not affected. The motility of the four types of A. hydrophila strains were as follows (as measured by swarming diameter): wild type, 30 mm; cyaA, 2 mm; cyaB, 27 mm; and cyaB cyaA, 2 mm.
We deduced that disruption of the cyaB gene had no effect on several cAMP-dependent processes, since the cyaB mutant and the wild-type strain exhibited the same phenotype. We therefore measured the presence of cAMP in A. hydrophila derivative cultures. Table 2 displays the results of cAMP concentration measurements. Clearly, disruption of the cyaA gene impaired cAMP synthesis, whereas disruption of cyaB had no effect. This indicates that under the physiological conditions tested the cyaB gene had no function in cAMP processes.
We then investigated whether modifying the growth conditions could have an effect on cAMP production. A variety of conditions were assayed; we found that neither changing the pH of the medium (from 5 to 10), the salt concentration range (from 0.1 to 1 M NaCl), or the temperature of growth (from 30°C to 50°C) nor testing under anaerobic conditions allowed restoration of cAMP-dependent growth or swarming for the cyaA exconjugant.
(iii) Complementation.
Despite the absence of classical cya-related phenotypes of cyaB mutants, restoration of the cAMP-dependent phenotype was obtained after introduction of a low-copy-number plasmid harboring the cyaB gene (pDIA6539) into the cyaA mutant or into the cyaA cyaB double mutant. In this construction, the cyaB gene was cloned in the direction opposite that of the Kmr gene promoter of pLA2917. This demonstrated that a cyaB promoter is located in the 350-bp DNA region located upstream from the start codon of the gene. Thus, one to three copies of pDIA6539 in the cyaA cyaB A. hydrophila derivative strain allowed cAMP synthesis (higher than in a wild-type strain [Table 2]). We showed that repeating the same experiment with a plasmid carrying an interrupted cyaB gene (pDIA6540) in the cyaA derivative as a control could not restore the cAMP-dependent phenotypes.
DISCUSSION
Cloning the cya genes from A. hydrophila to complement the defect of an E. coli cya mutant led to the identification of two distinct genes, namely, cyaA and cyaB. cyaA codes for an AC which is 73% similar to its E. coli counterpart. The gene shares the same locus organization (the same neighboring coding sequence, conserved promoter region, a poor ribosome binding site, and a TTG instead of ATG as a translation initiation codon), suggesting a similar mode of transcription and translation regulation (26). This AC belongs to the class I ACs. In contrast, cyaB has no homology with cyaA or with any identified cya gene from other organisms. In the present study, we investigated the biochemical characteristics and biological functions of the cyaB gene product. The purified cyaB-encoded protein, AC2, was shown in vitro to synthesize cAMP from ATP, demonstrating that it is an authentic AC. This 191-amino-acid 21-kDa protein exhibits unusual biochemical characteristics. The best conditions for maximal AC activity were found to be pH 9.5 in a 25 mM glycine buffer with 20 mM MgCl2. Moreover, a high temperature had an unexpected effect, increasing the cAMP synthesis eight times compared to the amount obtained at 30°C, which is the optimal temperature for A. hydrophila growth. The specific activity of AC2 under these conditions reaches 6,300 nmol/min/mg. This compares with the in vitro activity of the E. coli class I AC (700 nmol/min/mg) (28) or the yeast Saccharomyces cerevisiae class III AC (1,000 nmol/min/mg for the H-Ras and cyclase-associated-protein-activated form [22]). This is, nevertheless, not as efficient as the activity of class II ACs, such as the Bordetella pertussis toxic AC, which was shown to be 2,000 μmol/min/mg in the presence of calmodulin (6).
Whereas activities of AC1 and AC2 are comparable, their different optimal biochemical conditions for a maximal in vitro activity (37 to 45°C and pH 8.5 for a class I AC [28]; 65°C and pH 9.5 for AC2) led us to think that both enzymes may have different in vivo effective functions.
A. hydrophila is an opportunistic enteropathogen with at least 10 well-defined virulence factors, including enterotoxins, hemolysins, and proteases (11, 20). It is conceivable that the AC may play a role in pathogenicity like that of the calmodulin-activated toxic cyclases of the B. pertussis and B. anthracis pathogens. We did not find, however, significant homology between AC2 and the toxic class of ACs. Moreover, there was no evidence indicating that the protein is secreted, a general prerequisite for a toxic function. The sequence analysis of AC2 polypeptide excluded the presence of a signal peptide, and when screening for genes in the adjacent DNA regions of the cyaB gene, we could not detect any known system for secretion (9, 18). Finally, the presence of AC2 could not be detected in an A. hydrophila culture supernatant (data not shown). The hypothesis of a toxic role was therefore not retained.
To investigate the function of AC2 in vivo, cyaA, cyaB, or both genes were disrupted in the chromosome. Analysis of the phenotypes associated with cAMP-dependent functions demonstrated that only cyaA inactivation was effective, resulting in a total loss of cAMP synthesis. In contrast, cAMP production in the cyaB-inactivated mutant was similar to that in the wild type. This indicates that, instead of being a secondary AC, AC2 has either another function in A. hydrophila or, more likely, the protein is not expressed in the environmental context tested. cAMP synthesis resulting from AC2 expression was, however, obtained in the double cyaA cyaB A. hydrophila disruptant after the introduction of a low-copy-number plasmid containing the cyaB gene. This suggests that the cyaB gene expression is negatively regulated when in the chromosome. Whether this is due to a cis or trans effect could not be concluded from this experiment alone. Further experiments were undertaken to test whether a transcriptional repressor could have been titrated out by the introduction of a few copies of the cyaB promoter region (the 350 bp located upstream from the start codon) and would allow AC2 expression in the cyaA mutant. Our experimental data suggest that repression is due to the cyaB gene chromosomal location or that the putative negative regulation site is far upstream and thus not present in the constructions.
Finally, the fact that the best conditions for the in vitro cAMP production are 65°C and pH 9.5 suggests that the enzyme may have been recruited and expressed in particular biological situations. Attempts to reproduce some extreme growth conditions (e.g., alkaline growth media, high growth temperatures, and heat shock) to derepress cyaB gene expression failed to show any effect. Conditions in which activation of the gene expression occur therefore remain to be identified.
To date, the presence of a cyaB gene counterpart (detected by DNA hybridization) was found in only three Aeromonas species: A. hydrophila, A. caviae, and A. trota (data not shown). These bacteria have been isolated from aquatic ecosystems and/or were associated with intestinal and extraintestinal infections. No other phylogenetically related bacteria have been found to harbor a homologous gene. These genes, which seem to be exclusively found in Aeromonas species, define a new class of ACs. This gives a new start to the debate on the evolution of the cyclase’s function, since the discovery of such a new and different class of ACs illustrates a phenomenon of evolutionary convergence rather than divergence (10).
A review of the protein databases revealed that there is a significant homology between AC2 and the product of genes belonging to hyperthermophilic archaebacteria: M. thermoautotrophicum, A. fulgidus, and M. jannaschii. The hypothetical proteins encoded by these genes have sizes very similar to that of AC2. A sequence alignment comparison between the four gene products indicates a strong degree of similarity (Fig. 2). This is a particularly exciting finding since these genes were first classified with other archeal genes with no known function, providing evidence that the Archaea evolved separately from other organisms on Earth (8). Furthermore, considering the supposed absence of cAMP in Archaea together with AC2’s thermophilic properties, this aroused special interest.
FIG. 2.
Sequence alignment (as determined with the Multialign program) of the A. hydrophila AC2 protein (AHAC2) and the three homologous proteins found in the databases: protein from M. thermoautotrophicum (MTH1629), protein from M. jannaschii (MJ0240), and protein from A. fulgidus (AF1988).
The M. jannaschii gene was cloned into pBR322 and into a construction in which the coding sequence (MJ0240) is under pTrc99A transcriptional and translational control. We were, however, unable to determine whether the product of the gene has an AC activity. None of the constructions could complement an E. coli cya mutant, and no cAMP synthesis could be detected in the bacterial extracts under the optimal conditions for the AC2 activity (65°C, pH 9.5).
The presence of a similar gene in such phylogenetically divergent bacteria and its absence in other organisms led us to consider a probable horizontal gene transfer. The lack of correspondence between rRNA evolutionary patterns and metabolic gene patterns suggests that species frequently swap their genetic materials (17). However, considering the distant environments of these bacteria (fresh and estuarine cold water for Aeromonas sp., high temperature sewage digestor for M. thermoautotrophicum, geothermally heated sea floor for A. fulgidus, and deep sea marine hydrothermal vent for M. jannaschii), such a transfer appears somewhat unlikely. Moreover, according to a recent proposal of the evolutionary biologist Günter Wächtershäuser, “it is impossible for a hot organism to exchange genes with a cold organism, and vice versa” (16). Thus, such an exchange would have more likely occurred with an archaebacterium living in a similar environment, for example, Methanobacterium wolfei or Methanococcus voltae, which are found in river and estuarine sediments, respectively.
ACKNOWLEDGMENTS
We thank Benjamin Scott Gold and Chung-Kwun Wun for their critical reading and careful editing of the manuscript. We are grateful to Anne-Marie Gilles, Véronique Perrier, and Octavian Barzu for fruitful discussions and assistance during the biochemical analysis of the protein.
Financial support came from the Institut Pasteur, CNRS (UA1129), and the Université de Versailles Saint Quentin.
REFERENCES
- 1.Altschul W, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett D H, Frantz B B, Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequence and 5′ consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988;170:1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biville F, Guiso N. Evidence for the presence of cAMP, cAMP receptor and transcription termination factor rho in different gram-negative bacteria. J Gen Microbiol. 1985;131:2953–2960. doi: 10.1099/00221287-131-11-2953. [DOI] [PubMed] [Google Scholar]
- 5.Botsford J L, Harman J G. Cyclic AMP in procaryotes. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouhss A, Krin E, Munier H, Gilles A M, Danchin A, Glaser P, Barzu O. Cooperative phenomena in binding and activation of Bordetella pertussis adenylate-cyclase by calmodulin. J Biol Chem. 1993;268:1690–1694. [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye dinding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Granger G, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Coote J G. Structural and functional relationships among the RTX toxin determinants of gram-negative bacteria. FEMS Microbiol Rev. 1992;8:137–161. doi: 10.1111/j.1574-6968.1992.tb04961.x. [DOI] [PubMed] [Google Scholar]
- 10.Danchin A. Phylogeny of adenylyl cyclases. Adv Second Messenger Phosphoprotein Res. 1993;27:109–162. [PubMed] [Google Scholar]
- 11.Janda J M. Recent advances in the study of the taxonomy, pathogenicity and infectious syndromes associated with the genus Aeromonas. Clin Microbiol Rev. 1991;4:397–410. doi: 10.1128/cmr.4.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulfate-reducing archaeon Archaeglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 13.Ladant D, Brezin C, Alonso J M, Crenon I, Guiso N. Bordetella pertussis adenylate cyclase: purification, characterization and radioimmunoassay. J Biol Chem. 1986;261:16264–16269. [PubMed] [Google Scholar]
- 14.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 16.Morell V. Microbiology’s scarred revolutionary. Science. 1997;276:699–702. doi: 10.1126/science.276.5313.682b. [DOI] [PubMed] [Google Scholar]
- 17.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 18.Pugsley A, Francetic O, Possot O, Sauvonnet N, Hardie K R. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in gram-negative bacteria—a review. Gene. 1997;192:13–19. doi: 10.1016/s0378-1119(96)00803-7. [DOI] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sears C L, Kaper J B. Enteric bacterial toxins: mechanisms of action and linkage to intestinal secretion. Microbiol Rev. 1996;60:167–215. doi: 10.1128/mr.60.1.167-215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shepherd J C. Method to determine the reading frame of a protein from the purine/pyrimidine genome sequence and its possible evolutionary justification. Proc Natl Acad Sci USA. 1981;78:1596–1600. doi: 10.1073/pnas.78.3.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shima F, Yamawaki-Kataoka Y, Yanagihara C, Tamada M, Okada T, Karya K, Kataoka T. Effect of association with adenylyl cyclase-associated protein on the interaction of yeast adenylyl cyclase with Ras protein. Mol Cell Biol. 1997;17:1057–1064. doi: 10.1128/mcb.17.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposition mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 24.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steiner A L, Wehmann R E, Parker C W, Kipnis D M. Radioimmunoassay for the measurement of cyclic nucleotides. Adv Cyclic Nucleotide Res. 1972;2:51–61. [PubMed] [Google Scholar]
- 26.Trotot P, Sismeiro O, Vivares C, Glaser P, Bresson-Roy A, Danchin A. Comparative analysis of the cya locus in enterobacteria and related gram-negative facultative anaerobes. Biochimie. 1996;78:277–287. doi: 10.1016/0300-9084(96)82192-4. [DOI] [PubMed] [Google Scholar]
- 27.Ullmann A, Danchin A. Role of cyclic AMP in bacteria. Adv Cyclic Nucleotide Res. 1983;15:1–53. [Google Scholar]
- 28.Yang J K, Epstein W. Purification and characterization of adenylate cyclase of Escherichia coli K12. J Biol Chem. 1983;258:3750–3758. [PubMed] [Google Scholar]