Abstract
Background
The heterogeneity of lung adenocarcinoma (LUAD) plays a vital role in determining the development of cancer and therapeutic sensitivity and significantly hinders the clinical treatment of LUAD.
Objective
To elucidate the cellular composition and reveal previously uncharacterised tumour microenvironment in LUAD using single-cell RNA-sequencing (scRNA-seq).
Methods
Two scRNA-seq datasets with 106 829 high-quality cells from 34 patients including 11 normal, 9 early (stage I and II) and 14 advanced (stage III and IV) LUAD were integrated and clustered to explore diagnostic and therapeutic cell populations and their biomarkers for diverse stages of LUAD. Three independent bulk RNA-seq datasets were used to validate the results from scRNA-seq analysis. The expression of marker genes for specific cell types in early and advanced LUAD was verified by immunohistochemistry (IHC).
Results
Comprehensive cluster analysis identified that S100P+ epithelial and SPP1+ macrophage, positively related to poor outcomes, were preferentially enriched in advanced stage. Although the accumulation of KLRB1+CD8+ T cell and IGHA1+/IGHG1+ plasma cell both significantly associated the favourable prognosis, we also found KLRB1+CD8+ T cell decreased in advanced stage while IGHA1+/IGHG1+ plasma cells were increased. Cell-cell communication analysis showed that SPP1+ macrophage could interact with most of CD8+ subclusters through SPP1-CD44 axis. Furthermore, based on three independent bulk RNA-seq datasets, we built risk model with nine marker genes for specific cell subtypes and conducted deconvolution analysis, both supporting our results from scRNA-seq data. We finally validated the expression of four marker genes in early and advanced LUAD by IHC.
Conclusion
Our analyses highlight the molecular dynamics of LUAD epithelial and microenvironment and provide new targets to improve LUAD therapy.
Keywords: lung cancer
WHAT IS ALREADY KNOWN ON THIS TOPIC.
While a number of subpopulations of immune cells were previously identified to support cancer cell survival and metastatic dissemination in lung adenocarcinoma (LUAD), the data were mainly collected in a stage to study the specific cells.
WHAT THIS STUDY ADDS
We systematically characterised the four population cells with key markers (S100P+ epithelia, SPP1+ macrophage, KLRB1+CD8+ T cell and IGHA1+/IGHG1+ plasma cell) that potentially determine the development of LUAD from early to advanced stage.
A risk model with nine markers from above four subpopulations was built and showed efficient prediction for prognosis of LUAD in three independent cohorts.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Four population cells with key markers are important candidate targets for the therapy to improve outcomes of LUAD.
Introduction
Lung adenocarcinoma (LUAD), the leading cause of cancer death worldwide, is the one of the most common histological subtypes of lung cancer and accounts for ~40% of all patients with malignant lung.1–3 Most LUAD is often diagnosed at the advanced and even metastatic stage. Although surgery, targeting therapy and immunotherapy have been developed to improve the treatment and management of LUAD over the past 20 years, the 5-year overall survival rate is still low and <20%.4 5 The heterogeneity of composition of cells and the crosstalk between different cell types plays a key role in initiation and development of LUAD,6 7 and determines the response rate for drug therapies.8 Therefore, it is critical to improve fundamental understanding of the characteristics of tumour microenvironment (TME) in LUAD.
The epithelial cells, the origin of malignant cells, show a complex heterogonous and plasticity in LUAD. Generally, alveolar type 2 cells has been well known to give rise to LUAD.9–11 In addition, LUAD at some specific conditions would be derived by bronchioalveolar stem cells, ciliated cells and goblet cells.10 Whole genome sequencing data have revealed that the initiation and progression of LUAD is driven by the most highly somatic mutations in a diverse set of oncogenes, such as TP53, KRAS, EGFR, BRAF and MET.1 12 13 Each of the oncogenic mutations and/or combination of part of these driver mutations promoted tumourigenesis to result in different LUAD subtypes.14 15 The accumulation of diverse mutations in malignant cells then contributed to the heterogeneity of tumours. The major obstacle for LUAD treatment consequently was determined by the heterogeneity of malignant cells.16–18 Profiling of transcriptomic heterogeneity of malignant cells at single cell level could offer a novel perspective to understand the characterisation of malignant cells in LUAD.
The hallmark of LUAD TME is the predominant of immunosuppression, especially the suppression of cytotoxic CD8+ T cells.7 19 Cytotoxic T cell can kill tumour cells and plays a key role in antitumour immunity. However, the inactivation of cytotoxic CD8+ T cells with high expression of immune checkpoints, also known as the presence of exhausted CD8+ T cells, are characterised by progressive loss of cytokine production and killing functions, and the inhibitor of immunotherapy.20 21 Moreover, SPP1 in LUAD has been proposed to regulate macrophage polarisation and then act as an immune checkpoint to facilitate the suppression of T cell antitumour activation.22 Emerging evidence suggested that SPP1 promotes cell migration and invasion,23 and SPP1+ macrophage even can be a candidate biomarker for metastasis to early lymph node in LUAD.24 However, the whole profiling of composition of immune cells in LUAD TME has been largely unknown.
To better understanding of dynamic changes of malignant cell and TME in early and advanced LUAD, we integrated two independent single-cell RNA-sequencing (scRNA-seq) datasets including 11 normal, 9 early and 14 patients with advanced LUAD to reveal characterisation of epithelial and immune landscape at single-cell level. Furthermore, we used three independent bulk RNA-seq datasets to conduct deconvolution and survival analysis to validate the marker genes of each cluster and then to develop a risk model. The expression of these main marker genes was also validated by immunohistochemistry (IHC). Our results provide a comprehensive scRNA-seq profiling of LUAD across different stages and may be applied to improve diagnostics and prognosis in clinical practice.
Methods
Datasets
All scRNA-seq and bulk RNA-seq datasets analysed in this study were publicly available and collected from high-quality publications. The two independent scRNA-seq datasets19 25 were downloaded from the Gene Expression Omnibus (GEO; GSE131907 and GSE148071) with total 34 samples. The GSE131907 dataset included the 26 samples (11 normal, 9 early and 6 advanced stage) and other 8 advanced samples came from GSE148071 dataset. The other two bulk RNA-seq datasets26 27 were also obtained through the GEO series accession numbers (GSE31210, GSE72094) in Data Accessibility. The Cancer Genome Atlas (TCGA)-LUAD data with clinical information was downloaded from TCGA (https://portal.gdc.cancer.gov/projects/TCGA-LUAD).
Preprocessing for scRNA-seq data
We used the Python package Scanpy28 to conduct the quality control and preprocessing of two scRNA-seq data independently. Cells with <200 detected genes and genes detected in <3 cells were first excluded. We then filtered out cells with a percentage of expressed mitochondrial genes >10%. Finally, the samples with high-quality cells <1000 were excluded for further analysis.
Integration, clustering and annotation of scRNA-seq data
One obstacle of integration of scRNA-seq data is to efficiently remove the unwanted/unknown variations between different samples. The Python library scVI,29 offering a probabilistic analysis of single-cell omics data, could address the above limitation to integration of scRNA-seq data. The count matrix in data.raw object was used for scVI integration to perform model training and estimate the latent representation of each cell. We further conducted cell cluster analysis with ‘sc.pp.neighbors’ and ‘sc.tl.leiden’ function in Scanpy. The subclustering for each cell type had been performed by above pipeline.
To identify the differential gene expression for each cluster, we first normalised the raw counts using the function ‘normalize_total’ and then transformed the data using the function ‘log1p’. Wilcoxon rank-sum test in Scanpy was used to fund differential gene expression with log(fold change (FC)) >0.25 and adjusted p value <0.05. Combining the result of differential gene expression analysis and the known marker genes, we annotated the cell types clustered in our analysis. We also used a python library InferCNV (https://github.com/icbi-lab/infercnvpy) to infer copy number variation (CNV) from single-cell transcriptomics data for each cell.
Cell communication analysis
The CellChatDB is one of the signalling molecular databases including a comprehensive signalling ligand-receptor interactions, such as multimeric ligand-receptor complexes and stimulatory and inhibitory membrane-bound coreceptors. Here, we employed the CellChat package30 to perform cell-cell interaction analysis for LUAD scRNA-seq data. CellChat algorithm first identified differentially expressed signalling genes and calculated ensemble average expression for each cell cluster. With the calculation of intercellular communication probability, the statistically significant intercellular communications between two cell clusters had been determined to explore the differences in ligand and receptor interactions across the diverse cell clusters.
Trajectory inference for CD8+ T cells
The developmental trajectory of CD8+ T cells was reconstructed by partition-based graph abstraction (PAGA) in Scanpy. The PAGA graph was generated by the Leiden algorithm and visualised by denoising the graph. We defined the naïve cell cluster as root for estimating diffusion pseudotime. Finally, the developmental state of each CD8+ T cell was measured based on the computed diffusion pseudotime. All reconstructions of trajectory in this study were conducted with the default parameters.
Deconvolution of bulk transcriptome data
Due to the mixture of distinct cell types in TME, direct comparison of bulk transcriptomic profiles is ineffective to detect the cell type-specific transcriptome. The quantification of gene expression from bulk RNA-seq data is directly calculated by the mean abundance across cell types in tumour tissue. However, based on the reference expression matrix with a set of defined marker genes, deconvolution analysis could accurately infer cellular composition with specific marker genes for bulk RNA-seq data.31 In this study, we used a newly developed public method (BayesPrism31) with default parameters to predict cellular composition in TCGA-LUAD bulk RNA-seq dataset. Because of the significant infiltration of S100P+ epithelial cells and SPP1+ macrophage in LUAD and their key contribution to development of LUAD, we divided patients with LUAD in TCGA cohort into two groups with high and low proportion of S100P+ epithelial cells or SPP1+ macrophages for further survival analysis.
Construction prognostic model
To build the prognostic model, we first conducted univariate Cox analysis in R package survival (https://CRAN.R-project.org/package=survival) to filter out the marker genes of each cluster with p>0.05, suggesting these genes without significant association with the overall survival time. R package glmnet (https://CRAN.R-project.org/package=glmnet) was used to conduct the LASSO Cox regression analysis. We performed a 10-fold cross-validation to determine the penalty regularisation parameter λ and then obtain a suitable model. The coefficient for each gene in model was generated by multivariate Cox regression analysis. Based on the risk score value, patients with LUAD were classified into two groups (high risk and low risk).
Survival analysis
To determine the optimal cut-point for the gene expression value or risk score, we applied surv_cutpoint function in R package survminer (https://CRAN.R-project.org/package=survminer) to calculate a value that corresponds to the most significant outcome of bulk RNA-seq cohort in patients with LUAD. The Kaplan-Meier method with a log-rank test was used to generate survival curves (https://CRAN.R-project.org/package=survival). HR between two groups was estimated by the Cox regression model. To determine whether the gene expression level and risk score built by this study was an independent prognostic factor, the multivariate Cox regression analysis was performed in survival analysis. The significance threshold (p value) in the survival analysis was ≤0.05.
IHC staining
IHC was performed following the standard protocol. After dewaxing and rehydration, the samples were treated for antigen heat repair with sodium citrate solution for 20 min. Then treated for 10 min with 3% H2O2 in methanol to quench endogenous peroxidase activity, blocked with goat serum, incubated at 4°C overnight with primary antibodies against KLRB1 (Proteintech, 67537-1), IFITM3 (Proteintech, 66081-1), SPP1 (Proteintech, 25715-1-AP), anti-CD8a (Servicebio, GB114196), anti-CD68 (Servicebio, GB113109). After being washed and incubated with a horseradish peroxidase-conjugated secondary antibody for 1 hour at room temperature. The sections visualisation was performed using 3,3’-diaminobenzidine, counterstained with haematoxylin, dehydrated and mounted. Whole slides were scanned with an Aperio ScanScope system (Leica Biosystems).
Results
Single-cell profiling of tumour microenvironment in LUAD
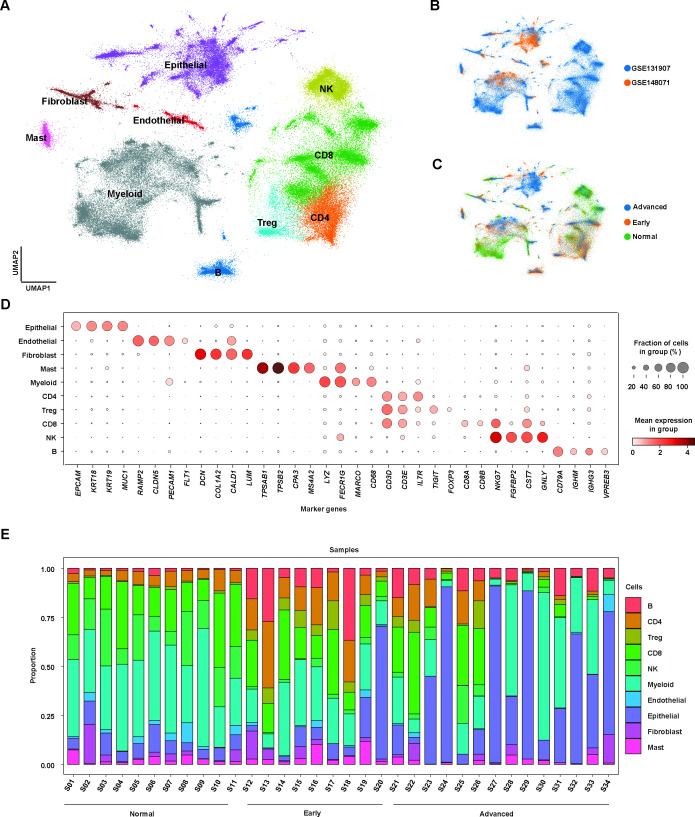
To evaluate the dynamic changes of the early and advanced LUAD TME, we integrated two independent scRNA-seq cohorts with 34 samples including 11 normal, 9 early and 14 advanced samples by Scanpy. All samples were obtained from primary tumours. After cell quality was filtered, we included a total of 106 829 single cells, comprising 40 630 normal cells, 32 221 early cells and 33 978 advanced cells (online supplemental table S1). Here, we defined early cell from patients with stage I, II LUAD, while advanced cell from patients with stage III, IV LUAD. ScVI tools were applied to remove batch effects and conduct single cell clustering analysis to identify cell types, including epithelial, endothelial, fibroblast, mast, myeloid and lymphoid cells (figure 1A, online supplemental figure 1). As shown in figure 1B,C, the cell populations we clustered were distributed in diverse cohorts or stages, suggesting that our integration was fair to perform batch correction.
Figure 1.
Single-cell atlas in early and advanced lung adenocarcinoma (LUAD). (A) Uniform manifold approximation and projection (UMAP) plot of integrated single-cell RNA-sequencing (scRNA-seq) data from 106 829 cells obtained from 34 samples with LUAD. Cell clusters found therein representing 10 cell types are shown. (B) UMAP plot of distribution of cell clusters by different cohorts. (C) UMAP plot of distribution of cell clusters by LUAD stages. (D) Dot plot showing expression levels of specific markers in each cell type. (E) Stacked plot showing the proportion of cells that contributed to each cell type by each sample.
bmjresp-2023-001878supp014.pdf (7.1MB, pdf)
bmjresp-2023-001878supp015.pdf (121.1KB, pdf)
bmjresp-2023-001878supp001.pdf (254.9KB, pdf)
We further used the Wilcoxon rank-sum test with Scanpy’s rank_genes_group function to estimate the differential gene expression and the expressions of marker genes for each cell types were visualised by dot plot (figure 1D, online supplemental table S2). Based on these annotated cells, we calculated the proportion of each cell type per patient, revealing the heterogenous changes of cell composition in LUAD TME, especially in advanced LUAD compared with normal and early patients (figure 1E).
Diverse malignant subpopulations enriched in advanced stage
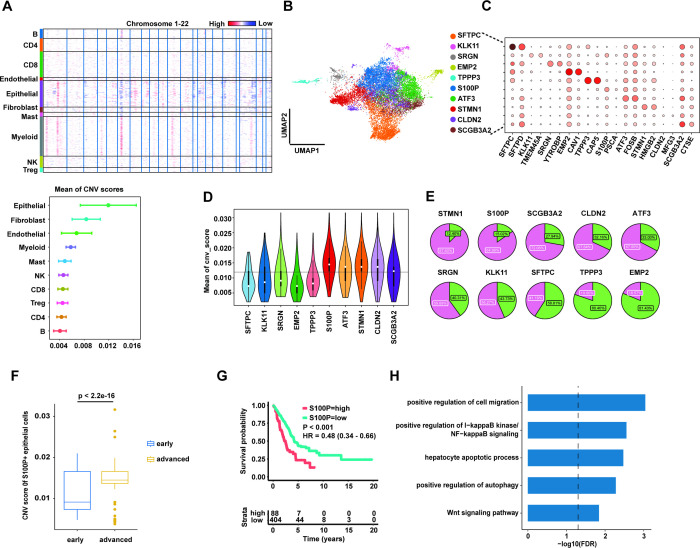
To investigate the characterisation of malignant cells in different LUAD stages, we first used the InferCNV to infer CNV events from single cell transcriptomics data with all B and T cells as the reference cells. As expected, the mean of CNV score was highest in epithelial cluster (figure 2A), supporting that malignant cells in LUAD were from the alteration of epithelial cell. Next, we conducted a subclustering analysis for all epithelial cells to identify the normal and malignant cells. Here, we only included the samples with number of epithelial cells ≥200.
Figure 2.
S100P+ epithelial cells with high copy number variation (CNV) score enriched in advanced stage and contributed to the poor prognosis. (A) Heatmap of CNV results across the genome (upper panel). The mean of CNV scores for each cell cluster (lower panel). (B) Uniform manifold approximation and projection (UMAP) plot of 10 subpopulations of epithelial cells. (C) Dot plot showing expression levels of specific markers in 10 epithelial subpopulations. (D) Violin plot showing the mean of CNV scores in each epithelial subpopulation. The horizontal black line indicates the mean of CNV scores of all epithelial cells. (E) Pie plot for each epithelial subpopulation consisting of cells from early and advanced patients with LUAD. Purple indicates the proportion of cells from advanced LUAD and green indicates the proportion of cells from early LUAD. (F) Box plot showing the CNV scores of S100P+ epithelial cells significantly higher in advanced LUAD than in early LUAD. Statistical significance is determined by Wilcoxon rank-sum test. (G) Kaplan-Meier survival analysis for two groups defined by patients in The Cancer Genome Atlas (TCGA)-LUAD cohort with high/low expression S100P. The sample numbers for each group are shown in brackets. Statistical significance is determined by log-rank test. (H) Gene Ontology pathway enrichment analysis of differential gene expression (log2 fold change ≥0.25 or ≤0.25 and false discovery rate ≤0.05) between S100P+ epithelial cells and the rest of epithelial cells.
The subclustering result revealed at least 10 subpopulations in LUAD (figure 2B). We further estimated the differential gene expression for each cluster to annotate the epithelial subpopulations. Notably, we identified >10 marker genes (SFTPC, KLK11, SRGN, EMP2, TPPP3, S100P, ATF3, STMN1, CLDN2 and SCGB3A2) that clearly distinguished each subpopulation (figure 2C). Among these marker genes, parts of gene play a key role in maintaining the normal function of lung. For example, SFTPC, secreted by the type II alveolar cells of the lung, regulates the homeostasis of pulmonary surfactant metabolism. On the other hand, other genes (such as S100P,32 STMN133 and SCGB3A234) had been reported to promote tumour progression in diverse cancers. Specially, the cells with high expression of SCGB3A2 and TPPP3 were also known as the club cell and ciliated cell, respectively. In addition, EMP2+ cells had significantly increased expression of AGER (log2FC=8.43, adjusted p value <0.0001), named as type I alveolar cell.
To identify the malignant cells enriched in advanced stage compared with the early stage, we integrated the estimation of CNV scores to subclustering analysis. We calculated the mean of CNV score for each subcluster (figure 2D), suggesting the mean values in potential malignant clusters were in top five, especially highest in S100P+ cluster. At the same time, the mean value was lowest in SFTPC+ cluster. We further counted the number of cells from advanced LUAD and non-advanced (normal and early) LUAD for each cluster (figure 2E). The results showed that most cells in malignant clusters mainly came from advanced patients, and supported the potentially predictable value of these marker genes for LUAD progression. Furthermore, we compared the mean of CNV score from S100P+ epithelial cells between early and advanced LUAD (figure 2F), showing that the CNV score was significantly higher in advanced stage.
To further validate the function of S100P in LUAD, we performed survival analysis to evaluate whether the increasing of S100P significantly contributed to unfavourable outcomes based on TCGA-LUAD cohort. Our data suggested that the high expression of S100P in epithelial cells was positively related to the shorter survival time in LUAD (figure 2G). Multivariate analysis further identified that the expression of S100P was an independent risk factor for survival (online supplemental figure 3). Based on significantly expressed genes in S100P+ epithelial cells compared with other epithelial cells, the Gene Ontology (GO) pathway enrichment analysis revealed that the elevated S100P probably affects cell migration, apoptosis and autophagy through the alteration of nuclear factor-κB and Wnt signalling pathway (figure 2H).
bmjresp-2023-001878supp003.pdf (264.4KB, pdf)
SPP1+ macrophage contributed LUAD development through binding CD44
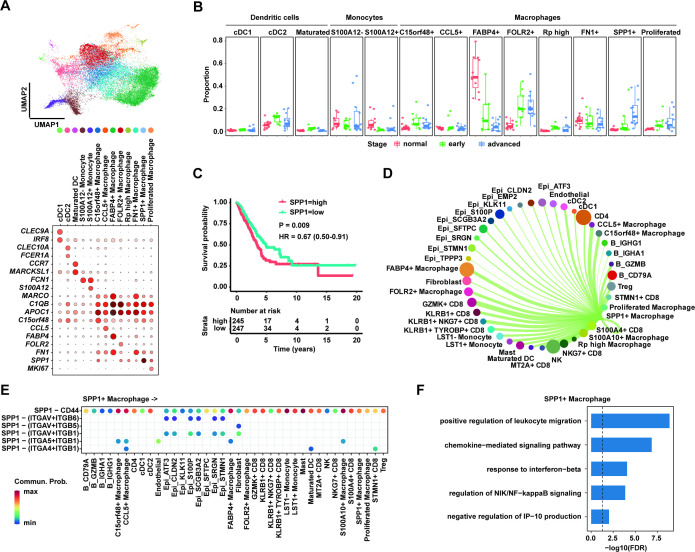
To elucidate the cellular composition of myeloid cells in LUAD TME, we reclustered myeloid cells to identify three main subtypes (dendritic cells, monocytes and macrophages) based on the expression of canonical gene markers (figure 3A). Classical dendritic cells 1 (cDC1), classical dendritic cells 2 (cDC2) and mature dendritic cells were characterised by CLEC9A, CLEC10A and CCR7, respectively. The monocytes-expressed FCN1 were clearly divided into two types with high expression of S100A12 and without high expression of S100A12. We next investigated the characteristics of macrophages and found at least eight subclusters in TME. These cells included C15orf48+ macrophages, CCL5+ macrophages, FABP4+ macrophages, FOLR2+ macrophages, FN1+ macrophages, SPP1+ macrophages, proliferated macrophages with Mki67 high expression and Rib high macrophages with high expression of ribosomal genes.
Figure 3.
SPP1+ macrophages enriched in advanced lung adenocarcinoma (LUAD). (A) Uniform manifold approximation and projection (UMAP) plot of 13 subpopulations of myeloid cells including three clusters of dendritic cells, two clusters of monocytes and eight clusters of macrophage cells (upper panel). Dot plot showing expression levels of specific markers in each cluster (lower panel). (B) Box plot showing the proportion of SPP1+ macrophages significantly increased in advanced LUAD compared with normal and early LUAD. (C) Kaplan-Meier survival analysis for two groups defined by patients in The Cancer Genome Atlas (TCGA)-LUAD cohort with high/low expression of SPP1. The sample numbers for each group are shown in brackets. Statistical significance is determined by log-rank test. (D) Circle plots showing putative ligand-receptor interactions between SPP1+ macrophages and other cell clusters, with the width of edges representing the strength of the communication. (E) The heatmap showing the interaction between SPP1 ligand in SPP1+ macrophages and six receptors of other clusters. Every row represents a ligand-receptor pair and every column defines a cell-cell interaction pair. (F) Gene Ontology pathway enrichment analysis of differential gene expression (log2 fold change ≥0.25 or ≤0.25 and FDR ≤0.05) between SPP1+ macrophage cells and the rest of macrophage cells.
To ask whether the changes of these myeloid subtype cells contribute to the progression of LUAD, we calculated the proportion of number of subtype cells to number of all myeloid cells per sample (figure 3B). The proportions of dendritic cells and monocytes were similar across the normal, early and advanced stages. However, the proportion of SPP1+ macrophages was significantly increased in advanced LUAD compared with normal and early stages. At the same time, the proportion of FABP4+ macrophages in advanced stage was significantly lower than in normal and early stages.
The SPP1+ macrophages were supposed to promote the progression of LUAD and be a risk predictor for metastasis of lung cancer. We used TCGA-LUAD cohort to test the association between SPP1 gene expression and clinical outcomes of patients, suggesting that high expression of SPP1 was an independent risk factor for poor survival outcomes (figure 3C, online supplemental figure 4). The median survival time (4.9 years/1790 days) of patients with low SPP1 expression was significantly longer than the time (3.45 years/1258 days) of patients with high SPP1 expression (p=0.009; HR 0.67, 95% CI 0.50 to 0.91) (figure 3C). Due to the fact that SPP1 encodes the chemokine osteopontin playing a key role in intercellular communication, CellChat was applied to explore ligand-receptor interactions between SPP1+ macrophages and other cell clusters in LUAD TME. Notably, we found that SPP1 in SPP1+ macrophages was mainly through CD44 to communicate with other cells (figure 3D,E). Except interacting with epithelial cells including malignant, SPP1+ macrophages and another immune cells, such as CD4+ T, CD8+ T, Tregs and B cells, had strong intercellular interaction with SPP1 and CD44. However, due to the low expression of CD44 in endothelial cells, the cell-cell communication between macrophages and endothelial cells through SPP1-CD44 axis was not identified in this study.
bmjresp-2023-001878supp004.pdf (244.4KB, pdf)
To gain insight into the role of SPP1+ macrophages in advanced LUAD, we conducted differential gene expression analysis between SPP1+ macrophage cells and the rest of macrophages and then performed GO pathway enrichment analysis. Our data showed that the high expression of SPP1 in macrophage cells contributes to the alteration of chemokine-mediated signalling pathway, response to interferon-beta and production of IP-10 (interferon gamma-induced protein 10) to regulate leucocyte migration (figure 3F), suggesting that the presence of SPP1+ macrophage probably remodels the LUAD TME to contribute to its tumourigenesis.
KLRB1+ CD8+ T cell candidate immunotherapy population
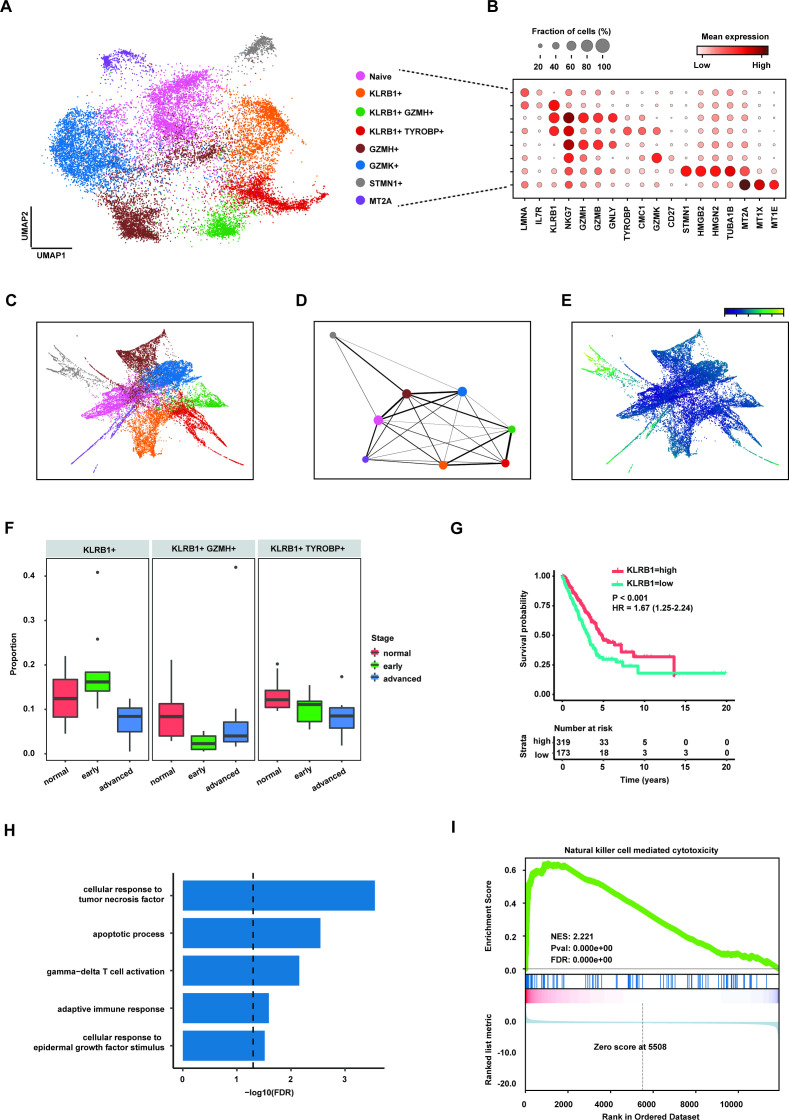
To reveal the cytotoxic character of CD8+ T cell in antitumour immunity, we identified six types of CD8+ T cells in eight clusters (figure 4A,B), including naïve CD8+ (IL7R+), KLRB1+CD8+, GZMH+CD8+, GZMK+CD8+, STMN1+CD8+ and MT2A+CD8+ T cells. KLRB1+CD8+ T cells were further divided into the subpopulations characterised by expression of GZMH and TYROBP (figure 4B). Moreover, trajectory inference method was used to reconstruct the differentiation paths of CD8+ T cells in LUAD TME. The result showed that three KLRB1+CD8+ T cells probably differentiated from GZMH+CD8+ T cells (figure 4C,D). At the same time, pseudotime analysis suggested that KLRB1+CD8+ T cells did not locate in the end of fate of CD8+ T cells (figure 4E). Taken together, our analyses supported previous studies showing that KLRB1+CD8+ T cells were one of the effector memory T cells and the cytotoxicity of them was probably mediated through granzymes.35
Figure 4.
CD8+KLRB1+ T cells decreased in advanced lung adenocarcinoma (LUAD). (A) Uniform manifold approximation and projection (UMAP) plot of subpopulations of CD8+ T cells. (B) Dot plot showing expression levels of specific markers in each CD8+ T subpopulation. (C) The partition-based graph abstraction (PAGA) embedding plot showing the trajectory inference of CD8+ T subpopulations. (D) PAGA graph showing the distribution of CD8+ T subpopulations. The width of line representing the connectivity adjacency between clusters. (E) Pseudotime plot of CD8+ T subpopulations. (F) Box plot showing the proportion of three CD8+KLRB1+ T cells in three stages of LUAD. (G) Kaplan-Meier survival analysis for two groups defined by patients in The Cancer Genome Atlas (TCGA)-LUAD cohort with high/low expression KLRB1. The sample numbers for each group are shown in brackets. Statistical significance is determined by log-rank test. (H) Gene Ontology pathway enrichment analysis of differential gene expression (log2 fold change (FC) ≥0.25 or ≤0.25 and FDR ≤0.05) between CD8+KLRB1+ T cells and the rest of CD8+ T cells. (I) Gene set enrichment analysis for the ranked gene lists according to FCs of gene expression in the CD8+KLRB1+ T cells compared with the rest of CD8+ T cells.
As KLRB1+CD8+ T cells may play a key role in tumour immunology, we calculated the proportion of number of KLRB1+CD8+ T cells to all CD8+ T cells, and found that the proportion of KLRB1+CD8+ T cells significantly decreased in advanced LUAD compared with normal and early stages (figure 4F). The survival analysis based on TCGA-LUAD cohort revealed that the low expression of KLRB1 was significantly related to poor prognosis in LUAD (figure 4G) and multivariate analysis further suggested KLRB1 expression as independent prognostic factor to predict the survival (online supplemental figure 5). These data suggested that the infiltration of KLRB1+CD8+ T into TME may improve the efficiency of antitumour immunity.
bmjresp-2023-001878supp005.pdf (244.6KB, pdf)
To investigate the potential function of KLRB1+CD8+ effector memory T cells, we identified differential gene expression between KLRB1+CD8+ and other CD8+ T cells and then conducted GO pathway enrichment and gene set enrichment analysis (GSEA). GO pathway analysis suggested that the function of KLRB1+CD8+ effector memory T cells may be regulated by the cytokine of tumour necrosis factor (figure 4H). In addition, result from GSEA showed the activated pathway of natural killer cell-mediated cytotoxicity in KLRB1+CD8+ T cells (figure 4I), supporting the potential cytotoxicity of this CD8+ subpopulation.
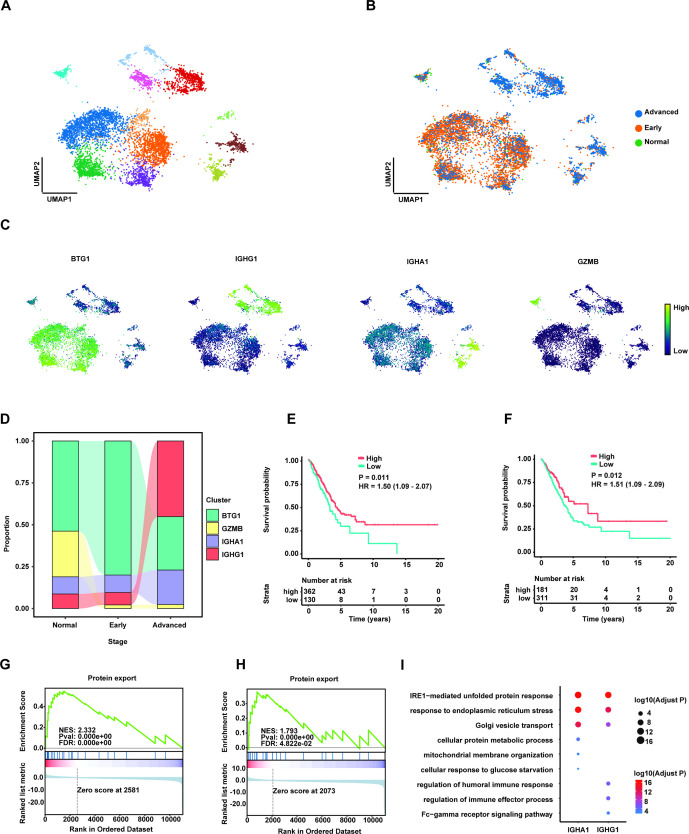
The presence of IGHG1+ and IGHA1+ plasma cells in advanced LUAD
Due to the fact that B cells are essential for immunotherapy with component of the adaptive immune, we next investigated the features in B cell subclusters in advanced LUAD to develop the potential therapeutic cell population and evaluated the distribution of each cluster across three stages. Notably, four subclusters of B cell were clearly identified by marker genes BTG1, IGHG1, IGHA1 and GZMB, respectively (figure 5A–C). We observed that proportion of IGHG1+ and IGHA1+ plasma cells both increased in advanced stage compared with the normal and early stages (figure 5D). After stimulated by the antigen, plasma cell could proliferate and secrete a specific antibody to play a significant role in the adaptive immune response. Our survival analysis supported that the high expression of mean of IGHA1 and CD79A (figure 5E), as well as IGHG1 and CD79A (figure 5F), were both significantly associated with good clinical outcomes, respectively. Further analysis revealed that high average of IGHA1 and CD79A (online supplemental figure 6) as well as IGHG1 and CD79A was an independent predictor of poor prognosis in LUAD (online supplemental figure 7).
Figure 5.
The increasing of IGHA1+/IGHG1+ plasma cells associated with favourable prognosis in advanced lung adenocarcinoma (LUAD). (A) Uniform manifold approximation and projection (UMAP) plot of subpopulations of B cells. (B) UMAP plot of distribution of B cell clusters by LUAD stages. (C) Expression of marker genes BTG1, IGHG1, IGHA1 and GZMB in UMAP. (D) The proportion of four B cell clusters in different stages. (E) Kaplan-Meier survival analysis for two groups defined by patients in The Cancer Genome Atlas (TCGA)-LUAD cohort with high/low expression of mean of CD79A and IGHA1. The sample numbers for each group are shown in brackets. Statistical significance is determined by log-rank test. (F) Kaplan-Meier survival analysis for two groups defined by patients in TCGA-LUAD cohort with high/low expression of mean of CD79A and IGHG1. The sample numbers for each group are shown in brackets. Statistical significance is determined by log-rank test. (G) Gene set enrichment analysis (GSEA) for the ranked gene lists according to fold changes (FCs) of gene expression in the IGHA1+ plasma cells compared with the rest of B cells. (H) GSEA for the ranked gene lists according to FCs of gene expression in the IGHG1+ plasma cells compared with the rest of B cells. (I) GO pathway enrichment analysis of differential gene expression (log2 FC ≥0.25 or ≤0.25 and FDR ≤0.05) between IGHA1+ or IGHG1+ plasma cells and the rest of B cells.
bmjresp-2023-001878supp006.pdf (245KB, pdf)
bmjresp-2023-001878supp007.pdf (234.2KB, pdf)
Next, we conducted the analysis of differential gene expression for each subcluster of B cells, specifically identified significantly expressed genes in IGHA1+ and IGHG1+ plasma cells compared with other B cells, respectively. GSEA result based on the differential gene expression showed that the function of protein export (KEGG protein export, hsa03060) was both activated in IGHG1+ (figure 5G) and IGHA1+ (figure 5H) plasma cell clusters. In addition, GO pathway enrichment analysis revealed that the pathway of IRE1-mediated unfolded protein response has been enriched in IGHG1+ and IGHA1+ plasma cells (figure 5I), respectively. However, the activation of metabolic process pathway was only found in IGHA1+ plasma cells while the humoral immune response was identified in IGHG1+ plasma cells.
Validation of marker genes in IHC and construction prediction model based on LUAD TME
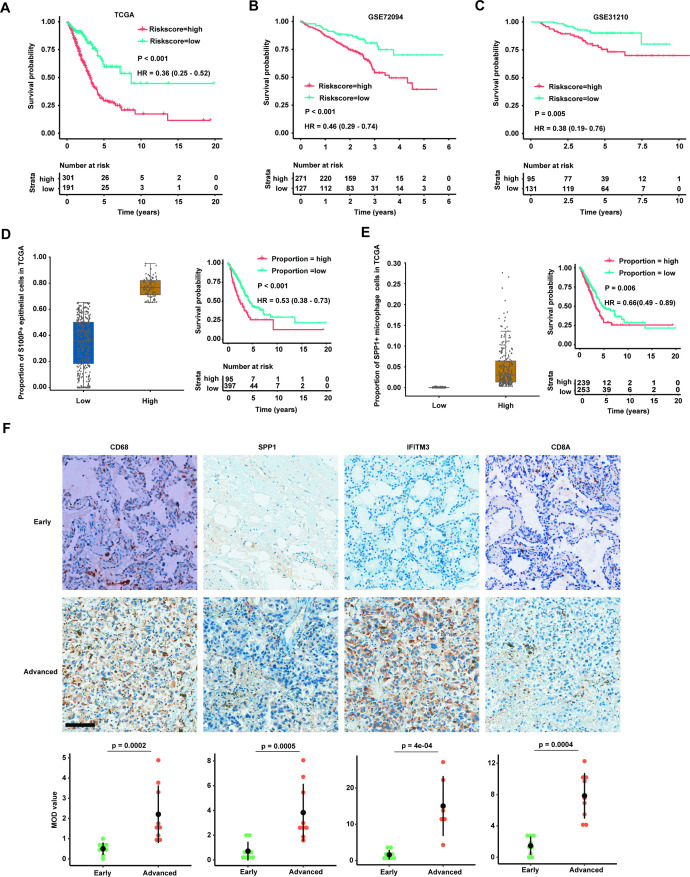
Given that the marker genes for certain cell type were free from the confounding effect of cell-type heterogeneity, we thought to develop the risk model based on the expression levels of these marker genes in bulk RNA-seq data from primary LUAD cancer tissue. Univariate Cox regression analysis was conducted to evaluate candidate marker genes in TGCA-LUAD cohort. Finally, we obtained nine genes with a threshold of p<0.05 and then performed multivariate Cox regression analysis to calculate the coefficients for each gene. Based on the gene expression values at RNA level and coefficients of these genes, a prognostic risk score for each patient with LUAD was estimated in TGCA cohort. The survival analysis for two groups with high-risk and low-risk scores showed that patients with high-risk score have unfavourable outcomes compared with patients with low-risk score (figure 6A), suggesting our risk score model based on the marker genes from scRNA-seq data could independently predict the prognosis of LUAD (online supplemental figure 8).
Figure 6.
Validation of marker genes for cell types by three independent bulk RNA-sequencing datasets and immunohistochemistry (IHC). (A) Kaplan-Meier survival analysis for two groups defined by patients in The Cancer Genome Atlas (TCGA)-LUAD cohort with high-risk/low-risk score. The sample numbers for each group are shown in brackets. Statistical significance is determined by log-rank test. (B) Kaplan-Meier survival analysis for two groups defined by patients in GSE72094 with high-risk/low-risk score. The sample numbers for each group are shown in brackets. Statistical significance is determined by log-rank test. (C) Kaplan-Meier survival analysis for two groups defined by patients in GSE31210 with high-risk/low-risk score. The sample numbers for each group are shown in brackets. Statistical significance is determined by log-rank test. (D) Patients with LUAD in TCGA cohort are divided into two groups based on the proportion of S100P+ epithelial cells estimated by deconvolution analysis (left panel). Kaplan-Meier survival analysis for two groups defined by patients in TCGA-LUAD cohort with high/low proportion of S100P+ epithelial cells (right panel). The sample numbers for each group are shown in brackets. Statistical significance is determined by log-rank test. (E) Patients with LUAD in TCGA cohort are divided into two groups based on the proportion of SPP1+ macrophage cells estimated by deconvolution analysis (left panel). Kaplan-Meier survival analysis for two groups defined by patients in TCGA-LUAD cohort with high/low proportion of SPP1+ macrophage cells (right panel). The sample numbers for each group are shown in brackets. Statistical significance is determined by log-rank test. (F) IHC staining of four marker genes (CD68, SPP1, IFITM3 and CD8A) in patients with early (upper panel) and advanced (middle panel) LUAD. The expression of four marker proteins (CD68, SPP1, IFITM3 and CD8A) are significantly increased in advanced stage compared with early stage (lower panel; n=18; scale bar=100 µm). Each IHC staining were replicated by six patients and three random regions of interest (ROI) for each marker per patient (total number=18) were selected for calculation. Mean optical density (MOD) is equal to the integrated optical density/measurement area.
bmjresp-2023-001878supp008.pdf (261.5KB, pdf)
To validate the risk score model, we constructed by TCGA-LUAD cohort, risk score for each patient was calculated in two independent LUAD cohorts and these patients were divided into high-risk group and low-risk group, respectively. We then combined clinical information to compare the survival time between the high-risk and low-risk groups. Notably, the clinical outcomes of patients with LUAD with high-risk scores were significantly worse than with low-risk scores in both cohorts (figure 6B,C), consistent with the result in TCGA-LUAD training cohort. Multivariable analysis further suggested that the risk score was independent predictor for LUAD prognosis in three cohorts (online supplemental figures 9 and 10).
bmjresp-2023-001878supp009.pdf (251.8KB, pdf)
bmjresp-2023-001878supp010.pdf (253KB, pdf)
Based on the expression of marker genes for each cell types in scRNA-seq, we furthermore used BayesPrism to conduct deconvolution of cell type for TCGA-LUAD bulk RNA-seq dataset to test the correlation between cell type composition and clinical outcomes. Given the large infiltration of S100P+ epithelial cells and SPP1+ macrophage cells in LUAD TME and their key role in LUAD progenesis, we calculated the proportion for each of these two cell populations and then classified patients into two groups with high and low proportion of cells, respectively. Kaplan-Meier survival curves further revealed that high compositions of S100P+ epithelial cells and SPP1+ macrophage cells could result in poor survival of LUAD (figure 6D,E), and the proportion of cells was independent factor for prognosis (online supplemental figures 11 and 12). Our survival analysis with more accurate estimation for S100P+ epithelial cells and SPP1+ macrophage cells was consistent with the result of marker genes. Finally, IHC results validated that the protein expressions of CD68, SPP1, IFITM3 and CD8A were increased in advanced LUAD (figure 6F). Each IHC staining was replicated by six patients (three early and three advanced stage) and three random regions of interest for each marker per patient were selected for calculation (total number=18). Taken together, our validation with bulk RNA-seq datasets supported the dynamic changes of cell subpopulations between early and advanced LUAD from scRNA-seq data.
bmjresp-2023-001878supp011.pdf (264.4KB, pdf)
bmjresp-2023-001878supp012.pdf (246.5KB, pdf)
Discussion
The TME is a complex and dynamic evolving ecosystem during the initiation, development and metastasis of tumour. The composition of TME and its interaction with surrounding tumour cells affect the efficiency of therapy and then determine the therapeutic resistance and tumour recurrence.8 36 37 Taking the advantage of cutting-edge sequencing technology, the understanding of TME characterisation in LUAD has significantly improved in recent years.7 16 19 25 However, the previous studies mainly focused on the certain stage of LUAD progression, and lacked the comprehensive comparison between different stages. In this study, we integrated two independent scRNA-seq cohorts to elucidate the dynamic changes of epithelial cell lineages and TME between early and advanced LUAD.
Compared with early LUAD, we found that three epithelial subclusters with high CNV score were significantly enriched in advanced stage. Specially, S100P, as a prognostic biomarker, has been reported to promote the migration and metastasis in diverse cancers, such as lung cancer,38 colorectal cancer,39 breast cancer40 and pancreatic cancer.41 Our survival analysis also revealed the upregulation of S100P predicting the poor survival outcomes in LUAD. Moreover, the mean of CNV scores was highest in S100P+ epithelial cells compared with other epithelial clusters. All evidence supported S100P+ epithelial cells could be a predictive biomarker for tumourigenesis of LUAD. Our functional analysis further uncovered the alteration of biological pathways in this subpopulation, which provides the insight into the newly therapeutic strategy for targeting S100P+ malignant cells.32 42 43
The fact that immunotherapy shows promising approach to significantly improve the clinical outcome of part of cancers, we also profiled the characterisation of TME during the progression of LUAD. The emerging studies and our result both revealed that SPP1+ macrophage, as the biomarker for metastasis, predicts poor prognosis in various tumours, especially in lung cancer. Currently, how osteopontin encoded by SPP1 interacts with TME still remains debated. Although SPP1+ macrophages have been generally reported to build intercellular networking with fibroblasts in colorectal cancer and lung cancer,23 44 there were few studies suggesting that SPP1 highly expressed in macrophages binds to its receptor CD44 on activated T cells to suppress cytotoxicity of T cells in TME.45 46 Here, our analysis supported that macrophages through SPP1-CD44 axis could interact with diverse subpopulations of immune cells in LUAD TME, such as CD8+ T cells, monocytes and mast cells. However, we had not observed crosstalk between macrophages and endothelial cells through SPP1 and CD44, suggesting the presence of other ligand-receptor interactions between these two cell clusters. Due to the critical function of SPP1+ macrophages and endothelial cells in metastases, the exploration of cell-cell communication between macrophages and endothelial cells should be useful to understand the tumour metastasis. In addition, the result from deconvolution analysis for bulk RNA-seq data showed that infiltration of SPP1+ macrophage cells into TME was significantly associated with poor outcome of LUAD. Taken together, these results suggested that SPP1 in macrophages may act as new immune checkpoint and inhibiting the interaction between macrophages and other cell populations in TME may be a promising method for LUAD treatment.
Moreover, we found other two immune cell clusters (KLRB1+CD8+ T cells and IGHG1+/IGHA1+ plasma cells) as potentially targeting cells to enhance the efficiency of antitumour. NK-cell receptor CD161 encoded by KLRB1 was one of the marked genes for conversing cytotoxicity of T cells (both CD4+ and CD8+ T cells).35 Inhibiting this receptor in infiltrating T cells activates the cytotoxic T cells to kill glioma tumour cells.47 Few studies further suggested that KLRB1+ T cells, as the cytotoxic memory cells, enriched in the gut48 and hepatocellular carcinoma49 TME, respectively, and could be high therapeutic potential cells to enhance the antitumour response. At the same time, plasma cells secrete antibodies to recognise tumour cell and then trigger the adaptive immune response.50 More and more evidence showed that tumour-infiltrated activated plasma cells were associated with improved prognosis in triple negative breast,51 prostate52 and colorectal cancers.53 The one probable reason to explore this better outcome was that tertiary lymphoid structures,54 55 normally consisting of the predominant plasma cells with high expression of IGHG1 and activation of T cells, facilitate the response to immunotherapy. However, the expression of marker genes and the presence of tertiary lymphoid structures in LUAD needed to be determined by further validation (such as IHC or multiple IHC) with sampling expansion.
Conclusions
Our findings highlight the key changes of composition of TME from LUAD early to advanced stage. Four cell populations (S100P+ epithelial, SPP1+ macrophage, KLRB1+CD8+ T cells and IGHG1+/IGHA1+ plasma cell) had significant changes in advance LUAD. The enriched S100P+ epithelial and SPP1+ macrophage are biomarkers of LUAD bad prognosis and even metastasis, while KLRB1+CD8+ T and IGHG1+/IGHA1+ plasma cells are candidates targeting populations to develop promising immunotherapy avenues to improve the clinical outcome.
bmjresp-2023-001878supp002.pdf (533.3KB, pdf)
bmjresp-2023-001878supp013.pdf (28.8KB, pdf)
Acknowledgments
The computations in this paper were run on the Siyuan-1 cluster supported by the Center for High Performance Computing at Shanghai Jiao Tong University.
Footnotes
HL and JQ contributed equally.
Contributors: CZ designed the study. HL, JQ, TC and CZ performed the data analysis. HL, LC and YS performed the experimental work. CZ, HL and TC wrote the manuscript. All authors have reviewed, corrected, read and approved the final manuscript. HL, TC and CZ are responsible for the overall content of the manuscript as the guarantor.
Funding: This work was supported by the Shanghai Pujiang Program(23PJ1411400), the National Natural Science Foundation of China (82303030), the Youth Foundation of the Shanghai Municipal Health Commission (20224Y0016), the Shanghai Sailing Programme (23YF1441100), and the Medical innovation project of Scientific and Technological innovation action plan of the Shanghai Committee of Science and Technology (21Y11913500).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. The h5ad file for integrated scRNA-seq data in the study is available at the figshare (https://figshare.com/articles/dataset/Lung_scVI_result_annotation_log1p_20230108_h5ad/22036547).
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Institutional Review Board of the Shanghai Chest Hospital, School of Medicine, Shanghai Jiao Tong University (ethical code: KS(Y)22013) and was conducted in accordance with the guidelines of the Declaration of Helsinki.
References
- 1.Shi J, Hua X, Zhu B, et al. Somatic Genomics and clinical features of lung adenocarcinoma: A retrospective study. PLoS Med 2016;13:e1002162. 10.1371/journal.pmed.1002162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender E. Epidemiology: the dominant malignancy. Nature 2014;513:S2–3. 10.1038/513S2a [DOI] [PubMed] [Google Scholar]
- 3.Testa U, Pelosi E, Castelli G. Molecular Charcterization of lung adenocarcinoma combining whole Exome sequencing, copy number analysis and gene expression profiling. Expert Rev Mol Diagn 2022;22:77–100. 10.1080/14737159.2022.2017774 [DOI] [PubMed] [Google Scholar]
- 4.Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363–85. 10.3322/caac.21565 [DOI] [PubMed] [Google Scholar]
- 5.Kumarakulasinghe NB, van Zanwijk N, Soo RA. Molecular targeted therapy in the treatment of advanced stage non-small cell lung cancer (NSCLC). Respirology 2015;20:370–8. 10.1111/resp.12490 Available: https://onlinelibrary.wiley.com/toc/14401843/20/3 [DOI] [PubMed] [Google Scholar]
- 6.Anderson NM, Simon MC. The tumor Microenvironment. Curr Biol 2020;30:R921–5. 10.1016/j.cub.2020.06.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altorki NK, Markowitz GJ, Gao D, et al. The lung Microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer 2019;19:9–31. 10.1038/s41568-018-0081-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune Microenvironment (TIME) for effective therapy. Nat Med 2018;24:541–50. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowbotham SP, Kim CF. Diverse cells at the origin of lung adenocarcinoma. Proc Natl Acad Sci U S A 2014;111:4745–6. 10.1073/pnas.1401955111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sainz de Aja J, Dost AFM, Kim CF. Alveolar progenitor cells and the origin of lung cancer. J Intern Med 2021;289:629–35. 10.1111/joim.13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim CF. Intersections of lung progenitor cells, lung disease and lung cancer. Eur Respir Rev 2017;26:170054. 10.1183/16000617.0054-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrot-Zhang J, Yao X, Devarakonda S, et al. Whole-genome characterization of lung adenocarcinomas lacking alterations in the RTK/RAS/RAF pathway. Cell Rep 2021;34:S2211-1247(21)00097-8. 10.1016/j.celrep.2021.108784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collisson EA, Campbell JD, Brooks AN, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkerson MD, Yin X, Walter V, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, Chromosomal instability, and methylation. PLoS ONE 2012;7:e36530. 10.1371/journal.pone.0036530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge X, Liu Z, Weng S, et al. Integrative Pharmacogenomics revealed three subtypes with different immune landscapes and specific therapeutic responses in lung adenocarcinoma. Comput Struct Biotechnol J 2022;20:3449–60. 10.1016/j.csbj.2022.06.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maynard A, McCoach CE, Rotow JK, et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell 2020;182:1232–51. 10.1016/j.cell.2020.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trefzer TB, Schneider MA, Jechow K, et al. Intratumoral heterogeneity and immune modulation in lung adenocarcinoma in female Smokers and never Smokers. Cancer Res 2022;82:3116–29. 10.1158/0008-5472.CAN-21-3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostman A. The tumor Microenvironment controls drug sensitivity. Nat Med 2012;18:1332–4. 10.1038/nm.2938 [DOI] [PubMed] [Google Scholar]
- 19.Wu F, Fan J, He Y, et al. Single-cell profiling of tumor heterogeneity and the Microenvironment in advanced non-small cell lung cancer. Nat Commun 2021;12:2540. 10.1038/s41467-021-22801-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otano I, Ucero AC, Zugazagoitia J, et al. At the crossroads of Immunotherapy for Oncogene-addicted Subsets of NSCLC. Nat Rev Clin Oncol 2023;20:143–59. 10.1038/s41571-022-00718-x Available: https://www.nature.com/articles/s41571-022-00718-x [DOI] [PubMed] [Google Scholar]
- 21.Liu B, Hu X, Feng K, et al. Temporal single-cell tracing reveals Clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat Cancer 2022;3:108–21. 10.1038/s43018-021-00292-8 [DOI] [PubMed] [Google Scholar]
- 22.Li S, Yang R, Sun X, et al. Identification of Spp1 as a promising biomarker to predict clinical outcome of lung adenocarcinoma individuals. Gene 2018;679:398–404. 10.1016/j.gene.2018.09.030 [DOI] [PubMed] [Google Scholar]
- 23.Yi X, Luo L, Zhu Y, et al. Spp1 facilitates cell migration and invasion by targeting Col11A1 in lung adenocarcinoma. Cancer Cell Int 2022;22:324. 10.1186/s12935-022-02749-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong B, Wu C, Huang L, et al. Macrophage-related Spp1 as a potential biomarker for early lymph node metastasis in lung adenocarcinoma. Front Cell Dev Biol 2021;9. 10.3389/fcell.2021.739358 Available: https://www.frontiersin.org/articles/10.3389/fcell.2021.739358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim N, Kim HK, Lee K, et al. Single-cell RNA sequencing demonstrates the molecular and cellular Reprogramming of metastatic lung adenocarcinoma. Nat Commun 2020;11. 10.1038/s41467-020-16164-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schabath MB, Welsh EA, Fulp WJ, et al. Differential Association of Stk11 and Tp53 with KRAS Mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2016;35:3209–16. 10.1038/onc.2015.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okayama H, Kohno T, Ishii Y, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res 2012;72:100–11. 10.1158/0008-5472.CAN-11-1403 [DOI] [PubMed] [Google Scholar]
- 28.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez R, Regier J, Cole MB, et al. Deep Generative modeling for single-cell Transcriptomics. Nat Methods 2018;15:1053–8. 10.1038/s41592-018-0229-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using Cellchat. Nat Commun 2021;12. 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu T, Wang Z, Pe’er D, et al. Cell type and gene expression Deconvolution with Bayesprism enables Bayesian integrative analysis across bulk and single-cell RNA sequencing in oncology. Nat Cancer 2022;3:505–17. 10.1038/s43018-022-00356-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer 2015;15:96–109. 10.1038/nrc3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Y, Fu Y, Liu C, et al. Stathmin 1 is a biomarker for diagnosis of Microvascular invasion to predict prognosis of early hepatocellular carcinoma. Cell Death Dis 2022;13:176. 10.1038/s41419-022-04625-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokoyama S, Nakayama S, Xu L, et al. Secretoglobin 3A2 eliminates human cancer cells through Pyroptosis. Cell Death Discov 2021;7:12. 10.1038/s41420-020-00385-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konduri V, Oyewole-Said D, Vazquez-Perez J, et al. Cd8+Cd161+ T-cells: cytotoxic memory cells with high therapeutic potential. Front Immunol 2021;11. 10.3389/fimmu.2020.613204 Available: 10.3389/fimmu.2020.613204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bejarano L, Jordāo MJC, Joyce JA. Therapeutic targeting of the tumor Microenvironment. Cancer Discov 2021;11:933–59. 10.1158/2159-8290.CD-20-1808 [DOI] [PubMed] [Google Scholar]
- 37.Obenauf AC. Mechanism-based combination therapies for metastatic cancer. Sci Transl Med 2022;14:eadd0887. 10.1126/scitranslmed.add0887 [DOI] [PubMed] [Google Scholar]
- 38.Wang T, Du G, Wang D. The S100 protein family in lung cancer. Clinica Chimica Acta 2021;520:67–70. 10.1016/j.cca.2021.05.028 [DOI] [PubMed] [Google Scholar]
- 39.Dong L, Wang F, Yin X, et al. Overexpression of S100P promotes colorectal cancer metastasis and decreases Chemosensitivity to 5-FU in vitro. Mol Cell Biochem 2014;389:257–64. 10.1007/s11010-013-1947-5 [DOI] [PubMed] [Google Scholar]
- 40.Cong Y, Cui Y, Wang S, et al. Calcium-binding protein S100P promotes tumor progression but enhances Chemosensitivity in breast cancer. Front Oncol 2020;10:566302. 10.3389/fonc.2020.566302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barry S, Chelala C, Lines K, et al. S100P is a metastasis-associated gene that facilitates Transendothelial migration of Pancreatic cancer cells. Clin Exp Metastasis 2013;30:251–64. 10.1007/s10585-012-9532-y [DOI] [PubMed] [Google Scholar]
- 42.Schmid F, Dahlmann M, Röhrich H, et al. Calcium-binding protein S100P is a new target gene of Macc1, drives colorectal cancer metastasis and serves as a Prognostic biomarker. Br J Cancer 2022;127:675–85. 10.1038/s41416-022-01833-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arumugam T, Logsdon CD. S100P: a novel therapeutic target for cancer. Amino Acids 2011;41:893–9. 10.1007/s00726-010-0496-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi J, Sun H, Zhang Y, et al. Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and Spp1+ Macrophages in colorectal cancer. Nat Commun 2022;13. 10.1038/s41467-022-29366-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng M, Liang G, Yin Z, et al. Immunosuppressive role of Spp1-Cd44 in the tumor Microenvironment of Intrahepatic Cholangiocarcinoma assessed by single-cell RNA sequencing. J Cancer Res Clin Oncol 2023;149:5497–512. 10.1007/s00432-022-04498-w Available: 10.1007/s00432-022-04498-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nallasamy P, Nimmakayala RK, Karmakar S, et al. Pancreatic tumor Microenvironment factor promotes cancer Stemness via Spp1-Cd44 axis. Gastroenterology 2021;161:1998–2013. 10.1053/j.gastro.2021.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inhibitory Cd161 receptor is expressed on glioma-infiltrating T cells. Cancer Discov 2021;11:OF19. 10.1158/2159-8290.CD-RW2021-027 Available: 10.1158/2159-8290.CD-RW2021-027 [DOI] [PubMed] [Google Scholar]
- 48.Billerbeck E, Kang Y-H, Walker L, et al. Analysis of Cd161 expression on human Cd8+ T cells defines a distinct functional subset with tissue-homing properties. Proc Natl Acad Sci U S A 2010;107:3006–11. 10.1073/pnas.0914839107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Y, Wu L, Zhong Y, et al. Single-cell landscape of the Ecosystem in early-relapse hepatocellular carcinoma. Cell 2021;184:404–21. 10.1016/j.cell.2020.11.041 [DOI] [PubMed] [Google Scholar]
- 50.Sharonov GV, Serebrovskaya EO, Yuzhakova DV, et al. B cells, plasma cells and antibody Repertoires in the tumour Microenvironment. Nat Rev Immunol 2020;20:294–307. 10.1038/s41577-019-0257-x [DOI] [PubMed] [Google Scholar]
- 51.Sakaguchi A, Horimoto Y, Onagi H, et al. Plasma cell infiltration and treatment effect in breast cancer patients treated with Neoadjuvant chemotherapy. Breast Cancer Res 2021;23:99. 10.1186/s13058-021-01477-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiner AB, Vidotto T, Liu Y, et al. Plasma cells are enriched in localized prostate cancer in black men and are associated with improved outcomes. Nat Commun 2021;12:935. 10.1038/s41467-021-21245-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia J, Xie Z, Niu G, et al. Single-cell landscape and clinical outcomes of infiltrating B cells in colorectal cancer. Immunology 2023;168:135–51. 10.1111/imm.13568 [DOI] [PubMed] [Google Scholar]
- 54.Cabrita R, Lauss M, Sanna A, et al. Author correction: tertiary Lymphoid structures improve Immunotherapy and survival in Melanoma. Nature 2020;580:E1. 10.1038/s41586-020-2155-6 [DOI] [PubMed] [Google Scholar]
- 55.Schumacher TN, Thommen DS. Tertiary Lymphoid structures in cancer. Science 2022;375:eabf9419. 10.1126/science.abf9419 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjresp-2023-001878supp014.pdf (7.1MB, pdf)
bmjresp-2023-001878supp015.pdf (121.1KB, pdf)
bmjresp-2023-001878supp001.pdf (254.9KB, pdf)
bmjresp-2023-001878supp003.pdf (264.4KB, pdf)
bmjresp-2023-001878supp004.pdf (244.4KB, pdf)
bmjresp-2023-001878supp005.pdf (244.6KB, pdf)
bmjresp-2023-001878supp006.pdf (245KB, pdf)
bmjresp-2023-001878supp007.pdf (234.2KB, pdf)
bmjresp-2023-001878supp008.pdf (261.5KB, pdf)
bmjresp-2023-001878supp009.pdf (251.8KB, pdf)
bmjresp-2023-001878supp010.pdf (253KB, pdf)
bmjresp-2023-001878supp011.pdf (264.4KB, pdf)
bmjresp-2023-001878supp012.pdf (246.5KB, pdf)
bmjresp-2023-001878supp002.pdf (533.3KB, pdf)
bmjresp-2023-001878supp013.pdf (28.8KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. The h5ad file for integrated scRNA-seq data in the study is available at the figshare (https://figshare.com/articles/dataset/Lung_scVI_result_annotation_log1p_20230108_h5ad/22036547).