Abstract
A 260-kDa structural cell wall protein was purified from sodium dodecyl sulfate-treated cell walls of Saccharomyces cerevisiae by incubation with Rarobacter faecitabidus protease I, which is a yeast-lytic enzyme. Amino acid sequence analysis revealed that this protein is the product of the SED1 gene. SED1 was formerly identified as a multicopy suppressor of erd2, which encodes a protein involved in retrieval of luminal endoplasmic reticulum proteins from the secretory pathway. Sed1p is very rich in threonine and serine and, like other structural cell wall proteins, contains a putative signal sequence for the addition of a glycosylphosphatidylinositol anchor. However, the fact that Sed1p, unlike other cell wall proteins, has six cysteines and seven putative N-glycosylation sites suggests that Sed1p belongs to a new family of cell wall proteins. Epitope-tagged Sed1p was detected in a β-1,3-glucanase extract of cell walls by immunoblot analysis, suggesting that Sed1p is a glucanase-extractable cell wall protein. The expression of Sed1p mRNA increased in the stationary phase and was accompanied by an increase in the Sed1p content of cell walls. Disruption of SED1 had no effect on exponentially growing cells but made stationary-phase cells sensitive to Zymolyase. These results indicate that Sed1p is a major structural cell wall protein in stationary-phase cells and is required for lytic enzyme resistance.
Yeast Saccharomyces cerevisiae has a rigid cell wall outside its cell membrane (21). The cell wall is the largest organelle of the yeast cell and protects the cell from mechanical injury, hypotonic lysis, and chemical substances that could damage the cell. The cell wall is composed of glucan, mannoproteins, and a small amount of chitin. Mannoproteins are proteins with a large amount of N- and/or O-linked mannoses and compose the outermost layer of the yeast cell wall (42). Mannoproteins in the yeast cell wall are grouped into two classes (9). The first class is proteins that can be extracted with sodium dodecyl sulfate (SDS). These proteins are considered to be noncovalently entrapped or associated in the cell wall. The other class is cell wall proteins that cannot be extracted with SDS (3, 36). These proteins are considered to be covalently bound to cell wall glucan and can be solubilized by incubation with β-1,3-glucanase (36). Among these proteins, α-agglutinin and Aga1p are involved in sexual agglutination (14) and Flo1p is related to flocculation (40), but there are many other proteins whose functions are not clear. These proteins are considered as structural cell wall proteins and some of them (Cwp1p, Cwp2p, and Tip1p) have been previously identified from a glucanase extract of SDS-treated cell walls (36). These proteins are rich in serine and threonine residues and contain signals for the addition of a glycosylphosphatidylinositol (GPI) anchor. This anchor is transferred to cell wall proteins in the endoplasmic reticulum, and GPI-anchored proteins are transported through the Golgi apparatus to the plasma membrane. Although some GPI-anchored proteins remain to bind to the plasma membrane (19), cell wall proteins are transferred to β-1,6-glucan that is bound to β-1,3-glucan by an unknown mechanism (7).
In previous studies, we have isolated cell wall proteins solubilized from SDS-treated cell walls with Rarobacter faecitabidus protease I (RPI) and characterized them as Cwp1p (31) and Tir1p (8). Cwp1p was coincidentally identified from a laminarinase extract of cell walls (36). Cwp1p is a putative GPI-anchored protein. The C-terminal hydrophobic sequence of Cwp1p is needed for attaching the molecule to cell walls because a mutant Cwp1p deficient in this sequence was secreted into the culture medium (31). Tir1p is a cell wall protein specifically expressed in cells cultured anaerobically (8) and also has a GPI anchor signal. Since these proteins are solubilized from cell walls with β-1,3-glucanase, these proteins were considered to be bound to cell wall glucan. RPI is a yeast-lytic protease that specifically recognizes mannose chains of mannoproteins and cleaves their peptide bonds (29, 30). Therefore, solubilization of cell wall proteins by digestion with RPI is very useful since extracted proteins show less heterogeneity in size than do cell wall proteins prepared by glucanase digestion (8, 31).
Here we report the isolation of another cell wall protein by using RPI. This protein was prepared from aerobic culture and was identified as Sed1p by amino acid sequence analysis. SED1 was highly expressed in the stationary phase, and its disruptant was more sensitive to Zymolyase than the wild-type cells in the stationary phase. We believe that Sed1p is required for stress resistance in stationary-phase cells.
MATERIALS AND METHODS
Yeast strains and media.
S. cerevisiae YPH499 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1), YPH501 (MAT a/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1) (32), and LB2134-3B (MATa mnn9) (1) were used in this study. YPAD medium (1% yeast extract, 2% Bacto Peptone, 40 mg of adenine per liter, 2% glucose) was used for yeast cultures. YPAD medium supplemented with 10% (wt/vol) sorbitol was used for the mnn9 mutant.
Preparation of cell walls and enzyme treatments.
RPI was prepared as described previously (31). Yeast cells were inoculated into the medium and cultured at 30°C for 48 h as a preculture. After 4 ml of the preculture was added to 200 ml of the main culture in a 500-ml culture flask, cells were incubated at 30°C for the appropriate time with shaking at 120 rpm in a rotary shaker. Cell walls were prepared as previously described (8). Briefly, cells were harvested from 1,000 ml of culture by centrifugation, washed, and disrupted by shaking with glass beads in a Braun homogenizer (B. Braun, Melsungen, Germany). The glass beads were removed by decantation, and the cell walls were pelleted by centrifugation and washed with 5 M LiCl. The cell walls were suspended in 20 ml of 50 mM Tris-HCl (pH 8.0) containing 2% SDS, 100 mM EDTA, and 40 mM dithiothreitol and then heated at 100°C for 10 min to remove noncovalently bound proteins and proteins bound by disulfide bridges. The SDS-treated cell walls were pelleted by centrifugation at 1,500 × g for 5 min, washed five times with 1 mM phenylmethylsulfonyl fluoride and one time with 50 mM Tris-HCl buffer (pH 8.0), and then digested with 5 μg of RPI per ml in 20 ml of 50 mM Tris-HCl buffer (pH 8.0) at 30°C for 6 h. The reaction mixture containing solubilized proteins was centrifuged at 15,000 × g for 10 min, and the supernatant was analyzed as RPI-extracted cell wall proteins. The SDS-treated cell walls were also digested with 50 mU of laminarinase (L5144; Sigma Chemical Co., St. Louis, Mo.) per ml in 20 ml of 50 mM sodium acetate buffer (pH 5.0) containing 1 mM phenylmethylsulfonyl fluoride at 37°C for 2 h. The reaction mixture was centrifuged at 15,000 × g for 10 min, and the supernatant was analyzed as glucanase-extracted cell wall proteins.
Purification of Sed1p.
Five-milliliter aliquots of RPI-extracted cell wall proteins were subjected to Superdex 200 gel filtration chromatography (26 by 600 mm; Pharmacia, Piscataway, N.J.). The elution buffer was 10 mM Tris-HCl (pH 8.0) containing 150 mM NaCl and 0.05% sodium azide with a flow rate of 1 ml/min. The chromatography was repeated, and fractions containing Sed1p were collected and lyophilized. These fractions was dissolved in 0.05% trifluoroacetic acid, applied to a TSKgel Phenyl-5PW RP reverse-phase column (Tosoh, Tokyo, Japan), and eluted with a linear gradient of 5 to 80% acetonitrile in 0.05% trifluoroacetic acid. The major peak containing Sed1p was collected and lyophilized.
Amino acid sequencing of Sed1p.
Purified protein (2 nmol) was dissolved in 10 mM Tris-HCl (pH 9.0) containing 10 pmol of Achromobacter protease I (Takara, Kyoto, Japan), and incubated at 37°C for 15 h. After the reaction was stopped by adding 0.1% trifluoroacetic acid, the produced peptides were subjected to reverse-phase chromatography (μBondasphere C18 100 Å; Waters, Milford, Mass.) and eluted with a linear gradient of 5 to 40% acetonitrile in 0.1% trifluoroacetic acid. The amino acid sequences of the purified peptides were determined by Edman degradation with an automated protein sequencer (491 Procise; Perkin-Elmer, Norwalk, Conn.). On the other hand, the purified protein (500 pmol) was blotted onto a Prosorb membrane (Perkin-Elmer) and incubated in 0.5% (wt/vol) polyvinylpyrrolidone-40–100 mM acetic acid at 37°C for 30 min. The membrane was washed 10 times with distilled water and incubated in 50 mM sodium phosphate (pH 7.0)–10 mU of pyroglutamate aminopeptidase (Takara) per ml–10 mM dithiothreitol at 50°C for 5 h. The membrane was washed three times with distilled water and applied to the protein sequencer.
PNGase F digestion.
The purified protein (10 μg) was denatured by boiling in 0.5% SDS for 10 min. After 1/10 volume of 0.5 M sodium phosphate buffer (pH 7.5) and 10% Nonidet P-40 were added, the sample was treated with 5,000 U of peptide N-glycosidase F (PNGase F; New England Biolabs, Beverly, Mass.) at 37°C for 15 h. Each sample was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and glycoproteins were detected by Western blotting analysis with concanavalin A as a probe.
SDS-PAGE and Western blotting analysis.
Protein samples were subjected to SDS-PAGE by the method of Laemmli (13) and stained with Coomassie brilliant blue R-250 (CBB; Bio-Rad, Richmond, Calif.) or electroblotted onto an Immobilon-P membrane (Millipore, Bedford, Mass.) in a solution of 25 mM Tris, 192 mM glycine, 20% methanol, and 0.05% SDS (34). The obtained blots were blocked for 30 min with 1% bovine serum albumin in Tris-buffered saline (TBS; 10 mM Tris-HCl buffer [pH 8.0]) containing 150 mM NaCl). For staining of glycoproteins, the membrane was probed with concanavalin A conjugated with biotin (Seikagakukogyou, Tokyo, Japan) diluted 1,000-fold in TBST (TBS with 0.05% Tween 20) and developed with avidin conjugated with alkaline phosphatase (Zymed, South San Francisco, Calif.). For staining of influenza virus hemagglutinin (HA) epitope-tagged proteins, the membrane was probed with 5 μg of anti-HA monoclonal antibody (Boehringer, Mannheim, Germany) per ml in TBST and developed with anti-mouse immunoglobulin G antibody conjugated with alkaline phosphatase (Promega, Madison, Wis.).
Disruption of the SED1 gene.
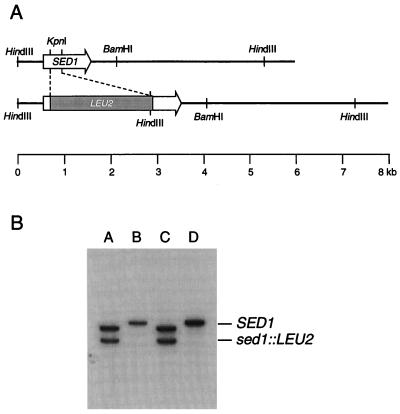
The SED1 gene was amplified by the PCR method (25) with 5′-TCATCTGTGTACACTAAGTAA-3′ and 5′-AGTCCATAACAAGGAAGGTAA-3′ as primers. The PCR product was digested with EcoRI and BamHI, and a 3.3-kb EcoRI/BamHI fragment was cloned into pUC118 that had been digested with EcoRI and BamHI. The resultant plasmid (pUC-SED1) was digested with KpnI, blunt ended with T4 polymerase, and ligated with a blunted BamHI fragment of pUC-LEU2 containing the LEU2 gene as a selectable marker (24). The resultant plasmid, in which the LEU2 gene was inserted in the middle of the SED1 open reading frame, was linearized by digestion with EcoRI and BamHI, and wild-type diploid strain YPH501 was transformed with it (2). Transformants with the LEU+ phenotype were recovered, and disruption of one of the two chromosomal SED1 genes was confirmed by Southern blot analysis. Chromosomal DNA was prepared as described previously (23) and then digested with HindIII, electrophoresed, blotted to a membrane, and hybridized with a 32P-labeled probe (the 3.3-kb EcoRI/BamHI fragment of pUC-SED1) at 65°C in a hybridization solution containing 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) 5× Denhardt’s solution, 0.5% SDS, and 20 μg of sonicated salmon sperm DNA per ml. After washing twice at 65°C for 20 min with 2× SSPE–0.1% SDS, the blot was used to expose X-ray film. The transformant with SED1/Δsed1::LEU2 was sporulated and dissected. Chromosome DNAs were prepared from a typical tetrad, and the gene disruption of SED1 was determined by Southern blotting analysis as described above.
Epitope tagging of Sed1p.
A single-strand DNA prepared from pUC-SED1 was annealed to a synthetic oligonucleotide containing a sequence coding for the influenza virus HA 12CA5 epitope (5′-TCGACTACTTTGGCCTACCCATACGACGTCCCAGACTACGCTCAATTTTCCAACAGT-3′; the inserted sequence is underlined) (11), and subsequent reactions were carried out (18) with a reagent kit (Amersham, Little Chalfont, England) according to the manufacturer’s instructions. Escherichia coli JM109 was transformed with the reaction product and screened for a transformant having a plasmid containing an AatII site that was present in the inserted sequence. The DNA of the candidate plasmid was sequenced (26) with 5′-CTTCTTCCACCGATGTCACTT-3′ as a primer to confirm that the appropriate mutation had been introduced. The EcoRI/BamHI fragment of the plasmid with the HA insertion was cloned into pRS426 that had been digested with EcoRI and BamHI. The resultant plasmid (pRS426-HA::SED1) was used for the transformation of YPH499.
Northern blot analysis.
Total RNA of yeast cells was prepared by the hot phenol extraction method (10). Five micrograms of total RNA was denatured at 65°C for 5 min in a mixture containing 50% formamide, MOPS [20 mM 3-(N-morpholino)propanesulfonic acid, 5 mM sodium acetate, 1 mM EDTA (pH 7.0)], and 0.16 volume of formaldehyde (37%, vol/vol). The samples were electrophoresed in a gel consisting of 1.2% agarose, MOPS, and 0.17 volume of formaldehyde (37%, vol/vol) and transferred to a nylon membrane (Hybond-N+; Amersham). The 3.3-kb EcoRI/BamHI fragment of pUC-SED1 was used as a probe. A probe for ACT1 was synthesized by the PCR method as described previously (8). The probes were labeled with 32P and hybridized to the blots for 16 h at 65°C in a buffer containing 50% formamide, 2× SSPE, 5× Denhardt’s solution, and 20 μg of denatured salmon sperm DNA per ml. After being washed twice at 65°C for 20 min with 2× SSPE–0.1% SDS, the blots were used to expose X-ray film.
Zymolyase sensitivity.
Yeast cells were cultured with shaking at 30°C for 6 and 48 h. Cells were harvested, washed with water, and suspended in 0.1 M sodium phosphate buffer (pH 7.5). After addition of 20 μg of Zymolyase 20T (Seikagakukogyou) per ml, the optical density at 660 nm was measured periodically.
RESULTS
Gel filtration analysis of cell wall proteins.
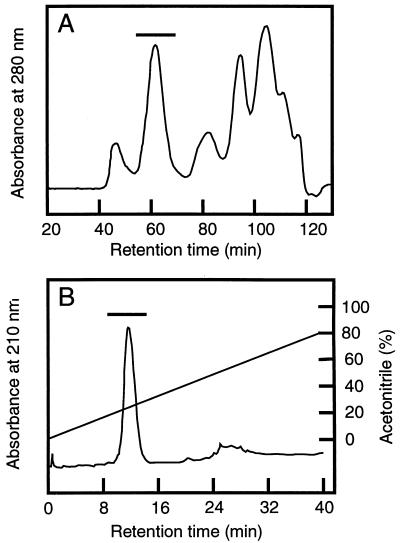
The mnn9 mutant was first used for analysis of cell wall proteins. The mnn9 mutant lacks the outer chains of N-linked sugars, which simplifies the analysis of cell wall proteins (1). Cell wall proteins were released from SDS-treated cell walls with RPI and analyzed by gel filtration chromatography. When the same sample was analyzed by SDS-PAGE and CBB staining, only Cwp1p could be observed clearly (31). However, the gel filtration chromatogram (Fig. 1A) shows the presence of several species of proteins. Among these proteins, the largest and most abundant, with a molecular mass of 260 kDa (gp260), was analyzed further.
FIG. 1.
Purification of a major cell wall protein by gel filtration and reverse-phase chromatographies. (A) Yeast cells (mnn9) were cultured in YPAD medium with shaking for 30 h. The cell wall fraction was prepared and treated with SDS to remove noncovalently bound proteins. Cell wall proteins were solubilized with RPI. After centrifugation, 5 ml of the supernatant was applied to a Superdex 200 gel filtration column. The column was eluted isocratically with 10 mM Tris-HCl (pH 8.0) containing 150 mM NaCl and 0.05% sodium azide with a flow rate of 1 ml/min. The protein peak indicated by the bar was collected and lyophilized. (B) The collected peak in panel A was further purified by TSKgel Phenyl-5PW RP reverse-phase chromatography with a linear gradient of 5 to 80% acetonitrile in 0.05% trifluoroacetic acid. The protein peak indicated by the bar (gp260) was collected and lyophilized.
Purification and identification of Sed1p.
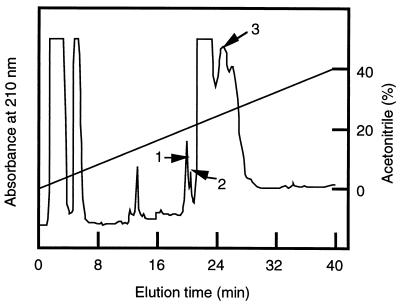
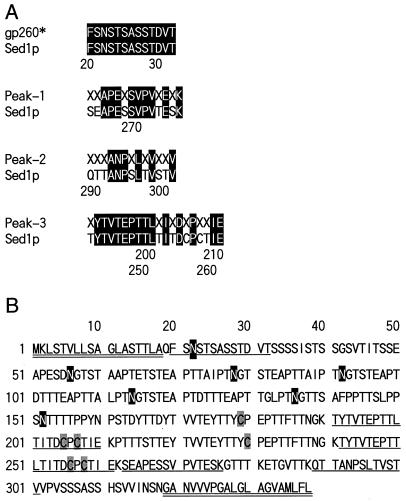
The peak fraction was applied to reverse-phase chromatography for purification. The chromatogram (Fig. 1B) contained essentially a single peak. The peak was collected, and 10 μg of the protein in this peak was applied to SDS-PAGE. The gel was stained with CBB, but no protein band was observed (data not shown). Hence, after electrophoresis the gel was blotted to a membrane and probed with concanavalin A. The protein migrated as a single but very smeared band ranging from 270 to 150 kDa (see Fig. 6, lane 1). The N-terminal amino acid residues of gp260 could not be sequenced, probably because of N-terminal blockage. Therefore, gp260 was digested with Achromobacter protease I and the resultant peptides were purified by reverse-phase chromatography (Fig. 2). The amino acid sequences of the purified peptides were analyzed, and the determined sequences are shown in Fig. 3A. The amino acid sequences of other peaks could not be determined. gp260 was also deblocked with pyroglutamate aminopeptidase and sequenced (Fig. 3A). All sequences that could be determined were homologous to sequences within Sed1p (Fig. 3), suggesting that gp260 was the SED1 gene product (6).
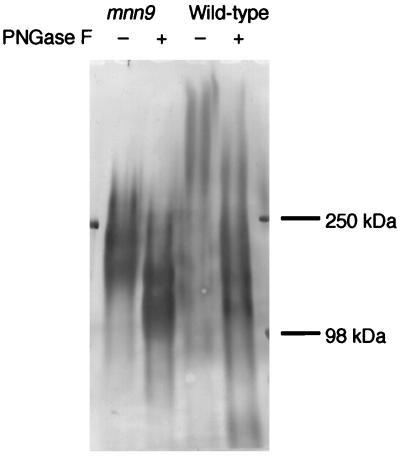
FIG. 6.
N-glycosylation of Sed1p. The mnn9 and wild-type cells (YPH499) were cultured in YPAD medium with shaking at 30°C for 30 h. The cell wall fractions were prepared and treated with SDS to remove noncovalently bound proteins. Cell wall proteins were solubilized with RPI, and the Sed1p fractions were purified by Superdex 200 gel filtration and TSKgel Phenyl-5PW RP reverse-phase chromatographies. The purified Sed1ps (10 μg) were treated with PNGase F as described in Materials and Methods. Untreated and PNGase F-treated Sed1ps from the mnn9 and wild-type cells were analyzed by SDS-PAGE (5 to 20% gel).
FIG. 2.
Reverse-phase chromatography of hydrolysates of gp260 with Achromobacter protease I. After gp260 and Achromobacter protease I (200:1 by molar ratio) were incubated at 37°C for 15 h, the hydrolysate was fractionated with a μBondasphere C18 reverse-phase chromatography column with a linear gradient of 5 to 40% acetonitrile in 0.1% trifluoroacetic acid. The numbered peaks were collected, and their amino acid sequences were determined.
FIG. 3.
Identification of gp260 as Sed1p. (A) Comparison of N-terminal and internal amino acid sequences of gp260 and Sed1p. The amino acid sequences of the numbered peaks in Fig. 2 were determined with a protein sequencer. X, unidentified amino acid; ∗, sequence identified after the N terminus of gp260 was deblocked with pyroglutamate aminopeptidase. Numbers indicate the residue numbers of Sed1p (6). (B) Amino acid sequence of Sed1p obtained from the literature (6). Underlines indicate sequences that were obtained from sequencing gp260 and its fragments. Sequences with double underlines are terminal hydrophobic sequences. Putative N-glycosylation sites are designated as shadowed letters. Cysteine residues are shaded.
Gene disruption of SED1.
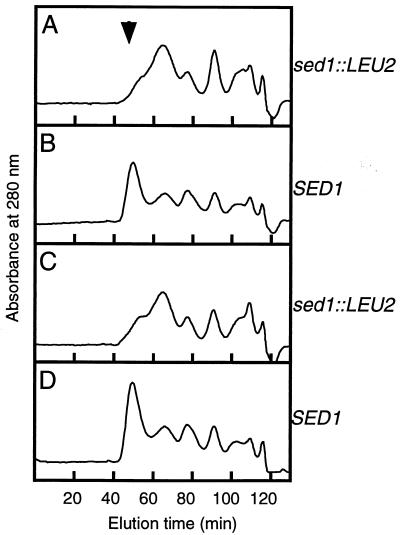
To confirm that gp260 is the SED1 gene product, gene disruption of SED1 was carried out and cell wall proteins were analyzed. A part of the coding region of SED1 (240 bp) was deleted and replaced with the LEU2 gene (Fig. 4A). The resultant sed1::LEU2 DNA was used to transform the wild-type diploid cells. Heterozygous SED1/sed1::LEU2 diploids were sporulated, and tetrads were dissected. All four spores produced viable cells on YPAD medium, indicating that SED1 is not required for viability in rich medium, in agreement with a previous report (6). Southern blotting analysis of four haploid cells from a representative ascus indicated that two of four haploids had the sed1::LEU2 allele (Fig. 4B). Cell wall proteins were solubilized by RPI treatment from the same four haploid cells and analyzed by gel filtration (Fig. 5). A major peak from cells with the SED1 allele eluted sooner than gp260 prepared from mnn9 cells (Fig. 5B and D). This protein, with a molecular mass of 300 kDa, was further purified by reverse-phase chromatography. We confirmed that this protein is Sed1p by amino acid sequencing after deblocking with pyroglutamate aminopeptidase (data not shown). The peak corresponding to Sed1p was not observed in RPI extract from cells with the sed1::LEU2 allele (Fig. 5A and C). These results clearly show that Sed1p is a major cell wall protein that can be released by RPI.
FIG. 4.
Gene disruption of SED1. (A) Construction of sed1::LEU2. The 240-bp KpnI fragment of SED1 was replaced with a DNA fragment containing LEU2 as a selectable marker. The linearized DNA containing sed1::LEU2 was used to transform a diploid strain (YPH501). (B) Southern blot analysis to confirm the gene disruption. Chromosomal DNAs from four haploid strains (A to D) from a representative ascus after sporulation of SED1/sed1::LEU2 heterozygous diploid were digested with HindIII and probed with a 3.3-kb EcoRI-BamHI fragment of pUC-SED1.
FIG. 5.
Analysis of cell wall proteins prepared from wild-type and sed1 disruptant cells. The same haploid strains (A to D) used for the Southern blot (Fig. 4B) were cultured in YPAD medium with shaking at 30°C for 30 h. The cell wall fractions were prepared and treated with SDS to remove noncovalently bound proteins. Cell wall proteins were solubilized with RPI, and the supernatants were analyzed by Superdex 200 gel filtration chromatography. The column was eluted isocratically with 10 mM Tris-HCl (pH 8.0) containing 150 mM NaCl and 0.05% sodium azide with a flow rate of 1 ml/min. The arrow indicates the position of Sed1p in all panels.
Sed1p is a highly glycosylated protein.
SED1 has previously been identified as a multicopy suppressor of erd2, which encodes a protein involved in retrieval of luminal endoplasmic reticulum proteins from the secretory pathway (27). The amino acid sequence of Sed1p is shown in Fig. 3B. Sed1p is composed of 338 amino acids, and its calculated molecular mass is 34,429 Da. Sed1p is very rich in threonine (29.3%) and serine (12.4%) and, like other cell wall proteins, contains a putative signal sequence for the addition of a GPI anchor (8, 31, 36). However, unlike other cell wall proteins such as Cwp1p, Tip1p, and Tir1p (36, 31, 8), Sed1p contains four cysteines and six putative N-glycosylation sites. The codon bias of the SED1 gene is 0.705 (22), which suggests that the SED1 gene is abundantly expressed.
The primary structure of Sed1p suggests that it is highly glycosylated by both N- and O-linked sugars. The purified Sed1ps from mnn9 and wild-type cells were analyzed by SDS-PAGE after PNGase F treatment, which eliminates N-linked sugars from proteins (Fig. 6). Sed1p from wild-type cells was very smeared and migrated more slowly than that from mnn9 cells. PNGase F treatment decreased the apparent molecular mass of Sed1p from both wild-type cells (from 810 to 360 kDa to 230 to 110 kDa) and mnn9 cells (from 270 to 150 kDa to 190 to 100 kDa). It was noteworthy that the Sed1p bands were very smeared even after PNGase F treatment. This may be due to the heterogeneity of O-linked sugars because Sed1p contains many serine and threonine residues. Considering that the molecular mass of the protein portion of Sed1p is only 34,429 Da, Sed1p must be heavily glycosylated by O- and N-linked sugars.
Sed1p is a glucanase-extractable cell wall protein.
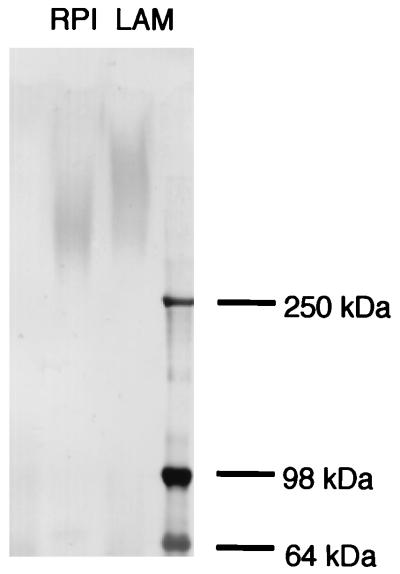
To characterize Sed1p localization, an influenza virus HA 12CA5 epitope was introduced just after the sequence coding for a putative signal peptide of the SED1 gene by oligonucleotide-directed in vitro mutagenesis. Cell walls were prepared from yeast cells harboring HA-tagged Sed1p, and Sed1p was purified from the RPI extract of the cell walls. The N-terminal amino acid sequence of this protein was YPYDVPDYAQFSNSTSASSTDV, indicating that the HA tag was correctly introduced into the N terminus of mature Sed1p and did not affect the synthesis of Sed1p. HA-tagged Sed1p was observed as a smeared band with a molecular mass of 600 to 300 kDa in Western blotting analysis of laminarinase-extracted cell wall proteins (Fig. 7, LAM), whereas RPI-extracted HA-tagged Sed1p migrated more quickly (Fig. 7, RPI). These results indicate that Sed1p is a structural cell wall mannoprotein that is covalently bound to the cell wall glucan and can be solubilized from SDS-treated cell walls by laminarinase or RPI digestion.
FIG. 7.
Analysis of HA-tagged Sed1p. The cells containing pRS426-HA::SED1 were cultured in YPAD medium with shaking at 30°C for 30 h. The cell wall fraction was prepared and treated with SDS to remove noncovalently bound proteins. Cell wall proteins were solubilized with RPI or laminarinase and applied to SDS-PAGE (5 to 20% gel). Proteins were transferred to a membrane and probed with anti-HA monoclonal antibody and anti-mouse immunoglobulin G secondary antibody conjugated with alkaline phosphatase. RPI, HA-Sed1p obtained by RPI extraction; LAM, HA-Sed1p obtained by laminarinase extraction.
Sed1p is highly expressed in stationary-phase cells.
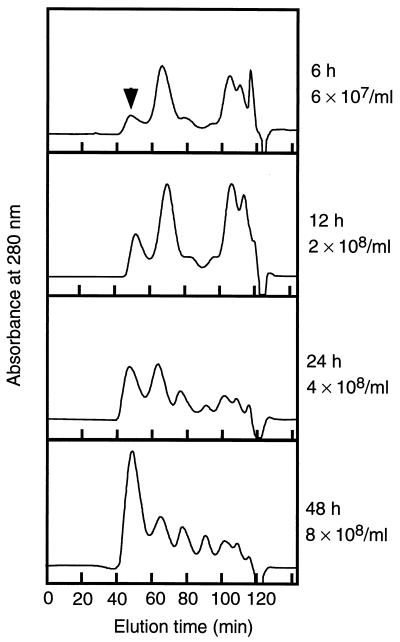
The amounts of Sed1p at various growth times were examined by RPI extraction of SDS-treated cell walls and gel filtration (Fig. 8). The amount of Sed1p was relatively minor in the exponential phase (Fig. 8, 6 and 12 h) but increased with culture time. After 48 h of culture, when the cells were in the stationary phase, Sed1p was the most abundant cell wall protein. In the stationary phase, Sed1p accounted for 30% of the RPI-extractable cell wall proteins.
FIG. 8.
Analysis of cell wall proteins with various culture times. Yeast cells (YPH499) were cultured in 1,000 ml of YPAD medium with shaking at 30°C for the indicated culture time. The cell wall fractions were prepared and treated with SDS to remove noncovalently bound proteins. Cell wall proteins were solubilized with RPI, and the supernatants were analyzed by Superdex 200 gel filtration chromatography. The column was eluted isocratically with 10 mM Tris-HCl (pH 8.0) containing 150 mM NaCl and 0.05% sodium azide with a flow rate of 1 ml/min. Each analyzed sample contained cell wall proteins extracted from approximately 2 × 1010 cells for each culture time. Cell numbers for each of the culture times are also indicated.
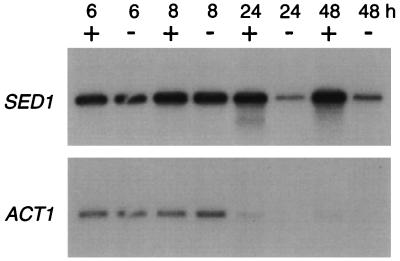
Expression of SED1 was also investigated by a Northern blotting analysis (Fig. 9). SED1 expression was moderate in the log phase (6 h) and 2.7-fold greater in the stationary phase (48 h) than at 6 h of culture, even though expression of ACT1 was weak in the stationary phase. SED1 was expressed in both shaking culture and static culture. Oxygen-dependent expression as seen in TIR1 was not observed (4, 8). These results indicate that Sed1p is highly expressed in the stationary phase in the shaking culture.
FIG. 9.
Northern blotting analysis of SED1. Wild-type yeast cells (YPH499) were cultured in YPAD medium with (+) or without (−) shaking at 30°C for the indicated time. Total RNA was extracted from the cells, and the expression of SED1 was analyzed by Northern blotting. The expression of ACT1 as a control gene was also analyzed. Numbers above panel show culture times in hours.
A SED1 disruptant is sensitive to Zymolyase in the stationary phase.
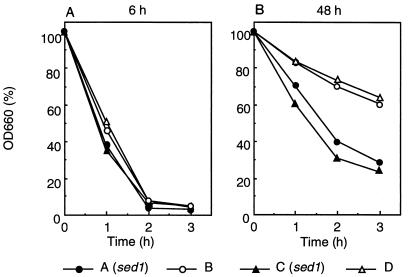
Stationary-phase cells are known to be resistant to lytic enzymes because their cell wall is thicker than that of exponentially growing cells (3). The increased expression of SED1 in the stationary phase led us to examine lytic enzyme resistance of sed1 disruptants. Four haploid strains from a representative ascus of a heterozygous diploid disruptant (SED1/sed1::LEU2), the same strains used for the Southern blot in Fig. 4B, were cultured and Zymolyase sensitivities were determined. The disruptant and wild-type cells in the exponential phase showed the same level of sensitivity to Zymolyase (Fig. 10A left), but the disruptant cells in the stationary phase were more sensitive to Zymolyase than wild-type cells (Fig. 10, right). These results indicate that Sed1p is required for Zymolyase resistance in the stationary phase.
FIG. 10.
Zymolyase sensitivity of the sed1 disruptant. The same haploid strains (A [sed1::LEU2], B [SED1], C [sed1::LEU2, and D [SED1]) used for the Southern blot (Fig. 4B) were cultured in YPAD medium with shaking at 30°C for 6 or 48 h. Cells were harvested, washed with water, and suspended in 0.1 M sodium phosphate buffer (pH 7.5). After addition of 20 μg of Zymolyase 20T per ml, the optical density at 660 nm (OD660) was measured periodically.
DISCUSSION
In this study, we used gel filtration analysis instead of SDS-PAGE to detect cell wall proteins. We isolated a major cell wall protein and identified it as Sed1p. Without the aid of a specific antibody, it is very difficult to identify cell wall proteins other than Cwp1p in SDS-PAGE (31). Poor staining of cell wall proteins with CBB in SDS-PAGE is probably because of their highly glycosylated nature (5). By this gel filtration method, we have already identified some other cell wall proteins, including Tir1p (8), Tip1p, and Cwp1p (our unpublished data). Gel filtration is also suitable for analytical profiling of cell wall proteins, as demonstrated by the experiment shown in Fig. 5.
Sed1p was predicted to be a glucanase-extractable cell wall protein because it has a putative GPI anchor signal that is a common character of such proteins (9). It was also shown that a fusion protein of a heterologous reporter protein and the C-terminal part of Sed1p was targeted to the cell walls (37, 38). However, there was no direct evidence showing that Sed1p is actually a structural cell wall protein bound to cell wall glucan. In this report, we have clearly shown that Sed1p is a cell wall protein covalently bound to cell wall glucan. First, Sed1p was extracted from SDS-treated cell walls by RPI, which is a protease that is specific to mannoproteins. Two other RPI-extractable proteins (Cwp1p and Tir1p) have already been determined to be cell wall proteins bound to cell wall glucan (8, 31). Second, HA-tagged Sed1p was extracted from SDS-treated cell walls by laminarinase, which is a mixture of β-1,3-glucanase and β-1,6-glucanase.
The N- and C-terminal hydrophobic sequences of Sed1p indicate that Sed1p is a putative GPI-anchored protein. Previous analysis of the GPI anchor attachment site revealed that the amino acid of the GPI anchor addition site (ω) of S. cerevisiae is Asn or Gly, and the amino acids at ω+1 and ω+2 are restricted to small amino acids (19, 20). According to this rule, Asn318 is the most likely candidate for the GPI attachment site in Sed1p. It has been proposed (7, 15, 16) that cell wall proteins with GPI anchor addition signals are transiently secreted to the plasma membrane as a GPI-anchored form and then are transferred to the cell walls by binding to the β-1, 6-glucan moiety of the cell walls, although the detailed mechanisms of relocation from the plasma membrane to the cell walls are unknown. The mechanisms that sorts GPI-anchored proteins between the plasma membrane and the cell wall is also unknown.
Since the molecular mass of the protein portion of mature Sed1p without the N- and C-terminal hydrophobic sequences is calculated to be 30,819 Da, the higher molecular mass obtained from gel filtration or SDS-PAGE is attributed to glycosylation of the protein. However, since highly glycosylated proteins might show anomalous electrophoretic mobility, the exact molecular mass of Sed1p should be evaluated by different methods. Based on the molecular masses estimated from SDS-PAGE, the protein content was calculated as only 5% in wild-type S. cerevisiae and only 14% in the mnn9 mutant, in which the outer chains of N-linked sugars are truncated (1). Sed1p can be considered a typical mannoprotein based on its high content of mannose. Sed1p may correspond to a cell wall mannoprotein described by Shibata et al. (28) and Frevert and Ballou (5) because all three proteins are similar in size, carbohydrate content, and amino acid composition.
Glucanase-extractable structural cell wall proteins that have been identified so far (Cwp1p, Cwp2p, Tip1p, and Tir1p) are mainly O-glycosylated proteins. Cwp1p has one putative N-glycosylation site in its amino acid sequence. However, the presence of an N-linked sugar in Cwp1p appears unlikely because no migration shift was observed by SDS-PAGE after the protein was treated with PNGase F (31). On the other hand, Sed1p has seven putative N-glycosylation sites and is known to be heavily glycosylated with N-linked sugars (Fig. 6). Furthermore, the O-glycosylated cell wall proteins mentioned above are serine rich but Sed1p is threonine rich. Sed1p also contains four cysteines, while the other proteins have no cysteine residues. Therefore, Sed1p belongs to another group of cell wall proteins due to its heavy N-glycosylation and its ability to form an intermolecular complex with other proteins by disulfide bonds.
SED1 is highly induced in the stationary phase, and Sed1p becomes the most abundant cell wall protein in stationary-phase cells (Fig. 8 and 9). It has been reported that the cell wall in the stationary phase is thicker and less porous than that in the exponential phase (3, 35). Since destruction of the mannoprotein layer by protease is a prerequisite for digestion of the glucan layer by glucanase, the thicker mannoprotein protects the glucan layer from lytic enzymes (3). Indeed, the sed1 disruptant cells in the stationary phase were more sensitive to Zymolyase than were wild-type cells (Fig. 10). This finding well agrees with the fact that Sed1p is a major cell wall protein in stationary-phase cells. However, the sed1 disruptant was more resistant to Zymolyase in the stationary phase than in the exponential phase. This means that the higher level of expression of Sed1p is one of the factors by which the stationary-phase cells are resistant to Zymolyase. Other genes expressed in the stationary phase may be involved in the Zymolyase resistance. TIP1 is one such gene because its expression was induced in the stationary phase (12). FKS2, a glucan synthase gene, is another possibility because its expression was induced during nutrient starvation (17).
Cells in the stationary phase are known to be resistant to various environmental stresses such as heat, ethanol, and lytic enzymes (41). Many factors are involved in the stress resistance of stationary-phase cells. The best-known mechanism is general stress responses caused by depletion of available nutrients. Reduction of the glucose concentration in the stationary phase results in inactivation of protein kinase A through the RAS-cyclic AMP signal transduction pathway (33). Inactivation of protein kinase A activates the expression of many stress-responsive element (STRE)-controlled genes. These genes have characteristic cis factors called STREs in their promoter regions (CCCCT or AGGGG) (39). In fact, the promoter region of SED1 contains AGGGG at −84 bases from the translation initiation codon and CCCCT at −912. Therefore, it is conceivable that SED1 is expressed during nutrient starvation in the stationary phase via STRE-mediated transcriptional activation.
In conclusion, Sed1p is a major structural cell wall protein in stationary-phase cells and plays an important role in cell defense mechanisms in the stationary phase. Our studies have revealed that structural cell wall proteins of yeast change dynamically in response to environmental changes, as shown by the expression of Tir1p in anaerobic culture (8) and by the expression of Sed1p in the stationary phase (this work). Further studies on the synthesis and control of expression of cell wall proteins, glucan, and chitin are needed to elucidate the detailed mechanism of cell wall reorganization that occurs in yeast proliferation and stress responses.
ACKNOWLEDGMENTS
We thank C. E. Ballou, University of California, Berkeley, for providing the mnn9 strain.
This work was supported by Special Coordination Funds for Promoting Science and Technology from the Science and Technology Agency of Japan.
REFERENCES
- 1.Ballou L, Cohen R E, Ballou C E. Saccharomyces cerevisiae mutants that make mannoproteins with a truncated carbohydrate outer chain. J Biol Chem. 1980;255:5986–5991. [PubMed] [Google Scholar]
- 2.Becker D M, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 3.De Nobel J G, Klis F M, Priem J, Munnik T, van den Ende H. The glucanase-soluble mannoproteins limit cell wall porosity in Saccharomyces cerevisiae. Yeast. 1990;6:491–499. doi: 10.1002/yea.320060606. [DOI] [PubMed] [Google Scholar]
- 4.Donzeau M, Bourdineaud J P, Lauquin G J M. Regulation by low temperatures and anaerobiosis of a yeast gene specifying a putative GPI-anchored plasma membrane protein. Mol Microbiol. 1996;20:449–459. doi: 10.1111/j.1365-2958.1996.tb02631.x. [DOI] [PubMed] [Google Scholar]
- 5.Frevert J, Ballou C E. Saccharomyces cerevisiae structural cell wall mannoprotein. Biochemistry. 1985;24:753–759. doi: 10.1021/bi00324a033. [DOI] [PubMed] [Google Scholar]
- 6.Hardwick K G, Boothroyd J C, Rudner A D, Pelham H R. Genes that allow yeast cells to grow in the absence of the HDEL receptor. EMBO J. 1992;11:4187–4195. doi: 10.1002/j.1460-2075.1992.tb05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapteyn J C, Montijn R C, Vink E, Cruz J, Llobell A, Douwes J E, Shimoi H, Lipke P N, Klis F M. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked β-1,3-/β-1,6-glucan heteropolymer. Glycobiology. 1996;6:337–345. doi: 10.1093/glycob/6.3.337. [DOI] [PubMed] [Google Scholar]
- 8.Kitagaki H, Shimoi H, Itoh K. Identification and analysis of a static culture-specific cell wall protein, Tir1p/Srp1p in Saccharomyces cerevisiae. Eur J Biochem. 1997;249:343–349. doi: 10.1111/j.1432-1033.1997.t01-1-00343.x. [DOI] [PubMed] [Google Scholar]
- 9.Klis F M. Cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 10.Köhrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 11.Kolodziej P A, Young R A. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- 12.Kondo K, Inouye M. TIP1, a cold shock-inducible gene of Saccharomyces cerevisiae. J Biol Chem. 1991;266:17537–17544. [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Lipke P N, Kurjan J. Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiol Rev. 1992;56:180–194. doi: 10.1128/mr.56.1.180-194.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu C-F, Kurjan J, Lipke P N. A pathway for cell wall anchorage of Saccharomyces cerevisiae α-agglutinin. Mol Cell Biol. 1994;14:4825–4833. doi: 10.1128/mcb.14.7.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C F, Montijn R C, Brown J L, Klis F, Kurjan J, Bussey H, Lipke P N. Glycosyl phosphatidylinositol-dependent cross-linking of α-agglutinin and β 1,6-glucan in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1995;128:333–340. doi: 10.1083/jcb.128.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazur P, Morin N, Baginsky W, el-Sherbeini M, Clemas J A, Nielsen J B, Foor F. Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol Cell Biol. 1995;15:5671–5681. doi: 10.1128/mcb.15.10.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamaye K, Eckstein F. Inhibition of restriction endonuclease Nci I cleavage by phosphorothioate groups and its application to oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1986;14:9679–9698. doi: 10.1093/nar/14.24.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuoffer C, Jeno P, Conzelmann A, Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol Cell Biol. 1991;11:27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nuoffer C, Horvath A, Riezman H. Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J Biol Chem. 1993;268:10558–10563. [PubMed] [Google Scholar]
- 21.Orlean P. Biogenesis of yeast wall and surface components. In: Broach J, Pringle J, Jones E, editors. The molecular and cellular biology of the yeast Saccharomyces. New York, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 229–362. [Google Scholar]
- 22.Payne W E, Garrels J I. Yeast Protein Database (YPD): a database for the complete proteome of Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:57–62. doi: 10.1093/nar/25.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philippsen P, Stotz A, Scherf C. DNA of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- 24.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 25.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semenza J C, Hardwick K G, Dean N, Pelham H R. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 1990;61:1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- 28.Shibata N, Mizugami K, Takano K, Suzuki S. Isolation of mannan-protein complexes from viable cells of Saccharomyces cerevisiae X2180-1A wild type and Saccharomyces cerevisiae X2180-1A-5 mutant strains by the action of Zymolyase-60,000. J Bacteriol. 1983;156:552–558. doi: 10.1128/jb.156.2.552-558.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimoi H, Tadenuma M. Characterization of Rarobacter faecitabidus protease I, a yeast-lytic serine protease having mannose-binding activity. J Biochem. 1991;110:608–613. doi: 10.1093/oxfordjournals.jbchem.a123628. [DOI] [PubMed] [Google Scholar]
- 30.Shimoi H, Iimura Y, Obata T, Tadenuma M. Molecular structure of Rarobacter faecitabidus protease I, a yeast-lytic serine protease having mannose-binding activity. J Biol Chem. 1992;267:25189–25195. [PubMed] [Google Scholar]
- 31.Shimoi H, Iimura Y, Obata T. Molecular cloning of CWP1: a gene encoding a Saccharomyces cerevisiae cell wall protein solubilized with Rarobacter faecitabidus protease I. J Biochem. 1995;118:302–311. doi: 10.1093/oxfordjournals.jbchem.a124907. [DOI] [PubMed] [Google Scholar]
- 32.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thevelein J M. Signal transduction in yeast. Yeast. 1994;10:1753–1790. doi: 10.1002/yea.320101308. [DOI] [PubMed] [Google Scholar]
- 34.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valentin E, Herrero E, Rico H, Miragall F, Sentandreu R. Cell wall mannoproteins during the population growth phases in Saccharomyces cerevisiae. Arch Microbiol. 1987;148:88–94. doi: 10.1007/BF00425354. [DOI] [PubMed] [Google Scholar]
- 36.Van Der Vaart J M, Caro L H P, Chapman J W, Klis F M, Verrips C T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Der Vaart J M, van Schagen F S, Mooren A T, Chapman J W, Klis F M, Verrips C T. The retention mechanism of cell wall proteins in Saccharomyces cerevisiae. Wall-bound Cwp2p is β-1,6-glucosylated. Biochim Biophys Acta. 1996;1291:206–214. doi: 10.1016/s0304-4165(96)00067-0. [DOI] [PubMed] [Google Scholar]
- 38.Van Der Vaart J M, Te Biesebeke R, Chapman J W, Toschka H Y, Klis F M, Verrips C T. Comparison of cell wall proteins of Saccharomyces cerevisiae as anchors for cell surface expression of heterologous proteins. Appl Environ Microbiol. 1997;63:615–620. doi: 10.1128/aem.63.2.615-620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varela J C, Praekelt U M, Meacock P A, Planta R J, Mager W H. The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol Cell Biol. 1995;15:6232–6245. doi: 10.1128/mcb.15.11.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watari J, Takata Y, Ogawa M, Sahara H, Koshino S, Onnela M L, Airaksinen U, Jaatinen R, Penttila M, Keranen S. Molecular cloning and analysis of the yeast flocculation gene FLO1. Yeast. 1994;10:211–225. doi: 10.1002/yea.320100208. [DOI] [PubMed] [Google Scholar]
- 41.Werner-Washburne M, Braun E, Johnston G C, Singer R A. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zlotnik H, Fernandez M P, Bowers B, Cabib E. Saccharomyces cerevisiae mannoproteins from an external cell wall layer that determines wall porosity. J Bacteriol. 1984;159:1018–1026. doi: 10.1128/jb.159.3.1018-1026.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]