Abstract
The genomic DNA of the BE strain of Escherichia coli has been scrutinized to detect porin genes that have not been identified so far. Southern blot analysis yielded two DNA segments which proved highly homologous to, yet distinct from, the ompC, ompF, and phoE porin genes. The two genes were cloned and sequenced. One of them, designated ompN, encodes a porin which, due to low levels of expression, has eluded prior identification. The functional properties (single-channel conductance) of the OmpN porin, purified to homogeneity, closely resemble those of the OmpC porin from E. coli K-12. The second DNA fragment detected corresponds to the nmpC gene, which, due to an insertion of an IS1 element in its coding region, is not expressed in E. coli BE.
Outer membranes of gram-negative bacteria are permeable to small (<600-Da), polar molecules that cross an otherwise impermeable lipid bilayer by diffusion through water-filled channel proteins, the porins (19). Escherichia coli K-12 encodes three major nonspecific proteins, the OmpC, OmpF, and PhoE porins, and several other channel-forming proteins with higher degrees of specificity. Since we are interested in the structural and functional characterization of nonspecific as well as specific porins to high resolution (6, 9, 25, 29, 30), it was of interest to know whether other, as yet unidentified genes coding for porin-like proteins were present in E. coli BE, a strain in which apparently a single porin is expressed (23, 25). During scrutiny of the genome of the BE strain for cross-hybridizing DNA, two additional genes were detected. One showed high similarity with the nmpC gene of E. coli K-12 and is not expressed in E. coli BE due to inactivation by an IS1 element. Cloning and overexpression of the other, which we call ompN, allowed characterization of the purified product, which reveals biochemical and functional properties highly similar to those of the OmpC porin.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
E. coli strains and plasmids used are listed in Table 1. Cells were grown aerobically at 37°C on 2× YT medium (16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl per liter). The antibiotics ampicillin (100 μg/ml) and kanamycin (50 μg/ml), where required, were included in the growth media.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Source (reference) |
|---|---|---|
| E. coli K-12 | ||
| CE1249 | F−thr leu thi pyrF thy ilvA his lacY argG fhuA rpsL cod dra vtr glpR ompB471 phoR69 proAB ΔphoE recA56 λvirr | J. Tomassen (32) |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG φ80lacZΔM15 | Invitrogen |
| E. coli BE | ||
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| BL21(DE3)omp8 | BL21(DE3), ΔlamB ompF::Tn5 ΔompA ΔompC | A. Prilipov (21) |
| BZB1107 | ompR ΔlamB ompF::Tn5 | This laboratory (15) |
| E. coli C JR301 | (r0 m0) Smr | J. Ryu (20) |
| Plasmids | ||
| pET-15b | E. coli T7 system expression vector, lacIq Apr | Novagen |
| pGEM-5Zf(+) | E. coli cloning vector, Apr | Promega |
| pJP29 | pACYC184 (vector) with cloned phoE gene | J. Tommassen (4) |
| pMY150 | pBR322 with cloned ompC gene | M. Inouye (18) |
| pMY222 | pBR322 with cloned ompF gene | M. Inouye (24) |
| pOmpN | pET-15b with cloned ompN gene | This study |
Standard DNA preparations, DNA labeling, and Southern blot analysis.
Standard DNA manipulations were performed as described previously (27). All DNA preparations were carried out by using available kits (Qiagen). To produce porin-specific gene probes, DNA fragments (a 900-bp HincII-BglII ompF fragment from pMY222, a 554-bp EcoRI-EcoRV ompC fragment from pMY150, and a 834-bp PstI-BglII phoE fragment from pJP29) were radioactively labeled with [α-32P]dATP (Amersham), using a Random Primer DNA labeling kit (Bio-Rad). For Southern blot analysis, performed as described by the manufacturer (Amersham), chromosomal DNAs of E. coli K-12 CE1249, BE BL21(DE3), and BE BZB1107 were digested with restriction endonuclease EcoRV and fractionated by electrophoresis on 0.8% agarose gels. The DNA was blotted to Hybond-N membranes (Amersham) and hybridized with the labeled porin gene probes overnight at 65°C.
Cloning, DNA sequencing, and PCR.
Chromosomal DNA (5 μg) from E. coli BE BL21(DE3) was digested with EcoRV and separated on an agarose gel (0.8%). Based on Southern blot analyses (see Results), DNA fragments of about 2.6 and 3.1 kb were isolated from gels and ligated with plasmid pGEM-5Zf(+) after linearization with EcoRV and dephosphorylation. Resulting plasmids were transformed into strain TOP10 and plated onto ampicillin–isopropyl-β-d-thio- galactopyranoside (IPTG)–5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates. White colonies were screened for recombinant plasmids harboring sequences homologous to the ompF gene by hybridization.
Cloned DNA was sequenced by the chain termination method (28), using a T7 sequencing kit (Pharmacia). To amplify the ompN gene from other E. coli strains by PCR (Pwo DNA polymerase [Boehringer Mannheim]; 30 cycles of amplification), we used oligonucleotides I (5′-TCTAGATATTTATCGGCTAACTGAACTTCT [XbaI site]) and II (5′-CAGGATCCTTTAGAACTGATAAACCAGACC [BamHI site]) (restriction sites used for cloning are underlined).
Amplified DNA from E. coli K-12 CE1249 was cloned as an XbaI-BamHI fragment into plasmid pET-15b, resulting in plasmid pOmpN.
Purification of the OmpN protein and N-terminal protein sequencing.
E. coli BE host strain BL21(DE3)omp8, which lacks all major porins but harbors the pOmpN expression plasmid, was used for OmpN protein overexpression. Cells were grown for 6 h (optical density at 600 nm of 0.6), followed by induction with IPTG (final concentration of 1 mM). After further growth for 4 h, cells (10 g [wet weight]) were suspended in 30 ml of breaking buffer (10). The suspension was passed through a French pressure cell (model FA-073; Aminco, Urbana, Ill.) at 162 MPa thrice. The outer membrane pellet was collected by two successive centrifugations, first at 8,000 × g for 10 min to remove unbroken cells and then at 75,000 × g for 30 min. The membrane pellet was extracted for 2 h at 37°C with 100 ml of extraction buffer (10) containing 0.125% octyl-polyoxyethylene (octyl-POE; Alexis, Läuflingen, Switzerland), followed by extraction buffer containing 3% octyl-POE. The latter extract was concentrated and applied to an ion-exchange DEAE-Sephacel (Pharmacia) column. It was washed with column buffer (10) containing 10 mM EDTA. The protein was eluted by using column buffer supplemented with 50 mM EDTA and 0.1 M NaCl and was then applied to a PBE94 chromatofocusing column (Pharmacia), followed by Sephadex G-150 gel filtration (Pharmacia), as described for OmpF (10). Purity of the protein was assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 12% acrylamide (16) and N-terminal protein sequencing (Applied Biosystems model 477A sequenator). Other porins (OmpF, OmpC, and PhoE) were purified as described previously (6).
Planar lipid bilayer experiments, liposome swelling assays, and CD spectroscopy.
Channel conductance properties of purified porins were measured with respect to single-channel conductance, critical voltage of closing, and ion selectivity, or by liposome swelling assays as described previously (26). The shift in electrophoretic mobility due to heat dissociation of trimers was monitored by SDS-PAGE (25). The circular dichroism (CD) spectrum of OmpN was recorded in a Jasco spectrometer (model J-720) at 25°C as described previously (25).
Homology searches.
Database BLASTP, BLASTN, and TBLASTN searches (2) were performed via the file server at blast@ncbi.nlm.nih.gov, using default parameters.
Nucleotide sequence accession numbers.
The sequences for E. coli BE nmpC and ompN have been deposited with GenBank under accession no. U91745 and AF035618, respectively.
RESULTS
Identification, cloning, and sequencing of the nmpC and ompN genes from E. coli BE.
The electrophoretic mobilities of various restriction fragments originating from the ompF, ompC, and phoE genes in E. coli BL21(DE3) were determined by Southern blot analyses (data not shown). To detect homologous genes, chromosomal DNA was purified from three different strains of E. coli [K-12 CE1249, BE BL21(DE3), and BE BZB1107], digested with restriction endonuclease EcoRV, and analyzed by Southern blot with the labeled ompF fragment as the probe (Fig. 1). In addition to the signals corresponding to ompF and ompC, two additional bands (I and II) were detected. The corresponding DNA fragments from strain BL21(DE3) were cloned and sequenced. They proved distinct from the phoE gene. BLASTN database analyses revealed the presence of an nmpC gene, described previously (3), in fragment I. This gene could encode a 360-residue polypeptide, including a 23-residue signal peptide, if it were not inactivated by an IS1 element insertion at a position corresponding to amino acid 170 of the mature protein. The sequence of fragment II revealed an open reading frame which encodes a polypeptide that is highly homologous to known porins (Fig. 2). We designate this gene, which has not been described previously, as ompN. To determine whether the same gene is also present in other E. coli strains, PCR was performed with the two ompN-specific oligonucleotides from the BE strain (see Materials and Methods). This resulted in the amplification of DNA fragments with the expected size both in E. coli C and the E. coli K-12 strains. The latter was cloned and sequenced. The predicted amino acid sequence revealed differences in two positions compared to the E. coli BE protein: a conservative change in the predicted β-strand 14 (V309I), and a substitution of Thr by Ala at 338 position in a predicted surface-exposed loop L8.
FIG. 1.
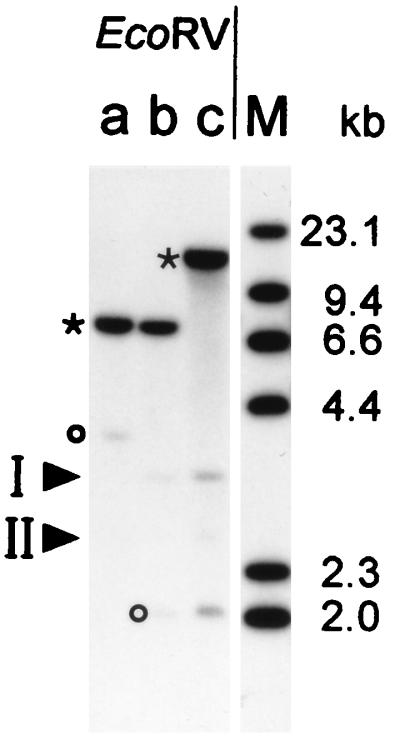
Southern blot analysis. Chromosomal DNAs were purified from E. coli CE1249 (lane a), BL21(DE3) (lane b), and BZB1107 (lane c), respectively, and digested with EcoRV. After fractionation of the DNA fragments by agarose gel electrophoresis, they were blotted onto a Hybond-N membrane and probed with [α-32P]dATP-labeled ompF-specific DNA. With strain BL21(DE3), two EcoRV fragments appeared in positions corresponding to ∼3.1 and 2.6 kb and are labeled by arrows as fragments I and II. Fragments corresponding to ompF (asterisk) and ompC (circle) are indicated. HindIII-digested λ DNA is shown in lane M.
FIG. 2.
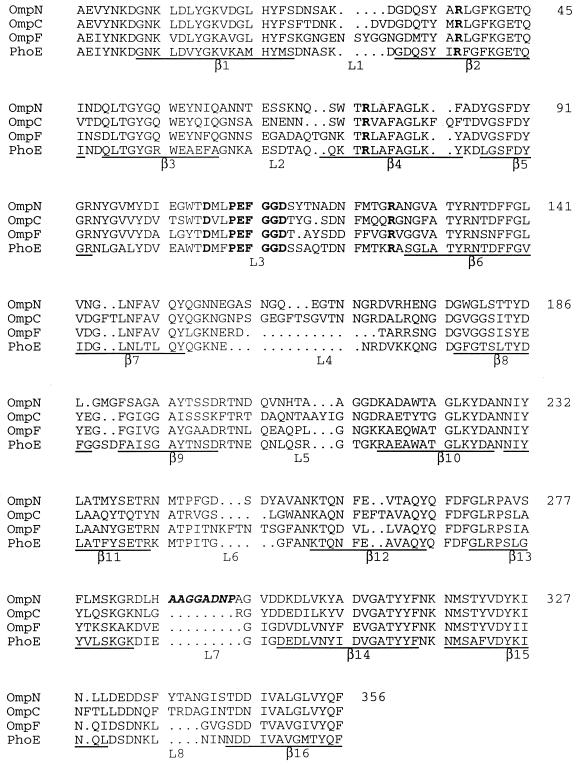
Comparison of the predicted amino acid sequence of OmpN with sequences of various known E. coli porins. Sequences underlined below each block correspond to the β strands in the three-dimensional structures of OmpF and PhoE (6). A highly conserved porin-specific sequence motif, PEFGGD (14), in loop L3, and five conserved charged residues (R37, R75, D106, E110, and R126) which form a strong transversal electrostatic field in the channel interior are shown in bold. The additional amino acid residues, present in the predicted surface-exposed loop L7 in OmpN, are shown in bold italics. The overall similarity of OmpN is highest to OmpC (65% identical residues), followed by PhoE (62% identical residues) and OmpF (58% identical residues).
Since the deduced amino acid sequence predicts the OmpN protein to be synthesized with a signal peptide, the processing site was confirmed by N-terminal protein sequencing. The mature OmpN protein consists of 356 amino acid residues, with a calculated mass of 39,152 Da. Comparison at the amino acid level with other porins (PhoE, OmpF, and OmpC from E. coli) revealed identity of the amino-terminal 24 residues of the mature OmpN with those of OmpC and a high degree of conservation with the other porins. The multiple alignment in Fig. 2 indicates the various degrees of identity. A highly conserved sequence motif, PEFGGD (14), and five charged residues (R37, R75, D106, E110, and R126) which form a strong transversal electrostatic field in the channel interior (6) are present at the same positions as in the superfamily of nonspecific porins. Altogether, the sequence and the characteristic β-sheet CD spectrum of the purified protein (absorption minimum at 218 nm [25]) suggest that OmpN porin also forms a 16-stranded antiparallel β barrel.
A standard BLASTP database search revealed numerous homologous proteins, among them several enterobacterial porins (13). Other closely related proteins, with an even higher score than OmpC, include the porins OmpS2 (GenBank accession no. X89756) and OmpS1 (7) from Salmonella typhi, OmpK36 from Klebsiella pneumoniae (1), and OpnP from Xenorhabdus nematophilus (8). The OmpN protein is also closely related to the bacteriophage-encoded Lc/NmpC proteins (3) and to the NmpC-like OmpD porin from Salmonella typhimurium (31). Interestingly, a TBLASTN database search identified a very similar sequence at 43.8 min of the E. coli K-12 chromosome. Closer scrutiny revealed that this sequence contains an internal stop codon and a frameshift.
Overexpression and characterization of the OmpN porin.
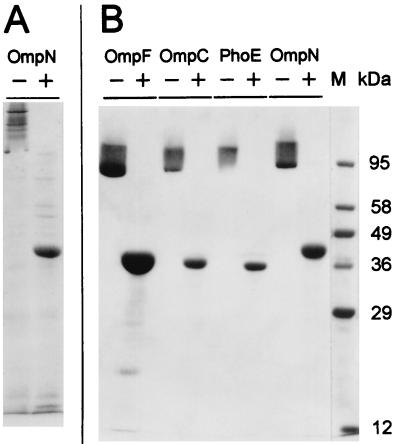
For the construction of an OmpN expression plasmid, the PCR-amplified ompN gene fragment from E. coli K-12 CE1249 was cloned as an XbaI-BamHI fragment (sites are present at the 5′ termini of the PCR primers) behind the T7-specific promoter of plasmid pET-15b. The resulting plasmid, pOmpN, allowed the overexpression of OmpN in E. coli BL21(DE3)omp8 (Fig. 3A), with its purification yielding 2 to 3 mg of OmpN protein per g (wet weight) of cell mass. The purity of the protein was established by SDS-PAGE (Fig. 3B) and N-terminal sequencing, which yielded the unique sequence AEVYNKDGNKLD. The apparent molecular mass of the polypeptide as determined by SDS-PAGE after 95°C heat treatment was 39 kDa (Fig. 3), which is in agreement with the calculated value from the deduced amino acid sequence. In samples not treated by heat prior to electrophoresis, the band revealed a significantly lower migration rate, characteristic for a trimeric state of association of porin monomers (25). The thermal stability with respect to the dissociation of the OmpN trimers into its monomers in the presence of 1% SDS occurred with a midpoint at 70°C. By comparison, the corresponding transition temperatures of OmpC, OmpF, and PhoE porins are at 80, 75, and 75°C.
FIG. 3.
SDS-PAGE analysis of E. coli porins. (A) Overexpression of OmpN porin in E. coli BL21(DE3)omp8. Membrane pellets were extracted with 3% octyl-POE as described in Materials and Methods. Protein samples (from 15 ml of culture) were applied to the gel without (−) or with (+) heat treatment for 10 min at 95°C in sample buffer. (B) Electrophoretic mobilities of purified porins (indicated at the top), which were used for functional analyses, represent monomers (high mobility) and trimers (low mobility). They were purified from E. coli outer membranes as described in the text. The protein samples (3 to 5 μg) were heated as described above. Lane M, molecular weight standard.
Reconstitution of the purified OmpN protein into planar lipid bilayers was as efficient as that of OmpF porin (29). The current traces were essentially free of noise and allowed the channel conductance properties to be monitored. Like other E. coli porins, OmpN porin showed high cooperativity in the initiation step, with conductance steps of 1.63 ± 0.06 nS (59 events), corresponding to the cooperative insertion of three monomers (single-channel conductances, 0.50 ± 0.03 nS; 286 events). Table 2 summarizes the measured values for single-channel conductances, critical voltage of channel closing, ion selectivities for various nonspecific porins. Similar to channels in the OmpC porin, OmpN channels are less sensitive to applied transmembrane potentials, requiring significantly higher voltages before the channels close (at ∼250 mV). The rates of sugar permeation through different porin channels, as determined by liposome swelling assays, exhibit a complex pattern (Table 3), as discussed below.
TABLE 2.
Channel conductance properties of various porins from E. coli outer membranes
| Porin | Mean ± SDa
|
||
|---|---|---|---|
| Single-channel conductance (nS) | Critical voltage of closing (mV) | Ion selectivity, pNa/pCl | |
| OmpN | 0.50 ± 0.03 (n = 286) | 243 ± 13 (n = 15) | 4.8 ± 0.8 (m = 6) |
| OmpC | 0.47 ± 0.04 (n = 138) | 267 ± 5 (n = 8) | 5.8 ± 0.3 (m = 5) |
| OmpF | 0.84 ± 0.06 (n = 156) | 145 ± 7 (n = 10) | 4.5 ± 0.8 (m = 6) |
| PhoE | 0.63 ± 0.06 (n = 170) | 135 ± 8 (n = 28) | 0.44 ± 0.05 (m = 2) |
n, number of events observed; m, number of independent experiments.
TABLE 3.
Rates of sugar permeation through various E. coli porins
| Sugar | Mean rate ± SDa
|
|||
|---|---|---|---|---|
| OmpN | OmpC | OmpF | PhoE | |
| Glucose | 74 ± 5 | 67 ± 5 | 73 ± 2 | 61 ± 6 |
| Galactose | 87 ± 6 | 84 ± 6 | 90 ± 6 | 69 ± 3 |
| Mannose | 79 ± 5 | 72 ± 5 | 79 ± 5 | 58 ± 3 |
| Fructose | 81 ± 5 | 71 ± 4 | 81 ± 4 | 70 ± 4 |
| GluNAc | 51 ± 6 | 44 ± 5 | 58 ± 7 | 32 ± 8 |
| Sucrose | 5 ± 3 | ND | 3 ± 3 | ND |
| Lactose | 6 ± 5 | ND | 7 ± 2 | 5 ± 4 |
| Melibiose | 5 ± 4 | ND | 2 ± 1 | ND |
| Maltose | 3 ± 3 | ND | 6 ± 2 | ND |
| Maltotriose | 4 ± 3 | ND | 4 ± 4 | ND |
Expressed as a percentage of the value for arabinose. Standard deviations correspond to at least 10 measurements with three different liposome preparations. ND, not detectable by the method used.
DISCUSSION
Initially, a single gene was found to encode a porin protein in E. coli BE (23, 25). Subsequently, an increasing number of porin variants, specific and nonspecific, were described. To determine how many further cryptic porins exist, we scrutinized the genome of E. coli BE, using a porin-specific gene probe. As described in this report, scrutiny of the E. coli BL21(DE3) chromosome revealed the presence of two other related genes.
The first gene identified in this study was very similar to the E. coli K-12 nmpC gene (91% identity at the amino acid level). The NmpC protein was first identified in pseudorevertants of E. coli K-12 strains which were impaired in the expression of both ompF and ompC porin genes (22). Apparently, this membrane protein was synthesized due to a precise excision of the IS5 element (3) and could functionally replace the OmpF or OmpC porin (12). In K-12 wild-type strains, the protein is not expressed, due to the presence of an IS5 element near the 3′ end of the coding sequence (3, 11). While this element does not exist at that position in B strains, the nmpC gene analog is not expressed in E. coli BE, as in this case an insertional inactivation by an IS1 element occurs. Thus, in both E. coli B and E. coli K-12, the nmpC genes are inactivated by insertion sequence elements, though their respective identities and locations are different.
The second gene, ompN, encodes a protein that is closely related to enterobacterial porins. Expression of the chromosomal ompN gene in strain BL21(DE3)omp8 was examined by comparison with an isogenic ompN knockout mutant. This revealed that under normal laboratory growth conditions in rich media, the gene product is found at levels in the outer membrane too low to be quantitated and partially overlapping another gel band. The ompN gene has also been found in E. coli C and K-12 strains. The demonstration that OmpN porin has properties which both in biochemical as well as in functional terms resemble those of the nonspecific porins brings their number to at least four: OmpC, OmpF, OmpN, and PhoE. An additional functional analog, the OmpG protein, has been identified and characterized (17). However, this protein does not exhibit substantial sequence homology to the four nonspecific porins mentioned, and its relationship remains to be determined.
Compared to OmpF porin from E. coli K-12, the OmpN protein contains an additional stretch of nine amino acid residues, predicted to be located in the surface-exposed loop L7. It is noteworthy that in terms of its functional properties, the OmpN and OmpC porins reveal very similar channel conductances (5), a finding that may be explained by the observation that these two porins both contain short inserts in regions of two loops (corresponding to L4 and L8) compared to the OmpF and PhoE porins (Fig. 2). Surprisingly, the differential uptake of mono- and disaccharides of the OmpN protein resembles that of the OmpF porin more closely than that in the OmpC protein (Table 3), differences which in the apparent absence of solute binding sites are rather interesting. Determinations of the structures of both OmpC and OmpN porins to high resolution, now in progress, should allow these differences to be explained.
ACKNOWLEDGMENTS
We thank P. Jenö, Department of Biochemistry, Biozentrum Basel, for N-terminal protein sequencing.
This study was supported by grant 31-36352 from the Swiss National Science Foundation. R.K. was supported by grant Ko1686/1-1 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Alberti S, Rodriquez-Quinones F, Schirmer T, Rummel G, Tomas J M, Rosenbusch J P, Benedi V J. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect Immun. 1995;63:903–910. doi: 10.1128/iai.63.3.903-910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Blasband A J, Marcotte W R, Schnaitman C A. Structure of the lc and nmpC outer membrane protein genes of lambdoid bacteriophages. J Biol Chem. 1986;261:12723–12732. [PubMed] [Google Scholar]
- 4.Bosch D, Leunissen J, Verbakel J, de Jong M, van Erp H, Tommassen J. Periplasmic accumulation of truncated forms of outer-membrane PhoE protein of Escherichia coli K-12. J Mol Biol. 1986;189:449–455. doi: 10.1016/0022-2836(86)90316-5. [DOI] [PubMed] [Google Scholar]
- 5.Buehler L K, Kusumoto S, Zhang H, Rosenbusch J P. Plasticity of Escherichia coli porin channels—dependence of their conductance on strain and lipid environment. J Biol Chem. 1991;266:24446–24450. [PubMed] [Google Scholar]
- 6.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 7.Fernández-Mora M, Oropeza R, Puente J L, Calva E. Isolation and characterization of ompS1, a novel Salmonella typhi outer membrane protein-encoding gene. Gene. 1995;158:67–72. doi: 10.1016/0378-1119(95)00171-2. [DOI] [PubMed] [Google Scholar]
- 8.Forst S, Waukau J, Leisman G, Exner M, Hancock R. Functional and regulatory analysis of the OmpF-like porin, OpnP, of the symbiotic bacterium Xenorhabdus nematophilus. Mol Microbiol. 1995;18:779–789. doi: 10.1111/j.1365-2958.1995.mmi_18040779.x. [DOI] [PubMed] [Google Scholar]
- 9.Garavito R M, Rosenbusch J P. Three-dimensional crystals of an integral membrane protein: an initial X-ray analysis. J Cell Biol. 1980;86:327–329. doi: 10.1083/jcb.86.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garavito R M, Rosenbusch J P. Isolation and crystallization of bacterial porin. Methods Enzymol. 1986;125:309–328. doi: 10.1016/s0076-6879(86)25027-2. [DOI] [PubMed] [Google Scholar]
- 11.Highton P J, Chang Y, Marcotte W R, Schnaitman C A. Evidence that the outer membrane protein gene nmpC of Escherichia coli K-12 lies within the defective qsr′ prophage. J Bacteriol. 1985;162:256–262. doi: 10.1128/jb.162.1.256-262.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hindahl M S, Crockford G W K, Hancock R E W. Outer membrane protein NmpC of Escherichia coli: pore-forming properties in black lipid bilayers. J Bacteriol. 1984;159:1053–1055. doi: 10.1128/jb.159.3.1053-1055.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutsul J-A, Worobec E. Molecular characterization of a 40 kDa OmpC-like porin from Serratia marcescens. Microbiology. 1994;140:379–387. doi: 10.1099/13500872-140-2-379. [DOI] [PubMed] [Google Scholar]
- 14.Jeanteur D, Lakey J H, Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 15.Jeanteur D, Schirmer T, Fourel D, Simonet V, Rummel G, Widmer C, Rosenbusch J P, Pattus F, Pagès J-M. Structural and functional alterations of a colicin-resistant mutant of porin (OmpF) from Escherichia coli. Proc Natl Acad Sci USA. 1994;91:10675–10679. doi: 10.1073/pnas.91.22.10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lugtenberg B, Meijers J, Peters R, van den Hoek P, van Alphen L. Electrophoretic resolution of the ‘major outer membrane protein’ of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 17.Misra R, Benson S A. A novel mutation, cog, which results in production of a new porin protein (OmpG) of Escherichia coli K-12. J Bacteriol. 1989;171:4105–4111. doi: 10.1128/jb.171.8.4105-4111.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuno T, Chou M-Y, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc Natl Acad Sci USA. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikaido H. Porins and specific channels of bacterial outer membranes. Mol Microbiol. 1992;6:435–442. doi: 10.1111/j.1365-2958.1992.tb01487.x. [DOI] [PubMed] [Google Scholar]
- 20.Prakash-Cheng A, Chung S S, Ryu J. The expression and regulation of hsdK genes after conjugative transfer. Mol Gen Genet. 1993;241:491–496. doi: 10.1007/BF00279890. [DOI] [PubMed] [Google Scholar]
- 21.Prilipov, A., P. S. Phale, P. Van Gelder, J. P. Rosenbusch, and R. Koebnik. Coupling site-directed mutagenesis with high-level expression: large scale production of porin mutants of E. coli. FEMS Microbiol. Lett., in press. [DOI] [PubMed]
- 22.Pugsley A P, Schnaitman C A. Identification of three genes controlling production of new outer membrane pore proteins in Escherichia coli K-12. J Bacteriol. 1978;135:1118–1129. doi: 10.1128/jb.135.3.1118-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugsley A P, Rosenbusch J P. OmpF porin synthesis in Escherichia coli strains B and K-12 carrying heterologous ompB and/or ompF loci. FEMS Microbiol Lett. 1983;16:143–147. [Google Scholar]
- 24.Ramakrishnan G, Ikenaka K, Inouye M. Uncoupling of osmoregulation of the Escherichia coli K-12 ompF gene from ompB-dependent transcription. J Bacteriol. 1985;163:82–87. doi: 10.1128/jb.163.1.82-87.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenbusch J P. Characterization of the major envelope protein from Escherichia coli. Regular arrangement on the peptidoglycan and unusual dodecyl sulfate binding. J Biol Chem. 1974;249:8019–8029. [PubMed] [Google Scholar]
- 26.Saint N, Lou K-L, Widmer C, Luckey M, Schirmer T, Rosenbusch J P. Structural and functional characterization of OmpF porin mutants selected for larger pore size. II. Functional characterization. J Biol Chem. 1996;271:20676–20680. [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schindler H, Rosenbusch J P. Matrix protein from Escherichia coli outer membranes forms voltage-controlled channels in lipid bilayers. Proc Natl Acad Sci USA. 1978;75:3751–3755. doi: 10.1073/pnas.75.8.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schirmer T, Keller T A, Wang Y F, Rosenbusch J P. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 31.Singh S P, Miller S, Williams Y U, Rudd K E, Nikaido H. Immunochemical structure of the OmpD porin from Salmonella typhimurium. Microbiology. 1996;142:3201–3210. doi: 10.1099/13500872-142-11-3201. [DOI] [PubMed] [Google Scholar]
- 32.Struyvé M, Visser J, Adriaanse H, Benz R, Tommassen J. Topology of PhoE porin: the ‘eyelet’ region. Mol Microbiol. 1993;7:131–140. doi: 10.1111/j.1365-2958.1993.tb01104.x. [DOI] [PubMed] [Google Scholar]