ABSTRACT
Universal PCR for bacteria, mycobacteria, and fungi can aid in the diagnosis of occult infections, especially in the case of fastidious organisms or when prior antimicrobial treatment compromises culture growth. However, the limitations of this technology, including lack of specificity, high cost, long turnaround time, and lack of susceptibility data, may limit its effect on clinical outcomes. We performed a retrospective analysis of all specimens sent for universal PCR over a 10-year period from 2013 to 2022, focusing on clinical indications for test utilization and patient outcomes. All specimens required approval by a microbiology laboratory director prior to testing. A total of 708 specimens were sent from 638 patients. Of these specimens, 163 were positive, with an overall positivity rate of 23%. Pre-analytic factors associated with a positive universal PCR result were the presence of organisms on Gram stain or histology, the presence of neutrophils on Gram stain, and growth on culture. Positivity rates varied significantly by specimen type. A total of 20% of all organisms detected were deemed clinically irrelevant by the clinical services. A positive universal PCR led to a change in antibiotic management in 29% of cases. Positive fungal universal PCR results sent from hospitalized patients were associated with worse outcomes, including increased hospital mortality. Our findings suggest that factors such as the presence of organisms or neutrophils on Gram stain, specimen source/clinical context, and anticipated changes in management based on results should be strongly considered when making stewardship decisions regarding the appropriateness of this testing modality.
IMPORTANCE
Our work provides a retrospective analysis of universal PCR orders for bacteria, mycobacteria, and fungi across our institution across a 10-year period. We assessed the positivity rates for this diagnostic tool by test type and specimen type and, critically, studied whether and how the results influenced the outcomes from treatment change, to readmission, to death.
KEYWORDS: universal PCR, molecular diagnostics, laboratory utilization, outcomes
INTRODUCTION
Universal PCR for bacteria, mycobacteria, and fungi is an attractive technology for the diagnosis of occult infections that may not be detected by routine microbiology laboratory methods (1). This technology involves sequencing of the 16S rRNA gene, the 65 kD heat-shock protein, or the 28S rRNA gene plus internally transcribed spacer regions to broadly detect and speciate bacterial, mycobacterial, and fungal DNA sequences, respectively. As molecular diagnostics such as these continue to evolve, laboratory stewardship is becoming increasingly important to ensure that these novel technologies are being utilized to the maximum benefit to the patient while minimizing unnecessary healthcare costs.
Universal PCR is best utilized in cases of culture-negative infections, especially in cases of fastidious organisms or when antimicrobials were given prior to specimen collection that compromise culture growth (2). While universal PCR has a fairly low sensitivity as a standalone assay (3), prospective studies have shown that when used in conjunction with culture, the overall detection rate may be enhanced over the use of culture alone (4 – 7). Various studies have supported the use of universal PCR in cases of meningitis (5, 8, 9), endophthalmitis (10, 11), joint and bone infections (6, 12 – 15), pleural effusions (16, 17), fungal sinusitis (18), and endocarditis (19 – 22). While positive universal PCR results have been reported to lead to changes in antimicrobial treatment, there is considerable variability between studies regarding the extent to which these results impact antimicrobial management (23 – 25).
Universal PCR is not without its limitations. Due to its non-specific nature, there is a high rate of false-positive results from contamination, especially in non-sterile body sites (26). Universal PCR also cannot provide antimicrobial susceptibility data, and even in the case of a positive result, clinicians must rely on epidemiologic data to guide coverage. In the absence of in-house universal PCR testing capacity, sending specimens to reference laboratories can take a week or longer to return results to the ordering institution, leaving clinicians to rely on empiric coverage of suspected organisms during a critical time in hospitalized patients’ care. Finally, this assay comes at a high cost compared to other routine microbiology testing methods, typically hundreds of dollars (27). Some studies have suggested that the combination of the lack of susceptibility data, high rate of contamination, sensitivity and faster turnaround time of concurrently submitted routine microbiology assays, and cost means that universal PCR provides little to no value to patient management or prognosis (28 – 30).
In this context, we aimed to further address the clinical utility of universal PCR, focusing on the patterns of utilization and patient outcomes. We performed a retrospective review of all universal PCR tests sent from our hospital network over a 10-year period and determined the cost of testing as well as the impact of positive universal PCR results on patient antimicrobial management and hospitalized patient outcomes. These data may impart diagnostic and prognostic significance for clinicians to determine the merit of testing for their patients and can ultimately guide stewardship decisions about the use of this technology for challenging diagnostic cases.
MATERIALS AND METHODS
We performed a retrospective chart review for all patients for whom universal PCR testing was ordered from our 743-bed adult tertiary-care medical center. These orders were placed by both outpatient and inpatient clinical teams, typically in conjunction with the infectious disease consult service. Clinicians had the option to order any combination of bacterial, mycobacterial, or fungal universal PCR based on their clinical judgement of potential etiologies of the patient’s infection. All universal PCR orders required review and approval by a clinical director of our institution’s microbiology laboratories, and approval was decided on a case-by-case basis without objective criteria. The specimens were sent for sequencing to the University of Washington Molecular Diagnosis Microbiology Section (Seattle, WA, USA). In the case of bacterial and fungal universal PCR, reflex testing to next-generation sequencing (NGS) was included in cases where multiple templates were present starting in late 2013.
We queried our institution’s clinical data repository to retrieve information on all specimens ordered for universal PCR from our electronic medical record network over the 10-year period from January 2013 through November 2022. A chart review was performed on all patients to determine patient factors and outcomes, including demographic data, relevant clinical laboratory data including concurrent Gram stain results and culture results, specimen type, clinical indications, antibiotic treatment decisions, inpatient status, and hospitalization outcomes. We considered other laboratory tests to be concurrent with the specimen sent for universal PCR if either the same specimen was utilized for both assays or if both specimens were retrieved from the same location during the same surgical procedure. We relied on clinical judgement by physicians treating the patient to determine if the detected organism was considered clinically relevant or if it may represent an irrelevant organism such as a commensal organism or a presumed contaminant.
All analyses were performed using R 3.5.1 (R Foundation for Statistical Computing, Vienna, Austria) and RStudio 1.1.463 (Posit, Boston, MA, USA). Population proportion confidence intervals were calculated to determine 95% confidence intervals for PCR positivity rates for different populations. A chi-squared test, odds ratio, or unpaired Student’s t-test was used for comparisons. P values were then corrected for multiple comparisons using the Benjamini-Hochberg method, and a corrected p value of < 0.05 was considered to be statistically significant. The Beth Israel Deaconess Medical Center Institutional Review Board reviewed the study protocol and determined it to be exempt (protocol no. 2017D000478).
RESULTS
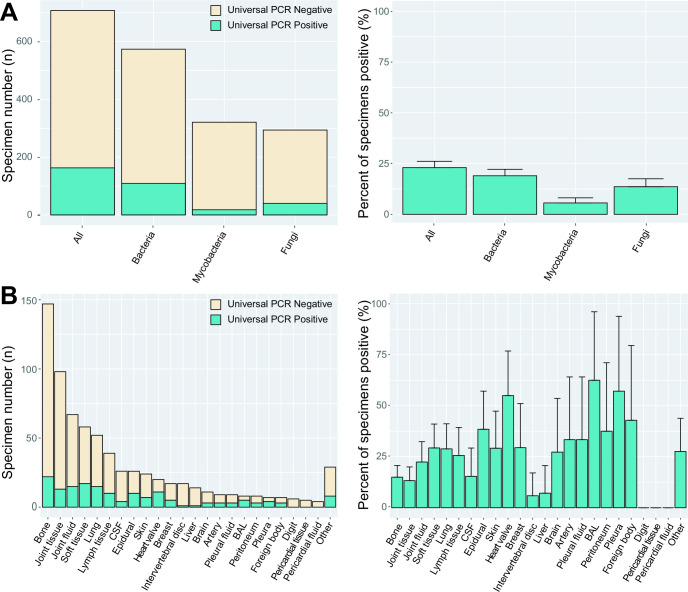
Over the 10-year period from January 2013 to November 2022, a total of 708 specimens from 638 patients (637 adult patients and 1 neonatal) were sent to our reference laboratory for universal PCR. Of those, 574 were tested for bacteria, 321 for mycobacteria, and 294 for fungi. A total of 54 patients had more than one specimen tested via universal PCR during the study period. Universal PCR specimens had a median turnaround time of 11 days from order to result (interquartile range of 8–14 days) regardless of the type of universal PCR testing. The overall positivity rate was 23% (163/708, 95% CI: 20%–26%). The positivity rate for those tested for bacteria was 19% (109/574, 95% CI: 16%–22%), for mycobacteria was 6% (18/321, 95% CI: 3.1%–8.1%), and for fungi was 14% (40/294, 95% CI: 9.7%–18%) (Fig. 1A).
Fig 1.
Total number of specimens sent for universal PCR and positivity rate by pathogen type (A) and specimen source (B). A single specimen could be sent for more than one universal PCR at a time. Error bars represent the 95% population proportion confidence intervals for positive results.
We examined whether other laboratory tests performed on specimens sent for universal PCR were associated with a positive PCR result (Table 1), including Gram stain results and results of concurrent histopathology slides examined in surgical pathology. The presence of organisms on Gram stain [23/44 positive, 52% (95% CI: 38%–67%)] was associated with positive universal PCR results (P < 0.05), and the absence of neutrophils on Gram stain [29/207 positive, 14% (95% CI: 9%–19%)] was negatively associated with positive universal PCR results (P < 0.05). We observed no significant difference in positivity rate between paraffin-embedded blocks and fresh frozen tissue. However, the presence of organisms seen on histology slides [34/73 positive, 47% (95% CI: 35%–58%)] was also significantly associated with a positive universal PCR result (P < 0.05). The presence of granulomata on histology was not associated with a positive PCR result. Having an organism detected on a prior specimen with universal PCR was significantly associated with a positive result [10/16 positive, 63% (95% CI: 39%–97%), P < 0.05], though the results of both PCR tests were concordant in only 40% of cases. Testing a new specimen following an initial specimen negative via universal PCR was not associated with a positive result.
TABLE 1.
Pre-analytic determinants of positive universal PCR results a
| Total specimens | Total positives by universal PCR | Percent positive (95% CIs) | Corrected P value | |
|---|---|---|---|---|
| Total | 708 | 163 | 23% (20%–26%) | -- |
| Female gender | 342 | 73 | 21% (17%–26%) | 0.79 |
| Age >50 | 498 | 113 | 23% (19%–26%) | 0.97 |
| Hardware present | 160 | 35 | 22% (15%–28%) | 0.97 |
| Gram stain organisms present | 44 | 23 | 52% (38%–67%) | 0.0001 |
| Neutrophils absent on gram stain | 178 | 29 | 16% (11%–22%) | 0.047 |
| Culture positive | 154 | 53 | 34% (27%–42%) | 0.018 |
| Prior universal PCR positive | 16 | 10 | 63% (39%–97%) | 0.013 |
| Organisms seen on histology | 73 | 34 | 47% (35%–58%) | 0.0001 |
| Granuloma on histology | 72 | 12 | 17% (8%–25%) | 0.62 |
| Formalin fixed tissue | 68 | 22 | 32% (21%–43%) | 0.40 |
| Outpatient | 115 | 27 | 23% (16%–31%) | 0.97 |
| Inpatient | 593 | 136 | 23% (20%–26%) | 0.97 |
Boldface indicates significant p values, -- indicates p value does not apply
Specimens sent for universal PCR had concurrent cultures taken in 656 cases (93%), with 154/656 (23%) of those cultures positive for an organism. Having a culture positive for any organism was significantly associated with positive universal PCR results [53/154 positive, 34% (95% CI: 27%–42%), P < 0.05]. Of those cases with a positive culture and a positive universal PCR result, there was concordance between those results for organism identification in 36/53 (68%) of cases. Positive cultures resulted sooner than universal PCR results in 32/53 (60%) of cases, including 19/36 (53%) of concordant cases.
A variety of specimen types were sent for universal PCR analysis. These included artery, bronchoalveolar lavage/sputum (BAL), bone, brain (including dura and meninges), breast, cerebrospinal fluid, digits, intervertebral disc, epidural tissues, foreign bodies, joints, liver, lungs, lymph node, pericardium, peritoneum, pleura, skin, soft tissues/muscles, heart valves, vitreous, esophagus, fallopian tube, bowel, bone marrow, uterus, and myocardium. Joint (including tissue and fluid) and bone tissue specimens were most frequent, with 165 and 148 specimens sent, respectively. Specimen types varied considerably in the overall positivity rates (Fig. 1B; Table S1). The highest positivity rates were seen in bronchoalveolar lavage/sputum specimens [5/8, 63% (95% CI: 29%–96%)] and heart valves [11/19, 58% (95% CI: 36%–80%)], while no positives were seen with digit or pericardial tissue/fluid specimens.
Likewise, clinical indications for testing varied considerably. The clinical indications for testing included osteomyelitis, septic arthritis, abscess, granulomata seen on surgical pathology, mass/lesion, endocarditis, pneumonia, cellulitis, lymphadenopathy, pericardial or pleural effusion, arteritis, meningitis, peritonitis, sinusitis, aplastic anemia, endophthalmitis, or a non-healing surgical wound. The highest positivity rate was noted for endocarditis [11/23 positive, 48%, (95% CI: 27%–68%)]. A complete table of positivity rates for specimen sources broken down by clinical indication and pathogen type is given in Table S1.
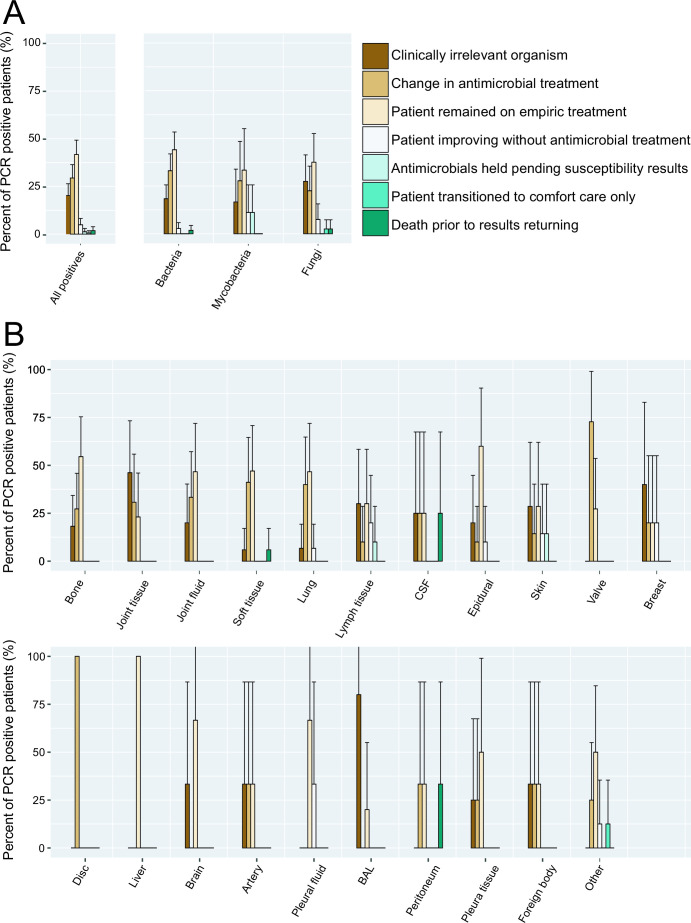
A total of 76% of all patients with positive universal PCR results were on empiric antimicrobial therapy prior to receiving results of the assay. Following the reporting of universal PCR results to the electronic medical record, clinicians considered the universal PCR result to be a clinically irrelevant organism in 33/163 (20%, 95% CI: 14%–26%) PCR positive cases (Fig. 2A). When broken down by specimen type, bronchoalveolar lavage/sputum specimens were considered to have the highest rate of clinical insignificance, with 4/5 (80%, 95% CI: 45%–100%) positive PCRs considered to be the result of a suspected contaminant or commensal organism (Fig. 2B). Then, 48/163 (29%, 95% CI: 22%–36%) positive results led to a change in antimicrobial management. Positive results led to a change in antibiotic management in 5/5 cases (100%) of intervertebral discs and 8/11 (72%, 95% CI: 46%–98%) of heart valves, while certain specimen types such as liver, brain, and BAL did not have any results that lead to a change in patient management. Reasons that the antimicrobial management was not changed include the patient already being on appropriate empiric therapy [68/163, 42% (95% CI: 34%–50%)], the patient improving without antibiotic intervention [8/163, 5% (95% CI: 2%–8%)], awaiting mycobacterial susceptibility results from a concurrently submitted culture [2/163, 2% (95% CI: 0%–4%)], or patient discharge to hospice or death prior to the return of the results [4/163, 3% (95% CI: 1%–5%)].
Fig 2.
Management decisions made by the clinical team based on positive universal PCR results by pathogen type (A) and specimen source (B). Error bars represent the 95% population proportion confidence intervals for positive results. Antimicrobial course was not changed in the case of death, transition to comfort care, patient improvement without intervention, satisfactory empiric treatment, or presumed contamination of the universal PCR sample.
We next determined whether a positive universal PCR result was associated with particular hospitalization outcomes. Of the 708 total specimens, 593 were from inpatients and 115 were from outpatients, with 136/593 (23%, 95% CI: 16%–31%) and 27/115 (23%, 95% CI: 20%–26%) being positive, respectively. From the time of specimen collection, universal PCR results from inpatients took a median of 11 days to return (interquartile range of 8–14 days) for all three types of universal PCR, regardless of positive or negative results. A total of 76% (95% CI: 72%–79%) of the results returned after the patient was discharged from the hospital. Among inpatients who were tested via universal PCR, 13/593 (2%, 95% CI: 1%–4%) died, 204/593 (34%, 95% CI: 31%–38%) required discharge to extended care facilities, including rehabilitation services and hospice care, and 123/593 (21%, 95% CI: 17%–24%) were readmitted to the hospital within 30 days of discharge. Universal PCR results returning prior to hospital discharge did not significantly affect hospitalization outcomes such as mortality and discharge to rehab services compared to patients whose results did not return until after discharge.
A positive universal PCR for fungi was significantly associated with increased hospital mortality [Odds ratio (OR) 7.65 (95% CI: 1.80–32.4), P < 0.05] (Fig. 3). A positive fungal PCR result was not significantly associated with discharge to rehabilitation or hospice care facilities [OR 2.38 (95% CI: 1.10–5.18, P = 0.052)] or readmission within 30 days following hospital discharge [OR 2.09 (95% CI 0.93–4.76), P = 0.07]. There were no significant differences in hospital mortality, discharge to rehabilitation or hospice facilities, or hospital readmission within 30 days among inpatients with positive bacterial or mycobacterial results. There were no significant differences in hospitalization length of stay associated with positive results for bacteria, mycobacteria, or fungi.
Fig 3.
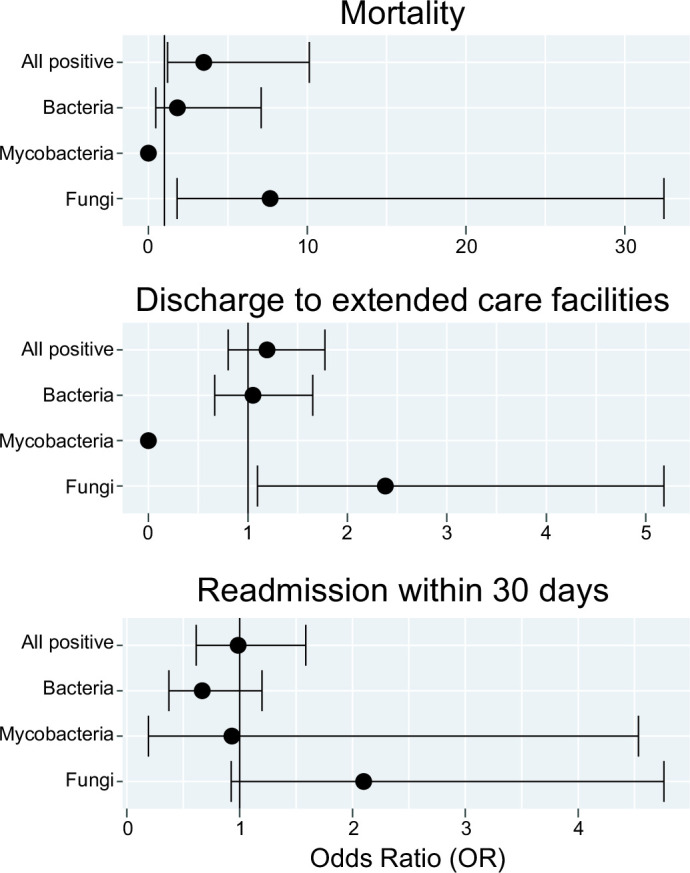
Hospitalization outcomes following positive universal PCR results compared to specimens which were negative via universal PCR. Error bars represent the 95% CI around the odds ratio.
Finally, we aimed to determine the cost associated with universal PCR testing by determining the budgetary cost of testing per positive result and per change in antibiotic management. Over the study period, our institution paid an average of $268 per bacterial PCR, $496 per mycobacterial PCR, and $439 per fungal PCR (Table 2). A reflex to NGS costs an additional $986, though an exact count of the number of specimens sent for reflex was not available and this cost was excluded from analysis. Because many specimens were ordered for more than one type of PCR, an average specimen costs $624 for overall testing. The average cost per positive PCR result was $1,411, $8,845, and $3,227 for bacteria, mycobacteria, and fungal PCR, respectively, for an overall cost per positive specimen of $2,712. For those positive results that led to a change in patient management, the cost per change in patient antibiotic regimen was $4,273, $31,843, and $14,341 for bacterial, mycobacterial, and fungal PCR, respectively, for an overall cost per change in antibiotic management of $9,211.
TABLE 2.
Budgetary cost associated with universal PCR testing
| Test type | Cost per test (USD) | Total tests sent (n) | Total positive tests (n) | Positive tests with a change in patient management (n) | Cost per positive test (USD) | Cost per change in patient management (USD) |
|---|---|---|---|---|---|---|
| Total | $624 | 708 | 163 | 48 | $2,712 | $9,211 |
| Bacteria | $268 | 574 | 109 | 36 | $1,411 | $4,273 |
| Mycobacteria | $496 | 321 | 18 | 5 | $8,845 | $31,843 |
| Fungi | $439 | 294 | 40 | 9 | $3,227 | $14,341 |
DISCUSSION
In this study, we examined the clinical factors associated with positive universal PCR results as well as associated hospitalization outcomes. In line with prior literature, we found that positive Gram stain results or histology results, neutrophils on Gram stain, and growth in culture were all significantly associated with positive universal PCR results (25, 31).
Our clinicians ordered universal PCR on a wide variety of specimen types, most frequently in the case of orthopedic infections. We found that although it was the most frequently ordered, orthopedic specimens such as joint tissues and fluids, bone, and intervertebral discs were among the least likely specimens to return positive. While less frequently ordered, heart valves were among the highest positivity rates and most frequently led to changes in antibiotic management. These findings are in line with the current literature demonstrating the great utility of universal PCR for endocarditis so much so that some studies have even suggested that universal PCR be incorporated as part of the modified Duke’s criteria for diagnosis of infective endocarditis (22).
In our cohort, 20% of all positive universal PCR results were considered clinically irrelevant by clinical teams. We relied on clinical judgement about whether a positive universal PCR result represented a true infection or clinically irrelevant organism. We believe that the fraction of results considered clinically irrelevant was actually underestimated in our data set due to clinicians potentially over-treating insignificant organisms detected on universal PCR out of an abundance of caution (Note that PCR disagreed with culture-based identification in 32% of cases). With a lack of gold standard in determining whether the resulting organism is truly causative of infection, universal PCR will likely continue to produce organism identifications that will present a clinical dilemma over whether to treat. Indeed, many of the changes in antibiotic regimens occurred when universal PCR results detected common commensals, such as Cutibacterium acnes or Corynebacterium species. While we did not evaluate the appropriateness of change in antimicrobials directly, these antibiotic changes may have had a negative impact on patient management, including drug side effects and IV infusion center costs, and antimicrobial stewardship programs.
Among patients with positive universal PCR results, only 29% of results led to a change in antibiotic management. The impact of positive universal PCR results on antibiotic management varies widely in the literature, with studies showing a change in management in 5% to 76% of cases (16, 24, 25, 32). The majority of our patients were on empiric treatment prior to universal PCR results, and so clinicians must consider the value that universal PCR brings to potentially changing or narrowing treatment and how strongly that will impact their patients’ care. A limitation of this study is that we were unable to perform a similar analysis to determine the changes in antibiotic management that may have resulted from a negative universal PCR result. While positive universal PCR results were invariably acknowledged in clinician notes, negative results were less frequently documented and therefore could not be directly correlated with treatment decisions. This may be a fruitful topic for future work.
We found that positive results from universal PCR for fungi were associated with worse hospital outcomes. A total of 4/30 (13%) positive cases in hospitalized inpatients were associated with patient death, and 15/30 (50%) cases required discharge to extended care facilities or hospice although such trends fell short of significance at the level of P < 0.05 after correction for multiple hypothesis testings. These findings likely reflect that patients with occult fungal infections are likely to be sicker with more severe systemic illness compared to patients without infections as the etiology of their disease.
There were no circumstances in which a positive result of universal PCR led to demonstrable improvements in hospitalization outcomes. Patients with positive universal PCR results for bacteria and mycobacteria did not have any change in hospital mortality, discharge to extended care facilities, readmission within 30 days, or length of stay. However, our universal PCR results took a median of 11 days from specimen collection to result, and 76% of hospitalized patients were discharged before the results returned. When considering the utility of this assay, turnaround time is a critical component. In rare instances, results took several months to return to the hospital, though these outliers were related to post-analytical reporting delays to our hospital. With improvements in turnaround time through the development of commercialized assays, it is possible that earlier time to appropriate antimicrobial intervention may be able to improve hospitalization outcomes (9, 13). However, many of these patients were treated empirically with broad-range antibiotics prior to results and it can be difficult to parse out specific improvements that occurred as a result of potentially narrowing coverage. It can be challenging to quantify other metrics of improper antibiotic use such as medication side effects, excess healthcare cost, and the promotion of resistance based on a retrospective analysis. With that said, universal PCR data may also prove valuable to infection control for epidemiologic studies; for example, in our hospital network, a cluster of four cases of Acinetobacter junii detected in orthopedic specimens in 2022 prompted an internal infection control investigation to identify potential sources of OR contamination to reduce false-positive rates. With further analysis, we were able to rule out orthopedic ORs as the source of contamination.
We found that with an average cost of $624 per specimen, ordering universal PCR costed an average of $2,712 to identify a single positive specimen and $9,210 to lead to a change in patient management following that positive result. Understanding the true contribution of a test cost to the overall healthcare cost for a patient can be complex. While the test cost to the microbiology laboratory associated with a single change in management may be greater than $30,000 as in the case of mycobacteria, when balanced against the cost of hospitalization, treatment regimens, ancillary testing, and other budgetary factors, these tests may still result in a net positive savings to the hospital at large.
Our findings here have provided a foundation of data for our institution to demonstrate to clinicians the costs and value of ordering this test for specific patient scenarios. Over-utilization of laboratory tests is at its highest for initial diagnostic testing with low-volume tests with subjective ordering criteria, as is currently the case of universal PCR (33). Given the significant variation in ordering and treatment practices currently among physicians, there will continue to be fluctuations in positivity rates and changes in antimicrobial practice from institution to institution until standardized objective guidelines are established for the use of this technology. To that end, a recent study assigning specimens a numeric score based on the amount of visualized inflammation or organisms on Gram stain and histopathology results found that low-scoring specimens tended to have lower positivity rates of universal PCR and lower rates of change in antibiotic management compared to those with a higher score (34). Our results complement such an approach by methodically examining clinical scenarios and specimen sources as well as outcome-based data to determine the pre-test probability of a positive assay and its potential to change management by objective criteria. Altering the ordering behaviors of clinical practices for these tests can be a complex and gradual process, but the foundation of standardizing ordering behaviors lies in a rigorous examination of the data from all relevant perspectives, including cost, positivity rates, and outcomes, to reassure clinical teams that stewardship decisions are made objectively in the best interests of their patients.
ACKNOWLEDGMENTS
We thank the University of Washington Molecular Diagnosis Microbiology Section (Seattle, WA, USA), our reference laboratory to whom we sent all samples to perform universal PCR studies. We also thank our clinical and laboratory staff for specimen submission and processing.
J.K. and S.R. conceived the studies and established parameters for data collection. A.M. and R.A. retrieved the data from our institution’s clinical data repository. Data analysis was performed by J.K. and A.K. Manuscript was written by J.K., R.A., and S.R. All authors reviewed the manuscript, data, and approved the final version.
Contributor Information
Ramy Arnaout, Email: rarnaout@bidmc.harvard.edu.
Stefan Riedel, Email: sriedel@bidmc.harvard.edu.
Daniel J. Diekema, Maine Medical Center Department of Medicine, Portland, Maine, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jcm.00952-23.
Table S1
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Church DL, Cerutti L, Gürtler A, Griener T, Zelazny A, Emler S. 2020. Performance and application of 16S rRNA gene cycle sequencing for routine identification of bacteria in the clinical microbiology laboratory. Clin Microbiol Rev 33:e00053-19. doi: 10.1128/CMR.00053-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sontakke S, Cadenas MB, Maggi RG, Diniz P, Breitschwerdt EB. 2009. Use of broad range16S rDNA PCR in clinical microbiology. J Microbiol Methods 76:217–225. doi: 10.1016/j.mimet.2008.11.002 [DOI] [PubMed] [Google Scholar]
- 3. Morel A-S, Dubourg G, Prudent E, Edouard S, Gouriet F, Casalta J-P, Fenollar F, Fournier PE, Drancourt M, Raoult D. 2015. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis 34:561–570. doi: 10.1007/s10096-014-2263-z [DOI] [PubMed] [Google Scholar]
- 4. Rampini SK, Bloemberg GV, Keller PM, Büchler AC, Dollenmaier G, Speck RF, Böttger EC. 2011. Broad-range 16S rRNA gene polymerase chain reaction for diagnosis of culture-negative bacterial infections. Clin Infect Dis 53:1245–1251. doi: 10.1093/cid/cir692 [DOI] [PubMed] [Google Scholar]
- 5. Schuurman T, de Boer RF, Kooistra-Smid AMD, van Zwet AA. 2004. Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from cerebrospinal fluid for diagnosis of bacterial meningitis in a clinical setting. J Clin Microbiol 42:734–740. doi: 10.1128/JCM.42.2.734-740.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tkadlec J, Peckova M, Sramkova L, Rohn V, Jahoda D, Raszka D, Berousek J, Mosna F, Vymazal T, Kvapil M, Drevinek P. 2019. The use of broad-range bacterial PCR in the diagnosis of infectious diseases: a prospective cohort study. Clin Microbiol Infect 25:747–752. doi: 10.1016/j.cmi.2018.10.001 [DOI] [PubMed] [Google Scholar]
- 7. Lass-Flörl C, Mutschlechner W, Aigner M, Grif K, Marth C, Girschikofsky M, Grander W, Greil R, Russ G, Cerkl P, Eller M, Kropshofer G, Eschertzhuber S, Kathrein H, Schmid S, Beer R, Lorenz I, Theurl I, Nachbaur D. 2013. Utility of PCR in diagnosis of invasive fungal infections: real-life data from a multicenter study. J Clin Microbiol 51:863–868. doi: 10.1128/JCM.02965-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deutch S, Pedersen LN, Pødenphant L, Olesen R, Schmidt MB, Møller JK, Ostergaard L. 2006. Broad-range real time PCR and DNA sequencing for the diagnosis of bacterial meningitis. Scand J Infect Dis 38:27–35. doi: 10.1080/00365540500372861 [DOI] [PubMed] [Google Scholar]
- 9. Welinder-Olsson C, Dotevall L, Hogevik H, Jungnelius R, Trollfors B, Wahl M, Larsson P. 2007. Comparison of broad-range bacterial PCR and culture of cerebrospinal fluid for diagnosis of community-acquired bacterial meningitis. Clin Microbiol Infect 13:879–886. doi: 10.1111/j.1469-0691.2007.01756.x [DOI] [PubMed] [Google Scholar]
- 10. Mishra D, Satpathy G, Chawla R, Venkatesh P, Ahmed NH, Panda SK. 2019. Utility of broad-range 16S rRNA PCR assay versus conventional methods for laboratory diagnosis of bacterial endophthalmitis in a tertiary care hospital. Br J Ophthalmol 103:152–156. doi: 10.1136/bjophthalmol-2018-312877 [DOI] [PubMed] [Google Scholar]
- 11. Ogawa M, Sugita S, Watanabe K, Shimizu N, Mochizuki M. 2012. Novel diagnosis of fungal endophthalmitis by broad-range real-time PCR detection of fungal 28S ribosomal DNA. Graefes Arch Clin Exp Ophthalmol 250:1877–1883. doi: 10.1007/s00417-012-2015-7 [DOI] [PubMed] [Google Scholar]
- 12. Fenollar F, Roux V, Stein A, Drancourt M, Raoult D. 2006. Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J Clin Microbiol 44:1018–1028. doi: 10.1128/JCM.44.3.1018-1028.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kühn C, Disqué C, Mühl H, Orszag P, Stiesch M, Haverich A. 2011. Evaluation of commercial universal rRNA gene PCR plus sequencing tests for identification of bacteria and fungi associated with infectious endocarditis. J Clin Microbiol 49:2919–2923. doi: 10.1128/JCM.00830-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gille J, Wallstabe S, Schulz A-P, Paech A, Gerlach U. 2012. Is non-union of tibial shaft fractures due to nonculturable bacterial pathogens? a clinical investigation using PCR and culture techniques. J Orthop Surg Res 7:20. doi: 10.1186/1749-799X-7-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qu X, Zhai Z, Li H, Li H, Liu X, Zhu Z, Wang Y, Liu G, Dai K. 2013. PCR-based diagnosis of prosthetic joint infection. J Clin Microbiol 51:2742–2746. doi: 10.1128/JCM.00657-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lampejo T, Ciesielczuk H, Lambourne J. 2021. Clinical utility of 16S rRNA PCR in pleural infection. J Med Microbiol 70. doi: 10.1099/jmm.0.001366 [DOI] [PubMed] [Google Scholar]
- 17. Insa R, Marín M, Martín A, Martín-Rabadán P, Alcalá L, Cercenado E, Calatayud L, Liñares J, Bouza E. 2012. Systematic use of universal 16S rRNA gene polymerase chain reaction (PCR) and sequencing for processing pleural effusions improves conventional culture techniques. Medicine (Baltimore) 91:103–110. doi: 10.1097/MD.0b013e31824dfdb0 [DOI] [PubMed] [Google Scholar]
- 18. Lieberman JA, Bryan A, Mays JA, Stephens K, Kurosawa K, Mathias PC, SenGupta D, Bourassa L, Salipante SJ, Cookson BT. 2021. High clinical impact of broad-range fungal PCR in suspected fungal sinusitis. J Clin Microbiol 59:e0095521. doi: 10.1128/JCM.00955-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Faraji R, Behjati-Ardakani M, Moshtaghioun SM, Kalantar SM, Namayandeh SM, Soltani M, Emami M, Zandi H, Firoozabadi AD, Kazeminasab M, Ahmadi N, Sarebanhassanabadi M. 2018. The diagnosis of microorganism involved in infective endocarditis (IE) by polymerase chain reaction (PCR) and real‐time PCR: a systematic review. Kaohsiung J Med Sci 34:71–78. doi: 10.1016/j.kjms.2017.09.011 [DOI] [PubMed] [Google Scholar]
- 20. Harris KA, Yam T, Jalili S, Williams OM, Alshafi K, Gouliouris T, Munthali P, NiRiain U, Hartley JC. 2014. Service evaluation to establish the sensitivity, specificity and additional value of broad-range 16S rDNA PCR for the diagnosis of infective endocarditis from resected endocardial material in patients from eight UK and Ireland hospitals. Eur J Clin Microbiol Infect Dis 33:2061–2066. doi: 10.1007/s10096-014-2145-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Armstrong C, Kuhn TC, Dufner M, Ehlermann P, Zimmermann S, Lichtenstern C, Soethoff J, Katus HA, Leuschner F, Heininger A. 2021. The diagnostic benefit of 16S rDNA PCR examination of infective endocarditis heart valves: a cohort study of 146 surgical cases confirmed by histopathology. Clin Res Cardiol 110:332–342. doi: 10.1007/s00392-020-01678-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marín M, Muñoz P, Sánchez M, Del Rosal M, Alcalá L, Rodríguez-Créixems M, Bouza E. 2007. Molecular diagnosis of infective endocarditis by real-time broad-range polymerase chain reaction (PCR) and sequencing directly from heart valve tissue. Medicine 86:195–202. doi: 10.1097/MD.0b013e31811f44ec [DOI] [PubMed] [Google Scholar]
- 23. Kerkhoff AD, Rutishauser RL, Miller S, Babik JM. 2020. Clinical utility of universal broad-range polymerase chain reaction amplicon sequencing for pathogen identification: a retrospective cohort study. Clin Infect Dis 71:1554–1557. doi: 10.1093/cid/ciz1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marbjerg LH, Holzknecht BJ, Dargis R, Dessau RB, Nielsen XC, Christensen JJ. 2020. Commercial bacterial and fungal broad-range PCR (micro-Dx) used on culture-negative specimens from normally sterile sites: diagnostic value and implications for antimicrobial treatment. Diagn Microbiol Infect Dis 97:115028. doi: 10.1016/j.diagmicrobio.2020.115028 [DOI] [PubMed] [Google Scholar]
- 25. Naureckas Li C, Nakamura MM. 2022. Utility of broad-range PCR sequencing for infectious diseases clinical decision making: a pediatric center experience. J Clin Microbiol 60:e0243721. doi: 10.1128/jcm.02437-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Renvoisé A, Brossier F, Sougakoff W, Jarlier V, Aubry A. 2013. Broad-range PCR: past, present, or future of bacteriology? Med Mal Infect 43:322–330. doi: 10.1016/j.medmal.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 27. Aggarwal D, Kanitkar T, Narouz M, Azadian BS, Moore LSP, Mughal N. 2020. Clinical utility and cost-effectiveness of bacterial 16S rRNA and targeted PCR based diagnostic testing in a UK microbiology laboratory network. Sci Rep 10:7965. doi: 10.1038/s41598-020-64739-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stempak LM, Vogel SA, Richter SS, Wyllie R, Procop GW. 2019. Routine broad-range fungal polymerase chain reaction with DNA sequencing in patients with suspected mycoses does not add value and is not cost-effective. Arch Pathol Lab Med 143:634–638. doi: 10.5858/arpa.2017-0299-OA [DOI] [PubMed] [Google Scholar]
- 29. Miller K, Harrington SM, Procop GW. 2015. Acid-fast smear and histopathology results provide guidance for the appropriate use of broad-range polymerase chain reaction and sequencing for mycobacteria. Arch Pathol Lab Med 139:1020–1023. doi: 10.5858/arpa.2013-0705-OA [DOI] [PubMed] [Google Scholar]
- 30. Panousis K, Grigoris P, Butcher I, Rana B, Reilly JH, Hamblen DL. 2005. Poor predictive value of broad-range PCR for the detection of arthroplasty infection in 92 cases. Acta Orthop 76:341–346. [PubMed] [Google Scholar]
- 31. Basein T, Gardiner B, Andujar-Vazquez GM, Chandranesan ASJ, Rabson A, Doron S, Snydman DR. 2017. Clinical utility of universal PCR and its real-world impact on patient management. Open Forum Infect Dis 4:S627–S627. doi: 10.1093/ofid/ofx163.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Racsa LD, DeLeon-Carnes M, Hiskey M, Guarner J. 2017. Identification of bacterial pathogens from formalin-fixed, paraffin-embedded tissues by using 16S sequencing: retrospective correlation of results to clinicians' responses. Hum Pathol 59:132–138. doi: 10.1016/j.humpath.2016.09.015 [DOI] [PubMed] [Google Scholar]
- 33. Zhi M, Ding EL, Theisen-Toupal J, Whelan J, Arnaout R. 2013. The landscape of inappropriate laboratory testing: A 15-year meta-analysis. PLoS One 8:e78962. doi: 10.1371/journal.pone.0078962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dumm RE, Glaser LJ, Rodino KG. 2023. Development of a scoring system to identify high-yield specimens for bacterial broad-range 16S rRNA gene PCR with sequencing at a tertiary care medical center. Am J Clin Pathol:aqad074. doi: 10.1093/ajcp/aqad074 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1