ABSTRACT
In 2017, the Centers for Disease Control and Prevention (CDC) established the Antimicrobial Resistance Laboratory Network to improve domestic detection of multidrug-resistant organisms. CDC and four laboratories evaluated a commercial broth microdilution panel. Antimicrobial susceptibility testing using the Sensititre GN7F (ThermoFisher Scientific, Lenexa, KS) was evaluated by testing 100 CDC and Food and Drug Administration AR Isolate Bank isolates [40 Enterobacterales (ENT), 30 Pseudomonas aeruginosa (PSA), and 30 Acinetobacter baumannii (ACB)]. We assessed multiple amounts of transfer volume (TV) between the inoculum and tubed 11-mL cation-adjusted Mueller-Hinton broth: 1 µL [tribe Proteeae (P-tribe) only] and 10, 30, and 50 µL, resulting in respective CFU per milliter of 1 × 104, 1 × 105, 3 × 105, and 5 × 105. Four TV combinations were analyzed: standard (STD) [1 µL (P-tribe) and 10 µL], enhanced standard (E-STD) [1 µL (P-tribe) and 30 µL], 30 µL, and 50 µL. Essential agreement (EA), categorical agreement, major error (ME), and very major error (VME) were analyzed by organism then TVs. For ENT, the average EA across laboratories was <90% for 7 of 15 β-lactams using STD and E-STD TVs. As TVs increased, EA increased (>90%), and VMEs decreased. For PSA, EA improved as TVs increased; however, MEs also increased. For ACB, increased TVs provided slight EA improvements; all TVs yielded multiple VMEs and MEs. For ENT and ACB, Minimum inhibitory concentrations (MICs) trended downward using a 1 or 10 µL TV; there were no obvious MIC trends by TV for PSA. The public health and clinical consequences of missing resistance warrant increased TV of 30 µL for the GN7F, particularly for P-tribe, despite being considered “off-label” use.
KEYWORDS: antimicrobial susceptibility testing, evaluation, commercial AST, broth microdilution, antimicrobial resistance
INTRODUCTION
In 2017, the Centers for Disease Control and Prevention (CDC) established the Antimicrobial Resistance Laboratory Network (AR Lab Network) to improve domestic detection and characterization of carbapenem-resistant (CR) Enterobacterales (ENT), Pseudomonas aeruginosa (PSA), and Acinetobacter baumannii (ACB) (1). These antimicrobial- resistant (AR) organisms are considered serious or urgent public health threats by CDC (2). Carbapenem resistance can be conferred by overexpression of efflux pumps, decreased permeability of membrane porins, and acquired resistance genes, including genes conferring the production of carbapenem hydrolyzing enzymes (carbapenemases), such as KPC, NDM, OXA-48-like, VIM, and IMP (3, 4). Carbapenemase genes pose a significant threat to patient safety due to their ability to transfer horizontally from organism to organism. The production of carbapenemases and concomitantly other β-lactamases (e.g., extended-spectrum β-lactamases and AmpCs) limits lifesaving treatment options for use against infections caused by these bacteria (5 – 7).
As part of the AR Lab Network’s infrastructure, antimicrobial susceptibility testing (AST) is performed to better characterize and monitor emerging AR threats (1). Many of the public health laboratories in the AR Lab Network previously validated the Sensititre GNX2F panel, a commercially available research-use only (RUO) broth microdilution (BMD) panel; however, the GNX2F panel lacks newer β-lactam/β-lactamase inhibitor combination agents which remain active against certain carbapenemase-producing organisms (CPOs) and lower dilutions for the updated fluoroquinolone breakpoints. The data generated from AST are key in monitoring for emerging resistance and providing critical information to aid in clinical decision making and informing isolate submission and laboratory workflows to target the detection of emerging CPOs (8).
The Sensititre GN7F panel is a BMD panel containing 24 antimicrobial agents, including key antimicrobials for Gram-negative bacteria such as the carbapenems, cephalosporins, monobactams, and newer β-lactam/β-lactamase inhibitor combination agents, such as ceftazidime/avibactam and ceftolozane/tazobactam. The Sensititre Food and Drug Administration (FDA)-cleared instructions for use (IFU) describes several options for transferring the bacterial suspension to inoculate the GN7F BMD panel, each of which results in a different final CFU per milliliter: a “standard” (STD) inoculum and an “enhanced standard” (E-STD) inoculum to aid in the detection of resistance, and an Enterobacterales tribe Proteeae (P-tribe)-specific method for preparing the inoculum. CDC and four AR Lab Network public health laboratories evaluated this panel by testing the different inoculum methods using a challenge set of well-characterized isolates obtained from the CDC & FDA AR Isolate Bank.
MATERIALS AND METHODS
Isolate selection
A total of 100 Gram-negative bacterial isolates from the CDC & FDA AR Isolate Bank (AR Bank, https://wwwn.cdc.gov/arisolatebank/) were used in this study (Table S1). Enterobacterales (n = 40, including 9 tribe Proteeae), Pseudomonas aeruginosa (n = 30), and Acinetobacter baumannii (n = 30). Isolates selected exhibited a range of resistance mechanisms with nearly 50% of isolates testing resistant to the carbapenems and having MICs that were on-scale (not less than or equal to and not greater than the MIC range of the GN7F panel). All AR Bank isolates had been characterized by the reference broth microdilution method for AST (9) and whole genome sequencing (WGS) (MiSeq platform; Illumina; San Diego, CA) for identifying resistance determinants. WGS gene detection was conducted using Resfinder and Antibiotic Resistance Gene-ANNOTation databases (accessed 6 August 2018) and thresholds were set at 99% identification and 100% coverage.
Phase I: multisite evaluation of GN7F using well-characterized isolates from the CDC & FDA AR Isolate Bank
Study design
In phase I, four AR Lab Network laboratories and CDC tested the same set of 100 isolates from the AR Bank. AST was performed with the Sensititre GN7F panel (ThermoFisher Scientific; Lenexa, KS) in accordance with the IFU (10) or established laboratory procedures as outlined in Table 1. Briefly, for the inoculum, a suspension of isolated colonies from a blood agar plate (trypticase soy agar + 5% sheep blood) was prepared and adjusted to the turbidity equivalent to a 0.5 McFarland standard by using either a MicroScan turbidity meter (Beckman-Coulter, Brea, CA) or the Sensititre Nephelometer (ThermoFisher Scientific). Next, an amount of the inoculum was transferred to a tube containing 11 mL cation-adjusted Mueller-Hinton broth (CAMHB) w/TES (ThermoFisher Scientific). According to the package insert, three different inoculum transfer volumes (TVs) could be used based on organism group or the need to enhance resistance detection; the final CFU per milliliter in the GN7F panel is directly affected by this TV. For the standard inoculum method, 10 µL of inoculum was transferred to the tube of 11 mL CAMHB (final: ~1 × 105 CFU/mL). For the enhanced inoculum method, as described in the IFU, 30 µL of inoculum was transferred (final: ~3 × 105 CFU/mL). The study group also tested a TV of 50 µL (final: ~5 × 105 CFU/mL), which is not supported in the IFU. For all P-tribe organisms (Proteus, Providencia, and Morganella spp.), 1 µL of inoculum is the only TV supported in the IFU (final: ~1 × 104 CFU/mL); nevertheless, the study group also assessed a 30 and a 50 µL TV for the P-tribe group of organisms. In summary, four TVs were used for testing with the GN7F panel: 1, 30, and 50 µL for P-tribe ENT, and 10, 30, and 50 µL for the non-P-tribe ENT, PSA and ACB. The Sensititre automated inoculating system (AIM) was used to dispense 50 µL of inoculated CAMHB into each well of the GN7F panel, which was then sealed, stacked three panels high, and incubated in ambient air or the Sensititre ARIS 2X at 35 ± 2°C for 16–20 hours (ENT and PSA) or 20–24 hours (ACB), then read by the Sensititre ARIS 2X with OptiRead, Sensititre Vizion (ThermoFisher Scientific), BioMIC (Giles Scientific USA; Santa Barbara, CA), or a mirror reader. Each study group laboratory’s specific workflow and the workflow’s regulatory status are described in Table 1.
TABLE 1.
GN7F AST methods and regulatory status by participating laboratory
| Lab | Inoculum media | Inoculum standardization | Read method | IFU status | Off-label reason |
|---|---|---|---|---|---|
| 1 | Sensititre dH2O a | MicroScan turbidity meter | Mirror stand with indirect light | Off-label | Inoculum standardization |
| 2 | Remel 0.85% sterile saline | MicroScan turbidity meter | Sensititre Vizion (manual) | Off-label | Inoculum media and standardization |
| 3 | Sensititre dH2O | Sensititre nephelometer | Sensititre Vizion (manual) | On-label | |
| Sensititre ARIS 2X with OptiRead (automated) | On-label | ||||
| 4 | Sensititre dH2O | Sensititre nephelometer | Sensititre Vizion (manual) | On-label | |
| 5 | Sensititre dH2O | Sensititre nephelometer | Giles Scientific BIOMIC V3 bottom-reader (with manual review) | Off-label | Read method |
dH2O, deionized water.
Quality control strains used were Escherichia coli American Type Culture Collection (ATCC) 35218, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Pseudomonas aeruginosa ATCC 27853, Enterococcus faecium ATCC 29212, and Staphylococcus aureus ATCC 29213 (ATCC, Manassas, VA).
Laboratories performed colony counts in duplicate on each day of testing for E. coli ATCC 25922, all P-tribe organisms, and a random selection of other organisms among the study isolates (at least one organism per laboratory’s defined batch) (9). Colony counts were performed by aspirating 10 µL from a positive control well of the GN7F panel and diluting into 10 mL of sterile water. From the diluted water suspension, 100 µL was spread onto a blood agar plate and incubated for 18–24 hours at 35 ± 2°C. Colony counts were averaged per isolate.
For each TV assessed, the interlaboratory reproducibility was assessed across all five laboratories by determining the mode, or median if a mode was unavailable, of the single MIC result generated by each laboratory, then determining essential agreement by comparing each laboratory’s result individually to the calculated modal or median MIC.
Data analysis
Data were collected from all sites in a custom Research Electronic Data Capture project hosted by CDC (11). Data were analyzed using Statistical Analysis Software (SAS) v.9.4 (Cary, NC) and Microsoft Excel. Accuracy was assessed by calculating essential agreement (EA), categorical agreement (CA), major errors (MEs, false resistant), and very major errors (VMEs, false susceptible) (12, 13). A few AR Bank isolates demonstrated MICs spanning a >4 dilution range for certain drugs by the reference BMD method; these specific isolate-drug combinations demonstrating this variability were not included in the analyses since many of the GN7F drug concentration ranges were ≤5 dilutions. Using combinations of the four inoculum TVs, up to four analytical categories describing procedures of inoculation were assessed for accuracy: STD (10 µL for non-P-tribe and 1 µL for P-tribe), E-STD (30 µL for non-P-tribe and 1 µL for P-tribe), 30 µL for all organisms (30-All), 50 µL for all organisms (50-All). The STD and E-STD are considered on-label; 30-All is considered on-label for non-P-tribe, but 30 µL TV for non-P-tribe and 50-All are considered off-label (Table 2).
TABLE 2.
Combinations of transfer volumes used for analysis
| Procedure group | Non-tribe Proteeae (µL) | Tribe Proteeae (µL) | IFU status |
|---|---|---|---|
| Standard (STD) | 10 | 1 | On-label |
| Enhanced standard (E-STD)-Enterobacterales only |
30 | 1 | On-label |
| 30 µL (30-All) | 30 | 30 | On-label for non-tribe Proteeae, off-label for tribe Proteeae |
| 50 µL (50-All) | 50 | 50 | Off-label |
The breakpoints assessed for all ENT included those in Clinical and Laboratory Standards Institute (CLSI) M100ed32 (14), as well as those identified in the GN7F IFU (037-NFAST FDA-USA Only–CID10253) if the antimicrobial agent was not currently FDA-cleared for use with current breakpoints on the FDA STIC website at the time of writing (15). Among the latter, the list of antimicrobial agents and current clearance for obsolete breakpoints (S/I/R in µg/mL) of the GN7F were as follows: aztreonam (8/16/32), cefazolin (8/16/32), cefepime (8/16/32), ceftazidime (8/16/32), ciprofloxacin (1/2/4), doripenem [≤0.5 (susceptible only)], imipenem (4/8/16), levofloxacin (2/4/8), and piperacillin/tazobactam (16/32–64/128). FDA interpretive criteria were used for tigecycline (15). Only CLSI M100ed32 breakpoints were assessed for PSA and ACB organisms, as the device had minimal FDA clearance pursued. For PSA, FDA clearance was obtained for only ceftazidime/avibactam and ceftolozane/tazobactam, doripenem (obsolete breakpoints), and piperacillin/tazobactam (obsolete breakpoints). For ACB, FDA clearance was obtained for only doripenem (obsolete breakpoints) and minocycline.
Bias, the proportion of GN7F MICs at least one dilution higher (positive) or lower (negative) than the reference BMD MICs, was also assessed per FDA guidance for 510(k) submissions (16) (Fig. S1). Briefly, MIC pairs were considered evaluable for trending when MICs generated from GN7F panel were one or more doubling dilutions higher or lower than MICs generated by the AR Bank irrespective of whether the MICs from the GN7F panel were on-scale (i.e., assessed when one or both MICs were on-scale and not assessed when both MICs were off-scale). Bias was calculated by the difference in proportion of MICs observed at +1 to −1 dilutions from the reference MICs. Confidence intervals (CIs) of bias were calculated using the Newcombe-Wilson method. An upward or downward trend was defined as when the bias was ≥±30% and significant (the CI did not include zero effect) (13, 16). Exact CIs were used to assess sensitivity and specificity of detecting carbapenem-resistant organisms (CROs).
Phase IIa: intermediary discrepancy testing by CDC
Study design
Due to limited resources and capacity within the study group (i.e., reagent supply and personnel-time), CDC alone conducted intermediary discrepancy testing of the subset of isolates where most or all partnering laboratories observed ME or VMEs. For these isolates, CDC performed side-by-side AST (using the same inoculum) with the GN7F panel and CDC’s in-house frozen reference BMD panels. Preparation of in-house reference BMD panels and subsequent AST were performed according to the CLSI M07 standard and as described previously (9, 17). In this study, we used 5 mL deionized water (ThermoFisher Scientific) when preparing the inoculum suspension, rather than 5 mL 0.85% sterile saline. This phase was used to determine whether all transfer volumes were required to undergo discrepancy testing by the entire study group.
Data analysis
Data were stored in an Excel file and analyzed using SAS (v.9.4). Each isolate was assessed for EA, ME, and VME for the four analytical TV categories. The comparator MIC was the MIC obtained by the in-house reference BMD panels tested alongside the GN7F from the same inoculum. Due to known inherent variability of AST and BMD, including the reference methods, these MICs may not reflect the exact modal MIC provided by the AR Bank.
Phase IIb: discrepancy testing by PHLs
Study design
Discrepancy testing of isolates with any ME or VME errors from 30 to 50 µL transfer volumes was conducted by all study group laboratories. For any ME or VME, regardless of occurrence in one or both of the 30 and 50 µL TVs, both TVs were retested (i.e., if a VME only occurred with 30 µL TV, both 30 and 50 µL TVs were repeated during discrepancy testing). For each day of discrepancy testing, laboratories also retested at least one non-discrepant isolate to avoid introducing bias to the analysis. Recommended QC strains were included in each test day.
Data analysis
Data collection and analysis for EA, CA, ME, and VMEs were collected and calculated as described in phase I.
RESULTS
Phase I: multisite evaluation of GN7F using well-characterized isolates from the CDC & FDA AR Isolate Bank
Accuracy
The results for phase I testing of all ENT are summarized in Tables 3 and 4. Overall, the STD and E-STD combinations of TVs performed poorly across the β-lactam agents. For the STD combination, all laboratories observed EA of <90% for doripenem (80.0%–82.5%), ertapenem (84.2%–86.8%), imipenem (86.8%–89.5%), meropenem (82.1–89.7%), cefepime (65.8%–76.3%), and piperacillin/tazobactam (79.5%–82.1%). For the E-STD combination, all laboratories observed EA of <90% for doripenem (85.0%–87.5%), ertapenem (84.2%–86.8%), cefepime (78.9%–81.6%), and piperacillin/tazobactam (84.6%–87.2%). The performance of these antimicrobial agents improved as the TV was increased to 30 µL (30-All) and 50 µL (50-All) across all Enterobacterales organisms, including P-tribe. Similarly, CA rates using CLSI M100 2022 breakpoints were lower for the STD and E-STD TV combinations but improved as the TV increased; using the obsolete FDA-cleared breakpoints outlined in the IFU generally resulted in lower CA (Table 3). The frequency of errors observed with STD and E-STD were higher for VMEs than MEs (Table 4). For STD combinations, all laboratories observed ≥1 VME for doripenem, ertapenem, imipenem, meropenem, aztreonam, ceftazidime, ceftolozane/tazobactam, piperacillin/tazobactam, and nitrofurantoin. VME frequency improved slightly with E-STD, and most VMEs were resolved with 30-All or 50-All.
TABLE 3.
Phase I summary of essential and categorical agreement across four laboratories a for 40 Enterobacterales isolates using 2022 CLSI and FDA-cleared breakpoints per IFU d
| Antimicrobial agent | N isolates | Essential agreement (%) b | Categorical agreement (%) b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STD d | E-STD | 30-All | 50-All | STD CLSI BPs |
STD IFU BPs |
E-STD CLSI BPs |
E-STD IFU BPs |
30-All CLSI BPs |
30-All IFU BPs |
50-All CLSI BPs |
50- IFU BPs |
||
| Doripenem | 40 | 81.3 (1.4) | 86.3 (1.4) | 97.5 (2) | 98.8 (1.4) | 77.5 (3.5) | 100 (0) c | 83.1 (2.4) | 100 (0) c | 92.5 (2) | 100 (0) c | 95 (2) | 99 (2.5) c |
| Ertapenem | 38 | 85.5 (1.5) | 85.5 (1.5) | 95.4 (1.3) | 94.7 (2.1) | 88.2 (1.5) | − | 88.2 (1.5) | − | 97.4 (2.1) | − | 99.3 (1.3) | − |
| Imipenem | 38 | 87.5 (1.3) | 89.5 (2.1) | 100 (0) | 98.7 (1.5) | 81.6 (2.1) | 73.7 (2.1) | 88.2 (2.6) | 78.9 (2.1) | 98.7 (1.5) | 90.8 (3.4) | 98 (1.3) | 90.1 (3.9) |
| Meropenem | 39 | 85.3 (3.2) | 89.1 (3.8) | 96.2 (3.3) | 93.6 (3.3) | 79.5 (0) | − | 83.3 (1.5) | − | 94.2 (1.3) | − | 95.5 (1.3) | − |
| Aztreonam | 40 | 90.6 (2.4) | 93.8 (1.4) | 98.8 (1.4) | 99.4 (1.3) | 90.6 (1.3) | 85.6 (1.3) | 95 (2) | 89.4 (2.4) | 97.5 (2) | 95.6 (2.4) | 96.9 (1.3) | 95 (2.9) |
| Ceftazidime | 40 | 95 (0) | 95 (0) | 98.1 (1.3) | 97.5 (2) | 95 (0) | 93.8 (1.4) | 95 (0) | 93.8 (1.4) | 96.3 (1.4) | 95 (1.4) | 97.5 (0) | 94.4 (1.4) |
| Ceftriaxone | 40 | 89.4 (1.3) | 90.6 (1.3) | 97.5 (2) | 99.4 (1.3) | 97.5 (0) | − | 97.5 (0) | − | 100 (0) | − | 100 (0) | − |
| Cefepime | 38 | 71.1 (4.3) | 80.3 (1.5) | 88.2 (3.4) | 92.8 (3.9) | 80.9 (3.3) | 69.7 (3.4) | 86.2 (2.5) | 75 (5) | 90.8 (3.4) | 81.6 (4.3) | 93.4 (3.4) | 90.1 (4.5) |
| Cefazolin | 19 | 100 (0) | 100 (0) | 100 (0) | 100 (0) | 88.2 (2.6) | NE | 92.1 (3) | NE | 92.1 (3) | NE | 90.8 (2.6) | NE |
| Ceftazidime-avibactam | 40 | 98.8 (1.4) | 100 (0) | 100 (0) | 99.4 (1.3) | 100 (0) | − | 100 (0) | − | 100 (0) | − | 100 (0) | − |
| Ceftolozane-tazobactam | 38 | 92.1 (2.1) | 94.7 (0) | 94.7 (0) | 95.4 (1.3) | 91.4 (3.3) | − | 92.1 (2.1) | − | 93.4 (1.5) | − | 94.7 (2.1) | − |
| Piperacillin-tazobactam | 39 | 81.4 (1.3) | 86.5 (1.3) | 94.2 (3.2) | 97.4 (2.1) | 85.3 (4.4) | 80.8 (1.5) | 88.5 (6.8) | 84.6 (2.1) | 93.6 (5.3) | 89.7 (3.6) | 94.9 (3.6) | 93.6 (1.5) |
| Ampicillin-sulbactam | 30 | 95.8 (1.7) | 95.8 (1.7) | 98.3 (3.3) | 100 (0) | 82.5 (3.2) | − | 85 (4.3) | − | 89.2 (6.3) | − | 87.5 (6.9) | − |
| Ampicillin | 10 | 100 (0) | 100 (0) | 100 (0) | 100 (0) | 100 (0) | − | 100 (0) | − | 100 (0) | − | 100 (0) | − |
| Ciprofloxacin | 40 | 100 (0) | 100 (0) | 100 (0) | 100 (0) | 97.5 (0) | 98.1 (2.4) | 97.5 (0) | 98.8 (1.4) | 97.5 (0) | 98.1 (1.3) | 96.3 (1.4) | 96.9 (2.4) |
| Levofloxacin | 40 | 100 (0) | 100 (0) | 100 (0) | 100 (0) | 95.6 (1.3) | 97.5 (0) | 95.6 (1.3) | 97.5 (0) | 97.5 (0) | 96.3 (1.4) | 96.9 (2.4) | 95.6 (0) |
| Gentamicin | 38 | 98.7 (1.5) | 100 (0) | 99.3 (1.3) | 99.3 (1.3) | 97.4 (2.1) | − | 97.4 (2.1) | − | 97.4 (2.1) | − | 98 (1.3) | − |
| Tobramycin | 38 | 98.7 (1.5) | 98.7 (1.5) | 100 (0) | 100 (0) | 94.1 (3.3) | − | 94.1 (3.3) | − | 96.1 (1.5) | − | 95.4 (1.3) | − |
| Amikacin | 40 | 100 (0) | 100 (0) | 100 (0) | 98.8 (1.4) | 98.8 (1.4) | − | 98.8 (1.4) | − | 96.3 (2.5) | − | 93.1 (2.4) | − |
| Tetracycline | 32 | 100 (0) | 100 (0) | 100 (0) | 99.2 (1.6) | 89.8 (3) | − | 90.6 (4.4) | − | 90.6 (4.4) | − | 89.8 (1.6) | − |
| Tigecycline | 29 | 96.6 (0) | 97.4 (1.7) | 97.4 (1.7) | 100 (0) | 89.7 (0) | − | 89.7 (2.8) | − | 89.7 (2.8) | − | 91.4 (2) | − |
| Nitrofurantoin | 19 | 94.7 (0) | 98.7 (2.6) | 98.7 (2.6) | 98.7 (2.6) | 75 (6.6) | − | 81.6 (9.1) | − | 81.6 (9.1) | − | 80.3 (2.6) | − |
| Trimethoprim-sulfamethoxazole | 40 | 96.3 (2.5) | 96.3 (1.4) | 97.5 (2) | 98.8 (1.4) | 96.3 (2.5) | − | 96.3 (1.4) | − | 97.5 (2) | − | 98.8 (1.4) | − |
Data from one study group laboratory were not included in this analysis due to being a technical outlier based on GN7F reading method.
Mean (standard deviation).
Susceptible-only breakpoint used (≤0.5 µg/mL). The denominator is the number of susceptible isolates tested.
STD, standard inoculum method [1-µL transfer volume (TV) for tribe Proteeae, 10-µL TV for other Enterobacterales]; E-STD, enhanced inoculum method (1 µL TV for tribe Proteeae, 30 µL TV for other Enterobacterales); 30-All, 30 µL TV for all Enterobacterales; 50-All, 50 µL TV for all Enterobacterales; CLSI BPs, categorical Agreement assessed using breakpoints from CLSI M100 32nd edition; IFU BPs, categorical agreement assessed using breakpoints FDA cleared for the device per instructions for use; Bold indicates percent where the mean EA or CA was <90%; NE: not evaluated, comparison using IFU BPs was not assessed due to incomplete dilution range on the reference BMD panel; – indicates the breakpoints FDA cleared for the device are the same as the breakpoints listed in CLSI M100 32nd edition.
TABLE 4.
Phase I number of errors across four laboratories a for 40 Enterobacterales isolates using 2022 CLSI and FDA-cleared breakpoints per IFU b
| Antimicrobial agent | N isolates | Major errors (N min–N max) | Very major errors (N min–N max) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N “S” | STD | E-STD | 30-All | 50-All | N “R” | STD | E-STD | 30-All | 50-All | ||||||||||||
| CLSI BPs | IFU BPs | CLSI BPs | IFU BPs | CLSI BPs | IFU BPs | CLSI BPs | IFU BPs | CLSI BPs | IFU BPs | CLSI BPs | IFU BPs | CLSI BPs | IFU BPs | CLSI BPs | IFU BPs | CLSI BPs | IFU BPs | CLSI BPs | IFU BPs | ||
| Doripenem | 40 | 21 | 20 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–1 | 16 | NA | 3–5 | NA | 3–4 | NA | 0–1 | NA | 0–0 | NA |
| Ertapenem | 38 | 18 | 0–0 | − | 0–0 | – | 0–0 | − | 0–0 | − | 19 | 3–4 | − | 3–4 | − | 0–0 | − | 0–0 | − | ||
| Imipenem | 38 | 18 | 23 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–1 | 19 | 9 | 1–3 | 3–3 | 1–3 | 3–3 | 0–0 | 0–0 | 0–0 | 0–0 |
| Meropenem | 39 | 22 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | 14 | 2–4 | − | 1–3 | − | 0–0 | − | 0–0 | − | ||
| Aztreonam | 40 | 18 | 20 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–1 | 20 | 19 | 1–2 | 1–2 | 1–1 | 1–2 | 0–0 | 0–1 | 0–0 | 0–0 |
| Ceftazidime | 40 | 16 | 17 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–1 | 0–0 | 0–1 | 23 | 22 | 1–1 | 0–0 | 1–1 | 0–0 | 0–0 | 0–1 | 0–0 | 0–0 |
| Ceftriaxone | 40 | 15 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | 25 | 0–1 | − | 0–1 | − | 0–0 | − | 0–0 | − | ||
| Cefepime | 38 | 19 | 20 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–1 | 0–1 | 18 | 15 | 0–1 | 3–6 | 0–1 | 2–3 | 0–0 | 0–1 | 0–0 | 0–0 |
| Cefazolin | 19 | 1 | NE | 0–0 | NE | 0–0 | NE | 0–0 | NE | 0–0 | NE | 17 | NE | 0–0 | NE | 0–0 | NE | 0–0 | NE | 0–0 | NE |
| Ceftazidime/avibactam | 40 | 33 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | 7 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | ||
| Ceftolozane/tazobactam | 38 | 16 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | 21 | 1–2 | − | 1–2 | − | 1–2 | − | 0–1 | − | ||
| Piperacillin/tazobactam | 39 | 16 | 18 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 21 | 17 | 0–1 | 1–4 | 0–0 | 1–4 | 0–1 | 0–0 | 0–1 | 0–0 |
| Ampicillin/ sulbactam | 30 | 4 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | 22 | 0–1 | − | 0–1 | − | 0–1 | − | 0–0 | − | ||
| Ampicillin | 10 | 1 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | 9 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | ||
| Ciprofloxacin | 40 | 16 | 18 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 23 | 20 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 |
| Levofloxacin | 40 | 16 | 20 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 22 | 19 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 | 0–0 |
| Gentamicin | 38 | 27 | 0–1 | − | 0–0 | − | 0–0 | − | 0–0 | − | 10 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | ||
| Tobramycin | 38 | 21 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | 16 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | ||
| Amikacin | 40 | 37 | 0–0 | − | 0–0 | − | 0–0 | − | 0–1 | − | 2 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | ||
| Tetracycline | 32 | 11 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | 19 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | ||
| Tigecycline | 29 | 26 | 0–0 | − | 0–1 | − | 0–1 | − | 0–0 | − | 0 | − | − | − | − | − | − | − | − | ||
| Nitrofurantoin | 19 | 7 | 0–0 | − | 0–0 | − | 0–0 | − | 0–0 | − | 9 | 1–1 | − | 0–1 | − | 0–1 | − | 0–1 | − | ||
| Trimethoprim/sulfamethoxazole | 40 | 19 | 0–1 | − | 0–1 | − | 0–2 | − | 0–1 | − | 21 | 0–2 | − | 0–1 | − | 0–0 | − | 0–0 | − | ||
Data from one study group laboratory was not included in this analysis due to being a technical outlier based on GN7F reading method.
STD, standard inoculum method [1 µL transfer volume (TV) for tribe Proteeae, 10 µL TV for other Enterobacterales]; E-STD, enhanced inoculum method (1 µL TV for tribe Proteeae, 30 µL TV for other Enterobacterales); 30-All, 30 µL TV for all Enterobacterales; 50-All: 50 µL TV for all Enterobacterales; CLSI BPs, categorical agreement assessed using breakpoints from CLSI M100 32nd edition; IFU BPs, categorical agreement assessed using breakpoints FDA cleared for the device per instructions for use. Bold indicates percent where the mean EA or CA was <90%. NA, not available for evaluation due to susceptible-only breakpoint evaluated; NE, not evaluated, comparison using IFU BPs was not assessed due to incomplete dilution range on the reference BMD panel; – indicates the breakpoints FDA cleared for the device are the same as the breakpoints listed in CLSI M100 32nd edition.
Data for Enterobacterales were excluded from one laboratory that read panels using a Sensititre ARIS 2X OptiRead auto reader. There were large discrepancies for the P-tribe results of the same panel between the MICs generated from the ARIS 2X automated readings and the manual readings (performed with the Sensititre Vizion). When increasing the TV to 30 or 50 µL (30-All and 50-All, respectively), the ARIS 2X would overcall MIC results, sometimes resulting MICs at opposite ends of the panel’s concentration range. This laboratory’s data were excluded due to these technical reasons. This phenomenon was not observed with PSA and ACB.
For PSA and ACB, EA was generally >90% for all five laboratories regardless of TVs used (Tables 5 and 6). For PSA, CA was <90% for the β-lactam agents (Table 5); for ACB, CA was <90% for cefepime, the aminoglycosides, ampicillin/sulbactam, and the tetracyclines (Table 6). MEs were found more frequently in PSA rather than ACB, with no discernible directional trend as TV increased; contrarily, VMEs were observed more frequently in ACB than PSA, and there was no obvious upward or downward trend as TV increased.
TABLE 5.
Phase I summary of essential and categorical agreement across five laboratories for 30 Pseudomonas aeruginosa isolates using 2022 CLSI breakpoints b
| Antimicrobial agent | Total N isolates | Essential agreement (%) a | Categorical agreement (%) a | Major errors (N min–N max) | Very major errors (N min–N max) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STD | 30-All | 50-All | STD | 30-All | 50-All | N “S” | STD | 30-All | 50-All | N “R” | STD | 30-All | 50-All | ||
| Doripenem | 30 | 94 (2.8) | 94 (1.5) | 96 (1.5) | 80.7 (2.8) | 83.3 (5.3) | 83.3 (6.2) | 17 | 0–0 | 0–0 | 0–0 | 11 | 0–0 | 0–0 | 0–0 |
| Imipenem | 30 | 92.7 (4.3) | 94.7 (1.8) | 94.7 (3.8) | 86.7 (6.7) | 86.7 (2.4) | 86.0 (4.9) | 14 | 0–2 | 0–1 | 0–2 | 14 | 0–0 | 0–0 | 0–0 |
| Meropenem | 30 | 94 (4.3) | 90.0 (2.4) | 92.7 (2.8) | 82.7 (3.7) | 80.7 (9.2) | 79.3 (8) | 16 | 0–0 | 0–2 | 0–1 | 10 | 0–0 | 0–0 | 0–0 |
| Aztreonam | 30 | 98.7 (1.8) | 97.3 (2.8) | 97.3 (2.8) | 90 (2.4) | 89.3 (4.3) | 88.7 (5.1) | 14 | 0–1 | 0–1 | 0–1 | 15 | 0–1 | 0–1 | 0–0 |
| Ceftazidime | 30 | 97.3 (2.8) | 96 (1.5) | 97.3 (4.3) | 98 (3) | 96 (3.7) | 97.3 (1.5) | 14 | 0–0 | 0–0 | 0–0 | 15 | 0–0 | 0–0 | 0–0 |
| Cefepime | 30 | 98 (1.8) | 97.3 (1.5) | 98 (1.8) | 82.7 (4.9) | 81.3 (7.7) | 82.7 (2.8) | 14 | 0–0 | 0–0 | 0–0 | 11 | 0–0 | 0–0 | 0–0 |
| Ceftazidime/avibactam | 29 | 97.2 (3.8) | 96.6 (4.9) | 95.2 (4.6) | 95.9 (3.8) | 93.1 (4.2) | 92.4 (4.5) | 24 | 0–3 | 0–3 | 0–3 | 5 | 0–1 | 0–0 | 0–0 |
| Ceftolozane/tazobactam | 30 | 96 (3.7) | 96 (1.5) | 95.3 (3) | 95.3 (1.8) | 96.7 (0) | 95.3 (3) | 23 | 0–0 | 0–0 | 0–1 | 6 | 0–0 | 0–0 | 0–0 |
| Piperacillin/tazobactam | 30 | 96.7 (2.4) | 99.3 (1.5) | 94.7 (3) | 87.3 (1.5) | 89.3 (3.7) | 86.7 (2.4) | 15 | 0–0 | 0–0 | 0–1 | 10 | 0–0 | 0–0 | 0–0 |
| Ciprofloxacin | 30 | 99.3 (1.5) | 99.3 (1.5) | 100 (0) | 94.7 (3) | 95.3 (4.5) | 97.3 (2.8) | 7 | 0–0 | 0–0 | 0–0 | 22 | 0–1 | 0–0 | 0–0 |
| Levofloxacin | 30 | 99.3 (1.5) | 100 (0) | 100 (0) | 94.7 (4.5) | 96 (1.5) | 95.3 (1.8) | 5 | 0–0 | 0–0 | 0–0 | 21 | 0–0 | 0–0 | 0–0 |
| Gentamicin | 30 | 98.7 (1.8) | 98.7 (1.8) | 99.3 (1.5) | 95.3 (3) | 97.3 (2.8) | 91.3 (4.5) | 17 | 0–0 | 0–0 | 0–0 | 9 | 0–0 | 0–0 | 0–0 |
| Tobramycin | 30 | 99.3 (1.5) | 100 (0) | 99.3 (1.5) | 99.3 (1.5) | 100 (0) | 99.3 (1.5) | 20 | 0–0 | 0–0 | 0–0 | 10 | 0–0 | 0–0 | 0–0 |
| Amikacin | 30 | 100 (0) | 97.3 (4.5) | 99.3 (1.5) | 98.6 (1.9) | 99.3 (1.5) | 100 (0) | 22 | 0–0 | 0–0 | 0–0 | 6 | 0–0 | 0–0 | 0–0 |
Mean (standard deviation).
STD, standard inoculum method, 10 µL transfer volume (TV); 30-All, 30 µL TV; 50-All, 50 µL TV. Bold indicates percent where the mean EA or CA was <90%.
TABLE 6.
Phase I summary of mean essential and categorical agreement and error rates across five laboratories for 30 Acinetobacter baumannii isolates using 2022 CLSI breakpoints b
| Antimicrobial agent | Total N
isolates |
Essential agreement (%) a | Categorical agreement (%) a | Major errors (N min–N max) | Very major errors (N min–N max) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STD | 30-All | 50-All | STD | 30-All | 50-All | N "S" | STD | 30-All | 50-All | N "R" | STD | 30-All | 50-All | ||
| Doripenem | 30 | 100 (0) | 100 (0) | 99.3 (1.5) | 89.3 (2.8) | 94.7 (3.8) | 94.7 (1.8) | 2 | 0–0 | 0–0 | 0–0 | 26 | 0–0 | 0–0 | 0–1 |
| Imipenem | 30 | 100 (0) | 99.3 (1.5) | 99.3 (1.5) | 99.3 (1.5) | 98 (3) | 96.7 (3.3) | 5 | 0–0 | 0–0 | 0–0 | 25 | 0–0 | 0–0 | 0–0 |
| Meropenem | 30 | 99.3 (1.5) | 100 (0) | 100 (0) | 90 (2.4) | 92.7 (2.8) | 92.7 (1.5) | 1 | 0–0 | 0–0 | 0–0 | 27 | 0–0 | 0–0 | 0–0 |
| Ceftazidime | 30 | 98 (3) | 98.7 (1.8) | 99.3 (1.5) | 94.7 (1.8) | 96.7 (3.3) | 95.3 (3) | 2 | 0–0 | 0–0 | 0–0 | 28 | 0–0 | 0–0 | 0–0 |
| Ceftriaxone | 30 | 100 (0) | 100 (0) | 99.3 (1.5) | 98 (3) | 99.3 (1.5) | 98.7 (1.8) | 0 | 0–0 | 0–0 | 0–0 | 30 | 0–0 | 0–0 | 0–1 |
| Cefepime | 30 | 92.7 (4.3) | 96 (5.5) | 96 (5.5) | 86 (8) | 89.3 (5.5) | 88.7 (3) | 0 | 0–0 | 0–0 | 0–0 | 25 | 0–2 | 0–1 | 0–2 |
| Ampicillin/sulbactam | 30 | 98 (1.8) | 98.7 (1.8) | 99.3 (1.5) | 75.3 (7.3) | 80 (9.7) | 83.3 (8.8) | 0 | 0–0 | 0–0 | 0–0 | 21 | 0–0 | 0–0 | 0–0 |
| Piperacillin/tazobactam | 30 | 100 (0) | 100 (0) | 100 (0) | 98.7 (1.8) | 99.3 (1.5) | 100 (0) | 0 | 0–0 | 0–0 | 0–0 | 30 | 0–0 | 0–0 | 0–0 |
| Ciprofloxacin | 30 | 99.3 (1.5) | 100 (0) | 100 (0) | 99.3 (1.5) | 100 (0) | 100 (0) | 0 | 0–0 | 0–0 | 0–0 | 30 | 0–1 | 0–0 | 0–0 |
| Levofloxacin | 30 | 95.3 (3.8) | 97.3 (2.8) | 97.3 (4.3) | 81.3 (6.1) | 86 (7.2) | 86.7 (6.7) | 0 | 0–0 | 0–0 | 0–0 | 27 | 0–3 | 0–2 | 0–3 |
| Gentamicin | 30 | 96.6 (3.4) | 97.2 (1.5) | 96.6 (2.4) | 87.6 (3.9) | 89.7 (2.4) | 89.7 (2.4) | 3 | 0–1 | 0–1 | 0–1 | 25 | 0–1 | 0–0 | 0–0 |
| Tobramycin | 30 | 98.7 (1.8) | 96 (2.8) | 96 (1.5) | 92 (1.8) | 86.7 (2.4) | 87.3 (2.8) | 8 | 0–1 | 0–2 | 0–2 | 22 | 0–0 | 0–0 | 0–1 |
| Amikacin | 30 | 96.7 (2.4) | 97.3 (1.5) | 98.7 (1.8) | 76 (3.7) | 81.3 (4.5) | 82.7 (5.5) | 12 | 0–0 | 0–0 | 0–0 | 11 | 0–0 | 0–0 | 0–0 |
| Tetracycline | 30 | 98 (3) | 97.3 (4.3) | 96 (3.7) | 87.3 (2.8) | 90 (6.2) | 89.3 (2.8) | 7 | 0–0 | 0–0 | 0–1 | 19 | 0–0 | 0–1 | 0–0 |
| Minocycline | 30 | 97.8 (5) | 96.7 (3) | 100 (0) | 80 (6.3) | 80 (5) | 78.9 (4.6) | 12 | 0–1 | 0–0 | 0–0 | 3 | 0–1 | 0–1 | 0–0 |
| Trimethoprim/sulfamethoxazole | 30 | 98.6 (1.9) | 95.9 (2.9) | 96.6 (2.4) | 98.6 (1.9) | 95.9 (2.9) | 96.6 (2.4) | 3 | 0–1 | 0–2 | 0–1 | 26 | 0–1 | 0–1 | 0–1 |
Mean (standard deviation).
STD, standard inoculum method, 10-µL transfer volume (TV); 30-All, 30 µL TV; 50-All, 50 µL TV. Bold indicates percent where the mean EA or CA was <90%.
Among all laboratories, each QC strain had >95% acceptable results regardless of TV used (data not shown).
Interlaboratory reproducibility
For all antimicrobial agents, organism group, and inoculum method, interlaboratory reproducibility was >95% (Table S2).
Colony counts
Across all five laboratories, 76 instances of colony counts for ATCC 25922 were performed to check the final inoculum delivery to the GN7F wells. The mean colony counts were as follows: 7.8 for 10 µL TV (~8 × 104 CFU/mL), 21.9 for 30 µL TV (~2.1 × 105 CFU/mL), and 35.5 for 50 µL TV (~3.6 × 105 CFU/mL). The mean colony counts for respective nephelometer and turbidity methods were as follows for the three TVs: 7.41 (~7 × 104 CFU/mL) and 8.7 (~9 × 104 CFU/mL) for 10 µL, 18.8 (~1.9 × 105 CFU/mL) and 24.8 (~2.5 × 105 CFU/mL) for 30 µL, and 33.1 (~3.3 × 105 CFU/mL) and 41.6 (~4.2 × 105 CFU/mL) for 50 µL. Thus, inoculum standardization by nephelometer resulted in slightly lower average colony counts.
Bias and trending
Trending was observed with STD for Enterobacterales (1 µL for P-tribe and 10 µL for non-P-tribe) (Table 7). When stratifying STD by P-tribe versus non-P-tribe organisms, the P-tribe organisms (1 µL TV) showed a bias of −83.6% (CI: −87.7% to −77.8%), while for the non-P-tribe, using a 10 µL TV showed a bias of −62.7% (CI: −67.0% to −57.9%). A downward trend was also observed for 30-All with Enterobacterales organisms (−31.7%, CI: −36.9% to −26.7%). Interestingly, PSA generally had a stronger negative bias (lower MICs) for non-β-lactam versus β-lactam agents, while the opposite was observed for Enterobacterales and ACB. When stratifying MIC results by inoculum standardization method, laboratories that used the Sensititre nephelometer tended to have a similar or larger negative bias than laboratories using a turbidity meter (Table 8). For PSA and ACB, no obvious trending was observed for any TV when assessing all drugs together. However, ACB generally undercalled MICs versus PSA. PSA had positive biases but no trend toward overcalling MICs; however, while stratifying by inoculum standardization method, a trend toward higher MICs was observed when utilizing the turbidity meter in 50-All TV. Interestingly, PSA generally had a negative bias for non-β-lactam versus β-lactam agents, while the opposite was observed for Enterobacterales and ACB.
TABLE 7.
Bias and trend assessment of organism group and the four inoculum transfer volumes d
| Reporting group | Transfer volume | Drug class | Labs | N evaluable MIC pairs | N (%) dilutions from reference MIC | Bias (%) | Ci (%) | Trend (direction) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| ≥ −1 | 0 | ≥ +1 | ||||||||
| Enterobacterales | 1 µL a | All | 4 c | 232 | 200 (86.2) | 26 (11.2) | 6 (2.6) | −83.6 | −87.7 to −77.8 | Yes (downward) |
| β-lactams | 201 | 180 (89.6) | 15 (7.5) | 6 (3.0) | −86.6 | −90.4 to −80.5 | Yes (downward) | |||
| Non-β-lactams | 31 | 20 (64.5) | 11 (35.5) | 0 (0.0) | −64.5 | −78.9 to −43.8 | Yes (downward) | |||
| 10 µL b | All | 4‡ | 499 | 344 (68.9) | 124 (24.8) | 31 (6.2) | −62.7 | −67.0 to −57.9 | Yes (downward) | |
| β-lactams | 331 | 242 (73.1) | 70 (21.1) | 19 (5.7) | −67.4 | −72.3 to −61.5 | Yes (downward) | |||
| Non-β-lactams | 168 | 102 (60.7) | 54 (32.1) | 12 (7.1) | −53.6 | −61.3 to −44.6 | Yes (downward) | |||
| 30 µL | All | 4‡ | 602 | 284 (47.2) | 225 (37.4) | 93 (15.4) | −31.7 | −36.5 to −26.7 | Yes (downward) | |
| β-lactams | 412 | 198 (48.1) | 156 (37.9) | 58 (14.1) | −34.0 | −39.7 to −27.9 | Yes (downward) | |||
| Non-β-lactams | 190 | 86 (45.3) | 69 (36.3) | 35 (18.4) | −26.8 | −35.5 to −17.6 | No | |||
| 50 µL | All | 4‡ | 585 | 207 (35.4) | 219 (37.4) | 159 (27.2) | −8.2 | −13.5 to −2.9 | No | |
| β-lactams | 384 | 134 (34.9) | 148 (38.5) | 102 (26.6) | −8.3 | −14.8 to −1.8 | No | |||
| Non-β-lactams | 201 | 73(36.3) | 71 (35.5) | 57 (28.4) | −8.0 | −16.9 to 1.2 | No | |||
| Acinetobacter baumannii | ||||||||||
| 10 µL | All | 5 | 455 | 218 (47.9) | 145 (31.9) | 92 (20.2) | −27.7 | −33.4 to −21.7 | No | |
| β-lactams | 206 | 123 (59.7) | 68 (33.0) | 15 (7.3) | −52.4 | −59.5 to −44.3 | Yes (downward) | |||
| Non-β-lactams | 249 | 95 (38.2) | 77 (30.9) | 77 (30.9) | −7.2 | −15.4 to 1.1 | No | |||
| 30 µL | All | 5 | 438 | 166 (37.9) | 164 (37.4) | 108 (24.7) | −13.2 | −19.2 to −7.1 | No | |
| β-lactams | 188 | 94 (50.0) | 80 (42.6) | 14 (7.4) | −42.6 | −50.2 to −31.1 | Yes (downward) | |||
| Non-β-lactams | 250 | 72 (28.8) | 84 (33.6) | 94 (37.6) | 8.8 | 0.5 to 16.9 | No | |||
| 50 µL | All | 5 | 424 | 122 (28.8) | 155 (36.6) | 147 (34.7) | 5.9 | −0.4 to 12.1 | No | |
| β-lactams | 177 | 65 (36.7) | 81 (45.8) | 31 (17.5) | −19.2 | −28.0 to −10.0 | No | |||
| Non-β-lactams | 247 | 57 (23.1) | 74 (30.0) | 116 (47.0) | 23.9 | 15.6 to 31.8 | No | |||
| Pseudomonas aeruginosa | ||||||||||
| 10 µL | All | 5 | 807 | 278 (34.4) | 344 (42.6) | 185 (22.9) | −11.5 | −15.9 to −7.1 | No | |
| β-lactams | 630 | 207 (32.9) | 255 (40.5) | 168 (26.7) | −6.2 | −11.2 to −1.1 | No | |||
| Non-β-lactams | 177 | 71 (40.1) | 89 (50.3) | 17 (9.6) | −30.5 | −38.7 to −21.8 | Yes (downward) | |||
| 30 µL | All | 5 | 793 | 184 (23.2) | 342 (43.1) | 267 (33.7) | 10.5 | 6.0 to 14.8 | No | |
| β-lactams | 612 | 141 (23.0) | 236 (38.6) | 235 (38.4) | 15.4 | 10.2 to 20.4 | No | |||
| Non-β-lactams | 181 | 43 (23.8) | 106 (58.6) | 32 (17.7) | −6.1 | −14.4 to 2.3 | No | |||
| 50 µL | All | 5 | 786 | 148 (18.8) | 318 (40.5) | 320 (40.7) | 21.9 | 17.4 to 26.2 | No | |
| β-lactams | 611 | 117 (19.1) | 215 (35.2) | 279 (45.7) | 26.5 | 21.4 to 31.4 | No | |||
| Non-β-lactams | 175 | 31 (17.7) | 103 (58.9) | 41 (23.4) | 5.7 | −2.8 to 14.1 | No | |||
tribe Proteeae organisms (Proteus, Providencia, and Morganella spp.).
Non-tribe Proteeae organisms.
Data from one study group laboratory were not included in this analysis due to being a technical outlier based on GN7F reading method.
MIC, minimum inhibitory concentration; CI, confidence interval (Newcombe); trend: trend is denoted “yes” when the absolute difference of ≥±1 is ≥30% and the confidence interval is significant.
TABLE 8.
Bias and trend assessment of inoculum transfer volumes stratified by inoculum standardization method and organism group c
| Transfer volume | Standardization method | N evaluable MIC pairs | N (%) dilutions from reference MIC | Bias (%) | CI (%) | Trend (direction) | ||
|---|---|---|---|---|---|---|---|---|
| ≥−1 | 0 | ≥+1 | ||||||
| Enterobacterales | ||||||||
| 1 µL a | Nephelometer | 111 | 94 (84.7) | 15 (13.5) | 2 (1.8) | −82.9 | −88.6 to −73.8 | Yes (downward) |
| Turbidity meter | 121 | 106 (87.6) | 11 (9.1) | 4 (3.3) | −84.3 | −89.5 to −75.7 | Yes (downward) | |
| 10 µL b | Nephelometer | 258 | 182 (70.5) | 65 (25.2) | 11 (4.3) | −66.3 | −71.8 to −59.6 | Yes (downward) |
| Turbidity meter | 241 | 162 (67.2) | 59 (24.5) | 20 (8.3) | −58.9 | −64.4 to −50.5 | Yes (downward) | |
| 30 µL | Nephelometer | 302 | 159 (52.6) | 111 (36.8) | 32 (10.6) | −42.1 | −48.4 to −35.2 | Yes (downward) |
| Turbidity meter | 300 | 125 (41.7) | 114 (38.0) | 61 (20.3) | −21.3 | −28.4 to −14.0 | No | |
| 50 µL | Nephelometer | 291 | 117 (40.2) | 114 (39.2) | 60 (20.6) | −19.6 | −26.7 to −12.2 | No |
| Turbidity meter | 294 | 90 (30.6) | 105 (35.7) | 99 (33.7) | 3.1 | −4.5 to 10.6 | No | |
| Acinetobacter baumannii | ||||||||
| 10 µL | Nephelometer | 277 | 145 (52.3) | 83 (30.0) | 49 (17.7) | −34.7 | −41.7 to −27.0 | Yes (downward) |
| Turbidity meter | 178 | 73 (41.0) | 62 (34.8) | 43 (24.2) | −16.9 | −26.2 to −7.1 | No | |
| 30 µL | Nephelometer | 271 | 129 (47.6) | 90 (33.2) | 52 (19.2) | −28.4 | −35.4 to −20.3 | No |
| Turbidity meter | 167 | 37 (22.2) | 74 (44.3) | 56 (33.5) | 11.4 | 1.8 to 20.7 | No | |
| 50 µL | Nephelometer | 258 | 87 (33.7) | 96 (37.2) | 75 (29.1) | −4.7 | −12.6 to 3.4 | No |
| Turbidity meter | 166 | 35 (21.1) | 59 (35.5) | 72 (43.4) | 22.3 | 12.3 to 31.7 | No | |
| Pseudomonas aeruginosa | ||||||||
| 10 µL | Nephelometer | 490 | 189 (38.6) | 198 (40.4) | 103 (21.0) | −17.6 | −23.1 to −11.9 | No |
| Turbidity meter | 317 | 89 (28.1) | 146 (46.1) | 82 (25.9) | −2.2 | −9.1 to 4.7 | No | |
| 30 µL | Nephelometer | 469 | 139 (29.6) | 203 (43.3) | 127 (27.1) | −2.6 | −8.3 to 3.2 | No |
| Turbidity meter | 324 | 45 (13.9) | 139 (42.9) | 140 (43.2) | 29.3 | 22.6 to 35.7 | No | |
| 50 µL | Nephelometer | 481 | 119 (24.7) | 193 (40.1) | 169 (35.1) | 10.4 | 4.6 to 16.1 | No |
| Turbidity meter | 305 | 29 (9.5) | 125 (41.0) | 151 (49.5) | 40.0 | 33.3 to 46.3 | Yes (upward) | |
tribe Proteeae organisms (Proteus, Providencia, and Morganella spp.).
Non-tribe Proteeae organisms.
MIC, minimum inhibitory concentration; CI, confidence interval (Newcombe-Wilson); Trend, trend is denoted “yes” when the absolute difference of ≥±1 is ≥30% and the confidence interval is significant.
Sensitivity for detecting carbapenem-resistant organisms
Data obtained for each TV were evaluated to determine whether they were sufficient for detection of carbapenem-resistant organisms (Table 9). Carbapenem-resistant organisms were defined as organisms resistant to one or more applicable carbapenem (imipenem, meropenem, and ertapenem for Enterobacterales; imipenem or meropenem for ACB and PSA). For all ENT, the STD or E-STD TV combinations yielded 18 and 16 false negative instances, respectively. These instances between STD and E-STD commonly involved five isolates (AR0059, AR0082, AR00133, AR0155, and AR0159), of which four were carbapenemase-producing P-tribe isolates. A non-P-tribe false negative with STD was a KPC-producing carbapenem-resistant E. coli isolate, AR0001, classified as non-CR by two laboratories. All false negatives were resolved when 30-All and 50-All were used for Enterobacterales organisms.
TABLE 9.
Detection of carbapenem-resistant organisms by reporting group, stratified by transfer volume combination analysis groups b , c
| Organism | TV analysis group | TP | TN | FP | FN | Sensitivity | Exact CI | Specificity | Exact CI |
|---|---|---|---|---|---|---|---|---|---|
| Enterobacterales (N = 160 datapoints; 40 isolates, four laboratories) a | STD | 66 | 76 | 0 | 18 | 0.7857 | 0.6826–0.8678 | 1 | 0.9526–1.0 |
| E-STD | 68 | 75 | 1 | 16 | 0.8095 | 0.7092–0.887 | 0.9868 | 0.9289–0.9997 | |
| 30-All | 84 | 75 | 1 | 0 | 1 | 0.957–1.0 | 0.9868 | 0.9289–0.9997 | |
| 50-All | 84 | 76 | 0 | 0 | 1 | 0.957–1.0 | 1 | 0.9526–1.0 | |
| Pseudomonas aeruginosa (N = 150 datapoints; 30 isolates, five laboratories) | STD | 88 | 58 | 2 | 2 | 0.9778 | 0.922–0.9973 | 0.9667 | 0.8847–0.9959 |
| 30-All | 86 | 54 | 6 | 4 | 0.9556 | 0.8901–0.9878 | 0.9000 | 0.7949–0.9624 | |
| 50-All | 89 | 52 | 8 | 1 | 0.9889 | 0.9396–0.9997 | 0.8667 | 0.7541–0.9406 | |
| Acinetobacter baumannii (N = 150 datapoints; 30 isolates, five laboratories) | STD | 133 | 14 | 2 | 1 | 0.9852 | 0.9475–0.9982 | 0.9333 | 0.6805–0.9983 |
| 30-All | 132 | 15 | 0 | 3 | 0.9778 | 0.9364–0.9954 | 1 | 0.782–1.0 | |
| 50-All | 134 | 14 | 1 | 1 | 0.9926 | 0.9594–0.9998 | 0.9333 | 0.6805–0.9983 |
One study group laboratory was not included in these data due to being a technical outlier based on GN7F reading method.
Datapoints are the total number of times the isolate was tested across laboratories. Each isolate and laboratory was 1 datapoint, and each isolate had 4 or 5 data points.Carbapenem-resistant organisms are defined as isolates resistant to at least 1 applicable carbapenem (ertapenem, meropenem, and imipenem) using CLSI M100 32nd edition breakpoints.
TV, transfer volume; TP, true positive; TN, true negative; FP, false positive; FN, false negative; CI, confidence interval.
Phase IIa results: intermediary discrepancy testing by CDC
To assess the subset of isolates yielding ME and VMEs among a majority of the study group (≥3 laboratories), CDC alone performed side-by-side AST of 12 isolates (Table 10) using CDC’s frozen reference BMD panel and the GN7F panel. As a result of this testing, MICs of P-tribe (n = 9) for the β-lactams, specifically the carbapenems, generated from lower transfer inoculum volume (1 µL), were notably lower than those generated from reference BMD panels derived from the same 0.5-McFarland equivalent inoculum suspension. Aside from the lower MICs resulting from using the 1 µL TV, the VMEs also reproduced. All 12 isolates were categorized as carbapenem-resistant strains when using reference BMD panels. However, when using either the 1 or 10 µL TV with the GN7F panel, only half of the strains were correctly identified as CRE. CRE classification for these isolates improved with 30-All (10 of 12) and 50-All (12 of 12) TVs.
TABLE 10.
Phase IIa: MIC results and carbapenem resistance classification for side-by-side testing with reference BMD and GN7F from CDC’s arbitration testing b
| AR bank isolate | Organism | β-lactam resistance genes | Ertapenem MIC a (µg/mL) | Imipenem MIC a (µg/mL) | Meropenem MIC a (µg/mL) | CRE classification | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref | 1 µL | 10 µL | 30 µL | 50 µL | Ref | 1 µL | 10 µL | 30 µL | 50 µL | Ref | 1 µL | 10 µL | 30 µL | 50 µL | Ref | 1 µL/10 µL | 30 µL | 50 µL | |||
| AR0001 | Escherichia coli | KPC-3 | 8 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 1 | 4 | 8 | Y | Y | Y | Y | |||
| AR0008 | Enterobacter cloacae | ACT-15 | >8 | 8 | 8 | 8 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | Y | Y | Y | Y | |||
| AR0029 | Proteus mirabilis | – | ≤0.12 | ≤0.25 | ≤0.25 | ≤0.25 | 4 | ≤ 1 | 2 | 4 | ≤0.12 | ≤0.5 | ≤ 0.5 | ≤ 0.5 | Y | N | N | Y | |||
| AR0057 | Morganella morganii | CTX-M-15, NDM-1, OXA-1 | 4 | 0.5 | 2 | 4 | 16 | 4 | 8 | >8 | 4 | 2 | 4 | 4 | Y | Y | Y | Y | |||
| AR0059 | Proteus mirabilis | TEM-1B | 0.5 | ≤0.25 | ≤0.25 | ≤0.25 | 8 | ≤ 1 | 4 | 8 | 0.5 | ≤0.5 | ≤0.5 | ≤0.5 | Y | N | Y | Y | |||
| AR0069 | Escherichia coli | NDM-1, CMY-6, TEM-1B | >8 | 4 | 8 | >8 | 8 | 4 | 8 | >8 | >8 | 4 | >8 | >8 | Y | Y | Y | Y | |||
| AR0082 | Providencia rettgeri | NDM-1 | 8 | ≤0.25 | 1 | 4 | 32 | 2 | >8 | >8 | 8 | ≤0.5 | 2 | 8 | Y | N | Y | Y | |||
| AR0133 | Morganella morganii | KPC-2 | 8 | 1 | 4 | 4 | 8 | 4 | 8 | 8 | 4 | 2 | 4 | 4 | Y | Y | Y | Y | |||
| AR0155 | Proteus mirabilis | KPC-6 | 2 | ≤0.25 | 4 | 2 | 16 | ≤ 1 | >8 | >8 | 2 | ≤ 0.5 | 2 | 4 | Y | N | Y | Y | |||
| AR0156 | Proteus mirabilis | KPC-2, OXA-10 | ≤0.12 | ≤0.25 | ≤0.25 | ≤0.25 | 4 | ≤ 1 | 2 | 4 | ≤0.12 | ≤ 0.5 | ≤0.5 | ≤0.5 | Y | N | N | Y | |||
| AR0159 | Proteus mirabilis | NDM-1 | 4 | 0.5 | 8 | 8 | 64 | ≤ 1 | >8 | >8 | 8 | ≤ 0.5 | 8 | 8 | Y | N | Y | Y | |||
| AR0519 | Morganella morganii | DHA-1 | >8 | >8 | >8 | >8 | 8 | 4 | 8 | 8 | 8 | 8 | >8 | >8 | Y | Y | Y | Y | |||
Breakpoints used were from CLSI M100 32nd edition. Doripenem was not assessed due to removal of the agent from the reference broth microdilution panel. No isolate was solely resistant to doripenem by the Sensititre GN7F.
MIC, minimum inhibitory concentration; CRE, carbapenem-resistant Enterobacterales; Ref (bolded) denotes MIC generated by CDC’s frozen reference broth microdilution panel; 1, 10, 30, and 50 µL are the transfer volumes used to produce the respective GN7F MIC result. Y, yes; N, no.
Phase IIb results: discrepancy testing by PHLs
The results of phase IIa showed that VMEs obtained when using the lower TVs of 1 and 10 µL were reproduced within an even stricter comparison with reference BMD and GN7F when tested side-by-side from the same inoculum. Therefore, the study group repeated testing of isolates having MEs and VMEs using the 30-All and 50-All TVs during testing in their laboratory in order to use data for study requirements to implement surveillance and/or Clinical Laboratory Improvement Amendments (CLIA)-compliant testing. Repeat testing did not reveal much change in either direction for EA, CA, and errors observed in phase I testing (data not shown).
DISCUSSION
Given the public health implications of carbapenem-resistant and particularly carbapenemase-producing Gram-negative bacteria, we evaluated the Sensititre GN7F broth microdilution panel, which offers AST for >20 antimicrobial agents, including newer combination agents like ceftazidime/avibactam and ceftolozane/tazobactam. Four AR Lab Network regional laboratories and CDC conducted a multisite evaluation of this newer panel, advertised as FDA-cleared, to better understand its performance and potential for use across public health laboratories in the AR Lab Network. Our objective was to determine which TV provided the most accurate AST results when compared to the reference broth microdilution method, particularly among the bacterial organisms targeted by the AR Lab Network. Our assessment of four TVs [STD (1 µL for P-tribe and 10 µL for other Enterobacterales), E-STD (1 µL for P-tribe and 30 µL for other Enterobacterales), 30-All, and 50-All] demonstrated that the optimal TV for the GN7F panel is 30 µL (30-All) for Enterobacterales (including P-tribe), Pseudomonas aeruginosa, and Acinetobacter baumannii.
We focused on EA to evaluate performance as the quantitative MIC data are a better determinant of device performance than the qualitative CA (18). Furthermore, AST is confounded by inherent variability which affects assessment of CA when MICs are near the breakpoint (19); this can lead to a false perception of poor performance. In phase I, for all ENTs, EAs of the manufacturer-recommended STD or E-STD inoculum methods were below acceptable criteria (90%) for multiple antimicrobial agents, commonly occurring with the β-lactams. For example, cefepime showed the lowest degree of agreement, with an averaged EA of 71%, 80%, 88%, and 92% for STD, E-STD, 30-All, and 50-All, respectively. The performance of GN7F greatly improved with 30-All and 50-All, with the average EA increasing from the mid-80% range to above 95% for many of the antimicrobial agents. This suggests the β-lactams are sensitive to the inoculum effect, likely driven by varying copy numbers and expression levels of β-lactamases or genes encoding them (20 – 24). Detection of β-lactam resistance in the P-tribe was most affected by changes in TV, which was illustrated by the substantial improvements observed in EA and CA from STD or E-STD to 30-All. When using the STD and E-STD TV, both of which indicate an inoculum of 1 µL for the P-tribe, sensitivity to detect carbapenem resistance was 78.6% and 80.1%, respectively. Using 30-All and 50-All TVs raised sensitivity to 100%. In contrast to the β-lactams, the non-β-lactams performed well in regard to EA, CA, and error rates across all organism groups regardless of TV used.
One study group laboratory revealed major discrepancies in results generated for organisms of the P-tribe when comparing the two Sensititre reading systems, the automated ARIS 2X with OptiRead and the manual Vizion reader. When the TV was increased to 30 or 50 µL, the results generated by the OptiRead considerably overcalled MICs, where sometimes the MIC results were oppositely off-scale from the manual Vizion read (data not shown), resulting in the MIC readings by the OptiRead being inaccurate when compared to the AR Bank MIC. It is likely that this phenomenon is the result of the “ghosting” effect (a very light and translucent growth in wells) associated with BMD of swarming organisms, which was observed only for Proteus and Providencia spp. (Fig. 1). When using reference BMD panels, this phenomenon can also be observed, and ghosting-type growth should be ignored; however, due to the fluorescence calibration settings and algorithms of the OptiRead, we hypothesize this ghosting can incorrectly be perceived as true growth, thus resulting in the overcalling of MICs.
Fig 1.
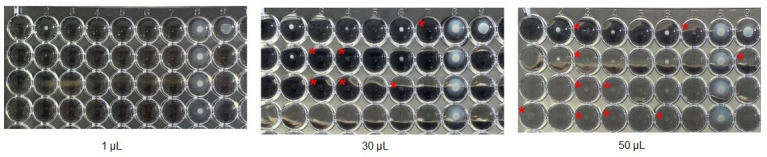
Isolate AR0059, a Proteus mirabilis, exhibiting faint growth pattern as transfer volume increases. The following antimicrobials and concentrations are included in the photo: 1a-c: amikacin (8–32 µg/mL); 1d: piperacillin/tazobactam (8/4 µg/mL); 2a-d: tigecycline (1–8 µg/mL); 3a-d: cefepime (2–16 µg/mL); 4a-d: doripenem (0.5–4 µg/mL); 5a-d: ertapenem (0.25–2 µg/mL); 7a-d: imipenem (1–8 µg/mL); 7a-d: meropenem (0.5–4 µg/mL); 8a-d: cefazolin (1–8 µg/mL); 9a-d: ceftazidime (1–8 µg/mL). For all antimicrobial agents, the concentrations increase from row A to D. Note: this photo does not capture the full concentration range for piperacillin/tazobactam, ertapenem, meropenem, cefazolin, and ceftazidime. Using the 1 µL TV, wells without growth are clear. When using the 30 or 50 µL TV, faint or “ghost-like” growth appears in wells marked with a red asterisk (*). This growth is hypothesized to be read as true growth by the OptiRead when they should be ignored.
For both PSA and ACB, there were no isolates for which multiple laboratories observed a major or very major error. CDC’s intermediary testing of 12 isolates that produced multiple errors across the study group laboratories further supported the initial phase I results obtained by the study group. For ACB and PSA, the sensitivity for detecting carbapenem resistance, a primary objective for AR Lab Network Gram-negative healthcare-associated infections activities, remained high regardless of TVs. Although EA was >90% for most antimicrobials no matter the TV, for ACB, the MICs generated by the GN7F panel had a slight bias, but not trend, toward producing lower MICs. The bias started to normalize around exact MICs as the TV increased. For PSA, 50 µL TVs produced MICs that tended to overcall resistance; thus, there were more false positive CR-PSA instances as TV increased.
In addition to evaluating the performance of four TVs, we also assessed whether procedural differences among study group laboratories influenced or biased the results. One such procedural difference among laboratories was the use of a nephelometer versus a turbidity meter. The manufacturer recommended use of a nephelometer, rather than a turbidity meter. In our study, we detected no inferiority in the turbidity meter inoculum suspension method. In fact, for ENT and ACB, results generated using a turbidity meter produced MICs closer to the reference MIC, while MICs generated using a nephelometer were lower. These differences in performance likely arise because the Sensititre nephelometer standardizes to the lower end of a 0.5 McFarland standard turbidity range (1.5 × 108; range 1–2 × 108 CFU/mL) compared with the MicroScan turbidity meter; the nephelometer stardardizes to 1.0 × 108 CFU/mL with the Sensititre 0.5-McFarland standard (from IFU v.GB V3.0-CID9104), while the turbidity meter standardizes to 1.5 × 108 CFU/mL with a Remel 0.5-McFarland standard (from IFU v.9020–7662, Rev. BB and Remel R20410 0.5 McFarland standard).
After the collaborating laboratories reviewed data from phase I (testing of 100 CDC & FDA AR Bank isolates) and conclusions from CDC’s intermediary testing, we halted additional assessment of the STD and E-STD TV combinations due to poor overall accuracy and high frequency of VMEs, including missed carbapenemase-producing CREs. Similarly, a study conducted by EUCAST and a subsequent EUCAST “Warnings!” also demonstrates that utilizing the 10 µL TV misses detecting resistance, particularly with meropenem (25). Because many antimicrobials on this device are currently either FDA-cleared for outdated FDA interpretive criteria (8 of 24 for ENT, 1 of 2 for PSA, and 2 of 4 for ACB) or lack FDA clearance altogether (7 of 9 for PSA and 10 of 14 for ACB lacked FDA-clearance when FDA interpretive criteria existed), along with the presence of many limitations for reporting resistant AST results, the study laboratories concluded that their use of this device would be mostly off-label and necessitate a validation study. In addition, overall CA and error rates improved using the updated breakpoints, further justifying the need for a validation study in order to use current breakpoints. Phase IIb thus focused on discrepancy testing so laboratories could use the data for implementation of surveillance and/or CLIA-compliant testing. Since FDA-clearance is granted per antimicrobial agent, not per panel configuration or name, these performance data could be extrapolated to other Sensititre non-RUO panels that contain shared antimicrobials of the same formulation. To complement this evaluation, we provided AR Lab Network laboratories with a concise refereed isolate list of 60 Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii across popular AR Bank panels to aid in validation studies (Table S3).
Our study had some limitations. First, discrepancy testing was not conducted for errors produced using the 1 and 10 µL TVs (affecting STD and E-STD) at each testing laboratory; however, CDC’s interim study served as arbitration to establish whether errors observed by the study group were due to chance or device performance. Second, we used a challenging set of organisms that did not reflect the populations of organisms or prevalence of resistant organisms encountered in a typical clinical laboratory, but rather those more relevant to the AR Lab Network public health laboratories’ isolate submissions. Despite the biases among the set of organisms tested (about half of which were resistant organisms and half were susceptible), their distribution provided more equality in the assessment of error types for both over- and undercalling of resistance, as well as biases. Third, we used on-scale MICs to adequately examine essential agreement; due to the shorter dilution ranges on the GN7F panel, many on-scale MICs were also straddling the breakpoint. The combination of the well-known inherent variance of AST and MICs straddling the breakpoint resulted in substandard performance for CA without adjustment (i.e., error rate bound method) (19, 20, 26, 27). Therefore, CA and minor errors had little influence on our conclusions. Nevertheless, we assessed VME and MEs because these types of errors are typically accompanied by MICs two or more dilutions from the comparator for most agents (except ceftazidime/avibactam). Fourth, we used a limited number of on-scale MIC pairs to adequately assess trending by individual laboratories and individual antimicrobial agents. Fifth, some laboratories deviated from the Sensititre FDA-cleared IFU procedure (Table 1). The use of a turbidity meter rather than of a nephelometer was the most significant deviation in the inoculum standardization procedure. Finally, because the study was conducted in 2020–2021, we applied 2022 CLSI breakpoints. Laboratories wishing to validate the GN7F panel using the 2023 CLSI breakpoints should be aware that for certain drugs, particularly the aminoglycosides, the panel lacks the lower drug concentrations necessary to be able to distinguish between susceptible and intermediate isolates.
Summary
Performance issues were identified in this evaluation of the Sensititre GN7F BMD panel even when the IFU procedure was followed, particularly attributed to the transfer volume used (from the 0.5 McFarland equivalent suspension to the 11 mL tube of CAMHB), which has a direct impact on the final CFU per milliter; the performance issues were driven predominately but not entirely by the P-tribe. Given the minimal performance differences between 30-All and 50-All, trending imbalances among reporting groups (50 µL for PSA had MICs biased higher, while other organism groups’ MICs trended lower), and the fact that 30-All is still considered partially on-label, study group laboratories determined that the most practical workflow would be to use 30 µL TV for all organism groups, including the P-tribe. Based on the results of one partnering laboratory’s comparison of MICs between read methods of the same panel, our data warrant caution for laboratories using the OptiRead automated reader for reading P-tribe when using the 30 or 50 µL TV. In December 2022, FDA issued a class 1 recall for this device, citing risk of false susceptible results when using the 1 µL TV for the P-tribe and certain antimicrobials (28). Together, the findings of this study group emphasize the value of continual post-market evaluation of commercial AST devices to assess their accuracy as bacterial populations evolve and resistance mechanisms emerge. Furthermore, where accessible, our findings support the use of contemporary isolates, including a variety of resistant strains with various underlying mechanisms of resistance, in verifications or validation studies, because testing of QC strains alone did not detect the performance problems described herein. In the era of the antimicrobial resistance crisis, it is imperative that AST devices continue to yield accurate and reliable results to ensure patient safety through informed therapeutic decision making and appropriate data-driven public health actions, policies, and initiatives.
ACKNOWLEDGMENTS
The authors wish to thank the Centers for Disease Control and Prevention (CDC) and FDA AR Isolate Bank staff for their contributions.
All authors reported no conflict of interest. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC, Wisconsin State Laboratory of Hygiene, Maryland Department of Health, Minnesota Department of Health, and Washington State Department of Health. Mention of any company or product does not constitute endorsement by CDC, Wisconsin State Laboratory of Hygiene, Maryland Department of Health, Minnesota Department of Health, and Washington State Department of Health.
This work was supported by the Centers for Disease Control and Prevention’s internal funding.
Contributor Information
Amelia S. Bhatnagar, Email: wmt7@cdc.gov.
Patricia J. Simner, Johns Hopkins University, Baltimore, Maryland, USA
ETHICS APPROVAL
This study was performed under General Protocol #7218 for ethical approval by the internal review board of the Centers for Disease Control and Prevention.
REFERENCES
- 1. Sabour S, Huang JY, Bhatnagar A, Gilbert SE, Karlsson M, Lonsway D, Lutgring JD, Rasheed JK, Halpin AL, Stanton RA, Gumbis S, Elkins CA, Brown AC. 2021. Detection and characterization of targeted carbapenem-resistant health care-associated threats: findings from the antibiotic resistance laboratory network, 2017 to 2019. Antimicrob Agents Chemother 65:e0110521. doi: 10.1128/AAC.01105-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. CDC . 2019. Antibiotic resistance threats in the United States, 2019
- 3. Meletis G. 2016. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis 3:15–21. doi: 10.1177/2049936115621709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nordmann P, Poirel L. 2019. Epidemiology and diagnostics of carbapenem resistance in Gram-negative bacteria. Clin Infect Dis 69:S521–S528. doi: 10.1093/cid/ciz824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. 2008. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol 29:1099–1106. doi: 10.1086/592412 [DOI] [PubMed] [Google Scholar]
- 6. Falcone M, Tiseo G, Antonelli A, Giordano C, Di Pilato V, Bertolucci P, Parisio EM, Leonildi A, Aiezza N, Baccani I, Tagliaferri E, Righi L, Forni S, Sani S, Mechi MT, Pieralli F, Barnini S, Rossolini GM, Menichetti F. 2020. Clinical features and outcomes of bloodstream infections caused by New Delhi metallo-beta-lactamase-producing enterobacterales during a regional outbreak. Open Forum Infect Dis 7:ofaa011. doi: 10.1093/ofid/ofaa011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ben-David D, Kordevani R, Keller N, Tal I, Marzel A, Gal-Mor O, Maor Y, Rahav G. 2012. Outcome of carbapenem resistant Klebsiella pneumoniae bloodstream infections. Clin Microbiol Infect 18:54–60. doi: 10.1111/j.1469-0691.2011.03478.x [DOI] [PubMed] [Google Scholar]
- 8. Vallabhaneni S, Huang JY, Grass JE, Bhatnagar A, Sabour S, Lutgring JD, Campbell D, Karlsson M, Kallen AJ, Nazarian E, Snavely EA, Morris S, Wang C, Lee R, Koag M, Lewis R, Garcia B, EIP Work Group, Brown AC, Walters MS. 2021. Antimicrobial susceptibility profiles to predict the presence of carbapenemase genes among carbapenem-resistant Pseudomonas aeruginosa isolates. J Clin Microbiol 59:e02874-20. doi: 10.1128/JCM.02874-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. CLSI . 2015. Methods for dilution antimicrobial susceptbility tests for bacteria that grow aerobically; approved standard. 10th ed. CLSI Document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10. ThermoScientific . 2019. Instructions for use: thermo scientific sensititre MIC susceptibility plates for testing non-fastidious Gram negative and Gram positive isolates, 037-NFAST FDA-USA only. CID10253 ed
- 11. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. 2009. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42:377–381. doi: 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. CLSI . 2015. Verification of commercial microbial identification and antimicrobial susceptibility testing systems. 1st ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13. FDA . 2009. Class II special controls guidance document: antimicrobial susceptibility test (AST) systems. Rockville, MD: [Google Scholar]
- 14. CLSI . 2022. M100 performance standards for antimicrobial susceptibility testing. 32nd ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15. FDA . 2022. Antibacterial susceptibility test interpretive criteria. Available from: https://www.fda.gov/drugs/development-resources/antibacterial-susceptibility-test-interpretive-criteria
- 16. FDA . 2021. 510(k) substantial equivalence determination decision summary (K203741). Available from: https://www.accessdata.fda.gov/cdrh_docs/reviews/K203741.pdf
- 17. Ransom E, Bhatnagar A, Patel JB, Machado M-J, Boyd S, Reese N, Lutgring JD, Lonsway D, Anderson K, Brown AC, Elkins CA, Rasheed JK, Karlsson M. 2020. Validation of aztreonam-avibactam susceptibility testing using digitally dispensed custom panels. J Clin Microbiol 58:e01944-19. doi: 10.1128/JCM.01944-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. ISO . 2021. ISO 20776-2:2021(E) clinical laboratory testing and in vitro diagnostic test systems-- susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices
- 19. Humphries RM, Ambler J, Mitchell SL, Castanheira M, Dingle T, Hindler JA, Koeth L, Sei K, CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing . 2018. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol 56:e01934-17. doi: 10.1128/JCM.01934-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith KP, Kirby JE. 2018. The inoculum effect in the era of multidrug resistance: minor differences in inoculum have dramatic effect on MIC determination. Antimicrob Agents Chemother 62:e00433-18. doi: 10.1128/AAC.00433-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thomson KS, Moland ES. 2001. Cefepime, piperacillin-tazobactam, and the inoculum effect in tests with extended-spectrum beta-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother 45:3548–3554. doi: 10.1128/AAC.45.12.3548-3554.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eng RH, Cherubin C, Smith SM, Buccini F. 1985. Inoculum effect of beta-lactam antibiotics on Enterobacteriaceae. Antimicrob Agents Chemother 28:601–606. doi: 10.1128/AAC.28.5.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Livermore DM. 1997. Beta-lactamases: quantity and resistance. Clin Microbiol Infect 3 Suppl 4:S10–S19. [PubMed] [Google Scholar]
- 24. Gould IM, MacKenzie FM. 1997. The response of Enterobacteriaceae to beta-lactam antibiotics--'round forms, filaments and the root of all evil’. J Antimicrob Chemother 40:495–499. doi: 10.1093/jac/40.4.495 [DOI] [PubMed] [Google Scholar]
- 25. EUCAST . 2018. Meropenem in a sensititre broth microdilution panel (DKMGN) from sensititre. Available from: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Warnings/Warnings_docs/Warning_-_meropenem_Sensititre_panel.pdf
- 26. Jorgensen JH. 1993. Selection criteria for an antimicrobial susceptibility testing system. J Clin Microbiol 31:2841–2844. doi: 10.1128/jcm.31.11.2841-2844.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhalodi AA, Oppermann N, Campeau SA, Humphries RM. 2021. Variability of beta-lactam broth microdilution for Pseudomonas aeruginosa. Antimicrob Agents Chemother 65:e0064021. doi: 10.1128/AAC.00640-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. FDA . 2022. Remel, inc recalls thermo scientific Gram negative IVD AST sensititre plate for risk of potential false susceptible results. Available from: https://www.fda.gov/medical-devices/medical-device-recalls/remel-inc-recalls-thermo-scientific-gram-negative-ivd-ast-sensititre-plate-risk-potential-false


