Abstract
A gene homologous to the rpoS gene of Escherichia coli was cloned from a Pseudomonas putida KT2440 gene bank by complementation of the rpoS-deficient strain E. coli ZK918. The rpoS gene of P. putida complemented the acid sensitivity and catalase deficiency of the rpoS mutant of E. coli and stimulated expression of the RpoS-controlled promoter, bolAp1. The gene was sequenced and found to be highly similar to the rpoS genes of other gram-negative bacteria. Like in other gram-negative bacteria, a homolog of the nlpD gene was found upstream to the rpoS gene. A transcriptional fusion of the promoter of the P. putida rpoS gene to the luxAB genes from Vibrio harveyi was constructed and used as an inactivated allele of rpoS for gene replacement of the wild-type copy in the chromosome of P. putida. The resultant rpoS mutant of P. putida, C1R1, showed reduced survival of carbon starvation and reduced cross-protection against other types of stress in cells starved for carbon, in particular after a challenge with ethanol. Survival in soil amended with m-methylbenzoate was also reduced in the mutant strain P. putida C1R1. The RpoS protein of P. putida controls the expression of more than 50 peptides, which are normally expressed in cells after a short period of carbon starvation.
Pseudomonas putida KT2442, a rifampin-resistant derivative of P. putida KT2440, has been studied under famine conditions, and its long-term viability after 1 month of carbon starvation was previously reported (13). When challenged with nongrowth conditions, P. putida develops a general cross-protection which enables the cells to survive various environmental stresses (13); in addition, P. putida exhibits a specific, temporal expression pattern of protein synthesis in response to starvation (12). P. putida KT2440 is a soil bacterium with significance in biodegradation and bioremediation. It is the natural host for several plasmids which confer the ability to mineralize toluene and other aromatic compounds (10), and its environmental application in bioremediation as an engineered, contained microorganism has been reported (35).
The rpoS gene encodes the transcription factor RpoS, which was identified as a central regulator during stationary phase in Escherichia coli (23); its role as the second principal sigma factor for this physiological state is known (28, 44). RpoS is involved in survival of famine conditions (23), in the transition from rod shape to coccus shape as cells reach stationary phase (22), and in cross-protection against stress (osmotic, acidic, and oxidative) (25); recently its role in osmoregulation has been studied (15). The rpoS gene encoding RpoS, also called ςS, has been described for other enteric bacteria and has been found to modulate virulence (7, 17, 41). Among nonenteric bacteria, the rpoS gene has been found in Pseudomonas aeruginosa, even though no phenotypic characteristic has been associated with the gene (43), and in P. fluorescens, in which case the gene was described as being responsible for osmoprotection, resistance to oxidative agents, and regulation of antibiotic synthesis (37).
The aims of this work were (i) to correlate the increased resistance of P. putida to general stress under starvation conditions with the transcription factor RpoS; (ii) to investigate whether this transcription factor is responsible for the protein synthesis program displayed as cells stop growing; and (iii) to generate a derivative strain easy to monitor under suboptimum conditions of growth (which are more similar to natural conditions in soil).
MATERIALS AND METHODS
Strains, plasmids, and growth and starvation conditions.
The strains used are listed in Table 1.
TABLE 1.
Bacteria and plasmids used
| Strain or plasmid | Relevant characteristicsa | Reference and/ or source |
|---|---|---|
| P. putida | ||
| KT2440 | hsdMR | 10 |
| KT2442 | P. putida KT2440 Rifr | V. de Lorenzo |
| R6C1 | Smr Sucs Lux+; P. putida KT2440 cointegrate containing pMIR592 | This study |
| C1R1 | Lux+; RpoS− derivative from KT2440 | This study |
| E. coli | ||
| ZK918 | W3110 ΔlacU169 tna-2 λMAV103 rpoS::kan | 3; G. W. Huisman |
| HB101 | F− Δ(gpt-proA)62 leuB6 supE44 ara-14 galK2 lacY1 Δ(mcrC-mrr) rpsL20 (Smr) xyl-5 mtl-1 recA13 | 4 |
| DH5αF′ | F′/endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15] | 48 |
| CC118(λpir) | λpir Rifr | 16 |
| Plasmids | ||
| pRK600 | Cmr; mob tra | V. de Lorenzo |
| pLAFR3 | Tcr derivative from the cosmid pLAFR1 (11) modified to include multiple cloning sites and the Plac promoter fused to the α peptide of ′LacZ | J. L. Ramos |
| pUTSm | Smr Apr; mini-Tn5/′Sm element inserted in pUT | 16 |
| pUC18 and pUC19 | Apr, multiple cloning site, Plac promoter fused to the α peptide of ′LacZ | 46 |
| pUNØ18 and pUNØ19 | Apr; identical to pUC18 and pUC19 except that the NheI site was changed to a NotI site and oriT was inserted as a 0.7-kb fragment in the new NotI site | Silvia Marqués |
| pUNØ19A | Apr; pUNØ19 after removing its unique AatII site | This study |
| pUJ20 | Tcr Apr; mini-Tn5/′luxABTc element inserted in pUT | V. de Lorenzo |
| pMIR0-1 | Tcr; chimeric cosmid of P. putida library bearing a gene homologous to the rpoS gene of E. coli | This study |
| pMIR1-2 | Tcr; chimeric cosmid of P. putida library bearing a gene homologous to the rpoS gene of E. coli | This study |
| pMIR1-34 | Tcr; chimeric cosmid of P. putida library bearing a gene homologous to the rpoS gene of E. coli | This study |
| pMIR2-9 | Tcr; chimeric cosmid of P. putida library bearing a gene homologous to the rpoS gene of E. coli | This study |
| pMIR13415 | Tcr Smr; mini-Tn5/′Sm element inserted in the P. putida rpoS gene on the 3.4-kb EcoRI fragment of pMIR1-34 | This study |
| pMIR13450 | Apr; 3.4-kb EcoRI fragment of pMIR1-34 was inserted in pUN19Ø | This study |
| pMIR11 | Apr; 7-kb BamHI fragment of pMIR2-9 was inserted in pUN19Ø | This study |
| pMIR61 | Apr; rpoS gene as a 2.5-kb BamHI/NheI P. putida DNA fragment inserted at the BamHI and XbaI sites of pUN19Ø | This study |
| pKNG101 | strAB mobRK2 oriR6K sacBR | 19 |
| pMIR492 | Apr; luxAB cassette as a SalI/BamHI fragment from pUJ20 inserted at AatII unique site of pMIR61 | This study |
| pMIR592 | Smr; PrpoS::luxAB fusion as KpnI/SphI fragment from pMIR492 inserted at the SmaI site of pKNG101 | This study |
| pGM112 | Kmr CmrbolAp1::lacZ | 26 |
| pGM115 | Kmr Cmrficp::lacZ | 26 |
| pGM118 | Kmr CmrlacZ promoterless | 26 |
Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin; Lux, light emission; Nal, nalidixic acid; Rif, rifampin; Sm, streptomycin; Suc, sucrose; Tc, tetracycline.
The minimal medium used for growth was either AB (6) supplemented with 0.01 mM Fe · EDTA (catalog no. E6760; Sigma, St. Louis, Mo.) and 10 mM sodium citrate or modified M9 minimal medium (36) supplemented with 10 mM sodium citrate. Alternatively, 1.5% (wt/vol) lactose-supplemented MacConkey agar (Difco) or rich Luria-Bertani (LB) medium (2) was used. The final concentrations (in micrograms per milliliter) of antibiotics, when required, were as follows: ampicillin, 100; carbenicillin, 500; chloramphenicol, 10; kanamycin, 25 (E. coli strains) and 50 (P. putida strains); nalidixic acid, 50; rifampin, 50; streptomycin, 100; and tetracycline, 10. The temperature under normal growth conditions was 30°C.
Carbon starvation was imposed as described by Givskov et al. (13), either by harvesting a growing culture (optical density at 450 nm [OD450] of 0.4) by centrifugation (preheated rotor and tubes at 30°C) followed by resuspension in preheated carbon-free minimal medium (AB or M9) or by exhaustion of the carbon source in AB or M9 medium supplemented with 1 mM sodium citrate (this condition resulted in starved cultures with an OD450 of 0.4). In all cases, the starvation temperature was 30°C.
Stress challenge protocol.
Growing or carbon-starved cultures (AB or M9) with a density of about 5 × 108 cells per ml were diluted 100- and 1,000-fold in either AB or M9 medium and subsequently diluted 1 to 10 in AB or M9 medium supplemented with either ethanol, H2O2, or NaCl to a final concentration of 18% (vol/vol), 200 μM, or 2.4 M, respectively. The ethanol treatment was performed at 25°C (13), whereas the peroxide and high-osmolarity treatments were performed at 30°C. Aliquots (0.1 ml), taken from the culture at different time points, were spread on LB plates, and after incubation for 16 to 24 h at 30°C, viable counts were determined.
In vitro DNA techniques.
Plasmid DNA was isolated by the alkaline lysis method (19), using Qiagen Plasmid Mini and Qiagen Plasmid Midi kits. All DNA manipulations, including restriction enzyme and alkaline phosphatase reactions, agarose gel electrophoresis, ligations, transformations, filling in, and digestion of protruding ends, were performed by using standard procedures (36). Electrotransformation of P. putida cells was performed with a Gene Pulser apparatus (Bio-Rad catalog no. 165-2098) according to the instruction manual. P. putida total DNA was isolated as described previously (14). DNA fragments were recovered from agarose gels with a GeneClean kit (Bio 101, Inc., Vista, Calif.), with suspension of silica to the glassmilk included in the kit used as an alternative (5). DNA sequencing was carried out on both strands by the dideoxy sequencing termination method (38), with 35S-labeled nucleotides, Sequenase version 1.0 T7 DNA polymerase, and universal or specific oligonucleotides to prime synthesis. DNA hybridization was performed basically as described previously (32), with positively charged nylon membranes from Boehringer.
Mobilization and transposition.
Plasmids were transferred by conjugation using a filter mating technique (32). Filters with a mixture of donor, recipient, and helper strain [E. coli HB101(pRK600)] in a 1:2:1 ratio were incubated overnight at 30°C on the surface of LB plates. The cells were washed and then suspended in 0.9% NaCl, and serial dilutions were plated on selective media. Delivery of minitransposons, either in the chromosome of the target strain or in the cosmids, was performed as described previously (32, 33).
2D-PAGE analysis of [35S]methionine-labeled cellular proteins.
Samples of 4 ml of P. putida in 10 mM sodium citrate-supplemented AB minimal medium with an OD450 of approximately 0.4 were labeled with 4 μl of [35S]methionine (5 mCi/ml; Amersham catalog no. SJ235) for 10 min, either during growth or after 1 h of carbon starvation, and for 15 min after 5 days of carbon starvation. The cultures were chased for 1 min with 4 μl of unlabeled methionine (10 mg/ml), after which 4 μl of chloramphenicol (50 mg/ml) was added; after 2 min, the cells were harvested by centrifugation at 0°C (10,000 × g for 5 min). Cells were lysed and proteins were precipitated with ice-cold acetone as described previously (12). The precipitated proteins were resuspended in 20 μl of sample buffer (50 mM Tris HCl, 0.3% sodium dodecyl sulfate [SDS], 0.6 M β-mercaptoethanol) and 80 μl of ampholyte solution (54% urea, 240 mM β-mercaptoethanol, 2% Pharmalyte pH 3-10 [Pharmacia Biotech], 0.5% Triton X-100) was added. Aliquots of 25 μl were analyzed with a Pharmacia two-dimensional (2-D) polyacrylamide gel electrophoresis (PAGE) system as recommended by the supplier, using 11-cm Immobiline dry strips (pH 4 to 7) and ExeGel XL SDS–12 to 14% precast gels. The protein sample was applied to the alkaline end of the Immobiline strip. After electrophoresis, the gels were fixed, dried, and monitored for radioactivity (1 day of exposure, except for gels of cells starved for 5 days, for which exposure times were prolonged to 6 days) as instructed by Pharmacia.
β-Galactosidase assays.
β-Galactosidase activity was measured as described previously (27), using Miller’s definition (OD420 per OD600 per minute) for specific activity units. Samples of stationary-phase cultures were diluted 10-fold in LB medium and then frozen and melted in ice before the cells were permeabilized with toluene; the same dilution factor was used with the samples for monitoring growth at OD600. β-Galactosidase activity, in Miller units, was corrected by the same dilution factor of 10.
Preparation of inocula and soil microcosm assays.
P. putida strains were grown to exponential phase (OD660 of about 1) (109 cells/ml) at 30°C with rotational shaking (200 rpm) in M9 minimal medium supplemented with 0.5% (wt/vol) glucose. Ten milliliters of the cultures (1010 cells) was washed in M9 minimal medium and resuspended in 1 ml of the same medium prior to introduction in the soil. For soil assays, a cambisol soil (0.63% [wt/wt] organic matter, 23.4% [wt/wt] CaCO3) was used (31). Before use, the freshly collected soils were sifted and sterilized under a vapor stream three times (31). Ninety grams of soil was placed in each jar. Survival of P. putida strains was tested in soils unamended and amended with 0.1% (wt/wt) m-methylbenzoate from a stock solution (0.5 M [pH 7.5]). One milliliter of cells, prepared as described above, was added to each jar containing the sterile soil to a density of about 108 CFU/g of soil. The soil microcosms (in duplicate) were kept at room temperature. To recover cells, 5 g of soil was added to 45 ml of 1× M9 medium and shaken at 30°C for 1 h. The number of viable cells was determined as described in legend to Fig. 9.
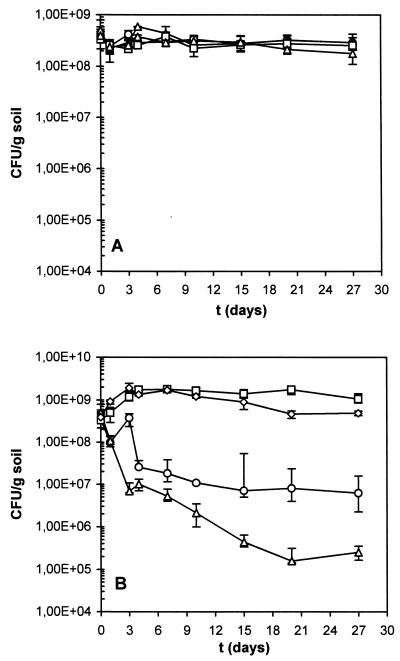
FIG. 9.
Survival of P. putida strains in soil. (A) P. putida KT2440 (○), its rpoS-deficient derivative strain P. putida C1R1 (▵), P. putida KT2440(pWWO) (□), and P. putida C1R1(pWWO) (◊) were introduced in unamended sterile soil in independent jars; (B) symbols as in panel A, but the microcosms were amended with 0.1% (wt/wt) m-methylbenzoate. For each determination, five different dilutions were plated by the drop-plating technique (20-μl drops were laid on selective plates). Mean values are shown, and maximum and minimum values are presented with error bars. Data are from a single experiment, although microcosms were run in duplicate, and typically the same result was observed. Selective medium for P. putida strains bearing the TOL plasmid was 5 mM m-methylbenzoate-supplemented M9; for strains without the TOL plasmid the same medium was supplemented with 5 mM benzoate.
Nucleotide sequence accession number.
The nucleotide sequence of the P. putida KT2440 rpoS gene is under accession no. X91654 (EMBL database).
RESULTS AND DISCUSSION
Isolation and characterization of an rpoS-homologous gene from P. putida KT2440.
A P. putida KT2440 gene bank was generated by random insertion of chromosomal DNA fragments in the cosmid vector pLAFR3, using E. coli HB101 as a host for transfection (31b). E. coli ZK918 (3) is an rpoS-deficient strain (due to a deletional insertion generated with an rpoS::km fusion); it also carries a chromosomal insertion of a transcriptional fusion of the bolAp1 promoter to lacZ at the bolA locus. Since bolAp1 is a promoter depending on RpoS (22), the phenotype of ZK918 is RpoS− LacZ−. Other activities depending on RpoS, such as catalase activity and acid resistance, are also missing in ZK918 (3). These features made ZK918 suitable as a reporter strain for complementing the activity of RpoS. The gene bank was transferred by mobilization to strain ZK918. Acid sensitivity is a phenotypic characteristic of rpoS-deficient E. coli strains (40). Based on this fact, the conjugation mix consisting of the P. putida library and E. coli ZK918 was acid treated before plating as follows. The cells were suspended in LB medium after the filter mating (see Materials and Methods) and cultivated to stationary phase. To the mating mixture was added 2 N HCl to pH 3.0; after acid shock for 30 min at room temperature, neutralization was achieved with 2 N NaOH, and serial dilutions were plated on MacConkey lactose agar containing kanamycin and tetracycline. This treatment reduced the survival of transconjugants 10,000-fold. About 1% of the transconjugants surviving the acid treatment were LacZ+, appearing as red colonies on MacConkey lactose agar, indicating stimulation of expression from the bolAp1::lacZ fusion controlled by RpoS. The catalase activity test of the LacZ+ clones (24- to 36-h colonies were scored for bubbling following the dropwise addition of H2O2) gave positive results as expected for RpoS+ cells. Among the acid-resistant, LacZ+, and catalase-positive transconjugants, four independent clones, harboring cosmids with different restriction patterns, were selected. The four cosmids from the P. putida library complementing the RpoS− phenotype of ZK918 were named pMIR0-1, pMIR1-2, pMIR1-34, and pMIR2-9. In all cases, the three RpoS-controlled phenotypic traits mentioned above were cotransferable with the resistance to tetracycline encoded by the cosmid vector. A 7-kb BamHI fragment and an internal 3.4-kb EcoRI fragment (Fig. 1) were present in all four cosmids. Mutagenesis of pMIR0-1, pMIR1-2, pMIR1-34, and pMIR2-9 with the mini-Tn5/′Sm contained in pUTSm (Table 1) generated mutant plasmids which were unable to complement the rpoS-deficient phenotype of E. coli ZK918. All of the mutant plasmids had insertions in the 3.4-kb EcoRI fragment (not shown). One of these mutants was randomly selected and named pMIR13415 (Fig. 1). The 3.4-kb EcoRI fragment of pMIR1-34 carrying the rpoS-complementing gene was further subcloned into pUNØ19 to yield pMIR13450, whose restriction map is shown in Fig. 1. The sequence of about 1.7 kb of DNA from the half part of the insert of pMIR13450 which contained the unique AatII site was determined and submitted to the EMBL database (accession no. X91654). The sequence was analyzed by the algorithm of Fickett (8) to detect open reading frames (ORFs) encoding polypeptides. Two ORFs were found: the C terminus of an ORF which ended at nucleotide 345 (ORF1) and a complete ORF of 1,008 nucleotides between positions 428 and 1435 (ORF2, whose translated sequence is shown in Fig. 2). The amino acid sequences corresponding to the ORFs were compared with all entries in the nonredundant GenBank CDS translations +PDB+SwissProt+PIR as described in the BLAST program (1) and with the SwissProt ALL library with the FASTA3 program (29). The data bank sequences that showed the most sequence identity with the partial ORF1 were seven precursor sequences encoding the lipoprotein B in gram-negative bacteria: P. aeruginosa (43), E. coli (42), Haemophilus somnus (45), H. influenzae (9), Salmonella typhimurium (21), Yersinia enterocolitica (17), and S. dublin (SwissProt entry 39700).
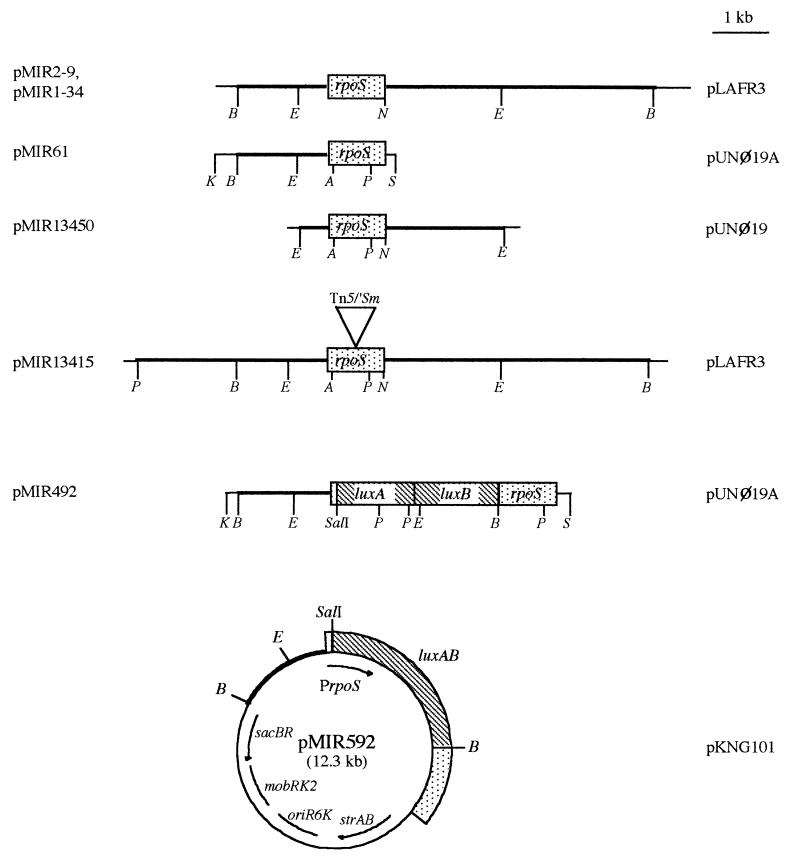
FIG. 1.
Restriction maps of plasmids harboring the P. putida rpoS gene. Plasmids pMIR2-9 and pMIR1-34 are two cosmids from a P. putida library carrying a gene homologous to the rpoS gene of E. coli. Plasmid pMIR61 contains the 2.5-kb BamHI/NheI fragment of pMIR11 (Table 1) inserted in the BamHI and XbaI sites of pUNØ19A (Table 1). Plasmid pMIR13450 carries the 3.4-kb EcoRI fragment of pMIR1-34 inserted in pUNØ19. Cosmid pMIR13415 harbors a mini-Tn5/′Sm element in the 3.4-kb EcoRI fragment of pMIR1-34 (Table 1). Plasmid pMIR492 is the result of inserting a luxAB cassette at position 43 inside the ORF rpoS. (The SalI/BamHI fragment from pUJ20 [Table 1], carrying the genes luxAB from Vibrio harveyi, after filling in the single-strand ends, was inserted in the unique AatII site of pMIR61 after removal of the single-strand protruding ends.) Plasmid pMIR592 was the result of inserting the fusion rpoSp::luxAB as a filled-in KpnI/SphI fragment from pMIR492 in the unique SmaI site of pKNG101 (Table 1). The plasmids listed on the right represent the relevant cloning vectors omitted from the maps. A, AatII; B, BamHI; E, EcoRI; K, KpnI; N, NheI; P, PstI; S, SphI.
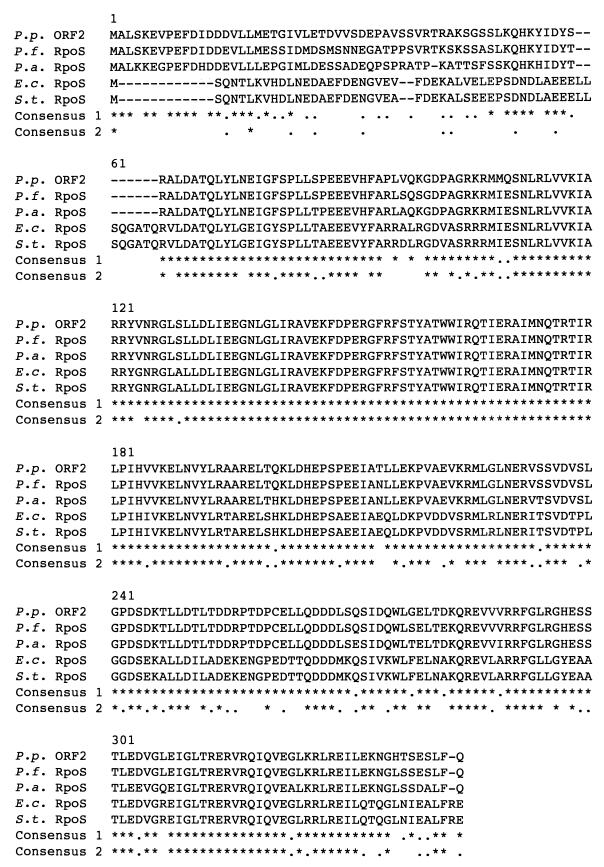
FIG. 2.
Multiple alignments of the P. putida ORF2 sequence with homologous sequences derived from the nonredundant GenBank CDS translations +PDB+SwissPot+PIR. An alignment with RpoS proteins from members of the pseudomonads and enteric bacteria is shown. The sequences of the RpoS proteins of P. fluorescens (P.f.), P. aeruginosa (P.a.), E. coli (E.c.), and S. typhimurium (S.t.) were derived from Samiguet et al. (37), Tanaka and Takahashi (43), Swiss-Prot entry 13445, and Swiss-Prot entry 37400, respectively. The P. putida (P.p.) RpoS sequence (ORF2) was determined in this study. ∗, amino acid conserved in all sequences; ., residue that belonged to the same group in all sequences (neutral changes). Changes of amino acids in the following groups were considered neutral: (i) A, G, P, S, and T; (ii) D, N, E, and Q; (iii) R, H, and K; (iv) I, L, M, and V; (v) F, Y, and W; and (vi) C. For consensus 1, pseudomonad RpoS was used as reference; for consensus 2, both pseudomonad RpoS and enteric RpoS were used.
The PROSITE search for conserved domains in the amino acid sequence derived from ORF2 produced two matches with ς70: sigma70_1 at residue 124 and sigma70_2 at residue 293. However, the sequences that showed the most extensive sequence identity with the complete ORF2, shown in Table 2, were the genes encoding RNA polymerase sigma factor RpoS from the pseudomonads P. fluorescens (37) and P. aeruginosa (43) and from the enteric bacteria E. coli (28), S. typhimurium (30), Y. enterocolitica (17), Serratia entomophila (accession no. U35777), and Shigella flexneri (41). The putative protein would be 38.1 kDa with a pI of 5.19. A potential ribosome binding sequence, AGGA (39), was found 12 bp upstream of the potential initiation ATG codon. Downstream of the rpoS-homologous gene (ORF2) and between nucleotides 1450 and 1480, a hairpin structure (ΔG° = −19.6 kcal/mol [47]) typical of a Rho-independent transcription terminator was identified.
TABLE 2.
Identities and similarities of P. putida KT2440 rpoS gene and RpoS sigma factor sequences with other rpoS gene and RpoS amino acid sequences from the database
| Organism | % DNA identity | Protein
|
|
|---|---|---|---|
| % Identity | % Similarity | ||
| P. fluorescens | 81 (1,009a) | 90 (335) | 96 (335) |
| P. aeruginosa | 84 (1,007) | 85 (335) | 95 (335) |
| E. coli | 70 (834) | 65 (327) | 84 (327) |
| S. typhimurium | 70 (834) | 65 (328) | 85 (328) |
| S. entomophila | 71 (824) | 65 (328) | 83 (328) |
| S. flexneri | 69 (828) | 64 (326) | 82 (326) |
| Y. enterocolitica | 66 (820) | 66 (325) | 81 (325) |
Extent of overlap (in base pairs).
A multiple alignment of ORF2 with two RpoS amino acid sequences of pseudomonads and two enterobacterial RpoS sequences was performed with the computer program SEQUENCE, and the results obtained are shown in Fig. 2. The overall identity between the deduced P. putida KT2440 ORF2 sequence and that of the known Pseudomonas RpoS proteins was about 87% (overall similarity of 95%); compared to the enterobacterial RpoS proteins, there was about 65% identity (overall similarity of 83%) (Table 2). On the basis of the nucleotide and predicted amino acid sequences, we hereafter refer to the ORF2 of P. putida KT2440 as the rpoS gene and to its gene product as RpoS. No cross-reaction with the P. putida RpoS protein was detected with a polyclonal antiserum against E. coli RpoS (not shown). A possible reason for this could be that the amino-terminal sequence of the P. putida RpoS was different from that of the E. coli RpoS (Fig. 2). In E. coli, the expression of RpoS is regulated at the levels of transcription, translation, and protein stability (24). It has been suggested that in E. coli, the amino-terminal part of the sequence might be involved in translational regulation of RpoS, through a mechanism where the Shine-Dalgarno sequence and the initiation codon are sequestered in a secondary structure of the mRNA; under inducing conditions, this structure may be altered and the frequency of translational initiation may be increased (15, 24). In this context, the 58 residues in the amino-terminal end of the protein should be considered of relevance in further studies in order to investigate the regulation of rpoS gene expression in pseudomonads.
Generation of an rpoS merodiploid strain and an rpoS-deficient strain.
Plasmid pMIR592 (Fig. 1) was introduced into P. putida KT2440 to obtain a replacement of the functional rpoS gene in its chromosome with a copy of the rpoS gene interrupted by luxAB at codon 15. The gene replacement was accomplished through homologous recombination. As a result of a single crossover event, one cointegrate containing isolate was selected and named R6C1; after the second crossover, one isolate which carried the mutant rpoS gene only, C1R1 (Fig. 3), was obtained. The selection of double crossovers, based on resistance to sucrose, was due to the conditional lethality which the sacBR genes, carried by pKNG101 and thus by pMIR592, conferred in the presence of sucrose. The genomes of P. putida R6C1 and C1R1 and other independent isolates similarly generated were examined by Southern blotting, and the analysis revealed correct single and double recombinant events, respectively, at the rpoS gene (not shown).
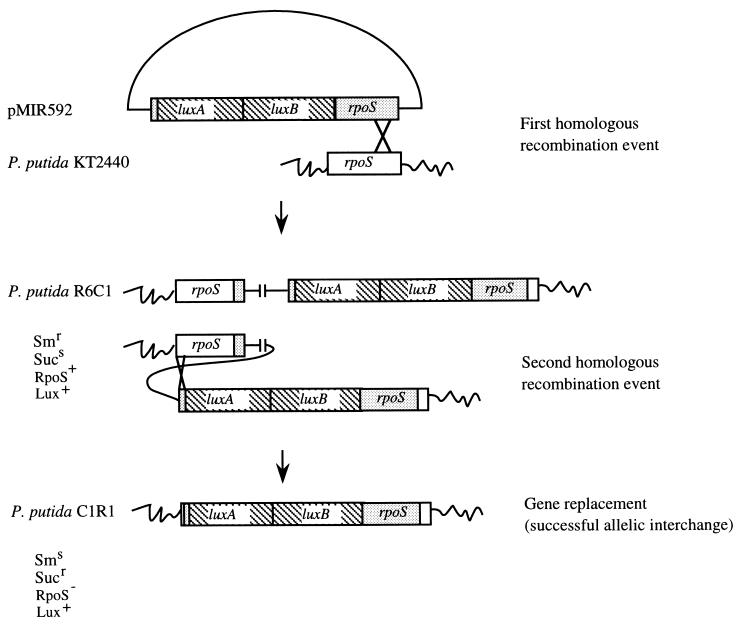
FIG. 3.
Replacements of the rpoS gene with the luxAB insertion mutant rpoS gene. A single homologous recombination event between the functional rpoS present on the chromosome of P. putida KT2440 and the inactivated rpoS present on pMIR592 was isolated by selection for resistance to streptomycin (pMIR592 [Fig. 1]). One of the Smr (see the footnote to Table 1 for abbreviations) transconjugants was selected and named R6C1; this merodiploid strain contained the entire plasmid pMIR592 integrated in the genome. A second crossover event at the rpoS locus was selected by cultivating R6C1 overnight in LB without streptomycin (about 10 generations) and subsequent plating on LB medium supplemented with 10% sucrose. Sucr colonies were analyzed by replica plating. One of the Sucr Sms Lux+ colonies was called C1R1. The genomes of three merodiploid isolates (from three independent matings) obtained as the result of the first recombination event and the genomes of six clones (two from each merodiploid) obtained as the result of the resolution of the merodiploids after the second recombination event were examined by Southern blot analysis, which revealed correct single and double recombination events, respectively, at the rpoS locus (not shown).
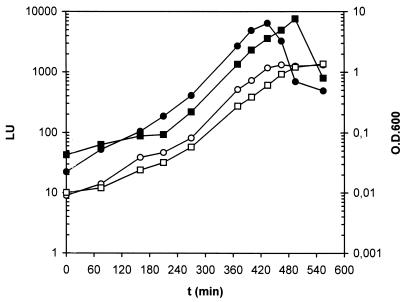
The special design of the above-described rpoS mutants allowed investigations of expression from the rpoS promoter due to the inserted luxAB reporter cassette. Monitoring of bioluminescence from strains R6C1 and C1R1 could in addition indicate if RpoS had any effect on its own expression. Expression from the fusion rpoSp::luxAB inserted in the chromosomes of P. putida R6C1 and C1R1 is shown in Fig. 4 as measurements of luminescence in relative light units (LU) (34). In both strains, the rpoS promoter was active throughout the growth phase, followed by a quick decay in light emission as the cells entered stationary phase. Our results show that rpoS transcription is not induced in cells at the end of the exponential phase of growth. Previously, Lange and Hengge-Aronis showed that in E. coli, rpoS transcription was not significantly induced in cells entering stationary phase in minimal media (24). Also, our data indicate that the rpoS gene does not seem to be involved in the control of its own transcription. The quick decay of light as cells stop growing, commonly observed in isolates bearing luxAB genes, likely reflects the requirement for an energy-generating activity in the cells in order for them to be bioluminescent, which makes the luxAB reporter system useful in monitoring of situations of low energy; however, it is a limitation when gene expression is studied under carbon starvation conditions.
FIG. 4.
Growth-dependent expression of the rpoS genes of P. putida R6C1 (○, •), and P. putida C1R1 (□, ▪). Exponentially growing cells in 10 mM citrate-supplemented AB minimal medium were diluted in the same medium, and growth (open symbols) and emission of light (solid symbols) were measured. Measurement of luminescence in liquid culture was carried out as described previously (34). LU values are not normalized per cell. Notice that the same range of log units, four, is plotted in both y axes, and therefore the curve slopes of light emission and optical density are comparable. Experiments were repeated with three cultures; results of a typical experiment from a single culture are presented. Duplicate measurements of LU in a single experiment yielded an average standard deviation of 10%.
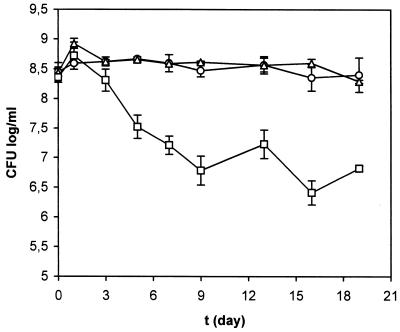
Phenotypic characterization of the P. putida rpoS mutant. (i) Effect of rpoS on survival of carbon starvation.
The P. putida rpoS-deficient strain C1R1 was investigated for its survival of carbon starvation compared to the P. putida parental strain KT2440. The results are shown in Fig. 5. The increase in cell number in the early starvation phase is consistent with previous findings for P. putida (13, 20). Viable counts of the rpoS-deficient strain C1R1 were reduced about 100-fold after 2 weeks of carbon starvation, whether starvation was accomplished by exhaustion of the citrate or by a shift to medium with no carbon source (not shown). Viable counts of the carbon-starved rpoS-proficient strain KT2440 were constant during the time of the experiment, and even after 3 to 4 weeks there was no significant change in viable counts compared to the prestarvation level (∼5 × 108 CFU). The same response to C starvation as for the wild-type strain was exhibited by the merodiploid strain, R6C1. The survival subpopulation of the mutant strain C1R1 was regrown, taken through a new cycle of carbon starvation, and found to be like the start population. To clarify the mechanisms which might be responsible for the survival of 1% of the bacterial population, more investigations are required. However, no extra copy of the rpoS gene was detected in either Southern blot or pulsed-field gel electrophoresis (PFGE) analysis (31a). The further generation of double mutants of the rpoS-deficient strain C1R1, which were unable to emit light (dark mutants produced by transposition, for example), will contribute to the identification of putative regulators which by controlling the expression of the rpoS gene might be the key to understanding the mechanisms responsible for survival of the subpopulation to C starvation.
FIG. 5.
Long-term survival of P. putida strains. Carbon starvation of P. putida KT2440 (○), R6C1 (▵), and C1R1 (□) cells was accomplished through exhaustion of 1 mM citrate present in AB minimal medium as described in Materials and Methods. Exponentially growing cells in 10 mM citrate-supplemented AB minimal medium were centrifuged, the supernatant was discarded, and the cells were resuspended in 1 mM citrate-supplemented AB medium up to 3 × 107 to 4 × 107 cells per ml. Time zero was defined in day 0 as cultures reached stationary phase. Survival of the starved cultures was monitored by determination of viable counts on LB plates, supplemented with streptomycin in the case of R6C1. Each starvation condition was repeated at least twice with two cultures each time. Means and standard deviations of duplicate experiments with the same cultures are plotted. Some of the error bars are too small to be distinguished.
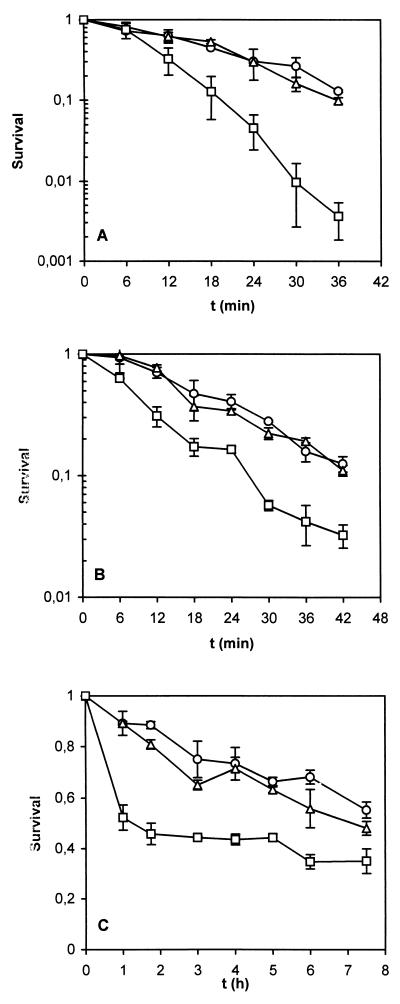
(ii) Effect of rpoS in cross-protection to stress.
Growing cells and carbon-starved cells were challenged with ethanol, hydrogen peroxide, and high medium osmolarity. Conditions (i.e., concentration, time, and temperature) were chosen such that a rapid decline in the survival of a growing culture was obtained (13). No difference between rpoS-proficient and rpoS-deficient exponentially growing cells was observed after challenge with these treatments. P. putida KT2440 and R6C1 cells carbon starved for 24 h or longer developed a high level of resistance to ethanol, resulting in at least 1,000-times-higher viable counts than was found for growing cells after 12 min of treatment (not shown). Carbon-starved rpoS-deficient C1R1 cells were significantly more sensitive to ethanol, showing 100-fold-lower viable counts than the wild type after 30 min of treatment (Fig. 6A). Carbon starvation induced a high level of resistance to H2O2 in the strains, including C1R1, which was only slightly more sensitive to the oxygen stress than the RpoS+ strains (Fig. 6B). Resistance to high osmolarity was also induced by carbon starvation (only 10% of growing RpoS+ cells survived after 30 min of osmotic stress, whereas up to 90% of the starved cells survived 60 min of stress). Again, starved C1R1 rpoS cells were slightly more sensitive than RpoS+ strains during the first hour of the osmotic stress (Fig. 6C).
FIG. 6.
Stress challenges of carbon-starved cultures of P. putida KT2440 (○), R6C1 (▵), and C1R1 (□). Starvation (for 48 h) was accomplished by exhaustion of 1 mM citrate from minimal medium M9. (A) Challenge with 18% (vol/vol) ethanol; (B) challenge with 200 μM H2O2; (C) challenge with 2.4 M NaCl. Viable counts present in each of the prechallenge samples (time zero) were normalized to 1. Survival was determined as relative viable counts. Each challenge experiment was repeated at least twice with two cultures each time. Means and standard deviations of duplicate experiments with the same cultures are plotted. Some of the error bars are too small to be distinguished.
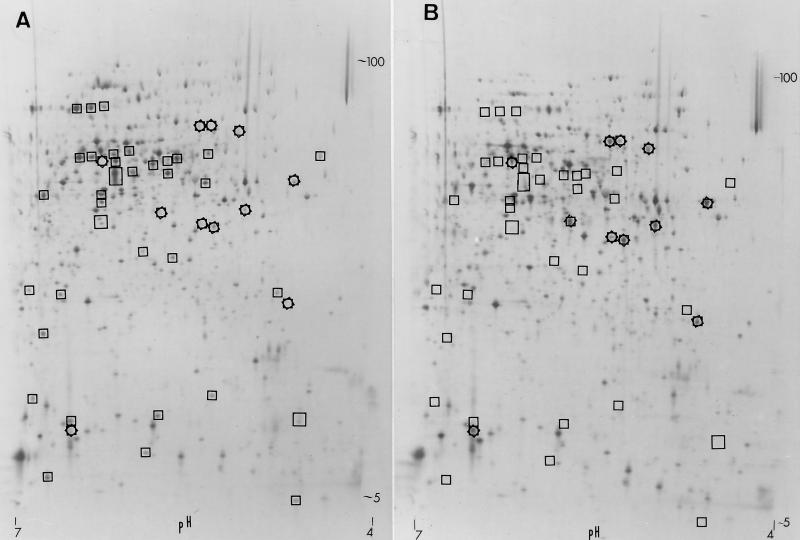
(iii) Synthesis of proteins after a shift to carbon starvation.
A 2D-PAGE system was used for the separation and analysis of [35S]methionine-labeled proteins from P. putida KT2440 and the rpoS derivative C1R1. [35S]methionine-labeled proteins from either growing cells, cells starved for carbon for 1 h, or cells starved for 5 days were analyzed. The different physiological states of the cells resulted in highly different patterns of labeled proteins, although it was possible to identify a common pool of background peptides from growing and 1-h starving cells. Identical patterns of protein synthesis were obtained from growing cells of the wild-type and rpoS mutant strains of P. putida (not shown). In contrast, significant differences were observed between carbon-starved cultures of the two strains: (i) for 1-h starvation, 39 polypeptide spots were missing in C1R1 (peptides positively dependent on RpoS [PPD]) and 13 new spots appeared which were not detected in the wild type (peptides negatively dependent on RpoS [PND]) (Fig. 7); (ii) for 5 days of starvation, 14 PPD were missing in C1R1 and 7 PND were not detected in the wild type (not shown). It was not possible to identify whether the PPD and PND observed after 5 days of carbon starvation corresponded to some of the PPD and PND observed after 1 h of carbon starvation because long-term C-starved cells resulted in a highly different pattern of labeled proteins. A program of protein synthesis as P. putida cells stop growing was reported previously (12). The differences between the wild-type and P. putida rpoS-deficient strains obtained in protein synthesis after the shift to carbon starvation indicate that RpoS acts as a central regulator of stationary-phase gene expression in P. putida. Similar differences in the pattern of protein synthesis between rpoS-proficient and -deficient strains have been reported for E. coli (23), and RpoS-controlled E. coli promoters have been identified (3, 22, 23).
FIG. 7.
2D-PAGE autoradiograms of carbon starvation-induced proteins of P. putida KT2440 (A) and P. putida C1R1 (B) 60 min after removal of the carbon source. Cells were cultivated in 10 mM citrate-supplemented AB minimal media. Spots positively (not present in panel B) and negatively (not present in panel A) dependent on rpoS are enclosed in boxes and stars, respectively. Molecular mass decreases from top to bottom (100 to 5 kDa), and pH decreases from left to right.
(iv) Expression of rpoS-controlled E. coli promoters in P. putida.
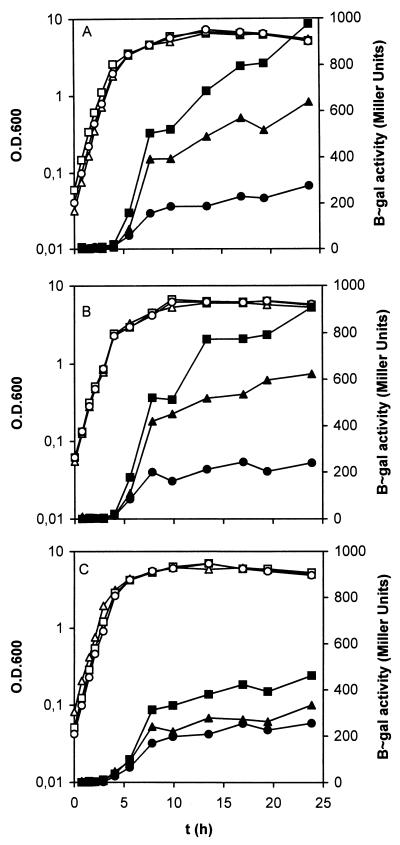
Expression of the growth-phase-dependent E. coli promoters bolAp1 and ficp, carried by plasmids pGM112 and pGM115, respectively, was monitored in growing cultures of P. putida KT2440, R6C1, and C1R1 (Fig. 8). Plasmid pGM118 is a control plasmid expressing the background transcription activity of the vector. No β-galactosidase activity was detected from any of these strains until the cultures were about to enter the stationary phase. The β-galactosidase activities measured in stationary-phase cells of P. putida KT2440(pGM112) and R6C1(pGM112) increased from a background level of 200 to 600 Miller units (Fig. 8A and B). Likewise, the β-galactosidase activity of the same two strains harboring pGM115 increased from 200 to 900 units (Fig. 8A and B). For the rpoS-deficient strain P. putida C1R1 (Fig. 8C), however, only marginal increases above the background level of gene expression were measured. It was recently reported that these promoters are induced in wild-type P. putida cells after entry into stationary phase, and the induction was associated with the detection of a chromosomal fragment hybridizing with the E. coli rpoS gene in Southern blot analysis (26). Thus, the lack of induction of ficp and bolAp1 in the rpoS-deficient background (C1R1 [Fig. 8C]) confirms that induction of these two genes, also in P. putida, is dependent on a functional rpoS gene. The increase in β-galactosidase activity in early stationary phase with the vector pGM118 alone without any promoter is RpoS independent since it was observed in the wild type and the mutant; it could be explained by an increase in plasmid copy number. The marginal induction of ficp expression at the entrance to stationary phase in C1R1 was not observed with the rpoS-deficient strain E. coli MC4100 (not shown). Further studies of expression with growth-dependent Pseudomonas promoters might be helpful in revealing whether sigma factors other than RpoS are involved in transcription at stationary phase in P. putida.
FIG. 8.
Expression from bolAp1 and ficp in P. putida KT2440 (A), P. putida R6C1 (B), and C1R1 (C) bearing either pGM112 (▵, ▴), pGM115 (□, ▪), or pGM118 (○, •). P. putida cells were electrotransformed with plasmids pGM112 (bolAp1), pGM115 (ficp), and pGM118 (control vector, no promoter), and expression from the plasmids carrying bolAp1 and ficp was studied in growing cultures. Exponentially growing LB cultures were used as inoculates, and cells were grown in the same LB medium supplemented with the appropriate antibiotics. OD600 (open symbols) and specific β-galactosidase (B∼gal) activities (solid symbols) were monitored. β-Galactosidase activities are not the result of subtracting the background levels that are synthesized from the vector pGM118 alone. Duplicate measurements of β-galactosidase activities in a single experiment yielded an average standard deviation of 5%.
(v) Effect of rpoS on survival in soil.
P. putida strains were introduced in cambisol soil at a density of 2 × 108 to 4 × 108 cells per g. P. putida KT2440 and C1R1 survived at densities higher than 108 cells per g for almost 1 month (27 days) in unamended soil, and the same result was obtained with cells that harbored the TOL plasmid pWWO (Fig. 9A). pWWO confers on P. putida the ability to degrade contaminants such as toluene and alkylbenzoates (10). In soils amended with m-methylbenzoate (Fig. 9B), the population (CFU) of P. putida KT2440 decreased by about 1 log after 3 days and remained more or less constant thereafter, with a slight tendency to decrease. However, the population of the rpoS-deficient strain, C1R1, decreased by about 1.5 logs after 3 days and by about 3 logs after 1 month, by the end of the experiment (Fig. 9B). Strains bearing the TOL plasmid in amended soils remained at levels above the inoculum size throughout the experiment. Levels of survival of the wild-type and mutant populations were the same in the unamended soil, probably because the bacterial cells were not carbon starved due to the organic matter present in the cambisol soil. In the absence of m-methylbenzoate, the presence of the TOL plasmid had no effect on survival. As the microcosms had been amended with the contaminant, pWWO played a major role in survival: the populations of P. putida KT2440(pWWO) and C1R1(pWWO) increased above 109 cells per g, whereas the populations of the same host strains, wild-type and C1R1, without TOL declined to 107 and 105 cells per g of soil, respectively. Thus m-methylbenzoate caused stress to cells without the TOL plasmid and supported the growth in soil of bacteria containing the TOL plasmid. The rpoS-deficient strain C1R1 was more sensitive than the wild type to contaminant stress with m-methylbenzoate. However, the biodegradation capability conferred by the TOL plasmid protected the rpoS mutant against the toxic effect of m-methylbenzoate.
ACKNOWLEDGMENTS
We thank G. W. Huisman for E. coli ZK918 and for advice on cloning of the rpoS gene from P. putida; we thank G. Miksch for plasmids pGM112, pGM115, and pGM118. We also thank Michael Givskov and Mogens Kilstrup for help with the 2D-PAGE and Flemming G. Hansen for use of his computer program SEQUENCE. We thank Silvia Marqués and J. L. Ramos for communicating unpublished results, and we thank M. A. Ramos-Díaz for useful discussions on PFGE.
This work was supported by grants to S.M. from the Danish Biotechnology Program and to M.I.R.-G., who held a Spanish Government postdoctoral research fellowship. The work was further supported by the Plasmid Foundation.
ADDENDUM
After this paper was submitted, the sequence of the Pseudomonas tolaasii rpoS gene became available in the DDBJ database. The identity between the P. putida RpoS protein and that of P. tolaasii was about 90%, and the similarity was 97%. As for the other known pseudomonad RpoS proteins, the amino-terminal sequence of P. tolaasii RpoS was more conserved than that of the enteric bacteria.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohannon D E, Connell N, Keener J, Tormo A, Espinosa-Urgel M, Zambrano M M, Kolter R. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of ς70. J Bacteriol. 1991;173:4482–4492. doi: 10.1128/jb.173.14.4482-4492.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer H W, Roulland-Dussoix D. A complementary analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 5.Boyle J S, Lew A M. An inexpensive alternative to glassmilk for DNA purification. Trends Genet. 1995;11:8. doi: 10.1016/s0168-9525(00)88977-5. [DOI] [PubMed] [Google Scholar]
- 6.Clark J D, Maaloe O. DNA replication and the cell cycle in Escherichia coli. J Mol Biol. 1967;23:99–112. [Google Scholar]
- 7.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative ς factor KatF (rpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fickett J W. Fickett, J. W. 1982. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 10.Franklin F C H, Bagdasarian M, Bagdasarian M M, Timmis K N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of the genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 12.Givskov M, Eberl L, Molin S. Responses to nutrient starvation in Pseudomonas putida KT2442: two-dimensional electrophoretic analysis of starvation- and stress-induced proteins. J Bacteriol. 1994;176:4816–4824. doi: 10.1128/jb.176.16.4816-4824.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Givskov M, Eberl L, Møller S, Poulsen K, Molin S. Responses to nutrient starvation in Pseudomonas putida KT2442: analysis of general cross-protection, cell shape, and macromolecular content. J Bacteriol. 1994;176:7–14. doi: 10.1128/jb.176.1.7-14.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimberg J, Maguire S, Belluscio L. A simple method for the preparation of plasmid and chromosomal E. coli DNA. Nucleic Acids Res. 1989;17:8893. doi: 10.1093/nar/17.21.8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hengge-Aronis R. Back to log phase: sigmaS as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 16.Herrero M, De Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iriarte M, Stainier I, Cornelis G R. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect Immun. 1995;63:1840–1847. doi: 10.1128/iai.63.5.1840-1847.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jouanin L, de la Judie P, Bagztoux S, Huguet T. DNA sequence homology in Rhizobium meliloti plasmids. Mol Gen Genet. 1981;182:189–195. doi: 10.1007/BF00269657. [DOI] [PubMed] [Google Scholar]
- 19.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y, Watrud L S, Matin A. A carbon starvation survival gene of Pseudomonas putida is regulated by ς54. J Bacteriol. 1995;177:1850–1859. doi: 10.1128/jb.177.7.1850-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowarz L, Coynault C, Robbe-Saule V, Norel F. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J Bacteriol. 1994;176:6852–6860. doi: 10.1128/jb.176.22.6852-6860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor ςS. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 24.Lange R, Hengge-Aronis R. The cellular concentration of the ςS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 25.Loewen P C, Hengge-Aronis R. The role of the sigmaS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 26.Miksch G, Dobrowolski P. Growth phase-dependent induction of stationary-phase promoters of Escherichia coli in different gram-negative bacteria. J Bacteriol. 1995;177:5374–5378. doi: 10.1128/jb.177.18.5374-5378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.Mulvey M R, Loewen P C. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel ς transcription factor. Nucleic Acids Res. 1989;17:9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prince R W, Fang F C, Libby S J. Cloning and sequencing of the RpoS (KatF) sigma factor from Salmonella typhimurium 14028s. Biochim Biophys Acta. 1994;1219:198–200. doi: 10.1016/0167-4781(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 31.Ramos J L, Duque E, Ramos-González M I. Survival in soils of an herbicide-resistant Pseudomonas putida strain bearing a recombinant TOL plasmid. Appl Environ Microbiol. 1991;57:260–266. doi: 10.1128/aem.57.1.260-266.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Ramos-Díaz, M. A., and J. L. Ramos. A combined physical and genetic map of the Pseudomonas putida KT2440 chromosome. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 31b.Ramos-González, M. I. Unpublished data.
- 32.Ramos-González M I, Duque E, Ramos J L. Conjugational transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmids. Appl Environ Microbiol. 1991;57:3020–3027. doi: 10.1128/aem.57.10.3020-3027.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramos-González M I, Ramos-Díaz M A, Ramos J L. Chromosomal gene capture mediated by the Pseudomonas putida TOL catabolic plasmid. J Bacteriol. 1994;176:4635–4641. doi: 10.1128/jb.176.15.4635-4641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rattray E A S, Prosser J I, Glover L A, Killham K. Characterization of rhizosphere colonization by luminescent Enterobacter cloacae at the population and single-cell levels. Appl Environ Microbiol. 1995;61:2950–2957. doi: 10.1128/aem.61.8.2950-2957.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronchel M C, Ramos C, Jensen L B, Molin S, Ramos J L. Construction and behavior of biologically contained bacteria for environmental applications in bioremediation. Appl Environ Microbiol. 1995;61:2990–2994. doi: 10.1128/aem.61.8.2990-2994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Samiguet A, Kraus J, Henkels M D, Muehlchen A M, Loper J E. The sigma factor sigmaS affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc Natl Acad Sci USA. 1995;92:12255–12259. doi: 10.1073/pnas.92.26.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shine J, Dalgarno L. The 3′-terminal sequence of E. coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegele D A, Kolter R. Life after log. J Bacteriol. 1992;174:345–348. doi: 10.1128/jb.174.2.345-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takayanagi Y, Tanaka K, Takahashi H. Structure of the 5′ upstream region and the regulation of the rpoS gene of Escherichia coli. Mol Gen Genet. 1994;243:525–531. doi: 10.1007/BF00284200. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka K, Takahashi H. Cloning, analysis and expression of an rpoS homologue gene from Pseudomonas aeruginosa PAO1. Gene. 1994;150:81–85. doi: 10.1016/0378-1119(94)90862-1. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. Heterogeneity of the principal ς factor in Escherichia coli: the rpoS gene product, ς38, is a second principal ς factor of RNA polymerase in stationary-phase Escherichia coli. Proc Natl Acad Sci USA. 1993;90:3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theisen M, Rioux C R, Potter A A. Molecular cloning, nucleotide sequence, and characterization of lppB, encoding an antigenic 40-kilodalton lipoprotein of Haemophilus somnus. Infect Immun. 1993;61:1793–1798. doi: 10.1128/iai.61.5.1793-1798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vieira J, Messing J. The pUC plasmid: an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 47.Walter A E, Turner D H, Kim J, Lyttle M H, Müller P, Mathews D H, Zucker M. Coaxial stacking of helices enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodcock D M. Quantitative evolution of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]