Abstract
Coenzyme M (CoM) is methylated during methanogenesis from monomethyamine in a reaction catalyzed by three proteins. Using monomethylamine, a 52-kDa polypeptide termed monomethylamine methyltransferase (MMAMT) methylates the corrinoid cofactor bound to a second polypeptide, monomethylamine corrinoid protein (MMCP). Methylated MMCP then serves as a substrate for MT2-A, which methylates CoM. The genes for these proteins are clustered on 6.8 kb of DNA in Methanosarcina barkeri MS. The gene encoding MMCP (mtmC) is located directly upstream of the gene encoding MMAMT (mtmB). The gene encoding MT2-A (mtbA) was found 1.1 kb upstream of mtmC, but no obvious open reading frame was found in the intergenic region between mtbA and mtmC. A single monocistronic transcript was found for mtbA that initiated 76 bp from the translational start. Separate transcripts of 2.4 and 4.7 kb were detected, both of which carried mtmCB. The larger transcript also encoded mtmP, which is homologous to the APC family of cationic amine permeases and may therefore encode a methylamine permease. A single transcriptional start site was found 447 bp upstream of the translational start of mtmC. MtmC possesses the corrinoid binding motif found in corrinoid proteins involved in dimethylsulfide- and methanol-dependent methanogenesis, as well as in methionine synthase. The open reading frame of mtmB was interrupted by a single in-frame, midframe, UAG codon which was also found in mtmB from M. barkeri NIH. A mechanism that circumvents UAG-directed termination of translation must operate during expression of mtmB in this methanogen.
The phylogenetic diversity of methanogens is evident from their dispersal throughout the Euryarchaeota kingdom of the domain Archaea (54). In this light it is ironic that most methanogens are capable of reducing a single substrate, carbon dioxide, to methane. Only one order of methanogenic archaea, the Methanosarcinales, has uniformly evolved the ability to reduce other compounds to methane (2). As a result, this highly successful group is found in a number of different environments. A representative species such as Methanosarcina barkeri can produce methane autotrophically, by the reduction of CO2; acetotrophically, by the cleavage of acetate to methane and carbon dioxide; or methylotrophically, by the dismutation of methanol, methylated thiols, or methylated amines to methane and carbon dioxide.
As with all methane precursors, methylotrophic substrates are first used to methylate coenzyme M (CoM) (9). Recently, three proteins required for CoM methylation from monomethylamine (MMA) were identified: a 170-kDa protein comprised of 52-kDa subunits, termed the MMA methyltransferase (MMAMT); a monomeric 29-kDa corrinoid-binding polypeptide designated the monomethylamine corrinoid protein (MMCP); and a monomeric 40-kDa MT2-type methylcobamide:CoM methyltransferase designated MT2-A (5, 6). These three proteins are sufficient to achieve in vitro methylation of CoM with MMA, but do not methylate CoM with other methylamine growth substrates such as trimethylamine (TMA) or dimethylamine (DMA). Although MMAMT and MMCP can be purified separately from cell extracts, they complex in solution and effect the methylation of the corrinoid bound to MMCP with MMA. Methyl-MMCP is demethylated and CoM is methylated by MT2-A. MMCP binds a single corrinoid cofactor per polypeptide (28), while MT2-A binds zinc as its only detectable prosthetic group (22, 30).
MT2-A is the predominant methylcobamide:CoM methyltransferase in cells grown on TMA (55) and is also involved in methanogenesis from DMA and TMA (19). In addition to MT2-A, TMA-dependent CoM methylation requires a 26-kDa corrinoid-binding polypeptide, designated the TMA corrinoid protein, and a 52-kDa polypeptide (17). The last two of these polypeptides copurify but form an unstable complex. These proteins do not catalyze CoM methylation with MMA or DMA. The N-termini of TMA corrinoid protein and its associated 52-kDa polypeptide differ from MMCP and MMAMT (18). DMA:CoM methyl transfer has not yet been reconstituted with highly purified proteins, but a corrinoid-containing protein supporting only DMA-dependent CoM methylation has been partially purified (52); MMCP is not involved in DMA metabolism (6). In short, methanogenesis from MMA, DMA, or TMA requires distinct methylamine methyltransferases and corrinoid-binding polypeptides but can use the same methylcobamide:CoM methyltransferase, MT2-A.
Two other MT2-type methylcobamide:CoM methyltransferases exist which are specific for methylotrophic methanogenesis from either methanol or methylated thiols. In the studies of Heltjens, van der Drift, Vogels, and coworkers, it was found that methanogenesis from methanol requires a corrinoid protein and the MT2-type enzyme predominant in methanol-grown cells (50–52). Similar to MMA and TMA metabolism, methanol is used to methylate a corrinoid cofactor bound to a 33-kDa protein which associates with a 52-kDa polypeptide (7, 43, 51). The two subunits purify in a tight complex termed MT1. The methylated corrinoid protein is then demethylated by a methanol-specific methylcobamide:CoM methyltransferase, MT2-M, (19, 52) which is a homolog of MT2-A (22, 23, 30). The 480-kDa methylthiol:CoM methyltransferase mediates CoM methylation with substrates such as dimethylsulfide or methylmercaptopropionate (47, 48) and is composed of the MT2 homolog MtsA, tightly bound to the corrinoid-binding polypeptide MtsB (38). Unlike CoM methylation with methanol or the methylated amines which require a minimum of three polypeptides, MtsB and MtsA are sufficient for CoM methylation by methylated thiols (48).
The three MT2 enzymes involved in methanogenesis from methylamines, methanol, or methylated thiols, show approximately 50% sequence similarity at the amino acid level (23, 30, 38). The genes encoding MT2-A (mtbA) and MT2-M (mtaA) are transcribed monocistronically (23). In contrast, the methylthiol-specific MT2 homolog MtsA is cotranscribed with MtsB, the corrinoid protein with which it mediates methylthiol:CoM methyl transfer (38). The sequence of MtsB revealed a corrinoid binding motif (38) typified by cobalamin-dependent methionine synthase (12). The two subunits comprising MT1 were found to be encoded on the genome contiguously and present on a single transcript (43). The smaller subunit (MtaC) was found to be homologous to MtsB and to share the corrinoid binding motif of methionine synthase, while the larger subunit of MT1 (MtaB) was unlike any other protein currently in the database.
With the exception of MT2-A (23, 30), none of the proteins specific for methanogenesis from the methylamines had been sequenced to date. We therefore undertook the cloning, sequencing, and transcript analysis of the region of the M. barkeri genome encoding the corrinoid protein specific for monomethylamine, MMCP (mtmC). Sequencing of the surrounding regions revealed the genes encoding MT2-A (mtbA) and MMAMT (mtmB). Cotranscribed with MMCP and MMAMT was a gene homologous to a large family of cationic amine permeases which could encode an MMA permease (mtmP). Surprisingly, the open reading frame of mtmB was interrupted by a single midframe canonical stop codon which does not appear to prevent translation of the full-length product.
MATERIALS AND METHODS
Organisms and culture.
M. barkeri MS (DSM 800) was cultured anaerobically in a phosphate-buffered medium and supplemented with MMA-HCl to a final concentration of 80 mM as described previously (5). Frozen cells of M. barkeri NIH were the generous gift of David A. Grahame. Escherichia coli DH5α (Gibco BRL, Gaithersburg, Md.) and LE392 (Promega Corp., Madison, Wis.) were cultured in Luria-Bertani (LB) broth (41) at 37°C.
Isolation of genomic, plasmid, and phage DNA.
M. barkeri genomic DNA was typically isolated from 2 g of frozen cell paste by resuspension in 10 ml of 1% (wt/vol) sodium dodecyl sulfate–45 mM EDTA–45 mM ammonium bicarbonate buffer (pH 8.0) and passage through a French pressure cell at 500 lb/in2. The DNA was then isolated by standard methods (41) and treated with RNase A (Boehringer-Mannheim Corp., Indianapolis, Ind.). pUC19 (Gibco BRL) and its derivatives propagated in E. coli DH5α were isolated with the Wizard Plasmid DNA Purification System (Promega).
Cloning techniques.
Unless specified otherwise, the molecular methods outlined by Sambrook et al. (41) were used throughout. It was hypothesized that MMCP would possess the corrinoid binding motif found in methylthiol:CoM methyltransferase (38) and MetH (12). Therefore, in order to generate a probe homologous to MMCP, PCR from was carried out with M. barkeri MS genomic DNA by using Taq polymerase (Gibco BRL) and primer 29A (5′-AA[C/T]CA[A/G]GA[A/G]AT[A/C/T]TT[C/T]GACAA-3′), designed with the N-terminal sequence of purified MMCP, as well as primer COB3 (5′-AT[A/G]TT[C/T]TTNCC[A/G/T]ATGTCATG-3′), designed from the HDIGKNIV corrinoid binding motif. The major 350-bp product was ligated with pGEM T-Vector (Promega) and propagated in E. coli DH5α. The DNA sequence of the insert was determined and compared to the N-terminal amino acid sequence of MMCP to confirm that the appropriate region of genomic DNA had been amplified and cloned. The positively identified insert was gel purified and labeled by the random primer method (15, 16) with the Prime-a-Gene System (Promega) using [α-32P]dATP (Amersham Corp., Arlington Heights, Ill.). This homologous probe was used to screen an M. barkeri MS Sau3AI genomic DNA library which was constructed in phage λ-GEM-11 (Promega) and propagated in E. coli LE392 as directed by the supplier. Plaque lifts were performed with Nytran nylon membranes (Schleicher & Schuell, Keene, N.H.) as recommended by the manufacturer. Hybridization and washing of membranes were performed as described by Hennigan and Reeve (25). Of 6,000 phage plaques tested, 20 clones hybridizing specifically to the probe were isolated and amplified in the host E. coli LE392, and the DNA was purified. The recombinant DNA preparations and M. barkeri MS genomic DNA were digested to completion with various restriction enzymes and examined for common restriction fragments by Southern hybridization with the oligonucleotide probe. The insert from one of four positive λ clones was isolated and subcloned with pUC19 as the vector and E. coli DH5α as the host.
A SacI-HindIII fragment (positions 2914 to 4437) (Fig. 1) containing part of the mtmB gene was cloned from M. barkeri MS genomic DNA with pUC19 as the vector and E. coli DH5α as the host. A similar fragment of mtmB was generated by PCR (see Fig. 4A) using the high-fidelity Vent DNA polymerase (New England Biolabs, Beverly, Mass.), either M. barkeri MS or M. barkeri NIH genomic DNA as the template, and the primers 29G (5′-TTGGAGGAGCTCCTGTATC-3′) and 52H (5′-GCTGTTCCCTTTGTAGTG-3′).
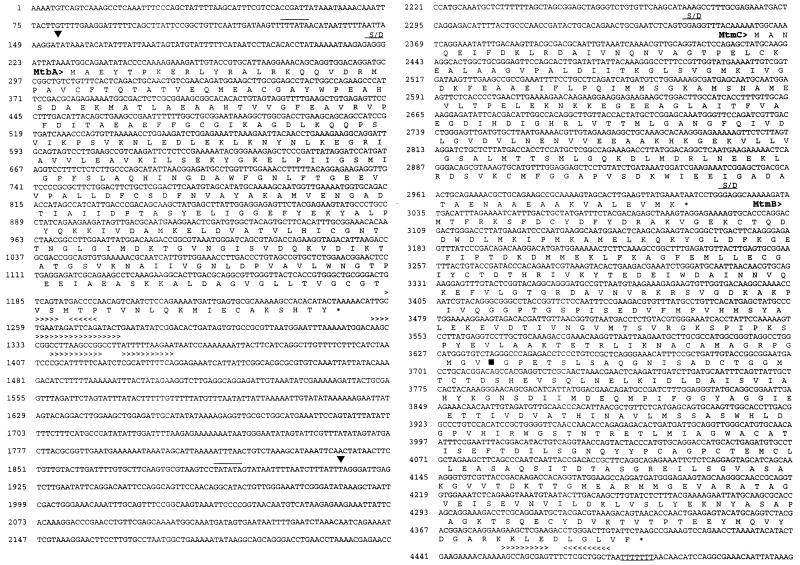
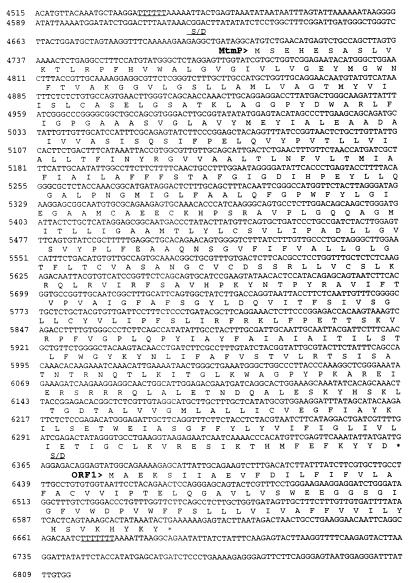
FIG. 1.
Nucleic acid sequences and predicted amino acid sequences of mtbA, mtmC, mtmB, mtmP, and orf1 of the mtm operon from M. barkeri. The predicted amino acid sequences are shown in single-letter code directly below the second base of each codon. Position numbers are given to the left of each row of the sequence. Overlined bases represent the predicted promoter sequences. Symbols and abbreviations: ▪, transcript start site determined from primer extension; ∗, predicted stop codon; ▾, position of UAG in reading frame of mtmB; > and < (shown above bases included in repeat sequences), direction of the repeat (indicated by the direction of the arrow head); S/D, predicted Shine-Dalgarno sequence.
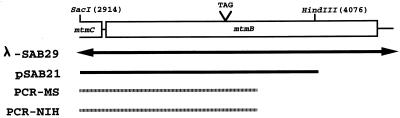
FIG. 4.
Confirmation of the mtmB sequence containing the in-frame UAG codon. The figure illustrates the regions that were cloned or amplified by PCR from M. barkeri genomic DNA and sequenced. λ-SAB29 is the original phage clone in which the amber codon was first observed. Nucleotide numbers are the same as those in Fig. 1. pSAB21 is a pUC19 derivative which contains the indicated 1.1-kb SacI-HindIII fragment cloned directly from M. barkeri MS genomic DNA. PCR-MS and PCR-NIH are fragments amplified from M. barkeri MS or NIH genomic DNA and directly sequenced.
DNA sequencing.
DNA sequences were determined by the dideoxynucleotide method (42) with Sequenase version 2.0 T7 DNA polymerase and 7-deaza-dGTP or dITP in place of dGTP (U.S. Biochemicals, Cleveland, Ohio). Alternatively, fluorescent tag sequencing using Dye Terminator Cycle Sequencing Reaction Mix (Perkin-Elmer Corp., Foster City, Calif.) supplemented with 5% (vol/vol) dimethyl sulfoxide (DMSO) and an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer Corp.) were employed.
Sequence analyses.
BLAST homology searches (1) were run with the nonredundant database maintained at the National Center for Biotechnology Information. Alignments of sequences were made with CLUSTALW (49) on the World Wide Web server maintained at the Baylor College of Medicine. Both of these programs employ the BLOSUM62 substitution matrix (24). Hydropathy profiles and transmembrane-spanning residue predictions from protein sequences were performed by the method of Kyte and Doolittle (29) and with the TMpred program (26) on the World Wide Web server maintained at the Swiss Institute for Experimental Cancer Research. Codon usage frequencies in protein coding sequences of Methanosarcina spp. in GenBank full flat file release version 102 were obtained by using CUTG (34) on the World Wide Web server maintained at the Kazusa DNA Research Institute. Additional files added in GenBank 104 were analyzed manually. Phylogenetic analysis of the relationship of MtmP with members of the APC family of cationic amine permeases (40) was performed as follows. A multiple sequence alignment of MtmP with protein sequences of APC family members was made with CLUSTALW. The distance data from the alignment were used to construct a phylogenetic tree by using the least-squares method of the TreeGen program (21) on the World Wide Web server maintained at the Eidgenössische Technische Hochschule in Zürich.
RNA isolation and Northern hybridizations.
Total RNA was isolated from M. barkeri MS grown on MMA to mid-log phase and treated with RNase-free DNase (Boehringer-Mannheim) as described previously (38). Samples were stored at −70°C as precipitates in isopropanol. Northern hybridizations were performed essentially as described by Hennigan and Reeve (25). Hybridizations and membrane washing conditions were determined empirically with each probe. Blots used for more than one hybridization were stripped of bound oligonucleotide probes as recommended by the membrane manufacturer (Schleicher & Schuell) and checked for complete removal by autoradiography.
Determination of transcript start sites.
The 5′ termini of the mtbA, mtmCB, and mtmCBP transcripts were determined by primer extension analysis with avian myeloblastosis virus reverse transcriptase (U.S. Biochemicals). The oligonucleotide primer 5′-GCAGTCTGAGTGAAACAG-3′ (complementary to bases 304 to 321 of the contiguous sequence [Fig. 1]) was used in the mapping of the 5′ end of the mtbA transcript, and the primers 5′-CAATTGCGTCGCGTAACTTG-3′ and 5′-CAGGTTCGGGTCCTTTGTG-3′ (complementary to bases 2384 to 2403 and 2072 to 2090) were used in determining the 5′ ends of the mtmCB and mtmCBPorf1 transcripts. Primers were 5′ end labeled (41) with [γ-32P]ATP (ICN Pharmaceuticals, Inc., Costa Mesa, Calif.) and T4 polynucleotide kinase (Boehringer-Mannheim). RNA samples (20 μg) isolated from cells growing exponentially on MMA were annealed with 3.0 pmol of phosphorylated primer by incubation at 90°C for 5 min followed by slow cooling of the mixture to 42°C over 45 min. One unit of reverse transcriptase and 0.5 mM deoxynucleoside triphosphates (Boehringer-Mannheim) were added, and the mixture was incubated at 42°C for 45 min. Extension products were purified by phenol-chloroform extraction and subjected to denaturing polyacrylamide gel electrophoresis alongside DNA sequencing ladders generated from plasmid clones with the same oligonucleotide primer.
Nucleotide sequence accession number.
The 6,814-nucleotide M. barkeri MS sequence (Fig. 1) and M. barkeri NIH mtmB partial sequence described in this article were deposited in GenBank under accession no. AF013713 and AF046875, respectively.
RESULTS
The genes for the initial steps of methanogenesis from MMA are clustered on the M. barkeri genome.
Using the approaches outlined in Materials and Methods, a phage clone, λ-SAB29, was identified which contained a 15-kb DNA insert that included mtmC (Fig. 1 and 2). The gene was positively identified by the N-terminal sequence of purified MMCP (ANQEIFDKLRDAIVNQNVAGTPELCKE). Further sequencing revealed an open reading frame immediately downstream of mtmC which was identified as encoding MMAMT by comparison to the N terminal sequence of the purified protein (TFRKSFDXYDF), which was designated mtmB. Located 1.1 kb upstream of mtmC was the gene encoding MT2-A (mtbA). Although mtbA has been previously sequenced from both M. barkeri NIH (30) and M. barkeri Fusaro (23), this is the first report of its proximity to mtmC. Interestingly, no open reading frame encoding a product larger than 17 amino acid residues was present in the 1.1-kb intergenic region of mtbA and mtmC.
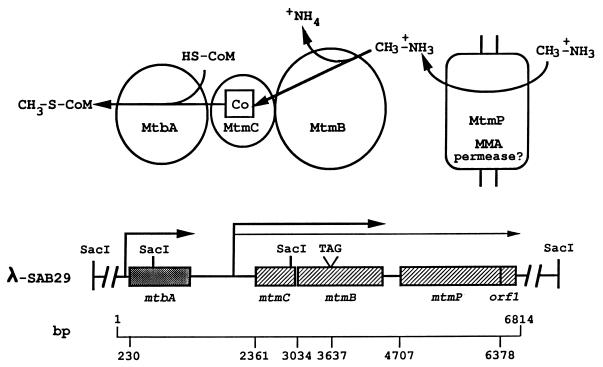
FIG. 2.
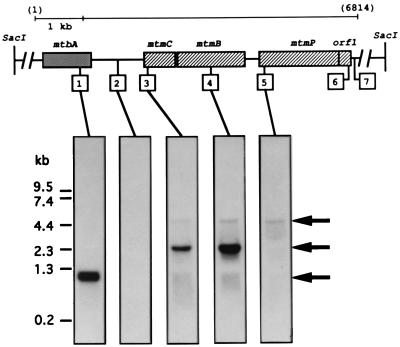
Organization of genes that mediate the initial steps of methanogenesis from MMA. The physical map of the 15-kb insert of M. barkeri genomic DNA from the λ-SAB29 clone shows the organization of the genes encoding the catalyzing polypeptides involved in the methylation of CoM with MMA. The transcripts containing these genes, designated by the arrows, are drawn to scale to indicate the identified 5′ termini and estimated 3′ termini. Base positions of initiation codons and the in-frame amber codon of mtmB are indicated below and correspond to the nucleic acid sequence shown in Fig. 1. The biochemical reactions and catalyzing polypeptides involved in CoM methylation with MMA are shown in the diagram at the top, along with the predicted function of the mtmP gene product.
Apparent ribosome-binding sites complementary to the 3′ terminus of the 16S rRNA of M. barkeri (Genbank accession number M59144) were located 8, 10, and 10 bases upstream from the respective start codons of mtbA, mtmC, and mtmB (Fig. 1). One inverted repeat sequence and two pairs of nonidentical direct repeats were located immediately downstream of the stop codon of mtbA, which may be involved in transcription termination (56). No obvious indication of a terminator was found in the intergenic region of mtmC and mtmB; however, an inverted repeat sequence followed by a polypyrimidine sequence, which could be involved in transcription termination (39, 56), was observed downstream of mtmB.
MtmC establishes a family of cobalamin-binding proteins involved in methylotrophic methanogenesis.
A BLASTP search of the nonredundant database maintained at the National Center for Biotechnology Information indicated significant homology of MtmC to a group of corrinoid-binding proteins which share the corrinoid binding motif exemplified by methionine synthase (MetH) from E. coli (12). The BLAST alignment of positions 27 to 152 of the MtmC sequence with MetH indicated 34% identity. Similarly, the alignment of positions 94 to 148 of MtmC with the MtsB subunit of methylthiol:CoM methyltransferase (38) and the MtaC subunit of MT1 from M. barkeri (43) revealed 50 and 52% identity, respectively. A CLUSTALW alignment of these proteins is shown in Fig. 3. Homology among all four sequences spans both the upper and lower domains demonstrated to contact the cofactor in the crystal structure of the 27-kDa cobalamin-binding tryptic fragment of MetH (12). MtmC contains all seven of the signature residues involved in B12 binding in MetH, including the histidyl residue which ligates the cobalt of the B12 cofactor.
FIG. 3.
Sequence alignment of the monomethylamine corrinoid protein (MtmC) with the corrinoid-binding subunits of methylthiol:CoM methyltransferase (MtsB) (38) and methyltransferase I (MtaC) (43) as well as the corrinoid-binding tryptic fragment of methionine synthase (MetH) (12). Residues shared between sequences are boxed. Position numbers are given to the left and right of each row of each sequence.
The sequence of mtmB contains an in-frame UAG codon.
Sequence analysis of mtmB revealed an in-frame amber codon located 603 bases into the coding sequence (nucleotide position 3637) (Fig. 1). This reading frame continues for an additional 770 bp before ending with a UAA stop. The other reading frames contain numerous UAA and UGA stop codons immediately upstream and downstream of the amber codon, indicating that the amber codon is not bypassed by +1 or −1 frameshift events. The purified gene product of mtmB has an approximate molecular mass of 52 kDa (6), and a much shorter 23-kDa product would result if the UAG codon was recognized as a stop codon. Therefore, the UAG codon was initially dismissed as a sequencing error or cloning error that occurred in E. coli during propagation of the λ genomic DNA library or pUC19 subclones. It was noted that the amber codon was located in a region (positions 3600 to 3680) (Fig. 1) with a GC ratio of 65 mol%, which was high relative to the rest of the sequence (46 mol%). Therefore, both strands of the DNA coding for mtmB were sequenced four times by a PCR-based fluorescent-tag sequencing reaction with added DMSO. The addition of DMSO has been shown to improve the quality of sequence data in regions prone to compressions (4). The same unambiguous sequence was obtained each time. Sequencing of the region was also repeated with a different methodology involving Sequenase 2.0 T7 DNA polymerase with 7-deaza-dGTP or dITP in place of dGTP. By either method, unambiguous sequence data indicating that the in-frame UAG codon was not a sequencing error was obtained, and this data confirmed the sequence in Fig. 1.
Further confirmation of the in-frame UAG codon of mtmB genomic DNA from two strains of M. barkeri.
To eliminate the possibility of a cloning error introduced during propagation of the λ library, a 1.1-kb SacI-HindIII fragment (positions 2914 to 4486) (Fig. 1) containing the majority of mtmB (Fig. 4) was isolated directly from genomic DNA of M. barkeri MS and cloned in pUC19. The sequence determined for this new clone was identical to that determined from λ-SAB29.
An additional approach was utilized to circumvent the propagation of M. barkeri DNA in a foreign host altogether. A fragment of the same region of M. barkeri MS genomic DNA (Fig. 4) was amplified by PCR with Vent DNA polymerase. The fragment was directly sequenced, and the outcome again confirmed the original sequence.
Finally, the same region of DNA was amplified by PCR from the genome of a separate strain, M. barkeri NIH, to determine if the midframe UAG codon was an anomaly of our laboratory strain. The result revealed several base changes throughout the sequence; however, the GC-rich region involving the in-frame amber codon and the codon itself, as well as the overall reading frame, were conserved. These data suggest that the in-frame UAG codon is not the result of a sequencing error, cloning abnormality, or strain variation.
The unusual occurrence of the in-frame UAG codons within mtmB from M. barkeri strains MS and NIH prompted an examination of other protein-encoding genes from Methanosarcina species to determine the apparent frequency of UAG usage as a stop codon. GenBank version 104 was examined and, disregarding known partial sequences, only 3 of 134 open reading frames were found to terminate with a UAG codon. Two of these were putative genes with no demonstrated products. This suggests that Methanosarcina terminates translation with the UAG codon in only 2% of genes. In comparison, analysis of the Methanococcus jannaschii genome (3) indicated UAG is used to terminate transcription in 10% of all ORFs.
mtbA, mtmCB, and mtmCBP transcripts.
Oligonucleotides complementary to the 1.1-kb region between mtbA and mtmC, as well as to mtbA, mtmC, and mtmB, were synthesized and used to probe Northern blots of RNA isolated from M. barkeri MS grown to mid-log phase on MMA. Probes for mtmC and mtmB (Fig. 5, probes 3 and 4) hybridized specifically to two mRNA species of approximately 2.4 and 4.7 kb. The predominant 2.4-kb transcript was within the expected size range to encompass both mtmC and mtmB, indicating cotranscription of the two genes. The much less predominant 4.7-kb message was the appropriate size to contain the entire region from mtbA to mtmB. However, only one transcript of approximately 1.1-kb was detected for mtbA (Fig. 5, probe 1), agreeing with the previous report by Harms and Thauer (23) of monocistronic transcription of mtbA in M. barkeri Fusaro. Additionally, no message was detected with a probe complementary to the region approximately 300 nucleotides downstream of mtbA (Fig. 5, probe 2). This indicated that the larger transcript includes sequence downstream of mtmB. Indeed, a probe targeted for a region approximately 400 bases downstream of mtmB (Fig. 5, probe 5) detected only the 4.7-kb message.
FIG. 5.
Identification of the mtbA, mtmCB, and mtmCBPorf1 transcripts. Northern analyses of total RNA isolated from M. barkeri grown on MMA were performed with the same blot and with probes complementary to the regions designated in the diagram by the boxed numbers. The probes were complementary to the following positions of the contiguous nucleotide sequence: probe 1, 933 to 1010; probe 2, 1775 to 1792; probe 3, 2384 to 2403; probe 4, 3723 to 3741; probe 5, 4777 to 4794; probe 6, 6549 to 6566; and probe 7, 6750 to 6768. Molecular size standards are indicated to the left, while arrows designating the migration positions of the three transcripts are located to the right. Probes 6 and 7 gave essentially the same autoradiograms as those indicated for probe 5 and 2, respectively.
Further sequencing downstream of mtmB revealed two open reading frames, designated mtmP and orf1 (Fig. 1 and 2). The former begins approximately 300 bp downstream of mtmB and is predicted to encode a very hydrophobic protein of 552 amino acids, with a predicted molecular mass of 60 kDa. The orf1 product is also predicted to be a hydrophobic protein; however, it is much smaller, with only 78 amino acid residues and a predicted molecular mass of 8.8 kDa. Apparent ribosome-binding sites were located nine and eight bases upstream of the initiation codons for mtmP and orf1, respectively (Fig. 1). Northern blot analysis confirmed that mtmP and orf1 (Fig. 5, probes 5 and 6) were contained in the 4.7-kb mtmCBPorf1 transcript (blot of probe 6 not shown). A polypyrimidine sequence was noted following the end of orf1 (Fig. 2, positions 6669 to 6675) which could be involved in termination of the transcript (56). The 4.7-kb transcript was not detected with a probe to the region following the putative terminator (Fig. 5, probe 7; blot not shown). No evidence was seen for a smaller transcript encoding only mtmP.
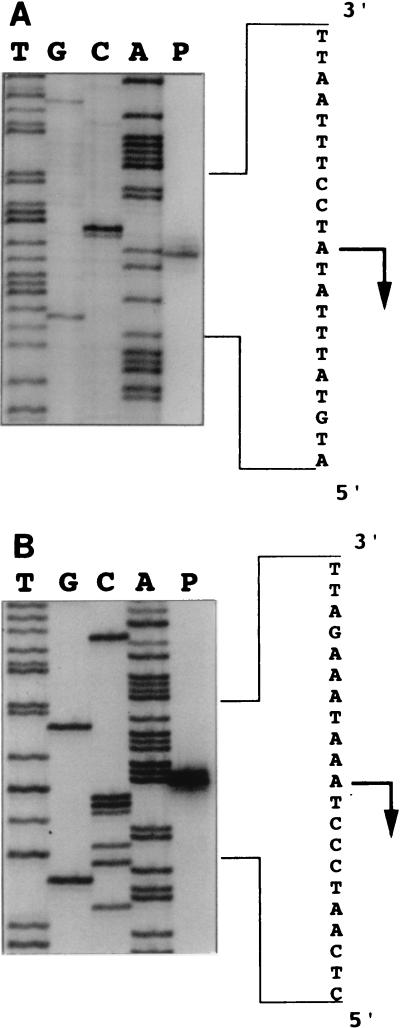
The locations of the 5′ termini of the mtbA and mtm transcripts were determined by primer extension analysis (Fig. 6; also Fig. 1). Transcription of mtbA was found to initiate 76 bp upstream of the translational start and 22 bp downstream of a sequence closely resembling the methanogen consensus promoter, TTTA(A/T)ATA (36, 39, 56). Interestingly, a single 5′ terminus was identified for both mtm transcripts, located 447 bp upstream of the translational start of MtmC. Although only one 5′ terminus was found, it was noted that other potential promoter sites which resembled the methanogen consensus promoter were found within the large leader region. Since the RNA samples utilized in these experiments were isolated from cells grown only on MMA, the possibility of alternate promoter sites which might be used under other growth conditions is not excluded.
FIG. 6.
Identification of initiation sites of mtbA, mtmCB, and mtmCBPorf1 transcripts. Primers were annealed with total RNA isolated from M. barkeri grown on MMA. The products of primer extension for the mtbA (A) and mtm (B) transcripts are shown adjacent to the DNA sequencing ladders generated with the same primers. The antisense nucleotide sequence is shown to the right with the 5′ terminus and direction of transcription indicated by an arrow. The positions of the transcriptional starts are indicated in Fig. 1.
Similarity of MtmP to the APC family of cationic amine permeases.
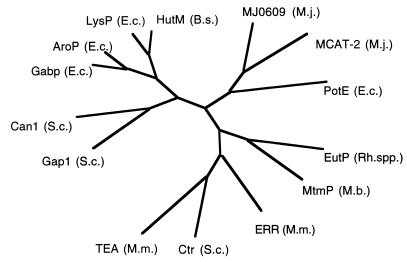
The mtmP gene product is predicted to have at least 12 membrane-spanning regions. A BLASTP search with the protein sequence of MtmP indicated strong similarity to the APC family of integral transport proteins (40). The most statistically significant alignment was to a putative ethanolamine permease (EutP) from Rhodococcus sp. strain NI86/21 (11). The BLAST alignment of positions 21 to 119 and 186 to 280 of the MtmP sequence with the EutP sequence gave scores of 177 and 173, respectively, and indicated 38% identity and 57% similarity in both regions. The number of sequences in the database that would randomly produce these scores when paired with MtmP over the given lengths was predicted to be 5.3 × 10−36. A phylogenetic tree constructed from a CLUSTALW alignment of MtmP with several members of this permease family, including two putative cation permeases from the genome of M. jannaschii (3), is shown in Fig. 7. Reizer et al. (40) have made the observation that the proteins within this family fall into major groups or clusters according to their substrate specificity. MtmP and EutP, although clearly related to this family, appear to be members of a new cluster. A transporter for MMA is to be expected, since the pKa of MMA is reported to be 10.79 at 20°C (10) and MMA is almost entirely ionized at physiological pH ranges. Cotranscription of this cationic amine transporter homolog with mtmCB indicates that MtmP could function as a methylammonium permease. No obvious similarity was detected between MtmP and the known ammonium/methylammonium transporters of E. coli (AmtA) (13) and Corynebacterium glutamicum (AmtA) (44) or the putative ammonium transporters from M. jannaschii (ORFs MJ0058 and MJ1343) (3).
FIG. 7.
MtmP belongs to the APC family of cationic amine permeases. A phylogenetic tree was constructed with the TreeGen program (21) from the CLUSTALW (49)-generated multiple sequence alignment of MtmP of M. barkeri with members of the APC family of transporters (40). The abbreviations for the gene products are as follows: EutP (Rh. spp.), putative ethanolamine permease from Rhodococcus sp. strain NI86/21 (GenBank accession no. L24492); Ctr (S.c.), choline transporter from Saccharomyces cerevisiae (J05603); ERR (M.m.), ecotropic retroviral receptor from Musculus musculus (M26687); TEA (M.m.), T-cell early activator from M. musculus (D83596); PotE (E.c.), putative putrescine/ornithine permease from E. coli (M64495); MJ0609 (M.j.) and MJ1196 [MCAT-2 (M.j.)], both putative cation transporters from M. jannaschii (3); Gap1 (S.c.), general amino acid permease from S. cerevisiae (X52633); Can1 (S.c.), arginine permease from S. cerevisiae (X03784); Gabp (E.c.), γ-aminobutyrate from E. coli (X65104); AroP (E.c.), general aromatic amino acid permease from E. coli (X17333); LysP (E.c.), lysine-specific permease from E. coli (M89774); HutM (B.s.), putative histidine permease from Bacillus subtilis (D31856).
DISCUSSION
The clustering of the genes encoding MMAMT, MMCP, and MT2-A provides further support of the working model of the initial steps of methanogenesis from MMA established in reconstitution studies. MMCP (MtmC) and MMAMT (MtmB) were shown to function in MMA metabolism following separate isolations (5, 6), yet they are encoded by genes in a single transcriptional unit. Likewise, the gene encoding the methylamine-specific CoM methylase MT2-A was located upstream and separated by a sequence which does not appear to code for protein. The monocistronic transcription of mtbA and cotranscription of mtmCB fit well with the evidence that MtmC and MtmB are specific to the MMA pathway, while MtbA functions in CoM methylation with all three methylamines.
MtmB interacts with MtmC to methylate the corrinoid cofactor of MtmC with MMA. MtaB interacts with MtaC to methylate the protein-bound corrinoid with methanol (43, 50, 51). The lack of sequence similarity between the functionally analogous MtaB and MtmB provides a rationale to the divergence of the homologous corrinoid proteins (MtaC and MtmC) and CoM methylases (MtaA and MtbA) used in the methanol and MMA pathways. Compensating changes in the homologous proteins may have been required to bind and maintain the correct orientations of active sites with nonhomologous methyltransferases.
A single promoter appears to direct synthesis of both the mtmCB and mtmCBPorf1 transcripts in cells grown on MMA. The sequence determinants for what appears to be an efficient terminator are present downstream of mtmB, as are putative promoter sites in front of mtmP. However, a monocistronic mtmP transcript was not observed. The ratio of the two mtm transcripts could be controlled by either RNA processing or antitermination events. Multiple transcripts from a methanogen gene cluster are not uncommon. Two transcripts are made from the fdhCAB operon of Methanobacterium thermoformicicum Z-254 (35), and multiple transcripts are made from the cdhABCDE operon of Methanosarcina thermophila (33). The relative ratios of the mtmCBP and mtmCB transcripts may reflect the need for an MMA transporter relative to the enzymes which initiate MMA metabolism.
To our knowledge, the 447-nucleotide leader sequences of the mtmCB and mtmCBPorf1 transcripts are the longest known for a methanogen. However, relatively long untranslated sequences appear to be a property of Methanosarcina transcripts encoding primary catabolic proteins. A leader of 370 bp was documented in the transcript encoding carbon monoxide dehydrogenase from M. thermophila (46), and a 296-bp leader begins the mtaCB transcript from M. barkeri (43).
Earlier it was suggested that the corrinoid proteins involved in methanogenesis from all classes of methylotrophic substrates would be homologous to the B12 binding domain of methionine synthase (38). This has been demonstrated for methylated thiols (38, 48), methanol (43), and now a methylamine. In essence, these corrinoid proteins act in methyl transfer as ferredoxins do in electron transfer, in that they are proteins of just sufficient size to bind the appropriate prosthetic group and act as methyl carriers between two different methyltransferases. Corrinoid-binding proteins homologous to the methanogen proteins have been found in the recently sequenced genomes of Methanobacterium thermoautotrophicum (45) and Archaeoglobus fulgidus (27). Although the function in these organisms is obscure, it is highly probable that these small corrinoid proteins will also act as methyl carriers, interacting with specific methyltransferases.
The sequence of mtmB contains an in-frame amber codon approximately midway through the predicted full-length coding sequence. The 52-kDa MMAMT polypeptide is one of the most predominate proteins in sodium dodecyl sulfate-polyacrylamide gels of MMA-grown cell extracts, and the predicted 23-kDa truncated product of the UAG-interrupted mtmB gene is not noticeably abundant (6). This raises the question as to how the ability of the UAG codon to direct translational termination is suppressed during MMAMT synthesis. We have obtained preliminary data that the mtmB transcript is not processed, indicating that termination is suppressed at the translational level. Natural populations of E. coli often contain amber suppressor tRNAs (32), and normal stop codon readthrough can occur at frequencies of 10−2 to 10−5 (37); neither event occurs at a frequency high enough to explain the abundant full-length mtmB product. Some protists use UAG to encode glutamine (20), but this would predict a fairly high occurrence of UAG in M. barkeri genes, which is not observed. The UAG codon could be bypassed by translational hopping (53) or a +1 or −1 frameshift followed by a compensating frameshift (14) which maintains the same reading frame. The UAG codon of mtmB might also direct the cotranslational insertion of a modified amino acid into MtmB, analogous to the well-known UGA-directed incorporation of selenocysteine (57, 58).
Members of the order Methanosarcinales are unique in having evolved the proteins to mediate methanogenesis from methylotrophic substrates. Molecular analysis of these proteins has shown that M. barkeri acquired this expanded substrate capability by adaptation of genes to new functions, followed by their subsequent duplication and dedication to specific methylotrophic pathways. The three methylotrophic methylcobamide:CoM methylases (MT2-type proteins) are specific for different substrates, but all have been noted to share an ancestor with uroporphyrinogen decarboxylase (23, 30, 38). The sequenced methyltrophic methanogenic corrinoid proteins are also specific for different substrates (38, 43, this work) but all share ancestory with the B12-binding domain of methionine synthase (31). Even transport of a methylotrophic substrate (MMA by MtmP) may be mediated by a protein most closely related to a Rhodococcus permease. It thus seems that the peculiar success of Methanosarcina spp. in adapting to methylotrophic growth is the result of recruitment of existing protein families to new functions and grafting them to the otherwise ancient process of methanogenesis.
ACKNOWLEDGMENTS
We thank D. J. Ferguson, Carey James, and Jack Presley for assistance on N-terminal sequences, and our colleagues at OSU Microbiology for their many helpful discussions and insights.
This work was supported by a grant from the Department of Energy (DE-FG02-91ER200042).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Boone D R, Whitman W B, Rouvière P. Methanogenesis. Ecology, physiology, biochemistry, and genetics. New York, N.Y: Chapman & Hall; 1993. Diversity and taxonomy of methanogens; pp. 35–80. [Google Scholar]
- 3.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodeck A, Scott J L, Georghagen N S M, Weidman J F, Fuhrman J L, Nguyen D, Utterback T R, Kelly J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C F, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 4.Burgett S G, Rosteck P R. Use of dimethylsulfoxide in fluorescent tag sequencing. In: Adams M D, Fields C, Venter J C, editors. Automated DNA sequencing and analysis. San Diego, Calif: Academic Press; 1994. pp. 211–215. [Google Scholar]
- 5.Burke S A, Krzycki J A. Involvement of the “A” isozyme of methyltransferase II and the 29-kilodalton corrinoid protein in methanogenesis from monomethylamine. J Bacteriol. 1995;177:4410–4416. doi: 10.1128/jb.177.15.4410-4416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke S A, Krzycki J A. Reconstitution of monomethylamine:coenzyme M methyl transfer with a corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J Biol Chem. 1997;272:16570–16577. doi: 10.1074/jbc.272.26.16570. [DOI] [PubMed] [Google Scholar]
- 7.Daas P J H, Gerrits K A A, Keltjens J T, van der Drift C, Vogels G D. Involvement of an activation protein in the methanol:2-mercaptoethanesulfonic acid methyltransferase reaction in Methanosarcina barkeri. J Bacteriol. 1993;175:1278–1283. doi: 10.1128/jb.175.5.1278-1283.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daas P J H, Wassenaar R W, Willemsen P, Theunissen R J, Keltjens J T, van der Drift C, Vogels G D. Purification and properties of an enzyme involved in the ATP-dependent activation of the methanol:2mercaptoethanesulfonic acid methyltransferase reaction in Methanosarcina barkeri. J Biol Chem. 1996;271:22339–22345. doi: 10.1074/jbc.271.37.22339. [DOI] [PubMed] [Google Scholar]
- 9.Daniels L. Biochemistry of methanogenesis. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of archaea. New York, N.Y: Elsevier; 1994. pp. 41–112. [Google Scholar]
- 10.Dawson R M C, Elliot D C, Elliot W H, Jones K M. Data for biochemical research. New York, N.Y: Oxford University Press; 1986. pp. 24–25. [Google Scholar]
- 11.De Mot R, Nagy I, Schoofs G, Vanderleyden J. Sequence of a Rhodococcus gene cluster encoding the subunits of ethanolamine ammonia-lyase and an APC-like permease. Can J Microbiol. 1994;40:403–407. doi: 10.1139/m94-066. [DOI] [PubMed] [Google Scholar]
- 12.Drennan C L, Huang S, Drummond J T, Matthews R G, Ludwig M L. How a protein binds B12: a 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 13.Fabiny J M, Jayakumar A, Chinault A C, Barnes E M., Jr Ammonium transport in Escherichia coli: localization and nucleotide sequence of the amtA gene. J Gen Microbiol. 1991;137:983–989. doi: 10.1099/00221287-137-4-983. [DOI] [PubMed] [Google Scholar]
- 14.Farabaugh P J. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg A P, Vogelstein B. Addendum: a technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson D J, Jr, Krzycki J A. Reconstitution of trimethylamine-dependent coenzyme M methylation with the trimethylamine corrinoid protein and the isozymes of methyltransferase II from Methanosarcina barkeri. J Bacteriol. 1997;179:846–852. doi: 10.1128/jb.179.3.846-852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson, D. J., and J. A. Krzycki. Unpublished data.
- 19.Ferguson D J, Krzycki J A, Grahame D A. Specific roles of methylcobamide:coenzyme M methyltransferase isozymes in metabolism of methanol and methylamines in Methanosarcina barkeri. J Biol Chem. 1996;271:5189–5194. doi: 10.1074/jbc.271.9.5189. [DOI] [PubMed] [Google Scholar]
- 20.Fox T D. Natural variation in the genetic code. Annu Rev Genet. 1987;21:67–91. doi: 10.1146/annurev.ge.21.120187.000435. [DOI] [PubMed] [Google Scholar]
- 21.Gonnet G H. New algorithms for the computation of evolutionary phylogenetic trees. In: Suhai S, editor. Computational methods in genome research. New York, N.Y: Plenum Press; 1994. [Google Scholar]
- 22.Grahame D A. Different isozymes of methylcobalamin:2-mercaptoethanesulfonate methyltransferase predominate in methanol-versus acetate-grown Methanosarcina barkeri. J Biol Chem. 1989;264:12890–12894. [PubMed] [Google Scholar]
- 23.Harms U, Thauer R K. Methylcobalamin:coenzyme M methyltransferase isoenzymes MtaA and MtbA from Methanosarcina barkeri. Eur J Biochem. 1996;235:653–659. doi: 10.1111/j.1432-1033.1996.00653.x. [DOI] [PubMed] [Google Scholar]
- 24.Henikoff S, Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hennigan A N, Reeve J N. mRNAs in the methanogenic archaeon Methanococcus vannielii: numbers, half-lives, and processing. Mol Microbiol. 1994;11:655–670. doi: 10.1111/j.1365-2958.1994.tb00344.x. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann R, Stoffel W. TMbase-a database of membrane-spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 27.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenny K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 28.Kremer J D, Cao X, Krzycki J. Isolation of two novel corrinoid proteins from acetate-grown Methanosarcina barkeri. J Bacteriol. 1993;175:4824–4833. doi: 10.1128/jb.175.15.4824-4833.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle R F. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:7217–7221. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Leclerc G M, Grahame D A. Methylcobamide:coenzyme M methyltransferase isozymes from Methanosarcina barkeri: physicochemical characterization, cloning, sequence analysis, and heterologous gene expression. J Biol Chem. 1996;271:18725–18731. doi: 10.1074/jbc.271.31.18725. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig M L, Matthews R G. Structure-based perspectives on B12-dependent enzymes. Annu Rev Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 32.Marshall B, Levy S B. Prevalence of amber suppressor-containing coliforms in the natural environment. Nature. 1980;286:524–525. doi: 10.1038/286524a0. [DOI] [PubMed] [Google Scholar]
- 33.Maupin-Furlow J A, Ferry J G. Analysis of the CO dehydrogenase/acetyl-coenzyme A sythase operon of Methanosarcina thermophila. J Bacteriol. 1996;178:6849–6856. doi: 10.1128/jb.178.23.6849-6856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1997;25:244–245. doi: 10.1093/nar/25.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nölling J, Reeve J N. Growth- and substrate-dependent transcription of the formate dehydrogenase (fdhCAB) operon in Methanobacterium thermoformicicum Z-245. J Bacteriol. 1997;179:899–908. doi: 10.1128/jb.179.3.899-908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer J R, Daniels C J. In vivo definition of an archael promoter. J Bacteriol. 1995;177:1844–1849. doi: 10.1128/jb.177.7.1844-1849.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1985;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul L, Krzycki J A. Sequence and transcript analysis of a novel Methanosarcina barkeri methyltransferase II homolog and its associated corrinoid protein homologous to methionine synthase. J Bacteriol. 1996;178:6599–6607. doi: 10.1128/jb.178.22.6599-6607.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeve J N. Molecular biology of methanogens. Annu Rev Microbiol. 1992;46:165–191. doi: 10.1146/annurev.mi.46.100192.001121. [DOI] [PubMed] [Google Scholar]
- 40.Reizer J, Finley K, Kakuda K, MacLeod C L, Reizer A, Saier M H., Jr Mammalian integral membrane receptors are homologous to facilitators and antiporters of yeast, fungi, and eubacteria. Protein Sci. 1993;2:20–30. doi: 10.1002/pro.5560020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. p. 1626. [Google Scholar]
- 42.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauer K, Harms U, Thauer R K. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri. Purification, properties and encoding genes of the corrinoid protein MT1. Eur J Biochem. 1997;243:670–677. doi: 10.1111/j.1432-1033.1997.t01-1-00670.x. [DOI] [PubMed] [Google Scholar]
- 44.Siewe R M, Weil B, Burkovski A, Eikmanns B J, Eikmanns M, Kramer R. Functional and genetic characterization of the (methyl)ammonium uptake carrier of Corynebacterium glutamicum. J Biol Chem. 1996;271:5398–5403. doi: 10.1074/jbc.271.10.5398. [DOI] [PubMed] [Google Scholar]
- 45.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qui D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sowers K R, Thai T T, Gunsalus R P. Transcriptional regulation of the carbon monoxide dehydrogenase gene (cdhA) in Methanosarcina thermophila. J Biol Chem. 1993;268:23172–23178. [PubMed] [Google Scholar]
- 47.Tallant T, Krzycki J. Coenzyme M methylase activity of the 480-kilodalton corrinoid protein from Methanosarcina barkeri. J Bacteriol. 1996;178:1295–1301. doi: 10.1128/jb.178.5.1295-1301.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tallant T C, Krzycki J A. Methylthiol:coenzyme M methyltransferase from Methanosarcina barkeri, an enzyme of methanogenesis from dimethylsulfide and methylmercaptopropionate. J Bacteriol. 1997;179:6902–6911. doi: 10.1128/jb.179.22.6902-6911.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Meijden P, Heythuysen H J, Pouwels A, Houwne F, van der Drift C, Vogels G D. Methyltransferases involved in methanol conversion by Methanosarcina barkeri. Arch Microbiol. 1983;134:238–242. doi: 10.1007/BF00407765. [DOI] [PubMed] [Google Scholar]
- 51.van der Meijden P, te Brömmelstroet B W, Poirot C M, van der Drift C, Vogels G D. Purification and properties of methanol:5-hydroxybenzimidazolylcobamide methyltransferase from Methanosarcina barkeri. J Bacteriol. 1984;160:629–635. doi: 10.1128/jb.160.2.629-635.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wassenaar R W, Daas P J H, Geerts W J, Keltjans J T, van der Drift C. Involvement of methyltransferase-activating protein and methyltransferase 2 isozyme II in methylamine:coenzyme M methyltransferase reactions in Methanosarcina barkeri Fusaro. J Bacteriol. 1996;178:6937–6944. doi: 10.1128/jb.178.23.6937-6944.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss R B. Ribosomal frameshifting, jumping, and readthrough. Curr Opin Cell Biol. 1991;3:1051–1055. doi: 10.1016/0955-0674(91)90128-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woese C R. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeliseev A, Gaertner P, Harms U, Linder D, Thauer R K. Function of methylcobalamin:coenzyme M methyltransferase isoenzyme II in Methanosarcina barkeri. Arch Microbiol. 1993;159:530–536. doi: 10.1007/BF00249031. [DOI] [PubMed] [Google Scholar]
- 56.Zillig W, Palm P, Klenk H-P, Langer D, Hudepohl U, Hain J, Landendorfer M, Holz I. Transcription in the archaea. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of archaea (Archaeabacteria). New York, N.Y: Elsevier Science Publishing; 1993. pp. 367–391. [Google Scholar]
- 57.Zionini F, Birkmann A, Leinfelder W, Bock A. Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UAG codon. Proc Natl Acad Sci USA. 1987;84:3156–3160. doi: 10.1073/pnas.84.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zionini F, Birkmann A, Stadtman T C, Bock A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci USA. 1986;83:4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]