Abstract
The gene for a novel endotype membrane-bound lytic transglycosylase, emtA, was mapped at 26.7 min of the E. coli chromosome. EmtA is a lipoprotein with an apparent molecular mass of 22 kDa. Overexpression of the emtA gene did not result in bacteriolysis in vivo, but the enzyme was shown to hydrolyze glycan strands isolated from murein by amidase treatment. The formation of tetra- and hexasaccharides, but no disaccharides, reflects the endospecificity of the enzyme. The products are characterized by the presence of 1,6-anhydromuramic acid, indicating a lytic transglycosylase reaction mechanism. EmtA may function as a formatting enzyme that trims the nascent murein strands produced by the murein synthesis machinery into proper sizes, or it may be involved in the formation of tightly controlled minor holes in the murein sacculus to facilitate the export of bulky compounds across the murein barrier.
Lysozymes are widely distributed among living organisms, including both pro- and eukaryotes (29). They cleave the β-1,4-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine in the murein (peptidoglycan) sacculus of bacteria. By degrading the sacculus, which serves as a bacterial exoskeleton (37, 43, 51), they cause bacteriolysis. Hence, lysozymes are among the most powerful antibacterial enzymes.
Despite their potential bacteriolytic power, lysozymes and other murein hydrolyzing enzymes, including glucosaminidases, amidases, and endopeptidases, are ubiquitous in bacteria themselves (43). This is due to the net-like structure of murein, which can only be enlarged by cleaving meshes to allow the insertion of new building blocks into the preexisting sacculus (23, 24, 43, 48, 51). Thus, murein hydrolases are likely to be involved in the enlargement and division of the sacculus during bacterial growth. In addition, murein hydrolases are believed to be needed for a localized opening of the murein net during the export of bulky compounds such as DNA, toxins, flagella, and fimbrial proteins (13).
The muramidases found in Escherichia coli differ from lysozymes in that they catalyze an intramolecular transglycosylation onto the C6 hydroxyl group following cleavage of the β-1,4-glycosidic bond. As a result of this reaction the products are marked by a 1,6-anhydromuramic acid (22, 45). The meaning of this peculiar reaction mechanism is still not understood. In a way similar to the action of lysozyme, murein is completely degraded by these enzymes. They are therefore called lytic transglycosylases. Four different proteins with this activity have been identified in E. coli (12, 39). Three of them, MltA, MltB, and MltC, each with a mass of about 38 kDa, are lipoproteins residing in the outer membrane (12, 15, 32). The fourth, Slt70, is a soluble enzyme with a mass of 70 kDa (6, 22). Although not a lipoprotein, Slt70 preferentially binds to the outer face of the murein sacculus (50). Hence, a specific topological localization of these enzymes in the cell wall is realized.
Unlike lysozyme, these lytic transglycosylases are exoglycosylases, which by starting at one end processively degrade murein strands releasing anhydrodisaccharide units. Unfortunately, it is not clear at which end these enzymes start. The crystal structure of the Slt70 molecule suggests that binding occurs at the 1,6-anhydromuramic acid end (47), whereas biochemical data show that the enzymes start at the nonreducing end, i.e., the N-acetylglucosamine terminus of the polysaccharide (4, 39), and release their products in the form of anhydromuropeptides. If true, the latter reaction mechanism raises a question: where are the anhydro groups present in the murein sacculus (18, 24) coming from?
Here we show that a lytic endotransglycosylase exists in E. coli that, by cleaving glycosidic bonds within the murein strands, produces shorter strands with 1,6-anhydromuramic acid ends in the murein sacculus.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and plasmids.
E. coli MC1061 (10) and the mltA mutant LT12 (mltA::cat) (32) were used as indicated below. The general cloning host was E. coli XL1-Blue (Stratagene, La Jolla, Calif.). Phage λ245 (7C10) from the Kohara phage library (30) was used as a DNA source for cloning the gene of interest. In order to construct an inducible expression system, the gene was cloned into plasmid pJFK118EH (7).
Media and growth conditions.
The bacteria were cultivated aerobically at 30°C in Luria-Bertani (LB) medium (34). The medium was supplemented when required with antibiotics at the following concentrations (per milliliter): kanamycin, 50 μg; tetracycline, 12.5 μg; and chloramphenicol, 20 μg. For strains carrying pAK5, overnight cultures and plates were supplemented with 0.2% glucose and 10 mM MgCl2.
DNA manipulations and PCR.
Standard techniques were used for manipulating the plasmid DNA (41), and E. coli was transformed as described by Inoue et al. (26). Restriction endonucleases were purchased from Boehringer (Mannheim, Germany), shrimp alkaline phosphatase was obtained from U.S. Biochemicals (Cleveland, Ohio), and the oligonucleotides came from Pharmacia (Uppsala, Sweden) or MWG-Biotech (Ebersberg, Germany).
PCR (40) was performed in a volume of 50 μl of 20 mM Tris-HCl (pH 8.8) containing 2 mM MgSO4, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100, 100 μg of bovine serum albumin per ml, 400 μM deoxynucleoside triphosphates, and 1.25 U of Pfu polymerase (Stratagene) with 0.5 μM concentrations of each primer and 2 μl of Kohara lambda phage 245 lysate (kindly provided by Guido Schiffer) as a template. After initial denaturation for 3 min at 92°C, touchdown PCR (14) was performed with 0.5 min of annealing, 1 min of extension at 72°C, and 0.5 min of denaturation at 92°C. The annealing temperatures were 65, 63, 61, 59, and 57°C for 3 cycles each and finally 55°C for another 15 cycles.
Construction of an inducible expression system.
The gene encoding the lytic endotransglycosylase was cloned into the plasmid pJFK118EH (7) to get an inducible expression system under the control of the tac promoter. The coding region was amplified from Kohara phage λ245 DNA by touchdown PCR by using the primers 5′-TAAGAATTCAAGAAATGAAATTGAGATGGTTTGCC-3′ and 5′-GAAGTCGACGCGCGACTGATTTACATCG-3′, introducing an EcoRI restriction site and a SalI restriction site at the ends. The first primer changed the start codon of the amplified coding sequence from the chromosomally encoded GTG to ATG. The amplified product was purified and digested with EcoRI and SalI and ligated into pJFK118EH cut with these enzymes and dephosphorylated with shrimp alkaline phosphatase. The resulting construct, pAK5, was amplified in E. coli XL1-Blue and transformed into E. coli LT12 for biochemical characterization of the protein overproduced upon induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
Preparation of membrane extracts.
To prepare membrane extracts, cells carrying either pJFK118EH or pAK5 were grown to an optical density at 578 nm (OD578) of 0.2 as described above. The expression of the cloned gene was induced by the addition of 1 mM IPTG, and the cells were cultivated for an additional 4 h. Cells from a 25-ml culture were harvested by centrifugation, washed with 1 ml of ice-cold buffer A (10 mM Tris-maleate, pH 6.8; 10 mM MgCl2; 0.1 mM dithioerythritol), and resuspended in 150 μl of buffer A containing 20 mg of DNase I (Boehringer). After breakage of the cells by shaking them with glass beads at 4°C, the beads were removed by low-speed centrifugation and the suspension was diluted with 800 μl of buffer A. The membranes were sedimented by ultracentrifugation (300,000 × g, 20 min, 4°C), extracted with 1 ml of buffer A containing 1 M NaCl for 90 min at 4°C, and sedimented again as described above. The salt-washed membranes were then resuspended in buffer A containing 500 mM NaCl and 2% Triton X-100 and extracted three more times at 4°C with a total volume of 1.4 ml of this buffer. The pooled Triton X-100 extracts were used for enzyme assays.
Murein hydrolase assays.
In one assay, isolated murein glycan chains radiolabeled with N-acetyl-d-1-[3H]glucosamine (259 GBq/mmol; Amersham) were used as a substrate to determine lytic transglycosylase activity (49). The glycan chains were separated by reversed-phase high-pressure liquid chromatography (HPLC) (20), and the fractions containing glycans with a defined length were desalted by gel filtration chromatography on Biogel P2 (Bio-Rad Laboratories, Hercules, Calif.). Membrane extracts were incubated with labeled glycan chains (10,000 cpm) in 110 μl of 10 mM Tris-maleate (pH 6.8) containing 10 mM MgCl2 and 0.2% Triton X-100 at 30°C for various times. The reaction was stopped by the addition of 4 μl of 2% phosphoric acid, and the samples were boiled for 3 min. After the addition of 55 μl of 100 mM sodium phosphate (pH 2.0), insoluble material was removed by centrifugation (13,000 × g, 5 min, 4°C), and the breakdown products were separated by reversed-phase HPLC as described by Harz et al. (20). Radioactivity in the eluate was detected with a Flo-One/Beta liquid scintillation counter (Canberra Packard, Frankfurt, Germany). The breakdown products were identified by comparison with standards of HPLC-separated murein glycan chains (20).
In a more general murein hydrolase assay, the enzyme activity was determined with murein sacculi (10,000 cpm, 5 μg) radiolabeled with meso-2,6-diamino-3,4,5-[3H]pimelic acid (18.5 MBq/mmol; CEA, Gif-sur-Yvette, France) as a substrate (31). The same buffer as described for the assay with glycan strands was used.
Zymogram analysis.
The detection of cell wall hydrolase activities after separation of the proteins by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed essentially as described by Potvin et al. (36). The SDS–12% polyacrylamide gel contained 0.2% (wt/vol) lyophilized Micrococcus lysodeikticus cells (Sigma, St. Louis, Mo.) and only 0.03% SDS. Prior to PAGE, the samples were boiled for 7 min in sample buffer (100 mM Tris-HCl, pH 6.8; 1% SDS; 6% sucrose; 10 mM dithioerythritol; 0.0025% bromophenol blue) (5). After PAGE, the gel was briefly rinsed with water and soaked for 30 min each with water and renaturation buffer (25 mM sodium phosphate [pH 7.0] containing 10 mM MgCl2 and 0.1% Triton X-100) with gentle agitation at room temperature. The gel was incubated overnight in 250 ml of renaturation buffer at 30°C and for another 10 h at 37°C. To obtain better contrast, the sacculi were stained with 0.1% methylene blue in 0.01% KOH as described by Bernadsky et al. (5). After they were destained with water, the unstained autolytic bands appeared in the blue background.
Palmitate labeling.
To label the lipoproteins, cells were grown aerobically in LB medium in the presence of 5 μCi of [3H]palmitate per ml (54 Ci/mmol; Amersham Buchler) from an OD578 of 0.08 to an OD578 of 0.4. Expression of the lytic endotransglycosylase was induced for 30 min by the addition of 1 mM IPTG in the presence or absence of 100 μg of globomycin per ml (Sankyo Co., Ltd., Tokyo, Japan) (25). Cells from a 10-ml culture were harvested by centrifugation, washed twice with 1 ml of ice-cold phosphate-buffered saline, and lysed by boiling in 120 μl of 20 mM Tris-HCl (pH 8.0)–1 mM EDTA–1% SDS for 10 min. After centrifugation (13,000 × g, 10 min, 4°C), the proteins in the supernatant were precipitated by the addition of 1.2 ml of ice-cold acetone, left overnight at 4°C, and pelleted by centrifugation as described above. The protein pellet was thoroughly resuspended in 50 μl of 1% SDS, and aliquots corresponding to 106 cpm were separated by SDS–15% PAGE (33). After electrophoresis, the gel was fixed overnight in 10% acetic acid, soaked in Amplify (Amersham Buchler), and dried. The labeled proteins were detected by fluorography with Hyperfilm MP (Amersham Buchler).
Murein structure analysis.
Murein sacculi were prepared and hydrolyzed with cellosyl (kindly provided by Hoechst-Roussel AG, Frankfurt, Germany) (17, 18). The resulting muropeptides were reduced with borohydride and fractionated by reversed-phase HPLC as described by Glauner (17).
RESULTS
Indication for an unknown membrane-bound lytic transglycosylase.
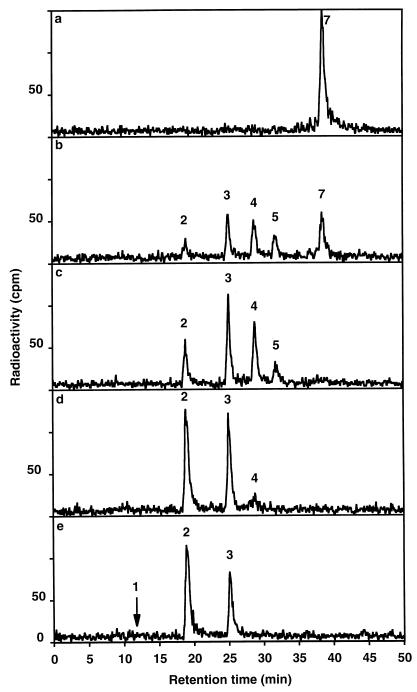
Of the four studied lytic transglycosylases of E. coli, Slt70, MltA, MltB, and MltC (24), only MltA accepts unsubstituted murein glycan chains as a substrate (39). Nevertheless, membrane extracts of an mltA mutant, E. coli LT12 (32), still showed significant enzymatic activity in degrading isolated murein glycan strands (Fig. 1a and c). As all resulting reaction products contained a 1,6-anhydromuramic acid chain end, the degrading enzyme must be a lytic transglycosylase. Partial chromatographic purification of this lytic transglycosylase activity resulted in the enrichment of a protein showing an apparent molecular mass of 22 kDa as determined by SDS-PAGE (data not shown). The isolated protein could not be sequenced N terminally, most probably due to the lack of a free amino terminus. As shown below, the protein indeed turned out to be a lipoprotein with its N-terminal cysteine residue replaced by a lipid moiety.
FIG. 1.
Murein glycan chain-degrading activities in membrane extracts of an mltA mutant. E. coli LT12 harboring pAK5 (b) or pJFK118EH as a control (a and c) were induced by the addition of 1 mM IPTG, and membrane extracts were prepared as described in Materials and Methods. Extracts corresponding to 1 μl (a and b) or 100 μl (c) of culture were incubated for 30 min at 30°C with radiolabeled murein glycan chains with a length of seven disaccharide units terminating with a 1,6-anhydromuramic acid residue. The breakdown products, all terminating with 1,6-anhydromuramic acid, were analyzed by reversed-phase HPLC. The numbers 2 to 7 indicate the degree of polymerization in the disaccharide units. Thus, for example, the “3” indicates a 1,6-anhydrohexasaccharide consisting of (GlcNAc-MurNAc)2–GlcNAc-(1,6-anhydro)MurNAc.
Identification and cloning of the gene encoding a novel lytic transglycosylase.
The indication for the presence of an unknown lytic transglycosylase prompted us to perform database searches. First, the protein sequences of the characterized lytic transglycosylases MltC and Slt70 were compared, and a sequence motif was defined by using the Block Maker program (1). This motif was used to search the complete genomic sequence of E. coli (8) translated into all possible reading frames. One score in the 26.7-min region of the genome seemed to be particularly interesting. A potential open reading frame containing the defined motif was identified. The possible protein would start with a signal peptide followed by a typical lipid modification site (9). The gene was thus expected to code for a lipoprotein with a calculated molecular mass of 21 kDa, which is consistent with the information obtained from the partial purification.
Therefore, the coding region was amplified by PCR and cloned behind the tac promoter of the pBR322 derivative pJFK118EH (7), resulting in plasmid pAK5. Induction of expression of the cloned gene in a mutant with a deletion of mltA (LT12) gave rise to a dramatic increase in glycan chain-degrading activity in membrane extracts (Fig. 1a and b). Therefore, the cloned gene encodes a novel membrane-bound lytic transglycosylase of E. coli.
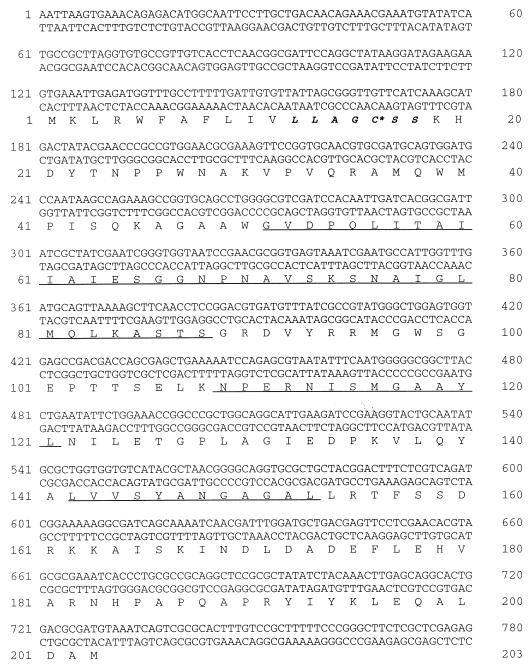
Nucleotide sequence analysis.
The novel lytic transglycosylase is encoded by an open reading frame of 609 bp starting with a GTG codon (Fig. 2). The coding sequence is found inside the reading frame o241 (GenBank entry AE000217), which predicts a 38-amino-acid-longer protein. Like the genes for MltA (32), MltB (15), and MltC (12), the coding region starts with a signal peptide followed by a lipoprotein processing site (9) and therefore is expected to encode a lipoprotein with the calculated molecular mass of 21 kDa. The residue following the modified cysteine of the bacterial lipoproteins determines the localization of the lipoprotein (52); the serine found in this protein predicts a localization in the outer membrane of E. coli. The deduced amino acid sequence shows significant similarity to other lytic transglycosylases. An alignment of protein sequences which score high in a database search (PSI-Blast algorithm, e values of <10−30 after one iteration) that was prepared with the BLOCKS program shows the conservation of three sequence motifs. These stretches (see Fig. 2) are actually found in the active center of the lytic transglycosylases, as shown for the Slt70 by X-ray crystallography (47).
FIG. 2.
Nucleotide sequence of the emtA gene and derived amino acid sequence. The consensus sequence of the lipoprotein processing site is shown in bold italics, with an asterisk indicating the modified cysteine. Regions of high similarity to other known lytic transglycosylases are underlined (12, 35).
Controlled overexpression of the cloned lytic transglycosylase.
Controlled overexpression of the novel lytic transglycosylase was achieved by the addition of 1 mM IPTG to E. coli LT12 harboring pAK5. Induction of expression at an OD578 of 0.2 caused only a negligible reduction in the growth rate compared to cells carrying the control vector pJFK118EH. Prolonged overexpression (2 to 4 h) led to the accumulation of chains of cells unable to separate properly. Overnight cultures of even uninduced E. coli XL1-Blue carrying pAK5 showed a slight tendency to lyse, probably due to the leakiness of the tac promoter. Therefore, all overnight cultures and plates were supplemented with 0.2% glucose to suppress expression of the cloned gene, and 10 mM MgCl2 was added to stabilize those cells that started to undergo lysis.
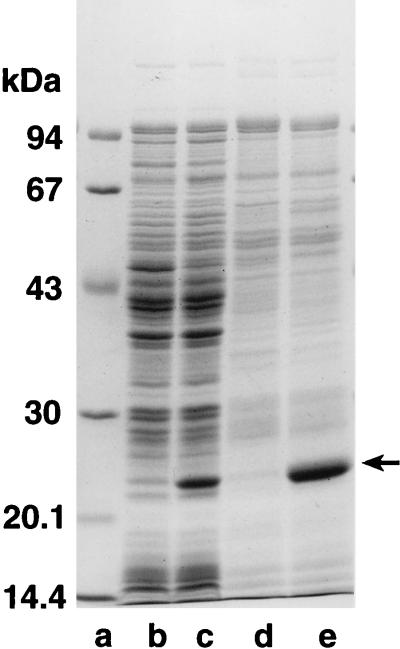
Analysis by SDS-PAGE of the cellular proteins in cells induced for the expression of the cloned gene revealed the appearance of a protein band with an apparent molecular mass of 22 kDa. As shown in Fig. 3, this band represents the major protein in Triton X-100 extracts of salt-washed membranes from induced cells. This finding is well in accordance with both the apparent molecular mass of the partially purified novel lytic transglycosylase and the size predicted from the sequence of the cloned gene.
FIG. 3.
Expression of the cloned lytic transglycosylase gene. E. coli LT12 harboring pJFK118EH (lanes b and d) or pAK5 (lanes c and e) was induced for 4 h by the addition of 1 mM IPTG. Cell homogenates (lanes b and c) or membrane extracts (lanes d and e) were separated by SDS–12% PAGE and stained with Coomassie brilliant blue. Lane a shows molecular mass markers. The position of the cloned lytic transglycosylase is indicated by an arrow.
Demonstration of the lipoprotein character of EmtA.
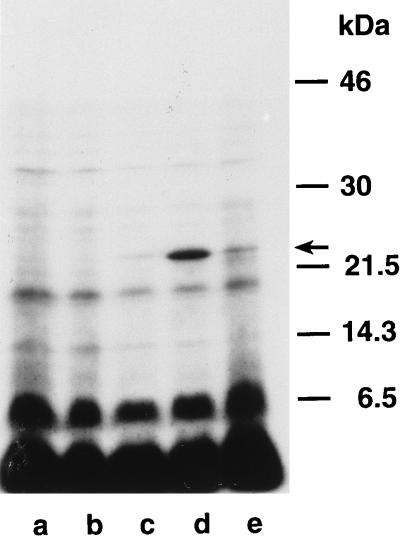
To test whether EmtA is indeed lipid modified, as indicated by the presence of a prokaryotic lipoprotein signal sequence and the fact that the protein isolated from the wild type could not be N-terminally sequenced (see above), the protein was expressed in cells grown in the presence of [3H]palmitate. After separation of the proteins by SDS-PAGE, a labeled band running at the appropriate molecular weight was detected in induced cells expressing EmtA but was hardly visible in cells harboring pJFK118EH (Fig. 4). The intensity of this band was greatly reduced when expression of the cloned gene was induced in the presence of globomycin, an inhibitor of signal peptidase II (25). No labeled band corresponding to a protein with a higher molecular mass could be detected, as one would expect in the presence of globomycin. Therefore, we assume that the unprocessed prolipoprotein is a rather unstable intermediate.
FIG. 4.
[3H]palmitate labeling of overexpressed EmtA. E. coli LT12 harboring pJFK118EH (lanes a and b) or pAK5 (lanes c to e) were labeled by growth in the presence of [3H]palmitate (5 mCi/ml). Cells were induced for 30 min by the addition of 1 mM IPTG (lanes b, d, and e). One culture (lane e) received 100 μg of globomycin per ml to inhibit the processing of lipoproteins. Separation of the proteins by SDS–15% PAGE and fluorography was performed as described in Materials and Methods. Molecular masses of prestained marker proteins and the position of EmtA are indicated on the right.
Endospecificity of the novel lytic transglycosylase.
Membrane extracts from induced cells of LT12(pJFK118EH) and LT12(pAK5) were used to further characterize the novel lytic transglycosylase activity. Studies on the kinetics of degradation of isolated murein glycan chains (Fig. 5) revealed that the enzyme preferentially cleaves at a distance of more than two disaccharide units from the ends of the glycan chain (it also cleaves at a distance of two disaccharide units, producing the anhydrotetrasaccharide, but prefers cleaving farther away from the ends of glycan chains). The end products of a complete glycan chain degradation by this lytic transglycosylase were fragments with lengths of two and three disaccharide units. Unlike MltA (49), an exospecific lytic transglycosylase, EmtA does not produce monomeric anhydrodisaccharide fragments. This indicates that the novel lytic transglycosylase is an endoglycosylase. We therefore propose to name it EmtA, for endo-type membrane-bound lytic transglycosylase.
FIG. 5.
Kinetics of glycan chain degradation by the lytic endotransglycosylase. E. coli LT12(pAK5) was induced by the addition of 1 mM IPTG, and membrane extracts were prepared as described in Materials and Methods. Extracts corresponding to 1 μl (b and c) or 100 μl (d and e) of culture were incubated with radiolabeled murein glycan chains with a length of seven disaccharide units terminating with 1,6-anhydromuramic acid (a) for 5 min (b and d) or 30 min (c and e) at 30°C. The breakdown products were analyzed by reversed-phase HPLC. Numbers 1 to 7, (GlcNAc-MurNAc)0–6–GlcNAc-(1,6-anhydro)MurNAc (as described in Fig. 1).
Changes in the murein structure upon overproduction of EmtA.
The morphological alterations observed when EmtA was overproduced (see above) point to a change in the structure of the murein sacculus. Indeed, analysis of the muropeptide composition of E. coli MC1061(pAK5) revealed a number of changes when noninduced cells were compared with cells overproducing EmtA (Table 1). Two major effects were seen. First, there was a threefold increase in all d,l-A2pm-A2pm cross-links and an increase in the relative amount of anhydromuropeptides, indicating a shortening of the glycan strands. The increase in anhydromuropeptides by a factor of about 2 indicates that the glycan strands were cleaved by the overproduced lytic transglycosylase, although without inducing bacteriolysis. Interestingly, only cross-linked anhydromuropeptides were increased, whereas the relative amounts of the monomeric anhydro compounds were not changed. It seems, therefore, that in vivo the enzyme has a preference for cross-linked murein.
TABLE 1.
Muropeptide composition of E. coli MC1061(pAK5)a
| Muropeptide | Trial I
|
Trial II
|
||||
|---|---|---|---|---|---|---|
| % in noninduced cells | % in induced cells | % Change | % in noninduced cells | % in induced cells | % Change | |
| Monomers | 54.39 | 47.17 | −13.3 | 55.75 | 48.26 | −13.4 |
| Tri | 7.83 | 10.96 | 39.9 | 7.61 | 11.17 | 46.8 |
| Tetra | 40.34 | 31.71 | −21.4 | 44.21 | 34.15 | −22.8 |
| Penta | 0.04 | 0.04 | 0 | 0.12 | 0.05 | −58.3 |
| Dimers | 41.69 | 47.30 | 13.5 | 37.65 | 42.57 | 13.1 |
| Ala-A2pm | 36.78 | 32.83 | −10.7 | 34.22 | 31.91 | −6.8 |
| A2pm-A2pm | 4.82 | 13.80 | 186.3 | 3.36 | 10.48 | 211.9 |
| Trimers | 3.81 | 5.30 | 39.1 | 6.91 | 8.82 | 27.6 |
| Ala-A2pm | 3.41 | 4.22 | 23.8 | 6.42 | 7.51 | 17.0 |
| A2pm-A2pm | 0.42 | 1.13 | 169.0 | 0.52 | 1.72 | 230.7 |
| Tetramers | 0.11 | 0.23 | 109.1 | 0.18 | 0.35 | 94.4 |
| Anhydros | 3.88 | 6.23 | 61.0 | 4.25 | 7.54 | 77.4 |
| Monomers | 0.93 | 1.09 | 17.2 | 1.38 | 1.87 | 33.5 |
| Dimers | 2.48 | 4.23 | 70.6 | 2.07 | 4.41 | 113.0 |
| Trimers | 0.46 | 0.91 | 97.8 | 0.81 | 1.61 | 98.8 |
| Cross-linked muropeptides | 23.47 | 27.35 | 16.5 | 23.57 | 27.43 | 16.4 |
| Ala-A2pm | 20.89 | 19.78 | −5.3 | 21.70 | 21.80 | 0.5 |
| A2pm-A2pm | 2.55 | 7.28 | 185.5 | 1.85 | 5.81 | 214.1 |
The abbreviations used for the muropeptides are as follows: Tri, GlcNAc-β-1,4-MurNAc-l-Ala-d-Glu-m-A2pm; Tetra, GlcNAc-β-1,4-MurNAc-l-Ala-d-Glu-m-A2pm-d-Ala; Penta, GlcNAc-β-1,4-MurNAc-l-Ala-d-Glu-m-A2pm-d-Ala-d-Ala; Ala-A2pm, cross-linkage via d-Ala and m-A2pm of two different stem peptides; A2pm-A2pm, cross-linkage via m-A2pm and m-A2pm of two different stem peptides; Anhydros, all muropeptides carrying a 1,6-anhydromuramic acid. The relative amounts of the muropeptides and the degree of cross-linkage were calculated as described by Glauner (17).
The increase in the number of d,l-A2pm-A2pm cross-links was found for all cross-linked muropeptides, dimers, trimers, and tetramers. By contrast, the cross-linkage via d,d-A2pm-Ala bonds was hardly changed. Formation of A2pm-A2pm cross-links has been reported to take place prior to the onset of bacteriolysis caused by various agents and growth conditions (18, 24). Thus, it has been speculated that an increase in A2pm-A2pm cross-bridges represents a kind of defense mechanism to avoid lysis (24). Consistent with this speculation, rapid lysis, even after prolonged expression of emtA, was not observed (see above).
Activity with intact murein sacculi.
Radiolabeled murein sacculi of E. coli were used as a substrate to determine in vitro murein degrading activity in membrane extracts from induced cells of LT12(pJFK118EH) and LT12. No increase in the release of murein breakdown products could be detected upon overexpression of the lytic endotransglycosylase.
In contrast, zymogram analysis revealed a double band with autolytic activity at approximately 22 kDa that was present in cell homogenates (Fig. 6) and membrane extracts (data not shown) of cells harboring pAK5 but not in control cells harboring pJFK118EH. The main, upper band most probably represents the native protein, whereas the weaker, lower band might be a proteolytic degradation product of EmtA. This shows that EmtA, first denatured and then renatured according to the zymogram procedure, can accept murein sacculi of Micrococcus lysodeikticus as a substrate.
FIG. 6.
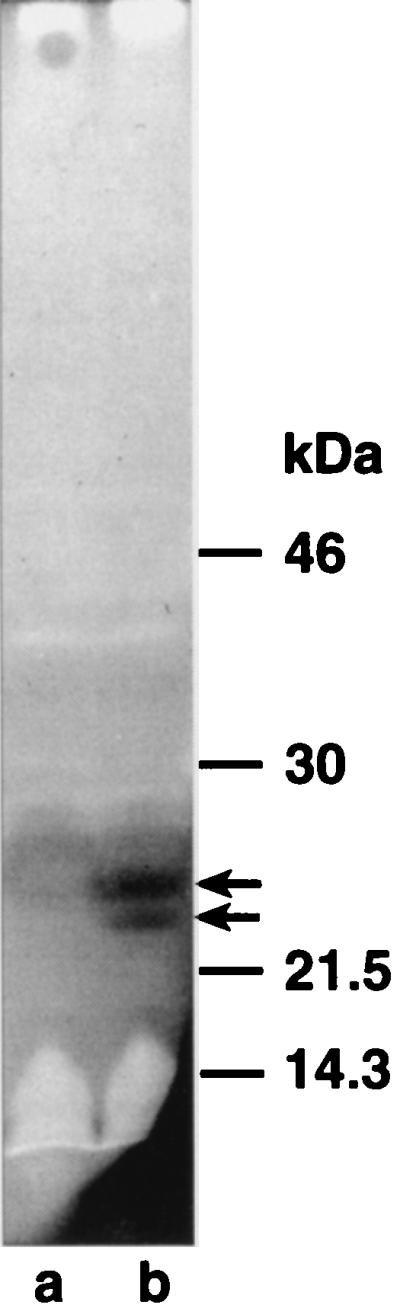
Zymogram analysis of overexpressed EmtA. E. coli LT12 harboring pJFK118EH (lane a) or pAK5 (lane b) was induced by the addition of 1 mM IPTG. Cell homogenates were separated by SDS–12% PAGE in gels containing 0.2% lyophilized cells of M. lysodeikticus. Renaturation of the separated proteins and staining of the gel were performed as described in Materials and Methods. The stained gel was scanned and inverted to obtain better contrast. Molecular masses of prestained marker proteins are indicated on the right.
DISCUSSION
Cooperation between exo- and endoenzymes is well known for the degradation of biopolymers such as chitin and cellulose (11, 16, 28). The advantage of producing more chain ends by endoenzymes for the exoenzymes to start their processive degradation process of the polymers is obvious. Therefore, it is not surprising to find that this strategy also seems to be used in the case of the cross-linked biopolymer murein.
An interesting feature of the endo-type membrane-bound lytic transglycosylase is its lipoprotein character, a feature EmtA shares with most of the other lytic transglycosylases, including MltA, MltB, and MltC (12, 15, 32). As demonstrated for MltA and MltB (15, 32), EmtA is also likely to be inserted into the outer membrane due to the presence of a serine at position 2 in the amino acid sequence (of the processed protein), which has been shown to direct lipoproteins to the outer membrane (52). It is tempting to speculate that the specific localization of the murein hydrolases in the outer membrane is a means to control these potentially suicidal enzymes. Interestingly, the synthetic enzymes, such as the bifunctional transpeptidase-transglycosylases PBP1A, PBP1B, and PBP1C, as well as the transpeptidases PBP2 and PBP3, are anchored to the cytoplasmic membrane (2, 44). Thus, hydrolases and synthases are localized in different layers of the complex cell wall. Organization of the murein hydrolases in the outer membrane is in accordance with the hypothesis of a three-for-one growth strategy of the stress-bearing murein sacculus, a hypothesis which assumes that old murein strands lined with new murein are removed from the side facing the outer membrane (23, 24).
The specific function of EmtA, however, is still uncertain. Although it cleaves isolated poly(MurNAc-GlcNAc) strands, the enzyme was unable to hydrolyze murein sacculi in vitro, and overproduction in vivo did not result in rapid bacteriolysis, although changes in the structure of the murein sacculus could be demonstrated (Table 1). It may well be that the enzyme has to cooperate with other proteins to carry out its specific function on murein sacculi, that is, on peptide-substituted glycan strands. Such a cooperation could also explain why the enzyme shows a preference for cross-linked murein in vivo. MltA, an exo-type lytic transglycosylase that, like EmtA, shows activity with unsubstituted glycan strands (49), also does not cause bacteriolysis when overproduced (32). Preliminary results suggest that MltA may cooperate with a novel amidase (46) that could supply the proper substrate, the murein glycan strands, for the lytic transglycosylase. A concerted action of two other murein specificities in E. coli has been demonstrated for the endopeptidase PBP7 and the lytic transglycosylase Slt70 (38).
Being an endo-type enzyme, EmtA would be an ideal enzyme to format the murein strands to proper sizes. As a matter of fact it is still unknown how the length distribution of the murein strands is determined (20, 24). Two main possibilities exist: either the strands are released from the bactoprenol carrier molecule by a specific termination reaction that forms a 1,6-anhydromuramic acid terminus, or the polymerization of the murein precursors by synthetic transglycosylases is a continuous reaction and the growing strands are formatted by lytic endotransglycosylases such as EmtA.
In addition to a general role in murein metabolism or even as its sole function, EmtA may be important for paving the way for bulky compounds to pass through the murein barrier. Preliminary results by Haigh and Williams at the University of Leicester, who isolated an emtA deletion mutant indicating that it is not an essential enzyme, suggest that this endoglycosylase may be involved in the production of pili in enteropathogenic E. coli strains (19). Indeed, several examples of the involvement of lytic transglycosylases in the transmurein export have been reported (3, 13, 35). The use of lytic transglycosylases rather than other specificities of murein hydrolases may have an advantage. Repair of the lesions in the murein net could make use of the energy stored in the 1,6-anhydro bond.
The strictly localized action of lytic transglycosylases in facilitating passage through the murein layer may be controlled by a specific interaction of the enzyme with other proteins of the export apparatus. This could explain the low autolytic activity found for EmtA. Studies of the control mechanisms for this specific function of murein hydrolases seem to be a promising experimental approach to gain more insights into the regulation of these potentially autolytic enzymes.
ACKNOWLEDGMENTS
We thank U. Schwarz for his interest and support and Y. Kohara for supplying the lambda phages. We are grateful to A. Ursinus for excellent assistance with muropeptide analyses. The plasmid pJFK118EH was kindly provided by R. Bishop, and globomycin was a generous gift from the Sankyo Co., Ltd., Tokyo, Japan.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbas J A, Diaz J, Rodriguez-Tebar A, Vazquez D. Specific location of penicillin-binding proteins within the cell envelope of Escherichia coli. J Bacteriol. 1986;165:269–275. doi: 10.1128/jb.165.1.269-275.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer M, Eferl R, Zellnig G, Teferle K, Dijkstra A, Koraimann G, Högenauer G. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J Bacteriol. 1995;177:4279–4288. doi: 10.1128/jb.177.15.4279-4288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beachey E H, Keck W, de Pedro M A, Schwarz U. Exoenzymatic activity of transglycosylase isolated from Escherichia coli. Eur J Biochem. 1981;116:355–358. doi: 10.1111/j.1432-1033.1981.tb05342.x. [DOI] [PubMed] [Google Scholar]
- 5.Bernadsky G, Beveridge T J, Clarke A J. Analysis of the sodium dodecyl sulfate-stable peptidoglycan autolysins of selected gram-negative pathogens by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1994;176:5225–5232. doi: 10.1128/jb.176.17.5225-5232.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betzner A S, Keck W. Molecular cloning, overexpression and mapping of the slt gene encoding the soluble lytic transglycosylase of Escherichia coli. Mol Gen Genet. 1989;219:489–491. doi: 10.1007/BF00259625. [DOI] [PubMed] [Google Scholar]
- 7.Bishop R E, Weiner J H. Complementation of growth defects in an ampC deletion mutant of Escherichia coli. FEMS Microbiol Lett. 1993;114:349–354. doi: 10.1111/j.1574-6968.1993.tb06597.x. [DOI] [PubMed] [Google Scholar]
- 8.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G, Gregor J, Davis N W, Kirkpatrick H, Goeden M, Rose D, Mau R, Shao Y. The complete genome sequence of Escherichia coli K12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 9.Braun V, Wu H C. Lipoproteins, structure, function, biosynthesis and model for protein export. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. New York, N.Y: Elsevier Science Publishing, Inc.; 1994. pp. 319–341. [Google Scholar]
- 10.Casabadan M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 11.Davies G, Henrissat B. Structure and mechanism of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 12.Dijkstra A J, Keck W. Identification of new members of the lytic transglycosylase family in Haemophilus influenzae and Escherichia coli. Microb Drug Resist. 1996;2:141–145. doi: 10.1089/mdr.1996.2.141. [DOI] [PubMed] [Google Scholar]
- 13.Dijkstra A J, Keck W. Peptidoglycan as a barrier to transenvelope transport. J Bacteriol. 1996;178:5555–5562. doi: 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehlert K, Höltje J-V, Templin M F. Cloning and expression of a murein hydrolase lipoprotein from Escherichia coli. Mol Microbiol. 1995;16:761–768. doi: 10.1111/j.1365-2958.1995.tb02437.x. [DOI] [PubMed] [Google Scholar]
- 16.Gilkes N R, Henrissat B, Kilburn D G, Miller R C, Jr, Warren R A. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 18.Glauner B, Höltje J-V. Growth pattern of the murein sacculus of Escherichia coli. J Biol Chem. 1990;265:18988–18996. [PubMed] [Google Scholar]
- 19.Haigh, R. D., and P. H. Williams. Personal communication.
- 20.Harz H, Burgdorf K, Höltje J-V. Isolation and separation of the glycan strands from murein of Escherichia coli by reversed-phase high-performance liquid chromatography. Anal Biochem. 1990;190:120–128. doi: 10.1016/0003-2697(90)90144-x. [DOI] [PubMed] [Google Scholar]
- 21.Henikoff S, Henikoff J G, Alford W J, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:GC17–26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- 22.Höltje J-V, Mirelman D, Sharon N, Schwarz U. Novel type of murein transglycosylase in Escherichia coli. J Bacteriol. 1975;124:1067–1076. doi: 10.1128/jb.124.3.1067-1076.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Höltje J-V. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology. 1996;142:1911–1918. doi: 10.1099/13500872-142-8-1911. [DOI] [PubMed] [Google Scholar]
- 24.Höltje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain M, Ichihara S, Mizushima S. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J Biol Chem. 1980;255:3707–3712. [PubMed] [Google Scholar]
- 26.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 27.Janson H, Hedén L-O, Forsgren A. Protein D, the immunoglobulin D-binding protein of Haemophilus influenzae, is a lipoprotein. Infect Immun. 1992;60:1336–1342. doi: 10.1128/iai.60.4.1336-1342.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeuniaux C. Chitinases. Methods Enzymol. 1966;8:644–650. [Google Scholar]
- 29.Jolles P. Lysozymes: model enzymes in biochemistry and biology. Berlin, Germany: Birkhäuser Verlag; 1996. [Google Scholar]
- 30.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 31.Kusser W, Schwarz U. Escherichia coli murein transglycosylase: purification by affinity chromatography and interaction with polynucleotides. Eur J Biochem. 1980;103:277–281. doi: 10.1111/j.1432-1033.1980.tb04312.x. [DOI] [PubMed] [Google Scholar]
- 32.Lommatzsch J, Templin M, Kraft A, Vollmer W, Höltje J-V. Outer membrane localization of murein hydrolases: MltA, a third lipoprotein lytic transglycosylase in Escherichia coli. J Bacteriol. 1997;179:5465–5470. doi: 10.1128/jb.179.17.5465-5470.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lugtenberg B, Meijers J, Peters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 34.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 35.Mushegian A R, Fullner K J, Koonin E V, Nester E W. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc Natl Acad Sci USA. 1996;93:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potvin C, Leclerc D, Tremblay G, Asselin A, Bellemare G. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol Gen Genet. 1988;21:241–248. doi: 10.1007/BF00337717. [DOI] [PubMed] [Google Scholar]
- 37.Rogers H J, Perkins H R, Ward J B. Microbial cell walls and membranes. London, England: Chapman and Hall, Ltd.; 1980. [Google Scholar]
- 38.Romeis T, Höltje J-V. Specific interaction of penicillin-binding proteins 3 and 7/8 with the soluble lytic transglycosylase in Escherichia coli. J Biol Chem. 1994;269:21603–21607. [PubMed] [Google Scholar]
- 39.Romeis T, Vollmer W, Höltje J-V. Characterization of three different lytic transglycosylases in Escherichia coli. FEMS Microbiol Lett. 1993;111:141–146. doi: 10.1111/j.1574-6968.1993.tb06376.x. [DOI] [PubMed] [Google Scholar]
- 40.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Schwarz U, Asmus A, Frank H. Autolytic enzymes and cell division of Escherichia coli. J Mol Biol. 1969;41:419–429. doi: 10.1016/0022-2836(69)90285-x. [DOI] [PubMed] [Google Scholar]
- 43.Shockman G D, Höltje J-V. Microbial peptidoglycan (murein) hydrolases. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1994. [Google Scholar]
- 44.Spratt B G. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur J Biochem. 1977;72:341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 45.Taylor A, Das B C, van Heijenoort J. Bacterial-cell wall peptidoglycan fragments produced by phage lambda or Vi II endolysin and containing 1,6-anhydro-N-acetylmuramic acid. J Biochem. 1975;53:47–54. [Google Scholar]
- 46.Templin, M. F. Unpublished results.
- 47.Thunnissen A-M, Rozeboom H J, Kalk K H, Dijkstra B W. Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. Implications for the enzymatic mechanism. Biochemistry. 1995;34:12729–12737. doi: 10.1021/bi00039a032. [DOI] [PubMed] [Google Scholar]
- 48.Tomasz A. Murein hydrolases: enzymes in search of a physiological function. In: Hakenbeck R, Höltje J-V, Labischinski H, editors. The target of penicillin. Berlin, Germany: Walter de Gruyter and Co.; 1983. pp. 155–172. [Google Scholar]
- 49.Ursinus A, Höltje J-V. Purification and properties of a membrane-bound lytic transglycosylase from Escherichia coli. J Bacteriol. 1994;176:338–343. doi: 10.1128/jb.176.2.338-343.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walderich B, Höltje J-V. Subcellular distribution of the soluble lytic transglycosylase in Escherichia coli. J Bacteriol. 1991;173:5668–5676. doi: 10.1128/jb.173.18.5668-5676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weidel W, Pelzer H. Bagshaped macromolecules: a new outlook on bacterial cell walls. Adv Enzymol. 1964;26:193–232. doi: 10.1002/9780470122716.ch5. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi K, Yu F, Inouye M. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell. 1988;53:423–432. doi: 10.1016/0092-8674(88)90162-6. [DOI] [PubMed] [Google Scholar]