ABSTRACT
Iron is required for the replication and growth of almost all bacterial species and in the production of myelin and neurotransmitters. Increasing clinical studies evidence that the gut microbiota plays a critical role in iron metabolism and cognition. However, the understanding of the complex iron-microbiome-cognition crosstalk remains elusive. In a recent study in the Aging Imageomics cohort (n = 1,030), we identified a positive association of serum ferritin (SF) with executive function (EF) as inferred from the semantic verbal fluency (SVF,) the total digit span (TDS) and the phonemic verbal fluency tests (PVF). Here, we explored the potential mechanisms by analyzing the gut microbiome and plasma metabolome using shotgun metagenomics and HPLC-ESI-MS/MS, respectively. Different bacterial species belonging to the Proteobacteria phylum (Klebsiella pneumoniae, Klebsiella michiganensis, Unclassified Escherichia) were negatively associated both with SF and executive function. At the functional level, an enrichment of microbial pathways involved in phenylalanine, arginine, and proline metabolism was identified. Consistently, phenylacetylglutamine, a metabolite derived from microbial catabolism of phenylalanine, was negatively associated with SF, EF, and semantic memory. Other metabolites such as ureidobutyric acid and 19,20-DiHDPA, a DHA-derived oxylipin, were also consistently and negatively associated with SF, EF, and semantic memory, while plasma eicosapentaenoic acid was positively associated. The associations of SF with cognition could be mediated by the gut microbiome through microbial-derived metabolites.
KEYWORDS: Cognition, iron stores, gut microbiota, ferritin, brain iron, epidemiology
Introduction
Iron is required for the replication and growth of almost all bacterial species. Approximately 5–20% of the iron is absorbed in the duodenum, and 80% of the iron ingested is used by the gut microbiota, mainly in the colon.1 The gut microbiota uses iron as a cofactor in proteins involved in metabolic pathways critical for its survival such as short-chain fatty acids production, DNA synthesis, redox reactions, and electron transport chain.1 In iron-deficient environments, the abundance of some bacterial species (Roseburia spp./Eubacterium rectale) decreases, while members of the Enterobacteriaceae (E. coli, Salmonella and Klebsiella pneumoniae) and Lactobacillaceae families increase. Conversely, iron-rich environments lead to decreased relative abundance of Bifidobacterium spp. and raised Enterobacteria/Lactobacilli ratio due to enhanced growth of the Escherichia/Shigella genus.1 The presence of intricate adaptive mechanisms, such as the secretion of modified forms of the siderophore enterobactin, empowers members of the Enterobacteriaceae family to thrive and proliferate not only in iron-deficient but also in iron-rich environments.1
Iron is also involved in the production of myelin and neurotransmitter synthesis in the central nervous system.2 It has been proposed that serum ferritin (SF) can transport iron into cells, constituting the main pathway of iron supply to oligodendrocytes for myelin production.3 In fact, in a recent study in young adults, higher levels of SF were found to be associated with enhanced working memory.4
There is an increased awareness that the gut microbiota plays a critical role in iron metabolism and cognition and recent observations linked impaired executive function (EF) to gut microbiota composition.5,6 To our knowledge, the potential relationships among SF, cognition, and gut microbiota composition and functionality in the elderly have not been explored.
In a recent study, we identified that higher levels of SF were associated with an enhanced EF and semantic memory (SM) in the Aging Imageomics cohort (n = 1,030). Therefore, our aim was to characterize the metagenomic and metabolomic profiles associated with SF, EF, and SM in the same cohort (Aging Imageomics). This study would be the first to analyze the microbiome-gut-brain-SF axis.
Results
We first assessed the relationships of microbial composition and SF in a discovery cohort (Aging Imageomics; n = 1,030). The subjects’ clinical features are shown in (Table 1). We identified 77 (Padj <0.1) species associated with SF in the entire cohort (Figure 1a, Table S1).
Table 1.
Clinical and neuropsychological data of the Aging Imageomics cohort.
| Characteristics | Total population (n = 1,030) |
|---|---|
| Age (years) | 67,05 ± 7,38 |
| Females n (%)/Males n (%) | 473 (45,9)/557 (54,1) |
| Education (years) | 8 [8–12] |
| BMI (kg/m2) | 27,55 [24,85–30,12] |
| Waist (cm) | 99 [91–107] |
| PVF (score) | 12 [9–15] |
| SVF (score) | 16 [13–20] |
| TDS (score) | 12 [10–14] |
| PHQ9 (score) | 3 [1–6] |
| FPG (mg/dL) | 102,0 [94,0–121,0] |
| HbA1c (%) | 5,7 [5,5–6,2] |
| Fasting Insulin (mU/L) | 8,7 [6,1–12,4] |
| HOMA-IR | 2,32 [1,48–3,58] |
| Serum creatinine (mg/dL) | 0,84 [0,71–0,97] |
| Serum urate (mg/dL) | 5,4 [4,5–6,4] |
| Total cholesterol (mg/dL) | 195,7 ± 35,04 |
| HDL-C (mg/dL) | 50 [42–61] |
| LDL-C (mg/dL) | 118,76 ± 31,18 |
| Fasting triglycerides (mg/dL) | 105,0 [77,5–146,0] |
| Hemoglobin (g/dL) | 14,4 [13,5–15,3] |
| Serum ferritin (ng/ml) | 115,0 [58,0–209,5] |
Results are expressed as number and frequencies for categorical variables, mean and standard deviation (SD) for normal distributed continuous variables and median and interquartile range [IQ] for non-normal distributed continuous variables. BMI, body mass index; PVF, phonemic Verbal Fluency; SVF, Semantic verbal fluency; FPG, fasting plasma glucose; HbA1c, glycated hemoglobin; HOMA-IR, homeostasis model assessment of insulin resistance; HDL-C, high density lipoprotein cholesterol; LDL-C low-density lipoprotein-cholesterol.
Figure 1.
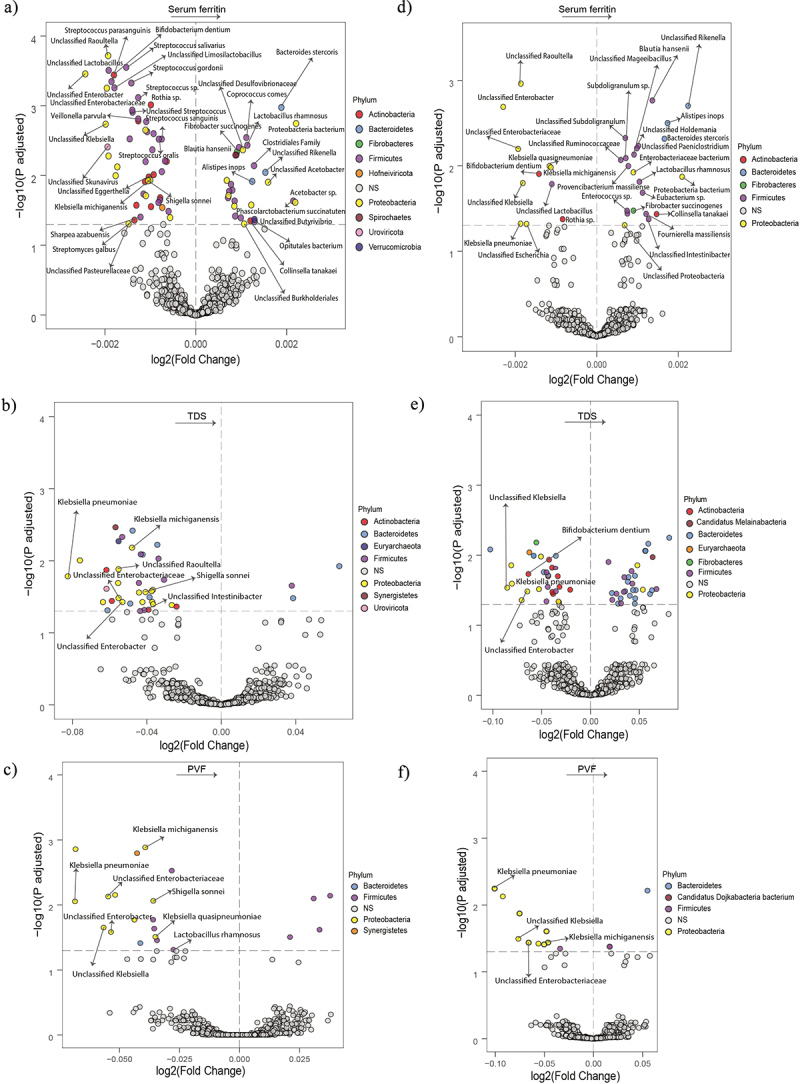
Association of the gut microbiota composition with serum ferritin and executive function in the entire cohort and in men. (a-c) Volcano plots of the gut microbiota associated (p adjusted < 0.05) with serum ferritin (a), TDS (b), and PVF (c) in the entire cohort as calculated by ANCOM-BC from shotgun metagenomic sequencing, adjusting all three models for age, BMI, and sex; and for years of education in TDS, and PVF models. Only the microbial species associated with serum ferritin, TDS (b) and serum ferritin, and PVF (c) are shown. (d-f) Volcano plots of the gut microbiota associated (p adjusted < 0.05) with serum ferritin (d), TDS (e), and PVF (f) in men as calculated by ANCOM-BC from shotgun metagenomic sequencing, adjusting for age and BMI (serum ferritin, TDS, and PVF), and years of education (TDS, and PVF). Only the microbial species associated with serum ferritin and TDS (e) and SF and PVF (f) are shown. The log2 fold change of the association with a unit change in the ANCOM-BC-transformed variable values and the log10 p values adjusted for multiple comparisons using a sequential goodness of fit were plotted for each taxon. Significantly different taxa are colored according to phylum. TDS, total digit span; PVF, phonemic verbal fluency; BMI, body mass index.
SF was negatively associated with species belonging to the Proteobacteria phylum (Klebsiella michiganensis, Klebsiella quasipneumoniae, Shigella sonnei, Unclassified Klebsiella,) and the Streptococcus genera (Streptococcus sanguinis, Streptococcus salivarius, Streptococcus oralis) (Figure 1a, Table S1).
We then explored the gut microbiota composition in relation to EF (Table S2, Table S3). Again, species from the Proteobacteria phylum (Klebsiella michiganensis, Shigella sonnei, Klebsiella pneumoniae, Unclassified Enterobacter, Unclassified Enterobacteriaceae, Unclassified Raoultella) were significantly and negatively associated with both SF and TDS score (Figure 1b). Another species of the Proteobacteria phylum (Klebsiella michiganensis, Shigella sonnei, Klebsiella pneumoniae, Unclassified Enterobacter, Unclassified Klebsiella, Unclassified Enterobacteriaceae,) were negatively associated with SF and PVF (Figure 1c).
When we stratified the population according to sex, we identified 53 species associated with SF in men (Padj <0.1) (Figure 1d, Table S4). As in the whole population, several bacteria belonging to the Proteobacteria phylum (Klebsiella pneumoniae, Klebsiella michiganensis, Klebsiella quasipneumoniae, Unclassified Escherichia,) were negatively associated with SF (Figure 1d, Table S4). Species from the phyla Proteobacteria were strongly negatively associated with both SF and TDS (Figure 1e). Similarly, species from the phylum Proteobacteria were negatively associated with SF and PVF (Figure 1f).
Next, we identified 49 species associated with SF in women (Table S5). In contrast to the observations in men, species belonging to Firmicutes (Veillonella atypica, Veillonella parvula, Veillonella tobetsuensis, and Streptococcus gordonii) were negatively associated with SF. Furthermore, species belonging to the phylum Firmicutes (Blautia producta, Lactobacillus ruminis, Lachnospiraceae bacterium), Bacteroidetes (Bacteroidetes mediterraneensis, Bacteroidetes acidifaciens), and Uroviricota (CrAssphage LMMB, CrAssphage ZA) were negatively associated with both SF and TDS (Figure 2).
Figure 2.
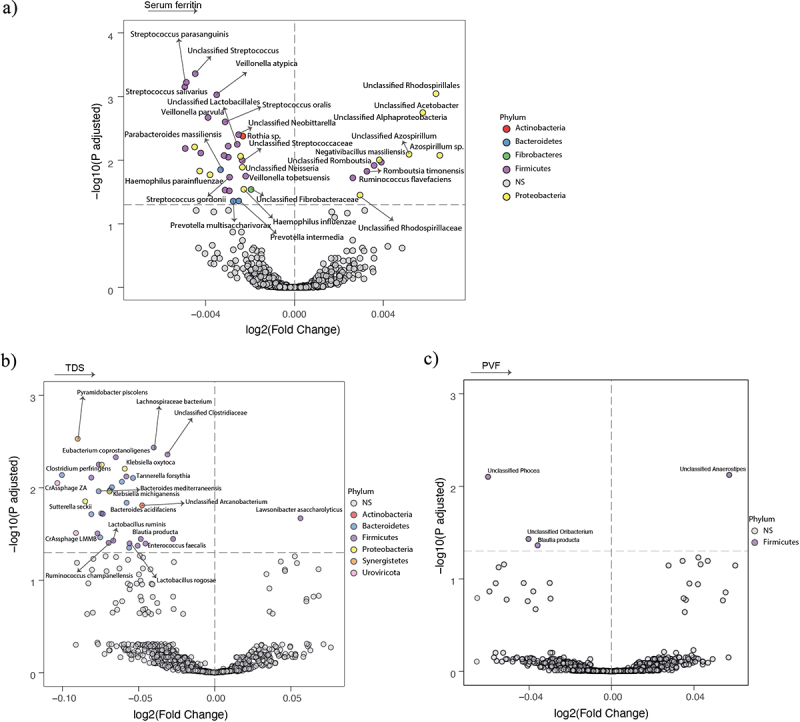
Association of the gut microbiota composition with serum ferritin and executive function in women. (a-c) Volcano plots of the gut microbiota associated (p adjusted < 0.05) with serum ferritin (a), TDS (b), and PVF (c) in women as calculated by ANCOM-BC from shotgun metagenomic sequencing, adjusting all three models for age and BMI; and for years of education in TDS, and PVF models. Only the microbial species associated with serum ferritin, TDS (b) and serum ferritin, and PVF (c) are shown. The log2 fold change of the association with a unit change in the ANCOM-BC-transformed variable values and the log10 p values adjusted for multiple comparisons using a sequential goodness of fit were plotted for each taxon. Significantly different taxa are colored according to phylum. TDS, total digit span; PVF, phonemic verbal fluency; BMI, body mass index.
To further assess the potential effects of the gut microbiome on SF and cognition, we performed functional analyses by mapping reads to KEGG orthologues. We identified microbiome molecular functions associated with SF after adjusting for age, sex, BMI, and years of education. We identified 1384 microbial functions (Padj <0.1) associated with SF, 832 with PVF, and 494 with TDS in the entire cohort, respectively (Figure 3a-c, Table S6–8). In men we identified 1180 microbial genes associated with SF, 829 with PVF, and 1282 with TDS, respectively (Figure 4, Table S9–11). We identified 296 microbial genes associated with SF, and 175 with PVF, respectively in women (Figure 5, Table S12–13).
Figure 3.
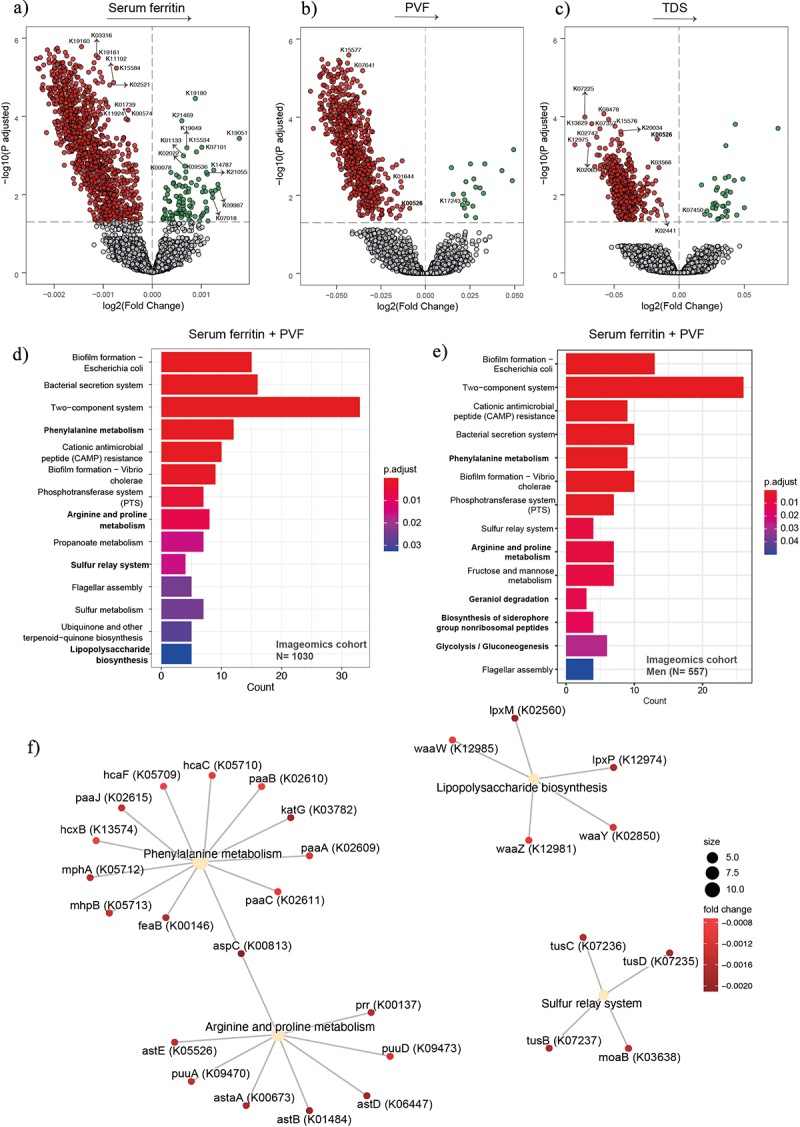
Association of serum ferritin and executive function with bacterial functionality in the Aging Imageomics cohort. (a-c) volcano plots of microbiome molecular functions associated (p adjusted < 0.05) with serum ferritin (a), PVF (b), and TDS (c) as calculated by ANCOM-BC from shotgun metagenomic sequencing, adjusting all three models for age, BMI, and sex; and for years of education in TDS, and PVF models. The log2 fold change of the association with a unit change in the ANCOM-BC-transformed variable values and the log10 p values adjusted for multiple comparisons using a sequential goodness of fit were plotted for each microbiome function. Significantly associated genes are colored in green (upregulated) or red (downregulated). d-e) Barplot plot of the KEGG pathway over-representation analyses (q value < 0.1) mapping the KEGG orthologues significantly associated with serum ferritin and PVF in the entire cohort (d) and in men (e). f) Gene-concept network depicting the linkage of significant KEGG orthologues participating in KEGG pathways related to lipopolysaccharide biosynthesis, sulfur relay system, and phenylalanine, arginine, and proline metabolism for serum ferritin and PVF in the entire cohort. TDS, total digit span; PVF, phonemic verbal fluency; BMI, body mass index.
Figure 4.
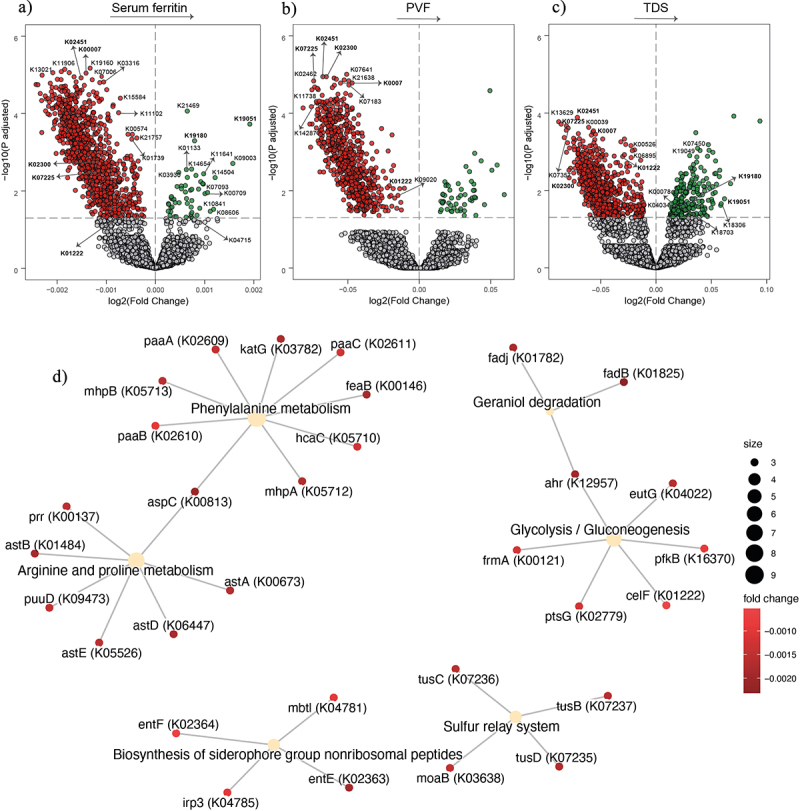
Association of serum ferritin and executive function with bacterial functionality in men. (a-c) Volcano plots of microbiome molecular functions associated (p adjusted < 0.05) with serum ferritin (a), PVF (b), and TDS (c) in men as calculated by ANCOM-BC from shotgun metagenomic sequencing, adjusting all three models for age and BMI; and for years of education in TDS, and PVF models. The log2 fold change of the association with a unit change in the ANCOM-BC-transformed variable values and the log10 p values adjusted for multiple comparisons using a sequential goodness of fit were plotted for each microbiome function. Significantly associated genes are colored in green (upregulated) or red (downregulated). d) Gene-concept network depicting the linkage of significant KEGG orthologues participating in KEGG pathways related to Glycolysis/Gluconeogenesis, biosynthesis of siderophore group nonribosomal peptides, geraniol degradation and phenylalanine, arginine, and proline metabolism for serum ferritin and PVF. TDS, total digit span; PVF, phonemic verbal fluency; BMI, body mass index.
Figure 5.
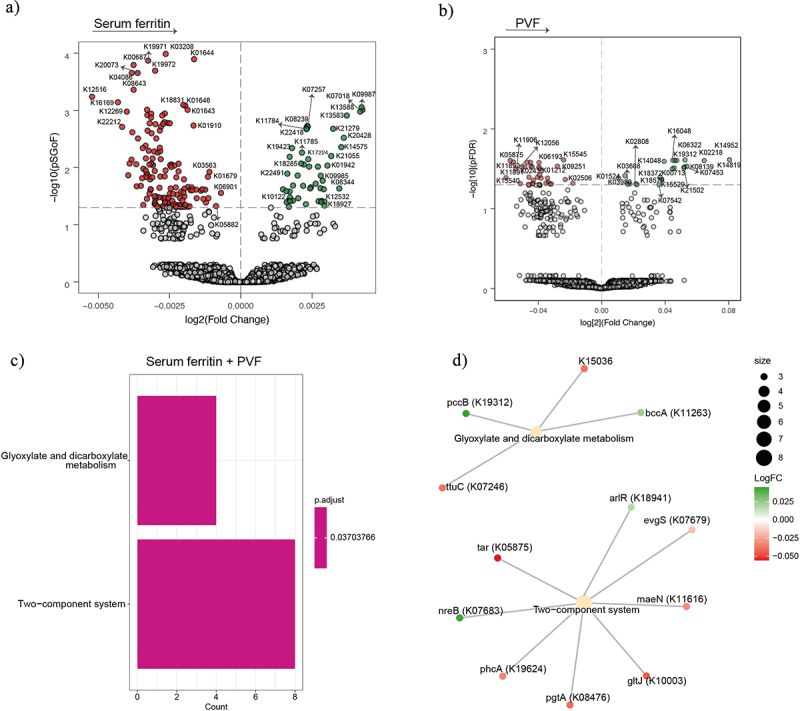
Association of serum ferritin and executive function with bacterial functionality in women. (a-b) Volcano plots of microbiome molecular functions associated (p adjusted < 0.05) with serum ferritin (a), and PVF (b), in women as calculated by ANCOM-BC from shotgun metagenomic sequencing, adjusting all models for age and BMI; and for years of education in PVF model. The log2 fold change of the association with a unit change in the ANCOM-BC-transformed variable values and the log10 p values adjusted for multiple comparisons using a sequential goodness of fit were plotted for each microbiome function. Significantly associated genes are colored in green (upregulated) or red (downregulated). c) Barplot plot of the KEGG pathway over-representation analyses (q value < 0.1) mapping the KEGG orthologues significantly associated with serum ferritin and PVF in women. d) Gene-concept network depicting the linkage of significant KEGG orthologues participating in KEGG pathways related to glyoxylate and dicarboxylate metabolism and two-component system for serum ferritin and PVF. PVF, phonemic verbal fluency; BMI, body mass index.
To obtain insights into the microbial pathways involved in these associations, we performed a pathway overrepresentation analysis of the KEGG orthologues associated with SF and PVF. Both in the entire cohort and in men, we identified a significant enrichment of pathways involved in phenylalanine, arginine, and proline metabolism (Figure 3de). Other significant pathways included the Geraniol degradation, the Biosynthesis of siderophore group nonribosomal peptides, and Glycolysis/Gluconeogenesis in men (Figure 3e). Additionally, a gene-concept network was performed to represent the linkage between significant KEGG orthologues involved in KEGG pathways associated with SF and PVF in the entire cohort (Figure 3f), in men (Figure 4) and in women (Figure S5).
Next, we analyzed the plasma metabolic profiles by HPLC-ESI-MS/MS and applied a machine learning variable selection strategy to identify metabolic signatures associated with SF (Figure 6a). After calculation of SHAP value, we found that SF was strongly associated with metabolites derived from the microbial catabolism of aromatic amino acids such as tryptophan (Indolylacryloylglycine, Indolealdehyde, Indoleacetaldeyde, Indole lactic acid) and tyrosine (cresol), and with LysoPI (20:5) (Figure 6b). In the negative mode, we found strong negative associations of SF with phenylacetylglutamine, derived from the microbial catabolism of phenylalanine (Figure 6c); and positive associations of SF with phosphatidylcholine plasmalogen (PC(P-36:3)) and Eicosapentaenoic acid (EPA). Similar results were found in the case of men (Figure 7). Of the metabolites associated with SF in the positive mode, only sphingomyelin species SM(d36:2) had positive associations with SVF and TDS, while Ubiquinone-2 had negative associations with the three tests (Figure 6e). Notably, EPA had strong positive associations with PVF, FVS, and TDS, while phenylacetylglutamine and ureidoisobutyric acid were strongly negatively associated with PVF, FVS, and TDS . In women, the results were different from those obtained in the entire cohort and in men. We found that SF was negatively associated with proline, linoleyl carnitine, and phosphatidylinositol phosphate (PIP (36:4)), among others, while it was positively associated with EPA, sphingomyelin(d36:2), and hydroxybutyric acid. PIP (36:4) had a strong negative association with PVF, FVS, and TDS, while linoleyl carnitine only with PVF and TDS (Figure 8).
Figure 6.
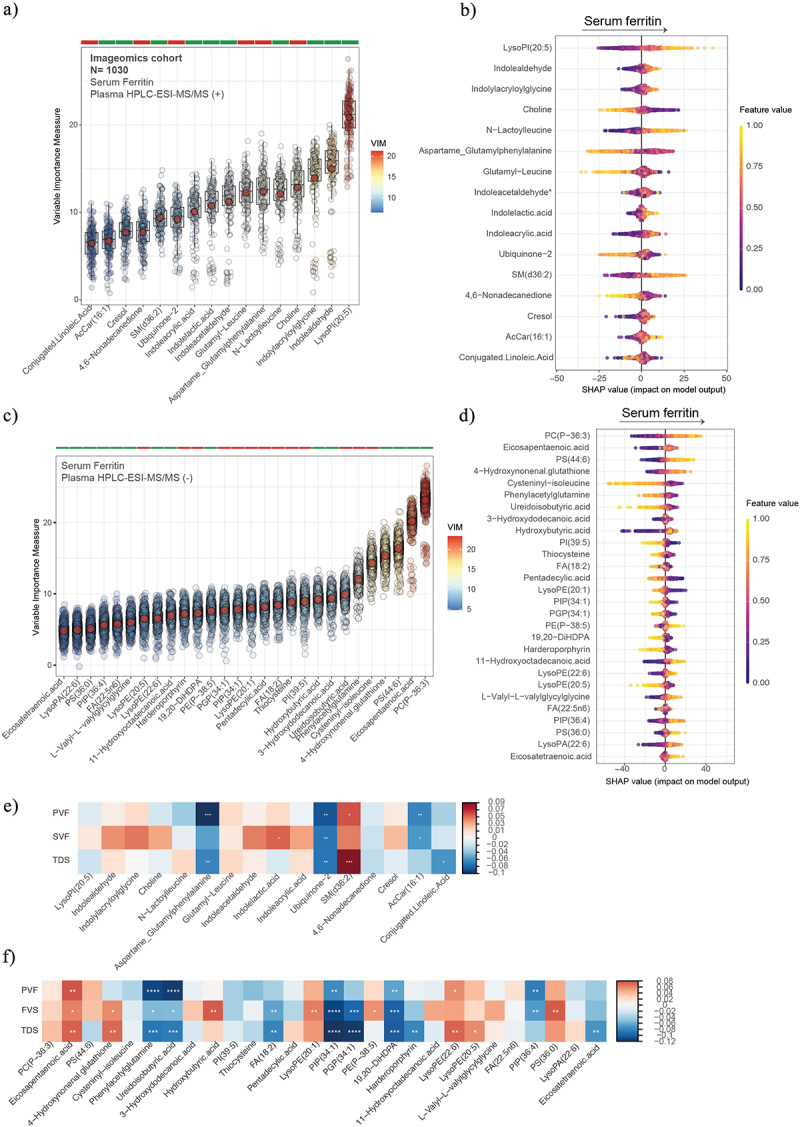
Plasma metabolomics associated with serum ferritin, executive function, and semantic memory in the entire cohort. Boxplots of the normalized variable importance measure (VIM) (a and c) and SHAP summary plots (b and d), for the metabolites associated with the SF levels measured by HPCL-ESI-MS/MS in positive (a) and negative (c) models. (e and f) Heatmap displaying the Spearman correlation (adjusted by age, sex, body mass index, and education years) between PVF, SVF, and TDS and the plasma metabolomics in positive mode (e) and negative mode (f). Significant associations are shown with a cross: **** <0.001, ***<0.01, **<0.05, *<0.1.
Figure 7.
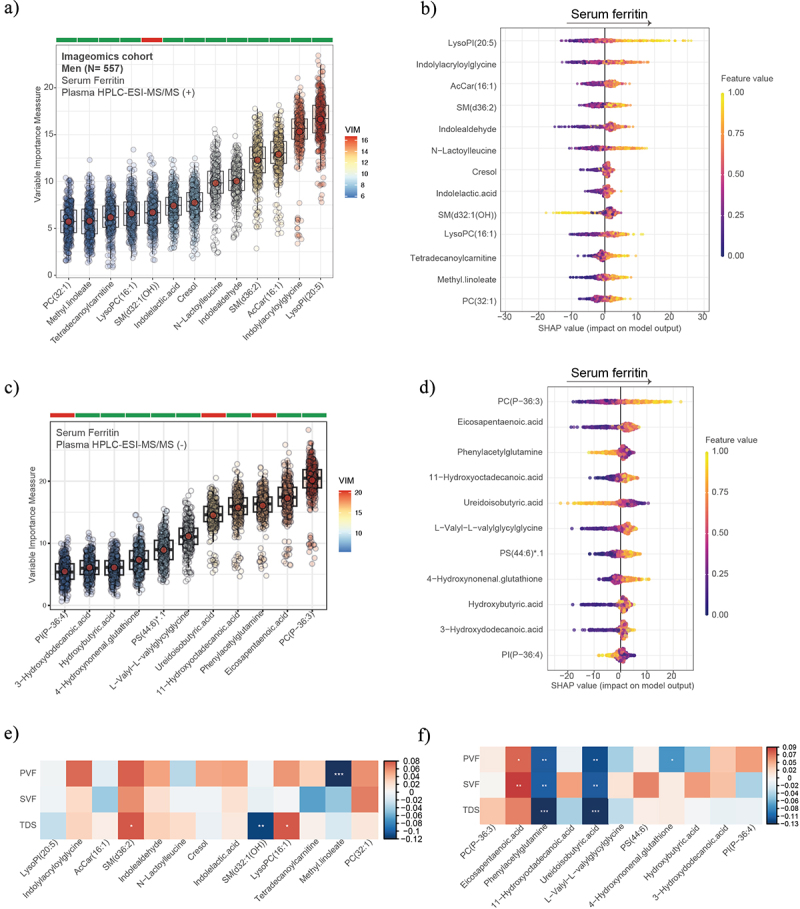
Plasma metabolomics associated with serum ferritin, executive function, and semantic memory in men. Boxplots of the normalized variable importance measure (VIM) (a and c) and SHAP summary plots (b and d), for the metabolites associated with the serum ferritin levels measured by HPCL-ESI-MS/MS in positive (a) and negative (c) models. (e and f) heatmap displaying the Spearman correlation (adjusted by age, body mass index, and education years) between PVF, SVF, and TDS and the plasma metabolomics in positive mode (e) and negative mode (f). Significant associations are shown with a cross: **** <0.001, ***<0.01, **<0.05, *<0.1. PVF, phonemic verbal fluency; SVF, semantic verbal fluency; TDS, total digit span.
Figure 8.
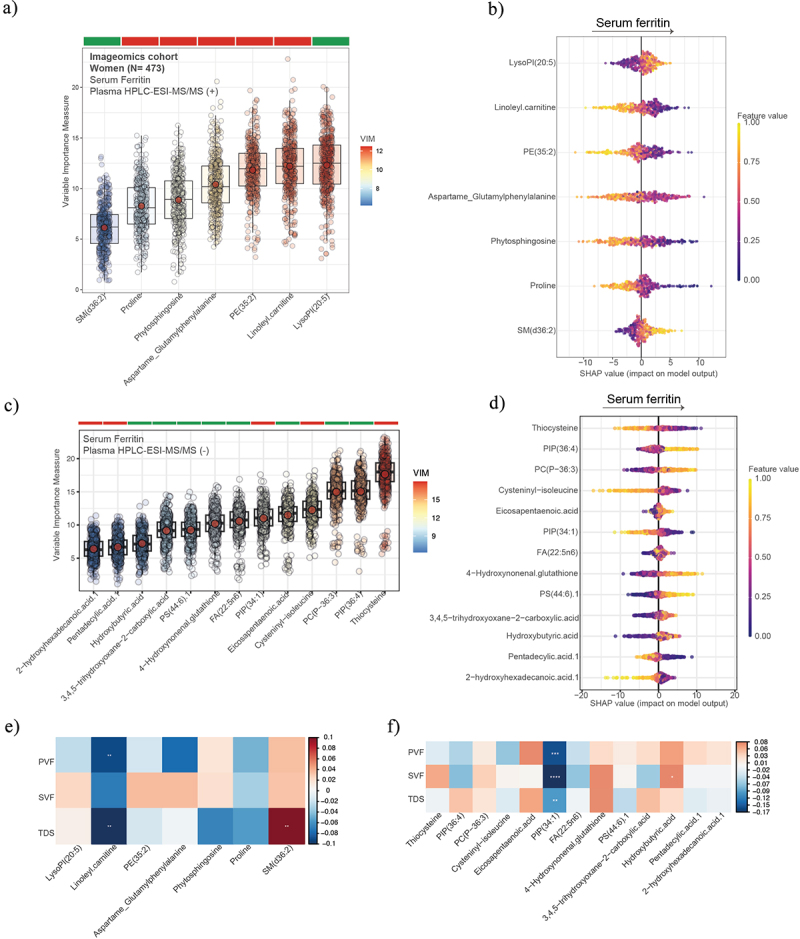
Plasma metabolomics associated with serum ferritin, executive function, and semantic memory in women. Boxplots of the normalized variable importance measure (VIM) (a and c) and SHAP summary plots (b and d), for the metabolites associated with the serum ferritin levels measured by HPCL-ESI-MS/MS in positive (a) and negative (c) models. (e and f) Heatmap displaying the Spearman correlation (adjusted by age, body mass index, and education years) between PVF, SVF, and TDS and the plasma metabolomics in positive mode (e) and negative mode (f). Significant associations are shown with a cross: **** <0.001, ***<0.01, **<0.05, *<0.1. PVF, phonemic verbal fluency; SVF, semantic verbal fluency; TDS, total digit span.
Finally, we also found positive associations of SF with several bacterial species belonging to the Firmicutes phylum (Phascolarctobacterium succinatutens, Coprococcus comes, and Lactobacillus rhamnosus) and with Collinsella tanakaei (Actinobacteria phylum) and Bacteroides stercoris (Bacteroidetes phylum). Similarly, in men, SF was positively associated with species belonging to the Firmicutes phylum such as Provencibacterium massiliensis, Blautia hansenii, Fournierella massiliensis, and Lactobacillus rhamnosus, Bacteroides stercoris (Bacteroidetes phylum) and Collinsella tanakaei (Actinobacteria phylum) (Figure 1). However, all these positive associations were not observed regarding cognitive test scores.
Discussion
Ferritin exists in both intracellular and extracellular compartments and has a major role in iron storage. Although extracellular ferritin or SF is poor in iron (compared to intracellular ferritin), it could be crucial in iron transport to oligodendrocytes for the synthesis of myelin.7,8 Our study examined metagenomic and metabolomic profiles associated with SF, SM, and EF.
In line with previous observations in which iron-deficient environments led to increased species of the Enterobacteriaceae and Lactobacillaceae families, higher SF levels were associated with a decrease in species belonging to the Proteobacteria phylum (Klebsiella pneumoniae, Klebsiella michiganensis, Klebsiella quasipneumoniae, Unclassified Escherichia). Of note, these same bacteria were also associated with worse scores in the EF tests (PVF and TDS). In humans with cognitive impairment in the context of systemic amyloidosis, an increased abundance of Escherichia/Shigella and proinflammatory cytokines (IL-6, CXCL2, NLRP3, and IL-1β) have been reported.9 Also, in line with current results, a study of 116 subjects negatively associated species belonging to the Proteobacteria phylum with memory and EF tests.5
We also observed relationships between microbial functionality, SF, and EF. We found an overrepresentation of genes involved in proline, arginine, and phenylalanine metabolism pathways in the gut microbiota associated with both SF and EF. Almost all microbial genes involved in arginine, proline, and phenylalanine metabolism were negatively associated with SF and EF. Thus, the higher the SF levels, the greater the downregulation of those genes, and the better the cognitive scores. These findings are similar to observations in subjects with mild cognitive impairment and Alzheimer’s disease, in whom increased levels of L-arginine were detected.10 In fact, L-arginine supplementation in older adults at high cardiovascular risk led to worsened EF.11 Proline metabolism has also been associated with depression12 and is also crucial in the progression of mild cognitive impairment in Alzheimer’s disease.13
Phenylalanine enters the brain using the amino acid transporter type (LAT1, SLC7A5). The affinity of LAT1 for phenylalanine is so high that even with small plasma elevations, it is preferentially transported to the brain over amino acids such as tryptophan and tyrosine. Reduced brain concentrations of tyrosine and tryptophan could lead to insufficient production of dopamine and serotonin, respectively.14 A study involving 19 early treated phenylketonuria patients showed that subjects with phenylketonuria had worse attention, memory, and EF compared to healthy controls.15 Remarkably, we found a strong negative association of phenylacetylglutamine with SF when analyzing the plasma metabolic profiles. Therefore, the higher the SF levels, the lower the phenylacetylglutamine levels. Furthermore, phenylacetylglutamine was associated with a worsening of EF and SM. Phenylacetylglutamine is derived from the microbial metabolism of phenylalanine. Altered phenylacetylglutamine levels were associated with EF in obese subjects.5 Another metabolite negatively associated with SF, SM, and EF was ureidoisobutyric acid, suggesting that lower ureidoisobutyric acid levels were associated with higher SF levels and enhanced SM and EF. Ureidoisobutyric acid is a degradation product of pyrimidine metabolism. Interestingly, two microbial functions (thyX and dut) involved in pyrimidine metabolism were strongly negatively associated with inhibitory control (executive function) in subjects without obesity.6
EPA levels were positively associated with SF, SM, and EF, indicating that higher EPA concentrations were associated with higher SF levels and improved SM and EF. EPA is an omega-3 polyunsaturated fatty acid with anti-inflammatory properties.16 Several studies have described that a low omega-3 intake predisposes to faster cognitive decline16 and has been positively associated with EF in obese subjects.6 EPA supplementation in older adults has been positively associated with EF.17 and memory.18
Lastly, SF was associated with increased Blautia hansenii, Fournierella massiliensis, and Lactobacillus rhamnosus. The genus Blautia is a well-known producer of short chain fatty acids (SCFAs), particularly acetic acid and butyric acid, and Blautia hansenii has been linked to less visceral fat accumulation.19 Fournierella massiliensis is also an acetic acid producer.20 SCFAs contribute to the integrity of the blood-brain barrier, inhibiting neuroinflammation and regulating the development and function of microglia.21 Lactobacillus rhamnosus has been associated with increased hippocampal neurogenesis, inhibiting the release of proinflammatory cytokines by the microglia, thus reducing hippocampal microgliosis and inflammation,22,23 which are known to impair learning and memory.24 In mice, supplementation with Lactobacillus rhamnosus led to improved memory and EF22,23 in mice and in subjects with mild cognitive impairment.25
To date, studies have focused on evaluating the composition of the gut microbiota in iron-deficient or iron-excess environments, or in subjects with associated diseases such as inflammatory bowel disease and nonalcoholic fatty liver disease. Therefore, the results described in the literature on iron and gut microbial composition are not comparable with those obtained in this study. Our results suggest that the beneficial associations of SF with EF and SM found in our recent article could be mediated by the gut microbiome and microbial-derived metabolites. However, these results should be interpreted with caution, as they are purely associative and represent a first step in human research that requires further validation in animal models. In addition, the data analyzed were obtained in a cross-sectional study. Longitudinal studies are required to validate and elucidate the complex interaction between the gut microbiota-brain axis and SF.
Materials and methods
Subjects
The Aging Imageomics Study is an observational study that included subjects from the province of Girona (Spain). The subjects originated from two independent cohorts: Maturity and Satisfactory Ageing in Girona study (MESGI50 study) and the Improving interMediAte RisK management study (MARK study). Data were collected at the Dr. Josep Trueta University Hospital facilities between November 14th, 2017, and June 19th, 2019. The study protocol was approved by the ethics committee of the Dr. Josep Trueta University Hospital.
The Aging Imageomics Study aimed to identify biomarkers of human aging by analyzing imaging, biopsychosocial, metabolomic, lipidomic, and microbiome variables. Consequently, the selection criteria were age ≥50 years, residing in the community, no infection within the last 15 days, and absence of contraindications to perform the magnetic resonance imaging (MRI). Participants were visited twice. Clinical history, physical examination, dietary evaluation, MRI, and neuropsychological evaluation of the participants were conducted. Blood, urine, and feces samples were obtained. Extensive information on the study protocol can be found elsewhere.26
Laboratory parameters
Anthropometric and clinical history were acquired by bioimpedance and ad hoc questionnaire, correspondingly. Fasting plasma glucose, lipids profile, serum creatinine, serum urate, hemoglobin and SF levels were obtained by standard laboratory methods using an analyzer (Cobas 8000 c702, Roche Diagnostics, Basel, Switzerland). Glycated hemoglobin was measured by performance liquid chromatography (ADAMA1c HA-8180 V, ARKRAY, Kyoto, Japan).
Neuropsychological assessment
A neuropsychological examination was performed to assess EF and SM. All tests were displayed as raw scores.
Executive function
Total Digits Span (TDS) is subtest of the Wechsler Adult Intelligence Scale-III (WAIS-III). TDS evaluates executive function, specifically working memory. TDS includes the Forward and Backward Digit Span tests. In the Forward Digit Span test, the subject repeats a series of numbers in the same order presented, whilst in the Backward Digit Span test, the subject repeats the sequence of numbers in reverse order. The Total Digit Span is the sum of the scores of the two previous tests. The higher the test score, the better the working memory.27
Phonemic verbal Fluency (PVF) evaluates the executive function and is influenced by processing speed. PVF is a spontaneous verbal production task that requires the production of words with a specific letter (P, M y R) for one minute each.28,29
Language function or semantic memory
Semantic verbal fluency (SVF) evaluates language. This is a test that requires producing as many words as possible in one minute for a specified category, typically “animals”.28,29
Extraction of fecal genomic DNA and whole-genome shotgun sequencing
Total DNA was obtained from frozen human stool using PowerSoil DNA extraction kit (MO BIO Laboratories). Illumina DNA Prep kit (Illumina) was used for Illumina sequencing library preparation with 400-500ng of total DNA. A TapeStation Highly Sensitive DNA kit (Agilent Technologies) was used to evaluate the library. Qubit (Invitrogen) was used in the quantification of the library. Equimolar amounts of validated libraries were pooled, then sequenced as a paired-end 150-cycle run on an Illumina NextSeq2000. Using an in-house python script raw readings were filtered for QV > 30.
FASTQ output files were first pre-processed using fastp for the taxonomic and functional diversity analysis of the microbiota.30 FASTQ is a data pre-processing tool for quality control, adapter trimming and quality filtering. To eliminate reads of human origin, the clean reads were mapped against Homo sapiens genome database (GRCh38.p13) using Bowtie2.31 SqueezeMeta v1.3.132 was used to execute unmapped reads in the co-assembly mode, which pools all samples into a single assembly. Contig assembly was performed with megahit.33 Mapping of reads in contigs was done with Bowtie2. Then, to anticipate codifying regions, Prodigal v2.6.342 was utilized.34 The functional subcategory, pathway, and annotation of genes were determined using HMMER35 against the 2016 version of the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.36 The statistical package R 4.2.237 was used for the selection of the best annotations and for assigning the orf annotation to each read. Also, R was utilized to count the aligned reads, to add the category and its coverage, and to construct abundance matrices. Kaiju v1.6.2 was used to implement taxonomic annotation on the human-free readings.38 Information on lineages, taxa count, and an abundance matrix was generated for all samples using R.
Metabolomics analyses
HPCL-ESI-MS/MS metabolomics analyses
Methanol was used to extract metabolites from plasma samples for non-targeted metabolomics analysis (containing phenylalanine-C13 as an internal standard).39 To each 10 μl of plasma, 30 μl of cold methanol was added, then vortexed for 1 minute and incubated for 1 hour at −20°C. FastPrep-24 (MP Biomedicals) was used to homogenize the samples and then were incubated overnight at 4°C in a rocker. Afterwards, all samples were centrifuged at 12.000 g for three minutes. The supernatant was separated and filtered through a 0.2 μm Eppendorf filter. Two μL of the extracted sample were put on a reversed-phase column (Zorbax SB-Aq 1.8 μm 2.1 × 50 mm; Agilent Technologies) equipped with a precolumn (Zorbax-SB-C8 Rapid Resolution Cartridge 2.1 × 30 mm 3.5 μm; Agilent Technologies) with a column temperature of 60°C. The flow rate was 0.6 mL/min. Solvent A was formed by water with 0.2% acetic acid and solvent B was formed by methanol with 0.2% acetic acid. The gradient increased from 2% B to 98% B in 13 minutes before remaining at 98% B for 6 minutes. Post-time was set in 5 min.
Using N2 as nebulizer gas (5 L/min, 350°C), data were acquired in positive and negative electrospray modes, time-of-flight operated in full scan mode at 50–3000 m/z over an extended dynamic range (2 GHz). A scan rate of 1 scan/s was used with a capillary voltage of 3500 V. For the continuous, low-level (10 L/min) introduction of reference mass compounds 121.050873 and 922.009798, the ESI source utilized a separate nebulizer. These compounds in turn were used for continuous, online mass calibration. The data analysis software MassHunter (Agilent Technologies, Barcelona, Spain) was used to obtain the results. MassHunter qualitative analysis software (Agilent Technologies, Barcelona, Spain) provided the molecular characteristics of the samples. Different ionic species co-migrating from a given molecular entity were represented using the Molecular Feature Extractor algorithm (Agilent Technologies, Barcelona, Spain). Samples with at least 2 ions were chosen. Multiple charge states were forbidden. Utilizing a mass window of 20.0 ppm 2.0 mDa and a retention time window of 0.1%± 0.25 minutes, compounds from various samples were aligned. Those corrected for individual bias and present in at minimum 50% of the samples were chosen.
Statistical analyses
Normal distribution and homogeneity of variances were determined. The results are presented as number and frequencies for categorical variables, mean and standard deviation for normally distributed continuous variables, and median and interquartile range for non-normally distributed continuous variables. These statistical analyses were conducted with SPSS, version 28.0 (IBM SPSS Statistics) or R Statistics. The statistics appear in the figures and legends. Analyses were performed on the entire population and then divided by sex, with findings being more consistent in men.
Metagenomics statistical analysis
Differential abundance analyses to identify bacterial species associated with the SF and cognitive test (Total Digit Span (TDS), PVF (phonemic verbal fluency) and semantic verbal fluency (SVF)) were performed using the microbiome compositional analysis methodology with bias correction (ANCOM-BC).40 By including a sample-specific offset to a linear regression model that is generated from the observed data, ANCOM-BC considers the bias resulting from differing sampling fractions between samples. To consider the compositional nature of metagenomics datasets, the linear regression model in log scale is equivalent to a log-ratio transformation and the offset term acts as the bias correction. All models were adjusted for age, sex, and body mass index (BMI). In the models including cognitive tests, we also adjusted for education years. P-values were corrected for multiple comparisons using a Sequential Goodness of Fit (SGoF). SGoF methods increase their power with a larger number of tests, in contrast to FDR methods which decrease their statistical power as the number of tests grows. In settings with a high number of tests and small sample size, as in large metagenomic datasets, SGoF has proven to perform better than FDR methods.41 Statistical significance was fixed at Padj < 0.1. Pathway overrepresentation analyses were conducted by mapping the KEGG orthologs to the KEGG pathways using the R package “ClusterProflier” (“enrichKEGG” function). A hypergeometric test was used to determine the significance of the pathway, and a Storey procedure (q-values) was adopted for multiple testing correction. Statistical significance was set at q-values <0.1.
Machine learning analysis (metabolomics)
A probabilistic quotient normalization was used to normalize the metabolomics data. Then, data from metabolomics were analyzed using Machine learning methods. We implemented an all-relevant machine learning variable selection strategy using multiple random forests as implemented in the Boruta algorithm.42 All models were adjusted for age, BMI, sex, and years of education. We performed the Boruta algorithm with 500 iterations, a confidence level of 0.005 for the Bonferroni adjusted p-values, and 5000 trees to grow the forest. To facilitate the interpretation of the models, we used the exact computation of Shapley Additive explanations (SHAP) scores, which leverages the internal structure of the random forest models. SHAP calculates the contribution of each metabolite to the predicted response variable.43 The SHAP scores were computed and plotted using the R packages “treeshap” and “SHAPforXGBoost.” The association between metabolomics and cognition (TDS, PVF, SVF) was explored using R, by Spearman correlation, adjusting the data for sex, BMI, age, and years of education.
Supplementary Material
Funding Statement
This work was partially funded by Instituto de Salud Carlos III (ISCIII, Madrid, Spain) through the project PI15/01934 and PI21/01361 to J.M.F-R and the project PI20/01090 (co-funded by the European Union under the European Regional Development Fund (FEDER). “A way to make Europe”) to J.M-P. Generalitat de Catalunya (2021 SGR 01263). M.R.D. is funded by Girona Biomedical Research Institute (Girona, Spain) through the Horizon 2020 Framework Programme of the European Union under the Marie Skłodowska-Curie Innovative Training Network grant agreement No 859890. The Aging Imageomics Study has been funded by Pla estratègic de recerca i innovació en salut 2016-2020 from Generalitat de Catalunya (reference number, SLT002/16/00250). The Health Imageomics has been funded by Health LivingLab operation of the Girona Healthy Region Program which was granted by the Projectes d’Especialització i Competitivitat Territorial (PECT) of the RIS3Cat and the Operative Programme of the European Regional Development Fund of Catalonia 2014-2020. Albert Einstein College of Medicine owns the copyright for the Memory Binding Test and makes it available as a service to the research community but charges for commercial use (for permission requests contact the AECOM at: biotech@einstein.yu.edu); the Spanish and Catalan adaptations used were provided by the BarcelonaBeta Brain Research Center and the Pasqual Maragall Foundation with the AECOM’s permission. For further information about these versions, contact Nina Gramunt at: ngramunt@pmaragall.org. IRBLleida is a CERCA Programme of the Government of Catalonia. IDIBGI is a CERCA Programme/Generalitat de Catalunya. M.J. and J. G. are ‘Serra-Hunter’ Fellows. J.M.-P. is funded by Instituto de Salud Carlos III (Madrid, Spain) through the Miguel Servet Program CP18/00009 (co-funded by European Regional Development Fund “Investing in your future”).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
M.R-D. researched the data, performed the statistical analysis, and wrote the manuscript. E.S-G. performed statistical analysis. A.M-A. contributed to the neuropsychological examination. J.P., J.G-O., R.R., and L.R-T. contributed to the discussion and reviewed the manuscript. J.M-P. and J.M.F.-R. carried out the conception and coordination of the study, performed the statistical analysis and wrote the manuscript.
Data availability statement
We will upload the data to Dryad.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2290318
References
- 1.Mayneris-Perxachs J, Moreno-Navarrete JM, Fernández-Real JM.. The role of iron in host–microbiota crosstalk and its effects on systemic glucose metabolism. Nat Rev Endocrinol [Internet]. 2022;18(11):683–17. doi: 10.1038/s41574-022-00721-3. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira A, Neves P, Gozzelino R. Multilevel impacts of iron in the brain: the cross talk between neurophysiological mechanisms, cognition, and social behavior. Pharmaceuticals [Internet]. 2019;12(3):126. https://www.mdpi.com/1424-8247/12/3/126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todorich B, Zhang X, Slagle-Webb B, Seaman WE, Connor JR. Tim-2 is the receptor for H-ferritin on oligodendrocytes. J Neurochem [Internet]. 2008;107(6):1495–1505. https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1471-4159.2008.05678.x. [DOI] [PubMed] [Google Scholar]
- 4.Raz S, Koren A, Levin C. Associations between red blood cell indices and iron status and neurocognitive function in young adults: evidence from memory and executive function tests and event-related potentials. Ann N Y Acad Sci [Internet]. 2022;1517(1):300–313. doi: 10.1111/nyas.14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnoriaga-Rodríguez M, Mayneris-Perxachs J, Burokas A, Contreras-Rodríguez O, Blasco G, Coll C, Biarnés C, Miranda-Olivos R, Latorre J, Moreno-Navarrete J-M, et al. Obesity impairs short-term and working memory through gut microbial metabolism of aromatic amino acids. Cell Metab [Internet]. 2020;32(4):548–560.e7. doi: 10.1016/j.cmet.2020.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Arnoriaga-Rodríguez M, Mayneris-Perxachs J, Contreras-Rodríguez O, Burokas A, Ortega-Sanchez J-A, Blasco G, Coll C, Biarnés C, Castells-Nobau A, Puig J, et al. Obesity-associated deficits in inhibitory control are phenocopied to mice through gut microbiota changes in one-carbon and aromatic amino acids metabolic pathways. Gut [Internet]. 2021;70(12):2283. http://gut.bmj.com/content/70/12/2283.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Knovich MA, Coffman LG, Torti FM, Torti SV. Serum ferritin: past, present and future. Biochim et Biophys Acta (BBA) - Gen Subj [Internet]. 2010;1800(8):760–769. https://www.sciencedirect.com/science/article/pii/S0304416510000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plays M, Müller S, Rodriguez R. Chemistry and biology of ferritin. Metallomics [Internet]. 2021;13(5):mfab021. doi: 10.1093/mtomcs/mfab021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattaneo A, Cattane N, Galluzzi S, Provasi S, Lopizzo N, Festari C, Ferrari C, Guerra UP, Paghera B, Muscio C, et al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging [Internet]. 2017;49:60–68. https://www.sciencedirect.com/science/article/pii/S019745801630197X. [DOI] [PubMed] [Google Scholar]
- 10.Graham SF, Chevallier OP, Elliott CT, Hölscher C, Johnston J, McGuinness B, Kehoe PG, Passmore AP, Green BD, Jacobs JM. Untargeted metabolomic analysis of human plasma indicates differentially affected polyamine and L-Arginine metabolism in mild cognitive impairment subjects converting to alzheimer’s disease. PLoS ONE [Internet]. 2015;10(3):e0119452. doi: 10.1371/journal.pone.0119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckman JA, Hurwitz S, Fisher NDL. Arginine impairs endothelial and executive function in older subjects with cardiovascular risk. J Am Soc Hypertens [Internet]. 2018;12(10):723–731. https://www.sciencedirect.com/science/article/pii/S1933171118302006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayneris-Perxachs J, Castells-Nobau A, Arnoriaga-Rodríguez M, Martin M, de la Vega-Correa L, Zapata C, Burokas A, Blasco G, Coll C, Escrichs A, et al. Microbiota alterations in proline metabolism impact depression. Cell Metab [Internet]. 2022;34(5):681–701.e10. doi: 10.1016/j.cmet.2022.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Xie K, Qin Q, Long Z, Yang Y, Peng C, Xi C, Li L, Wu Z, Daria V, Zhao Y, et al. High-throughput metabolomics for discovering potential biomarkers and identifying metabolic mechanisms in aging and alzheimer’s disease. Front Cell Dev Biol [Internet]. 2021;9. doi: 10.3389/fcell.2021.602887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Spronsen FJ, Hoeksma M, Reijngoud D-J. Brain dysfunction in phenylketonuria: is phenylalanine toxicity the only possible cause? J Inherit Metab Dis. [Internet]. 2009;32(1):46. doi: 10.1007/s10545-008-0946-2. [DOI] [PubMed] [Google Scholar]
- 15.Pilotto A, Zipser CM, Leks E, Haas D, Gramer G, Freisinger P, Schaeffer E, Liepelt-Scarfone I, Brockmann K, Maetzler W, et al. Phenylalanine effects on brain function in adult phenylketonuria. Neurology [Internet]. 2021;96(3):e399. http://n.neurology.org/content/96/3/e399.abstract. [DOI] [PubMed] [Google Scholar]
- 16.von Schacky C. Importance of EPA and DHA blood levels in brain structure and function. Nutrients [Internet]. 2021;13(4):1074. https://www.mdpi.com/2072-6643/13/4/1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Power R, Nolan JM, Prado-Cabrero A, Roche W, Coen R, Power T, Mulcahy R. Omega-3 fatty acid, carotenoid and vitamin E supplementation improves working memory in older adults: a randomised clinical trial. Clin Nutr [Internet]. 2022;41(2):405–414. doi: 10.1016/j.clnu.2021.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Külzow N, Witte AV, Kerti L, Grittner U, Schuchardt JP, Hahn A, Flöel A, Anstey K. Impact of omega-3 fatty acid supplementation on memory functions in healthy older adults. J Alzheimer’s Dis. 2016;51(3):713–725. doi: 10.3233/JAD-150886. [DOI] [PubMed] [Google Scholar]
- 19.Ozato N, Yamaguchi T, Mori K, Katashima M, Kumagai M, Murashita K, Katsuragi Y, Tamada Y, Kakuta M, Imoto S, et al. Two blautia species associated with visceral fat accumulation: a one-year longitudinal study. Biology (Basel) [Internet]. 2022;11(2):11. https://www.mdpi.com/2079-7737/11/2/318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Togo AH, Durand G, Khelaifia S, Armstrong N, Robert C, Cadoret F, di Pinto F, Delerce J, Levasseur A, Raoult D, et al. Fournierella massiliensis gen. nov., sp. nov., a new human-associated member of the family ruminococcaceae. Int J Syst Evol Microbiol [Internet]. 2017;67(5):1393–1399. https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijsem.0.001826. [DOI] [PubMed] [Google Scholar]
- 21.O’Riordan KJ, Collins MK, Moloney GM, Knox EG, Aburto MR, Fülling C, Morley SJ, Clarke G, Schellekens H, Cryan JF. Short chain fatty acids: microbial metabolites for gut-brain axis signalling. Mol Cell Endocrinol [Internet]. 2022;546:111572. https://www.sciencedirect.com/science/article/pii/S0303720722000193. [DOI] [PubMed] [Google Scholar]
- 22.Gu X, Bi N, Wang T, Huang C, Wang R, Xu Y, Wang H-L. Probiotic Lactobacillus rhamnosus GR-1 supplementation attenuates Pb-induced learning and memory deficits by reshaping the gut microbiota. Front Nutr [Internet]. 2022;9. doi: 10.3389/fnut.2022.934118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel C, Pande S, Acharya S. Potentiation of anti-Alzheimer activity of curcumin by probiotic Lactobacillus rhamnosus UBLR-58 against scopolamine-induced memory impairment in mice. Naunyn Schmiedebergs Arch Pharmacol [Internet]. 2020;393(10):1955–1962. doi: 10.1007/s00210-020-01904-3. [DOI] [PubMed] [Google Scholar]
- 24.Ehsanifar M, Jafari AJ, Montazeri Z, Kalantari RR, Gholami M, Ashtarinezhad A. Learning and memory disorders related to hippocampal inflammation following exposure to air pollution. J Environ Health Sci Eng [Internet]. 2021;19(1):261–272. doi: 10.1007/s40201-020-00600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aljumaah MR, Bhatia U, Roach J, Gunstad J, Azcarate Peril MA. The gut microbiome, mild cognitive impairment, and probiotics: a randomized clinical trial in middle-aged and older adults. Clin Nutr [Internet]. 2022;41(11):2565–2576. https://www.sciencedirect.com/science/article/pii/S0261561422003442. [DOI] [PubMed] [Google Scholar]
- 26.Puig J, Biarnes C, Pedraza S, Vilanova JC, Pamplona R, Fernández-Real JM, Brugada R, Ramos R, Coll-de-Tuero G, Calvo-Perxas L, et al. The Aging imageomics study: rationale, design and baseline characteristics of the study population. Mech Ageing Dev [Internet]. 2020;189:111257. https://www.sciencedirect.com/science/article/pii/S0047637420300531. [DOI] [PubMed] [Google Scholar]
- 27.Wechsler D. WAIS-III: administration and scoring manual: Wechsler adult intelligence scale. [San Antonio Tex]: Psychological Corporation. 1997. [Google Scholar]
- 28.Casals-Coll M, Sánchez-Benavides G, Quintana M, Manero RM, Rognoni T, Calvo L, Palomo R, Aranciva F, Tamayo F, Peña-Casanova J. Estudios normativos españoles en población adulta joven (proyecto NEURONORMA jóvenes): normas para los test de fluencia verbal. Neurología [Internet]. 2013;28(1):33–40. https://www.elsevier.es/es-revista-neurologia-295-articulo-estudios-normativos-espanoles-poblacion-adulta-S0213485312000783.22652141 [Google Scholar]
- 29.Lezak MD, Howieson DB, Bigler ED, Tranel D. Neuropsychological assessment. 5th ed. New York, NY, US: Oxford University Press.[Google Scholar]; 2012. [Google Scholar]
- 30.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics [Internet]. 2018;34(17):i884–90. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods [Internet]. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamames J, Puente-Sánchez F. SqueezeMeta, a highly portable, fully automatic metagenomic analysis pipeline. Front Microbiol [Internet]. 2019;9. doi: 10.3389/fmicb.2018.03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics [Internet]. 2015;31(10):1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 34.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform [Internet]. 2010;11(1):119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durbin R, Eddy SR, Krogh A, Mitchison G. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge: Cambridge University Press; 1998. https://www.cambridge.org/core/books/biological-sequence-analysis/921BB7B78B745198829EF96BC7E0F29D. [Google Scholar]
- 36.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res [Internet]. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Team RC . R: a language and environment for statistical computing. 2013.
- 38.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun [Internet]. 2016;7(1):11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wikoff WR, Pendyala G, Siuzdak G, Fox HS. Metabolomic analysis of the cerebrospinal fluid reveals changes in phospholipase expression in the CNS of SIV-infected macaques. J Clin Invest [Internet]. 2008;118(7):2661–2669. doi: 10.1172/JCI34138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat Commun [Internet]. 2020;11:3514. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carvajal-Rodríguez A, de Uña-Alvarez J, Rolán-Alvarez E. A new multitest correction (SGoF) that increases its statistical power when increasing the number of tests. BMC Bioinform [Internet]. 2009;10(1):209. doi: 10.1186/1471-2105-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kursa MB, Rudnicki WR. Feature selection with the Boruta package. J Stat Softw [Internet]. 2010;36(11):1–13. https://www.jstatsoft.org/index.php/jss/article/view/v036i11. [Google Scholar]
- 43.Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, Nair B, Katz R, Himmelfarb J, Bansal N, Lee S-I. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell [Internet]. 2020;2(1):56–67. [accessed 2022 Sep 7]. https://pubmed.ncbi.nlm.nih.gov/32607472/. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We will upload the data to Dryad.


