Abstract
Protein phosphorylation is an important post-translational modification (PTM), which is involved in many important cellular functions. Understanding protein phosphorylation at the molecular level is critical to decipher its relevant biological processes and signaling networks. Mass spectrometry (MS) has become a powerful tool for comprehensive profiling of protein phosphorylation. Yet the low ionization efficiency and low abundance of phosphopeptides among complex biological samples make its MS analysis challenging; an enrichment strategy with high efficiency and selectivity is always necessary prior to MS analysis. In this study, we developed a phosphorylated cotton fiber-based Ti(IV)-IMAC material (termed as: Cotton Ti-IMAC) that can serve as a novel platform for phosphopeptide enrichment. The cotton fiber can be effectively grafted with phosphate groups covalently in a single step, where the titanium ions can then be immobilized to enable capturing phosphopeptides. The material can be prepared using cost-effective reagents within only 4 hours. Benefiting from the flexibility and filterability of cotton fibers, the material can be easily packed as a spin-tip and make the enrichment process convenient. Cotton Ti-IMAC successfully enriched phosphopeptides from protein standard digests and exhibited a high selectivity (BSA/β-casein = 1000:1) and excellent sensitivity (0.1 fmol/μL). Moreover, 2354 phosphopeptides were profiled in one LC-MS/MS injection after enriching from only 100 μg HeLa cell digests with an enrichment specificity of up to 97.51%. Taken together, we believe that Cotton Ti-IMAC can serve as a widely applicable and robust platform for achieving large-scale phosphopeptide enrichment and expanding our knowledge of phosphoproteomics in complex biological systems.
Keywords: Phosphoproteomics, Enrichment, Immobilized metal affinity chromatography, Cotton fiber, LC-MS/MS, Protein PTM analysis
Graphical Abstract
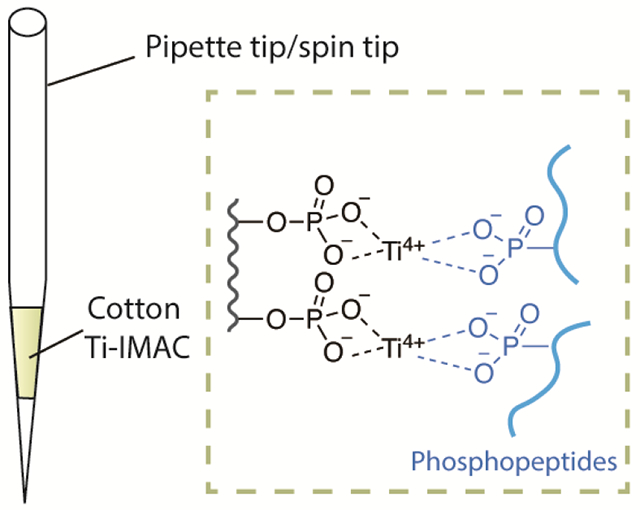
Introduction
Protein phosphorylation is one of the most prevalent protein post-translational modifications (PTMs), as one-third of all proteins in mammalian cells are dynamically phosphorylated.1 This reversible modification is involved in many biological processes, such as protein synthesis, cell signaling, apoptosis, and enzymatic regulation.2–4 Consequently, detailed analyses of protein phosphorylation at a molecular level are critical to decipher its relevant biological processes and signaling networks. Mass spectrometry (MS)-based methods have been widely applied to study phosphoproteome owing to the high sensitivity and high throughput capability.5 However, the low ionization efficiency and low abundance of phosphopeptides pose challenges for their direct analysis, especially in complex biological samples. To address this issue, an efficient enrichment method is necessary to separate the phosphopeptides from the bulk of unmodified peptides before MS analysis.
Various techniques have been established for phosphopeptide enrichment so far.6,7 Among them, immobilized metal affinity chromatography (IMAC) has become the most competent and widely used one. IMAC was first developed in 1975 by Porath et al. to fractionate protein solutions based on the transition metal ion affinity.8 Since then the technique has been successfully adapted to phosphopeptide enrichment, where robust, specific affinity between phosphate groups on phosphopeptides and metal ions separate the phosphopeptides from other peptides.9 Commonly immobilized metal ions include Fe3+, Ga3+ and Ni2+, which are bound to chelating ligands including iminodiacetic acid (IDA) and nitrilotriacetic acid (NTA).9–12 However, the enrichment still suffer from non-specific capturing of acidic peptides as the metal ion also binds to the carboxylic acid groups.13 To enhance the enrichment selectivity, Zhou et al. discovered that using phosphate groups as binding ligands and Ti4+ as metal ions provides stronger metal-phosphate affinity, as inside the MO6 octahedral structure of metal(IV) ions, each metal ion interacts with multiple phosphate groups and vice versa.14 Many Ti(IV)-IMAC materials have been reported since then.15–20 Despite their excellent enrichment specificity, the synthesis and phosphate group functionalization of these nanomaterials or mesoporous polymeric materials are often expensive or require specialized expertise in material science, potentially limiting their applicability in a MS lab.
Cellulose, the most common natural polymer and an eco-sustainable resource, has been utilized for various materials and applications.21 Examples include phosphorylated cellulose in flame retardant materials.22,23, drug delivery matrices.24, and metal ion adsorbents for wastewater purification.25,26 Phosphorylation of cellulose material can be achieved through both chemical modification.23 and enzymatic phosphorylation.27 Although both methods can effectively substitute hydroxyl groups on the cellulose surface, chemical methods are generally more cost-effective and faster.28,29 The phosphorylation reaction of cellulose can be realized with various reagents, e.g., phosphoric acid, phosphorus pentoxide, ammonium phosphate, polyphosphate, and phosphoryl chloride with the assistance of urea in one step.30–37 Inspired by the abundance of phosphate groups on the material surface, phosphorylated cellulose has also been used as IMAC material to enrich phosphopeptides.38,39 However, the preparation process of cellulose itself is still complicated, which hinders its application for large-scale MS-based analysis.
On the other hand, cotton fiber is the most common source of cellulose with good flexibility, strength, chemical resistance, and hydrophilicity. It has been used as the plug material in pipette tip-based solid-phase extractions.40,41, and as hydrophilic stationary phase for glycans and glycopeptides enrichment through hydrophilic interaction liquid chromatography (HILIC).42–44 Cotton fiber has also been reported to be grafted with carboxyl groups or phosphate groups to function as Ti-IMAC material for phosphopeptide enrichment.45,46 However, the reported chemical modification steps are still relatively complicated and time-consuming, and the enrichment specificity is limited in these studies.
To address the above limitations and develop a more accessible IMAC material, we have developed a phosphorylated cotton fiber-based Ti(IV)-IMAC material (termed as: Cotton Ti-IMAC) as a novel platform for phosphopeptide enrichment. Taking advantage of the convenient phosphorylation modification of cellulose materials with phosphoric acid and urea, the cotton fiber can be effectively grafted with phosphate groups in a single step. Titanium ions can then be immobilized onto the surface of the cotton fiber to capture phosphopeptides. In addition to the monophosphate groups, polyphosphate groups can also be introduced to the phosphorylated cotton, which facilitate to chelate more Ti4+ ions.47,48 Moreover, these modifications maintain the cotton’s properties, such as flexibility and filterability, enabling the material to be easily packed into a spin-tip and making the phosphopeptide enrichment procedures more convenient. Overall, the material can be prepared in just two steps within 4 hours with the significantly simplified process and common reagents, and do not require any specialized procedures or equipment. Through the evaluation with standard protein digests, Cotton Ti-IMAC has been proven to have excellent selectivity and high sensitivity. Our method has also been employed for the enrichment of phosphopeptides from HeLa cells, achieving an enrichment specificity as high as 97.51%. Taken together, we have developed a widely applicable, cost-effective, and robust platform for phosphopeptide enrichment that is capable of large-scale LC-MS/MS profiling of complex biological samples.
Experimental Section
Chemicals.
Sodium dodecyl sulfate (SDS), phosphoric acid, trifluoroacetic acid (TFA), triethylammonium bicarbonate (TEAB), iodoacetamide (IAA), and penicillin-streptomycin, as well as protein standards including bovine serum albumin (BSA), α-casein from bovine milk, and β-casein from bovine milk were all ordered from Sigma-Aldrich (St. Louis, MO). Sequencing grade trypsin and dithiothreitol (DTT) were purchased from Promega (Madison, WI). Dulbecco’s Modified Eagle Medium (DMEM) was purchased from Cytiva (Marlborough, MA). Titanium sulfate (Ti(SO4)2) was procured from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). The sterile cotton was obtained from First Aid Only (Shelton, CT) and the empty TopTips (200 μL) came from Glygen Corp (Columbia, MD). Centrifuge-assisted extraction Ti-IMAC (CAE-Ti-IMAC) microspheres were ordered from J&K Scientific Ltd. (Beijing, China). All other chemicals and LC-MS grade solvents were purchased from Fisher Scientific (Pittsburgh, PA).
Preparation of Cotton Ti-IMAC.
The phosphorylation of cotton fiber was performed based on previous literature with slight modification.31 A total of 200 mg cotton fiber was pre-soaked in a solution containing 49.6% urea (w/v), 18.4% phosphoric acid, and 32% water. Following saturation, the excess solution was squeezed out of the cotton. The cotton fiber was then transferred to a flask and heated at 150 °C for 60 minutes, followed by thorough washing with distilled water. After squeezing out the extra water, the cotton fiber was incubated in a 100 mM Ti(SO4)2 solution prepared with 0.1% TFA for 2 h. The cotton fiber was then washed with the 0.1% TFA solution for three times to remove unbound Ti4+ and stored at 4 °C for future use.
Characterization.
Attenuated total reflectance Fourier-transform infrared spectroscopy (ATR FT-IR) analysis was performed on an Equinox 55/S FT-IR spectrophotometer (Bruker, Germany). The surface morphologies were measured by a Zeiss Gemini 450 field emission scanning electron microscope (SEM) (Carl Zeiss, Germany). Energy-dispersive spectroscopy (EDS) elemental mapping were performed using the Thermo Noran Energy dispersive X-ray microanalysis system (Thermo Fisher Scientific, San Jose, CA).
Cell Lysate.
The samples were prepared following a protocol adapted from previous work.40 Briefly, HeLa cells were cultured in DMEM with 10% fetal bovine serum and 1% penicillin-streptomycin, under conditions of 37°C in a humidified chamber with 5% CO2. After harvest, cell pellets were obtained and lysed in a buffer containing 50 mM Tris base (pH 7.4), 4% SDS, 65 mM DTT, and 175 mM NaCl. To every 10 mL of the lysis buffer, 1 phosphatase inhibitor tablet and 1 protease inhibitor tablet (Roche, Mannheim, Germany) were added. The lysis buffer was added to the cell pellet at a ratio of 10:1 (v/v). The lysate was sonicated with a probe sonicator in ice water bath at 50% power with a 5s on/5s off pulse for 12 cycles. Cell lysates were centrifuged, and the resulting supernatant was mixed with a pre-chilled precipitation buffer (composed of acetone, ethanol, and acetic acid in a 50: 50: 0.1 ratio, by volume) at a 1:5 ratio. The mixture was left to precipitate overnight at −20 °C. Protein pellets were collected by centrifugation at 18000 g and washed twice with the same volume of ice-cold precipitation buffer. The pellets were dried in the fume hood for 10 min and then redissolved in 8M urea/50 mM TEAB buffer (pH 8.0). Protein concentrations were measured using a BCA protein assay kit (Thermo Fisher Scientific, San Jose, CA).
Protein Digestion.
Standard proteins or protein extracts from cell lysate were dissolved in 8M urea/50 mM TEAB buffer, reduced by adding 0.4 M DTT solution to a final concentration of 20 mM and incubated at 37 °C for 2h. For protein alkylation, 0.8 M IAA solution was added to reach a final concentration of 40 mM. The mixture was then incubated in the dark at room temperature for 30 min. Alkylation was quenched by adding the same amount of DTT and incubating for another 10 min. The urea concentration in the solution was brought down to 1 M using 50 mM TEAB buffer. Initial protein digestion was conducted with trypsin at a protein-to-enzyme ratio of 100:1 and incubated at 37 °C for 12 h. The same amount of trypsin was added again followed by another 4h incubation at 37 °C to achieve a final protein-to-enzyme ratio of 50:1. The digestion was stopped by adding TFA to a final concentration of 1%. The samples were stored at −80 °C for future use.
Phosphopeptide Enrichment.
Cotton Ti-IMAC material (3 mg) was packed into an empty TopTip, which was then attached to a 2 mL microcentrifuge tube through an adapter unit. The Cotton Ti-IMAC tip was equilibrated by adding 300 μL of 0.1% TFA solution and centrifuging at 200 g three times. Peptide samples were dissolved in 100 μL of loading buffer (40% ACN, 3% TFA) and loaded onto the Cotton Ti-IMAC tip, followed by centrifugation at 200 g for 2 min. The flow-through was re-loaded to the Cotton Ti-IMAC tip four more times to ensure complete retention. After sample loading, the Cotton Ti-IMAC tip was washed three times each with 300 μL of washing buffer I (50% ACN, 6% TFA and 200 mM NaCl) and 300 μL of washing buffer II (30% ACN, 0.1% TFA) at 200 g for 2 min. Finally, phosphopeptides were eluted with consecutive additions of 150 μL of 1% NH4OH (v/v), 150 μL of 5% NH4OH (v/v), and 150 μL of 10% NH4OH (v/v) with a centrifugation speed of 200 g. The phosphopeptides enrichment procedures of CAE-Ti-IMAC followed the manufacturer’s instructions. The collected eluate was combined and dried down in vacuo prior to MS analysis.
MALDI-TOF and NanoLC-MS/MS analysis.
The samples were analyzed by MALDI-MS and LC-MS/MS using previously described instrument parameters.49 Briefly, for MALDI-MS, standard protein samples were reconstituted in 0.1% FA and spotted on a plate with DHB matrix (25 mg/mL in 50%ACN/1%H3PO4 (v/v)). The dried spots were subjected to analysis using a Bruker Rapiflex MALDI-TOF/TOF instrument (Bruker Daltonik, Bremen, Germany). The LC-MS/MS analysis of HeLa digests samples was performed on an Orbitrap Fusion Lumos mass spectrometer connected to a Dionex Ultimate 3000 UPLC system (Thermo Fisher Scientific, San Jose, CA). Peptides were dissolved in 20 mM citric acid/1% FA solution and one fifth of each sample was finally loaded for LC-MS/MS analysis. LC separation was performed on a 15 cm long, in-house packed BEH C18 (1.7 μm, 130 Å, Waters) capillary column using a 96-min gradient (0-30% mobile phase B containing 0.1% FA in 100% ACN) and a flow rate of 0.3 μL/min. MS1 scans of peptides were taken at a resolution of 120 K, with the AGC target set to 2E5 and a maximum injection time set to 50 ms. Stepped higher-energy collision dissociation (HCD) fragmentation (30 ± 8%) and top 20 data-dependent acquisition (DDA) mode were applied for MS/MS data acquisition. Other MS/MS scan settings were performed as follows: 30K resolution, AGC target of 5E4, and maximum injection time of 150 ms.
Data Analysis.
LC-MS/MS raw data files were processed using MaxQuant software (Version 1.6.7.0) following previously described settings.49,50 In brief, UniProt Homo sapiens reviewed database (August 2020, 20311 entries) was used as the protein database. Mass tolerance for precursor ions and fragment ions were specified as 4.5 ppm and 20 ppm, respectively. Two miss cleavages were allowed for trypsin digestion. Fixed modification included carbamidomethylation and variable modifications included oxidation on methionine and phosphorylation on serine, threonine, and tyrosine. False discovery rates (FDR) were controlled less than 1% for both peptide and protein identifications. The MS data were submitted to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD037549.51 Gene ontology (GO) annotations of identified phosphoproteins were performed on the DAVID Bioinformatics Resources following the website instructions.52
Results and Discussion
Synthesis and Characterization of Cotton Ti-IMAC material.
The Cotton Ti-IMAC material was prepared as illustrated in Figure 1A. In the presence of urea and phosphoric acid, the cellulose of the cotton fiber underwent phosphorylation through a dehydration condensation reaction involving the phosphoric acid and hydroxy groups, which primarily occurred at the C2 and C6 positions of glucose.47 Consequently, the reaction product may contain the following functional groups: PO32−, P-O-P, and HPO2−.48
Figure 1.
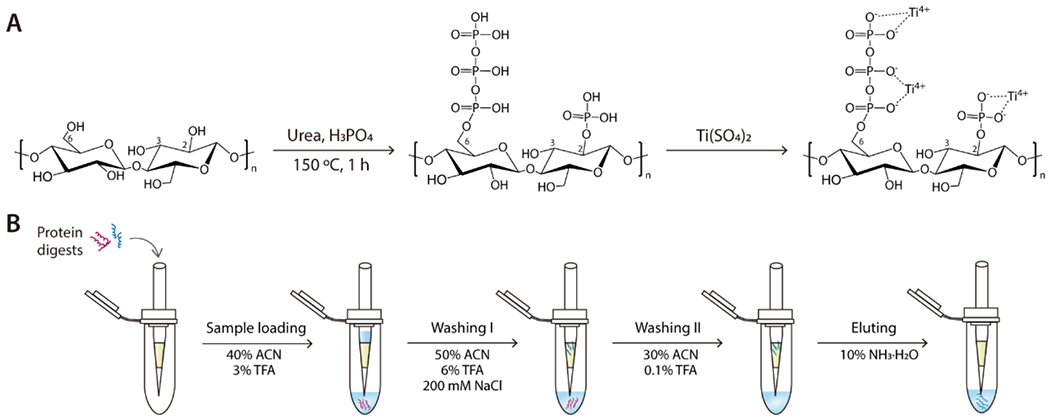
(A) Schematic illustration of the procedure for Cotton Ti-IMAC preparation; (B) Workflow of phosphopeptide enrichment by Cotton Ti-IMAC on a spin-tip.
To confirm the phosphorylation of cotton fiber, both unmodified cotton fiber and phosphorylated cotton fiber was characterized using ATR-FTIR. As shown in Figure 2A, the spectrum of the phosphorylated cotton exhibited a peak of 2366 cm−1 which was attributed to P-H stretching in a phosphite group.32 The peak at 1220 cm−1 was attributed to P-O-P vibrations. Peaks observed at 970 cm−1 and 1003 cm−1, as well as the broader shoulder from 900cm−1 to 942 cm−1 on the main 1030 cm−1 peak, were evidence of P-O stretching vibrations.35,53 Within the 3200-3600 cm−1 range, the OH stretching vibration band exhibited increased asymmetry due to the addition of the more acidic OH groups from phosphoric acids.36 The presence of these characteristic bands indicate the successful reaction between phosphoric acid and cotton fiber, with phosphate groups chemically bonding to the cellulose structure. Owing to the abundant phosphate groups in the modified cotton fiber, Ti4+ ions were easily immobilized onto the surface of the material via ionic interaction at room temperature.
Figure 2.
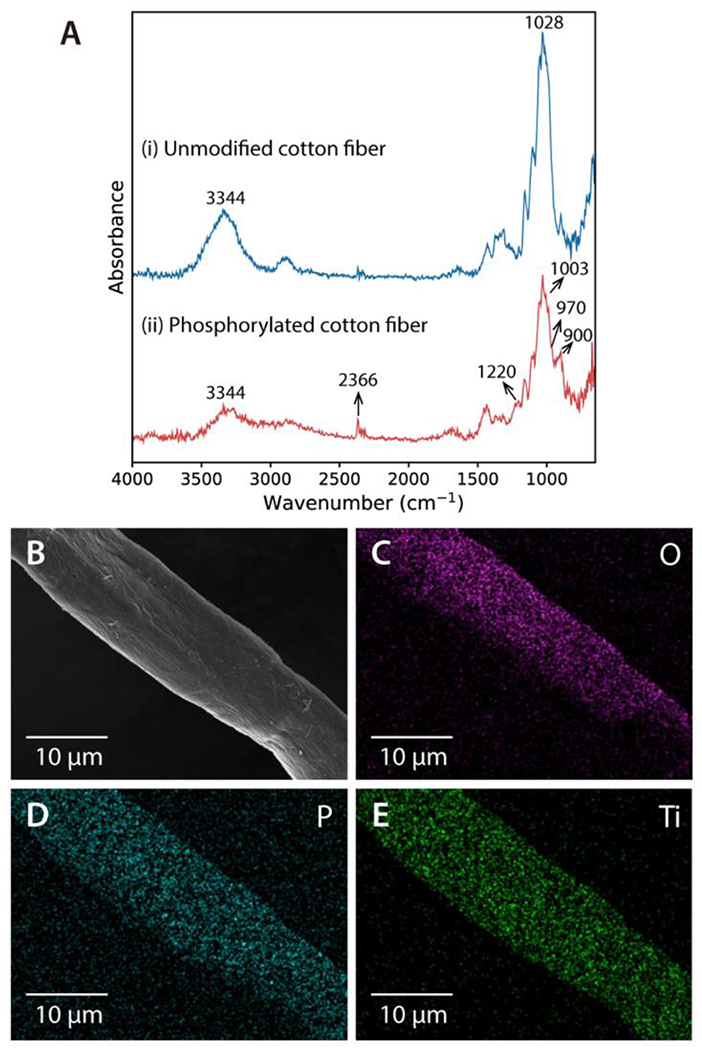
Characterization of Cotton Ti-IMAC. (A) FT-IR spectra of the cotton fiber before (i) and after (ii) the phosphorylation modification. EDS elemental mapping images of Cotton Ti-IMAC: (B) SEM image, (C) O element, (D) P element, and (E) Ti element.
Furthermore, SEM analysis was carried out to characterize the effect of these modifications on the cotton morphology. The SEM images (Figure S1) revealed that the fibrous morphology of the cotton fiber was not significantly affected during each step of the reaction, and the diameter of the cotton fiber remained around 15 μm. To examine the elemental composition, the Cotton Ti-IMAC fiber was characterized by EDS spectroscopy. As depicted in Figure 2B-E, the Cotton Ti-IMAC fiber contained evenly distributed O, P, and Ti elements. The detailed composition of each element and the corresponding EDS spectrum are included as Figure S2. It should be noted that the atom percentages of P and Ti were measured as 12.76% and 35.03%, respectively, demonstrating that the Ti4+ ions were successfully chelated to the cotton fiber, and the Cotton Ti-IMAC material had high Ti content that would contribute to excellent binding capacity for phosphopeptides.
Evaluating the Phosphopeptide Enrichment Performance of Cotton Ti-IMAC.
After the successful fabrication of Cotton Ti-IMAC material, it was initially tested to capture phosphopeptides from tryptic digests prepared from α-casein, a phosphoprotein standard. The protein digests were loaded onto the Cotton Ti-IMAC spin-tip with an acidic loading buffer (40% ACN, 3% TFA), and washed several times by solutions containing 50% ACN, 6% TFA and 200 mM NaCl (washing buffer I) and 30% ACN, 0.1% TFA (washing buffer II ) to remove non-specific bindings based on previous reported workflow.18 The retained phosphopeptides were eventually eluted in an ammonia solution. Figure 3A showed that the signals in the MALDI-TOF spectrum were dominated by unmodified peptides before enrichment, where only two mono-phosphorylated peptides were detected. After the Cotton Ti-IMAC enrichment, most unmodified peptides were removed, and phosphopeptides could be detected with enhanced signals (Figure 3B). In total, 15 phosphopeptides were detected from the α-casein digests with greatly improved signal-to-noise (S/N) ratios, including 5 mono-phosphorylated and 10 multi-phosphorylated peptides. Our identification results covered all phosphorylation sites reported in α-casein that were enriched by other IMAC or commercial TiO2 materials.17,20,54 Details of identified phosphopeptides can be found in Table S1. The results demonstrate the ability of the modified cotton fibers to enrich phosphopeptides as an IMAC material.
Figure 3.
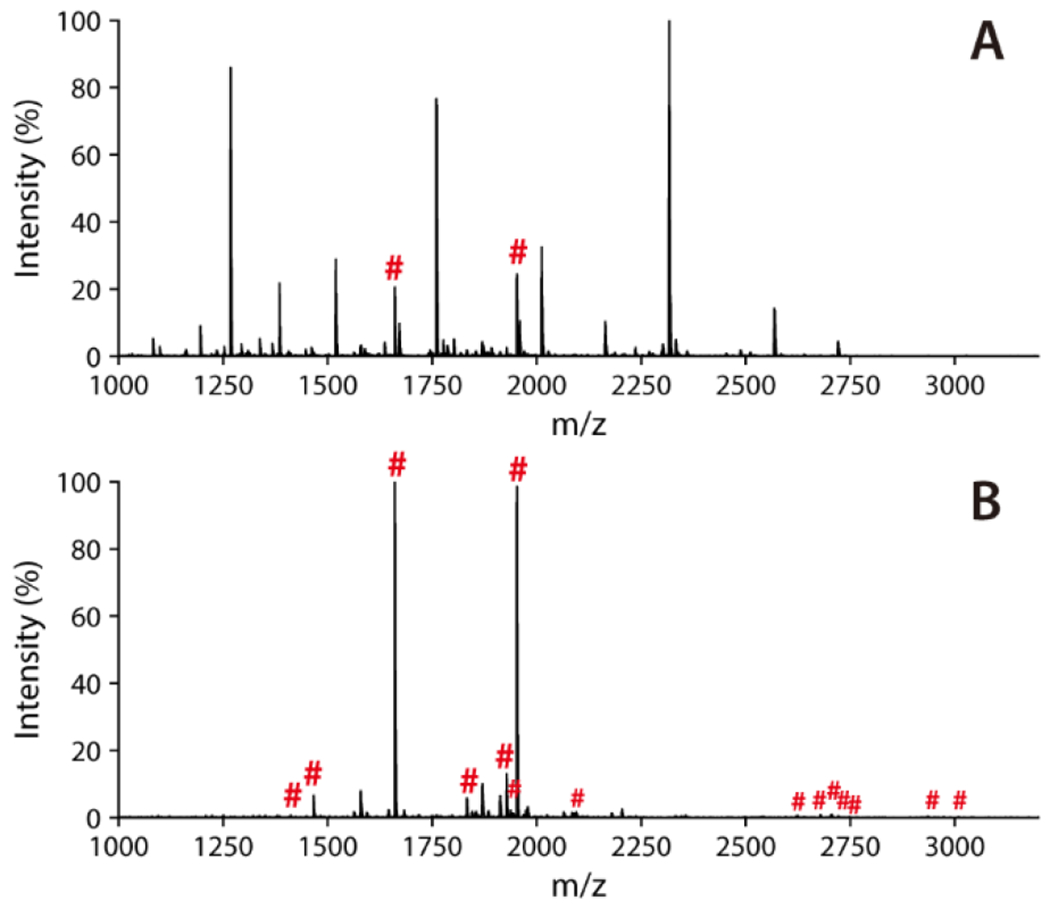
MALDI-TOF MS spectra of α-casein tryptic digest (10 μg). (A) Direct analysis; (B) After phosphopeptide enrichment by Cotton Ti-IMAC. Phosphopeptides are marked with red “#”.
As phosphopeptides are often present in very low abundance in real samples, it is essential to prepare Ti-IMAC material that possesses high enrichment selectivity and sensitivity for selective enrichment of these peptides. A mixture of BSA and β-casein tryptic digests were used for examining the enrichment selectivity of Cotton Ti-IMAC material. β-casein is another commonly used phosphoprotein standard that contains multiple phosphorylation sites. Figure S3 shows that three phosphopeptides including one mono- phosphorylated and two tetra-phosphorylated peptides can be clearly detected after enrichment (Table S2). BSA is a protein without any phosphorylation modification, so its tryptic digests can be used as the background interference sample during the enrichment of β-casein tryptic digests. Upon mixing BSA and β-casein tryptic digests at a molar ratio of 100:1, it was hard to find any signals of phosphopeptides in the MALDI MS spectrum which were all suppressed by the high abundance unmodified peptides (Figure 4A). After enrichment, the signals from phosphopeptides were significantly improved with practically no interference from other peptides (Figure 4B). All three phosphopeptides were successfully separated from the mixture of peptides and detected with appreciable signal intensities. The same experiment was also performed at higher molar ratios; in contrast to the highly interfered direct analyses (Figure S4), the material exhibited remarkable selectivity even at molar ratios of 500:1 and 1000:1 (Figure 4C, 4D) where signals from both mono- and multi-phosphorylated peptides could still be clearly observed.
Figure 4.
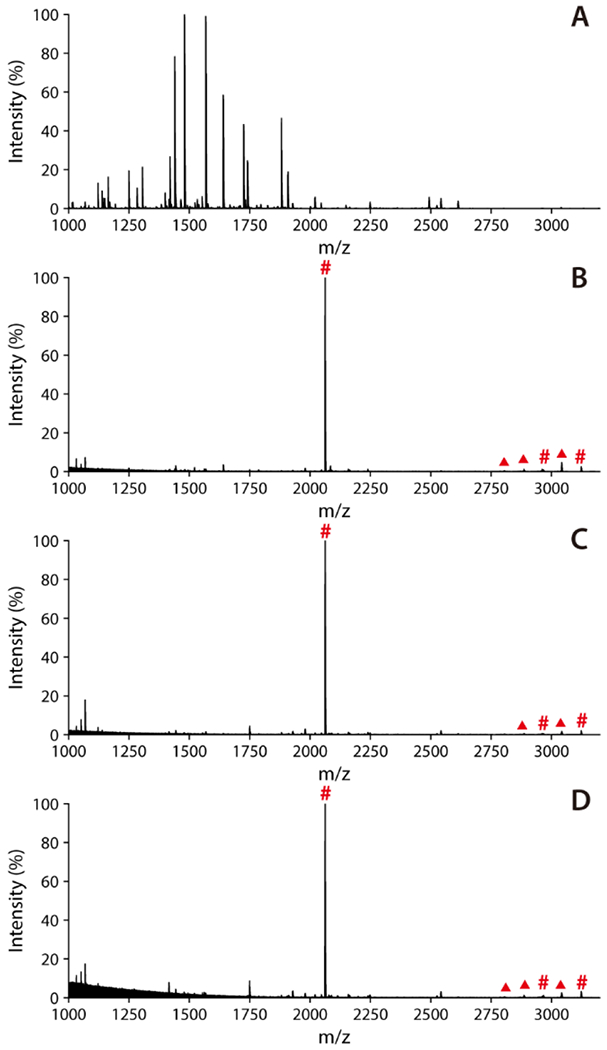
MALDI-TOF MS spectra of the mixture of BSA and β-casein digests at different molar ratios. (A) Direct analysis at 100:1 ratio; Post-enrichment at ratios of (B) 100:1, (C) 500:1 and (D) 1000:1. Phosphopeptides are marked with red “#”, while neutral loss peaks are marked with red “▲”.
To further investigate the sensitivity of the Cotton Ti-IMAC enrichment, this approach was applied to enrich phosphopeptides from diluted β-casein tryptic digests, with sample concentrations starting to decrease from 10 fmol/μL. As depicted in the MALDI-MS spectra in Figure 5, signals from the phosphopeptides were still distinctively evident with an S/N over 3, even at concentrations as low as 0.1 fmol/μL. The results highlighted that the abundant phosphate groups on the modified cotton fiber ensured potent chelation with Ti4+ ions, which in turn contributed to the robust affinity towards phosphopeptides, thus greatly enhancing the sensitivity of phosphopeptide enrichment and subsequent detection. Collectively, the remarkable selectivity and sensitivity suggest the promising capabilities of Cotton Ti-IMAC for isolating and enriching phosphopeptides from more complicated biological samples.
Figure 5.
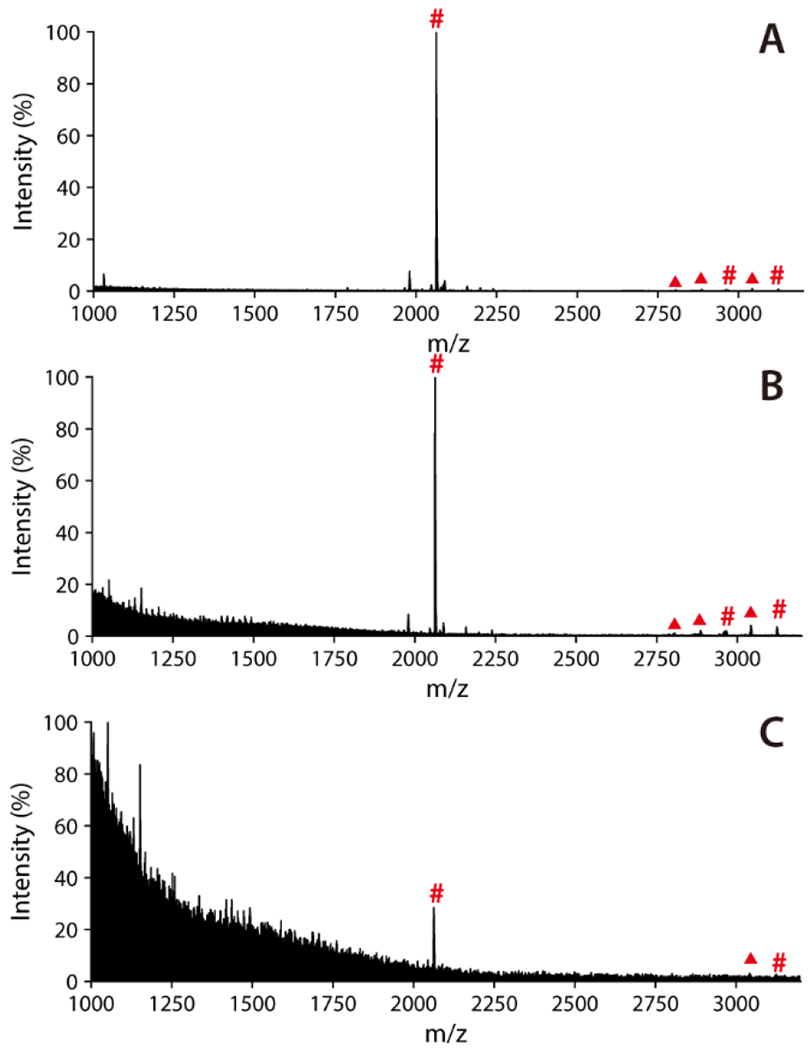
MALDI-TOF MS spectra of phosphopeptides captured from β-casein tryptic digest enriched by Cotton Ti-IMAC at different concentrations: (A) 10 fmol/μL, (B) 1 fmol/μL, and (C) 0.1 fmol/μL. Phosphopeptides are marked with red “#”, while neutral loss peaks are marked with red “▲”.
Phosphopeptide Enrichment from HeLa Cell Lysate.
In view of the outstanding performance of phosphopeptide enrichment in standard samples, the Cotton Ti-IMAC spin-tip approach was used to enrich phosphopeptides from HeLa cell protein digests. The enrichment procedures were the same as the one performed using the standard samples. As a comparison, we also performed phosphopeptide enrichment from the same aliquots of 100 μg HeLa cell protein digests with conventional CAE-Ti-IMAC material following the manufacturer’s instructions. The enriched phosphopeptides underwent LC-MS/MS analysis, with the detailed dentification results listed in Table S3. In general, Cotton Ti-IMAC and CAE-Ti-IMAC enabled identification of 2354 and 2029 phosphopeptides, respectively, as an average of three technical replicates (Figure 6A). This comparison demonstrated that the Cotton Ti-IMAC method could identify 300 more phosphopeptides in a single injection compared to the conventional enrichment approach. It is also worth noting that the numbers of multi-phosphopeptides identified by the two methods were comparable, suggesting that the Cotton-Ti-IMAC method identified more mono-phosphopeptides. Mono-phosphopeptides generally have a lower affinity for IMAC enrichment than multi-phosphopeptides. With more mono-phosphopeptides being identified, Cotton Ti-IMAC demonstrated greater affinity for phosphopeptides than the conventional CAE-Ti-IMAC. Moreover, although both methods exhibited very high enrichment specificity, the Cotton Ti-IMAC method (97.51%) still slightly outperformed the CAE-Ti-IMAC method (96.56%). Besides CAE-Ti IMAC, the results were also compared to previously reported methods in the literature for phosphopeptide enrichment in HeLa cell digests (Table S4). As summarized in this table, Cotton Ti-IMAC turned out to achieve the highest identification number and enrichment specificity with least sample starting amount.
Figure 6.
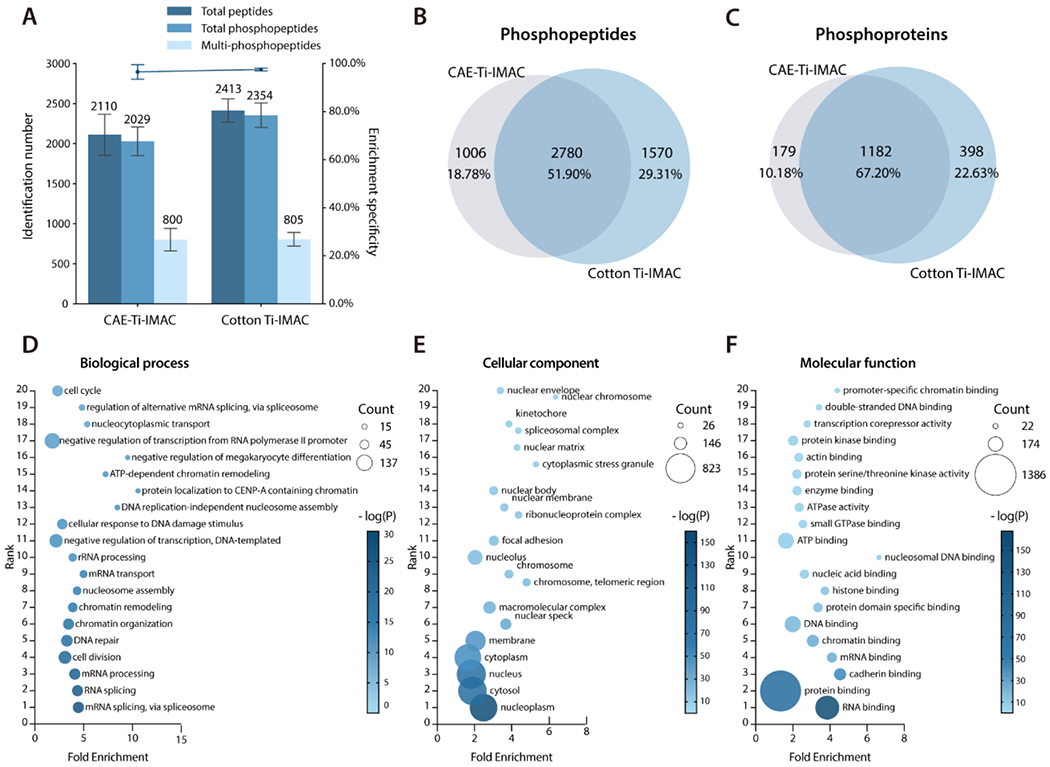
LC-MS/MS analysis of phosphopeptides enriched from HeLa cell tryptic digests. (A) Comparison of phosphopeptide identification number and enrichment specificity by CAE-Ti-IMAC and Cotton Ti-IMAC enrichment, (B) Overlap of phosphopeptides enriched by the two methods, (C) Overlap of phosphoproteins identified by the two methods. Gene ontology analysis of phosphoproteins identified in the Cotton Ti-IMAC method based on (D) biological process, (E) cellular component, and (F) molecular function. Top 20 most significant categories were plotted.
The Cotton Ti-IMAC also showed a better reproducibility due to its simpler enrichment procedure. The complementarity of the phosphopeptides and phosphoproteins identified by the two approaches was also investigated. The combined results from three replicates, as illustrated in Figures 6B and 6C, revealed that the two methods enabled profiling of 5356 phosphopeptides in total corresponding to 1759 phosphoproteins, with an overlap of 2780 phosphopeptides (51.90%), and 1182 phosphoproteins (67.20%), respectively. According to Figure 6B, 1570 phosphopeptides were uniquely identified in the Cotton Ti-IMAC method while only 1006 unique phosphopeptides were present in the CAE-Ti-IMAC dataset. The distribution of the phosphoproteins closely followed the pattern of the phosphopeptides, with 398 phosphoproteins uniquely identified in the Cotton Ti-IMAC dataset and 179 identified in the CAE-Ti-IMAC dataset, respectively (Figure 6C). These datasets clearly showed that the majority of phosphopeptides found in the CAE-Ti-IMAC dataset could be identified using the Cotton Ti-IMAC, and more importantly, the Cotton Ti-IMAC approach enabled profiling of more phosphopeptides and phosphoproteins.
To functionally categorize the profiled phosphoproteins, GO annotation of phosphoproteins identified in the Cotton Ti-IMAC method was performed based on three categories: biological process (Figure 6D), cellular component (Figure 6E), and molecular function (Figure 6F). The top 20 most significant categories were plotted. The results of biological processes showed that the most significant terms were associated with translation and transcription, representing major biological processes of cancer cells. The top 5 significant cellular component annotation showed that the phosphoproteins were evenly distributed in the cell nucleus, cytosol, and membrane, which indicated that the phosphoprotein enrichment was unbiased for all cellular components. Molecular functional analysis of the identified phosphoproteins showed that the majority of the phosphoproteins were binding associated proteins, especially for protein and RNA binding, which was consistent with the fact that phosphorylation is an important signaling modification in cells.
Conclusions
In this study, a novel Cotton Ti-IMAC platform for phosphopeptide enrichment was developed based on the phosphorylation modification of cotton fibers. The synthesis of Cotton Ti-IMAC was facile, rapid, and cost-effective, as only sterile cotton, urea, phosphoric acid, and titanium sulfate were involved during the synthesis. The whole synthesis only requires two reaction steps which can be completed within a short time without any requirement for specialized procedures or equipment, indicating its broad applicability to any analytical or MS lab. The Cotton Ti-IMAC has been demonstrated to have excellent phosphopeptide enrichment performance on both standard protein digests and cell lysate digests, including extremely high selectivity and sensitivity. Owing to its good biocompatibility and high titanium content, the enrichment specificity could achieve 97.51% when analyzing complex samples. In addition, benefiting from the physical property of cotton fiber, the enrichment can be performed in various forms. Depending on the sample type or amount, the enrichment can be achieved either in a packed spin-tip, a packed pipette tip, or even just in-solution mode, which offers great flexibility to bioanalysis. Given the numerous merits of the material, it is anticipated to serve as a widely applicable and robust platform for large-scale phosphopeptide enrichment, which can provide deeper understanding of phosphoproteome in any complex biological or clinical samples. By taking advantage of the intrinsic hydrophilicity of cotton fiber, future implementation of Cotton Ti-IMAC could also be extended to enrich other PTMs simultaneously with phosphorylation.40,49,55
Supplementary Material
Acknowledgements
The authors express gratitude to Gary Girdaukas from the Analytical Instrumentation Center (AIC) at the School of Pharmacy, University of Wisconsin-Madison for his assistance with the FT-IR analysis. This research received partial financial support from the National Institutes of Health (NIH) under grants U01CA231081, R01AG052324, R01AG078794, and R01 DK071801 (to L.L). The mass spectrometers were obtained through the support of NIH shared instrument grants S10 RR029531 and S10 OD025084 (to L.L.), as well as the Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. L.L. acknowledges the Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Distinguished Chair Professorship, funded by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website:
Data availability; SEM images; EDS spectrum and chemical composition of Cotton Ti-IMAC; MALDI-MS spectra of β-casein phosphopeptides; MALDI-MS direct analysis of the mixture of BSA and β-casein digests; Tables of identified phosphopeptides from α-casein and β-casein; Tables of identified phosphopeptides from HeLa cell; Table of HeLa cell phosphopeptide enrichment performance comparison.
The authors declare no conflict of interest.
References
- (1).Zolnierowicz S; Bollen M Protein Phosphorylation and Protein Phosphatases. EMBO J. 2000, 19 (4), 483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Gjertsen BT; Døskeland SO Protein Phosphorylation in Apoptosis. Biochim. Biophys. Acta 1995, 1269 (2), 187–199. [DOI] [PubMed] [Google Scholar]
- (3).Pawson T; Scott JD Protein Phosphorylation in Signaling – 50 Years and Counting. Trends Biochem. Sci 2005, 30 (6), 286–290. [DOI] [PubMed] [Google Scholar]
- (4).Ardito F; Giuliani M; Perrone D; Troiano G; Muzio LL The Crucial Role of Protein Phosphorylation in Cell Signaling and Its Use as Targeted Therapy (Review). Int. J. Mol. Med 2017, 40 (2), 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Yang C; Zhong X; Li L Recent Advances in Enrichment and Separation Strategies for Mass Spectrometry‐based Phosphoproteomics. Electrophoresis 2014, 35 (24), 3418–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Li X-S; Yuan B-F; Feng Y-Q Recent Advances in Phosphopeptide Enrichment: Strategies and Techniques. TrAC - Trends Anal. Chem 2016, 78, 70–83. [Google Scholar]
- (7).Zhao Y; Xu W; Zheng H; Jia Q Light, PH, and Temperature Triple-Responsive Magnetic Composites for Highly Efficient Phosphopeptide Enrichment. Anal. Chem 2023, 95 (23), 9043–9051. [DOI] [PubMed] [Google Scholar]
- (8).Porath J; Carlsson J; OLSSON I; Belfrage G Metal Chelate Affinity Chromatography, a New Approach to Protein Fractionation. Nature 1975, 258 (5536), 598–599. [DOI] [PubMed] [Google Scholar]
- (9).Han G; Ye M; Zou H Development of Phosphopeptide Enrichment Techniques for Phosphoproteome Analysis. Analyst 2008, 133 (9), 1128–1138. [DOI] [PubMed] [Google Scholar]
- (10).Neville DCA; Townsend RR; Rozanas CR; Verkman AS; Price EM; Gruis DB Evidence for Phosphorylation of Serine 753 in CFTR Using a Novel Metal‐ion Affinity Resin and Matrix‐assisted Laser Desorption Mass Spectrometry. Protein Sci. 1997, 6 (11), 2436–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Posewitz MC; Tempst P Immobilized Gallium(III) Affinity Chromatography of Phosphopeptides. Anal. Chem. 1999, 71 (14), 2883–2892. [DOI] [PubMed] [Google Scholar]
- (12).Trojer L; Stecher G; Feuerstein I; Lubbad S; Bonn GK Characterisation and Evaluation of Metal-Loaded Iminodiacetic Acid–Silica of Different Porosity for the Selective Enrichment of Phosphopeptides. J. Chromatogr. A 2005, 1079 (1–2), 197–207. [DOI] [PubMed] [Google Scholar]
- (13).Ficarro SB; McCleland ML; Stukenberg PT; Burke DJ; Ross MM; Shabanowitz J; Hunt DF; White FM Phosphoproteome Analysis by Mass Spectrometry and Its Application to Saccharomyces Cerevisiae. Nat. Biotechnol 2002, 20 (3), 301–305. [DOI] [PubMed] [Google Scholar]
- (14).Zhou H; Ye M; Dong J; Han G; Jiang X; Wu R; Zou H Specific Phosphopeptide Enrichment with Immobilized Titanium Ion Affinity Chromatography Adsorbent for Phosphoproteome Analysis. J. Proteome Res 2008, 7 (9), 3957–3967. [DOI] [PubMed] [Google Scholar]
- (15).Yu Z; Han G; Sun S; Jiang X; Chen R; Wang F; Wu R; Ye M; Zou H Preparation of Monodisperse Immobilized Ti4+ Affinity Chromatography Microspheres for Specific Enrichment of Phosphopeptides. Anal. Chim. Acta 2009, 636 (1), 34–41. [DOI] [PubMed] [Google Scholar]
- (16).Zhang L; Zhao Q; Liang Z; Yang K; Sun L; Zhang L; Zhang Y Synthesis of Adenosine Functionalized Metal Immobilized Magnetic Nanoparticles for Highly Selective and Sensitive Enrichment of Phosphopeptides. Chem. Commun 2012, 48 (50), 6274–6276. [DOI] [PubMed] [Google Scholar]
- (17).Zhao L; Qin H; Hu Z; Zhang Y; Wu R; Zou H A Poly(Ethylene Glycol)-Brush Decorated Magnetic Polymer for Highly Specific Enrichment of Phosphopeptides. Chem. Sci 2012, 3 (9), 2828–2838. [Google Scholar]
- (18).Zhou H; Ye M; Dong J; Corradini E; Cristobal A; Heck AJR; Zou H; Mohammed S Robust Phosphoproteome Enrichment Using Monodisperse Microsphere–Based Immobilized Titanium (IV) Ion Affinity Chromatography. Nat. Protoc 2013, 8 (3), 461–480. [DOI] [PubMed] [Google Scholar]
- (19).Su J; He X; Chen L; Zhang Y Adenosine Phosphate Functionalized Magnetic Mesoporous Graphene Oxide Nanocomposite for Highly Selective Enrichment of Phosphopeptides. ACS Sustain. Chem. Eng 2018, 6 (2), 2188–2196. [Google Scholar]
- (20).Hong Y; Zhao H; Pu C; Zhan Q; Sheng Q; Lan M Hydrophilic Phytic Acid-Coated Magnetic Graphene for Titanium(IV) Immobilization as a Novel Hydrophilic Interaction Liquid Chromatography–Immobilized Metal Affinity Chromatography Platform for Glyco- and Phosphopeptide Enrichment with Controllable Selectivity. Anal. Chem 2018, 90 (18), 11008–11015. [DOI] [PubMed] [Google Scholar]
- (21).Wang X; Yao C; Wang F; Li Z Cellulose‐Based Nanomaterials for Energy Applications. Small 2017, 13 (42), 1702240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ghanadpour M; Carosio F; Larsson PT; Wågberg L Phosphorylated Cellulose Nanofibrils: A Renewable Nanomaterial for the Preparation of Intrinsically Flame-Retardant Materials. Biomacromolecules 2015, 16 (10), 3399–3410. [DOI] [PubMed] [Google Scholar]
- (23).Fiss BG; Hatherly L; Stein RS; Friščicć T; Moores A Mechanochemical Phosphorylation of Polymers and Synthesis of Flame-Retardant Cellulose Nanocrystals. ACS Sustain. Chem. Eng 2019, 7 (8), 7951–7959. [Google Scholar]
- (24).Wu S; Gong Y; Liu S; Pei Y; Luo X Functionalized Phosphorylated Cellulose Microspheres: Design, Characterization and Ciprofloxacin Loading and Releasing Properties. Carbohydr. Polym 2021, 254, 117421. [DOI] [PubMed] [Google Scholar]
- (25).Lehtonen J; Hassinen J; Kumar AA; Johansson L-S; Mäenpää R; Pahimanolis N; Pradeep T; Ikkala O; Rojas OJ Phosphorylated Cellulose Nanofibers Exhibit Exceptional Capacity for Uranium Capture. Cellulose 2020, 27 (18), 10719–10732. [Google Scholar]
- (26).Liu P; Borrell PF; Božič M; Kokol V; Oksman K; Mathew AP Nanocelluloses and Their Phosphorylated Derivatives for Selective Adsorption of Ag+, Cu2+ and Fe3+ from Industrial Effluents. J. Hazard Mater 2015, 294, 177–185. [DOI] [PubMed] [Google Scholar]
- (27).Božič M; Liu P; Mathew AP; Kokol V Enzymatic Phosphorylation of Cellulose Nanofibers to New Highly-Ions Adsorbing, Flame-Retardant and Hydroxyapatite-Growth Induced Natural Nanoparticles. Cellulose 2014, 21 (4), 2713–2726. [Google Scholar]
- (28).Illy N; Fache M; Ménard R; Negrell C; Caillol S; David G Phosphorylation of Bio-Based Compounds: The State of the Art. Polym. Chem 2015, 6 (35), 6257–6291. [Google Scholar]
- (29).Noguchi Y; Homma I; Matsubara Y Complete Nanofibrillation of Cellulose Prepared by Phosphorylation. Cellulose 2017, 24 (3), 1295–1305. [Google Scholar]
- (30).Jurgens JF; Reid JD; Guthie JD Phosphorylated Cotton Cellulose as a Cation-Exchange Material. Text. Res. J 1948, 18 (1), 42–44. [Google Scholar]
- (31).Guthrie JD Ion Exchange Cottons. Industrial Eng. Chem 1952, 44 (9), 2187–2189. [Google Scholar]
- (32).Mucalo MR; Yokogawa Y; Toriyama M; Suzuki T; Kawamoto Y; Nagata F; Nishizawa K Growth of Calcium Phosphate on Surface-Modified Cotton. J. Mater. Sci.: Mater. Med 1995, 6 (10), 597–605. [DOI] [PubMed] [Google Scholar]
- (33).Mucalo MR; Yokogawa Y; Suzuki T; Kawamoto Y; Nagata F; Nishizawa K Further Studies of Calcium Phosphate Growth on Phosphorylated Cotton Fibres. J. Mater. Sci.: Mater. Med 1995, 6 (11), 658–669. [Google Scholar]
- (34).Suflet DM; Chitanu GC; Popa VI Phosphorylation of Polysaccharides: New Results on Synthesis and Characterisation of Phosphorylated Cellulose. React. Funct. Polym 2006, 66 (11), 1240–1249. [Google Scholar]
- (35).Mucalo MR; Kato K; Yokogawa Y Phosphorylated, Cellulose-Based Substrates as Potential Adsorbents for Bone Morphogenetic Proteins in Biomedical Applications: A Protein Adsorption Screening Study Using Cytochrome C as a Bone Morphogenetic Protein Mimic. Colloids Surf. B: Biointerfaces 2009, 71 (1), 52–58. [DOI] [PubMed] [Google Scholar]
- (36).Luneva NK; Ezovitova TI Cellulose Phosphorylation with a Mixture of Orthophosphoric Acid and Ammonium Polyphosphate in Urea Medium. Russ. J. Appl. Chem 2014, 87 (10), 1558–1565. [Google Scholar]
- (37).Kokol V; Božič M; Vogrinčič R; Mathew AP Characterisation and Properties of Homo- and Heterogenously Phosphorylated Nanocellulose. Carbohyd. Polym 2015, 125, 301–313. [DOI] [PubMed] [Google Scholar]
- (38).Shen F; Hu Y; Guan P; Ren X Ti4+-Phosphate Functionalized Cellulose for Phosphopeptides Enrichment and Its Application in Rice Phosphoproteome Analysis. J. Chromatogr. B 2012, 902, 108–115. [DOI] [PubMed] [Google Scholar]
- (39).Zhang L; Wang Y; Pan L; Tang R; Asoh T-A; Ou J; Uyama H Fabrication of a Reusable Bifunctional Biomimetic Ti 4+ -Phosphorylated Cellulose Monolith with a Coral-like Structure for Enrichment of Phosphorylated and Glycosylated Peptides. Green Chem. 2021, 23 (19), 7674–7684. [Google Scholar]
- (40).Huang J; Liu X; Wang D; Cui Y; Shi X; Dong J; Ye M; Li L Dual-Functional Ti(IV)-IMAC Material Enables Simultaneous Enrichment and Separation of Diverse Glycopeptides and Phosphopeptides. Anal. Chem. 2021, 93 (24), 8568–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wang D; Ma M; Huang J; Gu T-J; Cui Y; Li M; Wang Z; Zetterberg H; Li L Boost-DiLeu: Enhanced Isobaric N,N-Dimethyl Leucine Tagging Strategy for a Comprehensive Quantitative Glycoproteomic Analysis. Anal. Chem 2022, 94 (34), 11773–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Selman MHJ; Hemayatkar M; Deelder AM; Wuhrer M Cotton HILIC SPE Microtips for Microscale Purification and Enrichment of Glycans and Glycopeptides. Anal. Chem 2011, 83 (7), 2492–2499. [DOI] [PubMed] [Google Scholar]
- (43).Peng Y; Lv J; Yang L; Wang D; Zhang Y; Lu H A Streamlined Strategy for Rapid and Selective Analysis of Serum N-Glycome. Anal. Chim. Acta 2019, 1050, 80–87. [DOI] [PubMed] [Google Scholar]
- (44).Han J; Chen Q; Jin W; Zou M; Lu Y; Liu Y; Wang C; Wang Z; Huang L Purification of N- and O-Glycans and Their Derivatives from Biological Samples by the Absorbent Cotton Hydrophilic Chromatographic Column. J. Chromatogr. A 2020, 1620, 461001. [DOI] [PubMed] [Google Scholar]
- (45).He X-M; Chen X; Zhu G-T; Wang Q; Yuan B-F; Feng Y-Q Hydrophilic Carboxyl Cotton Chelator for Titanium(IV) Immobilization and Its Application as Novel Fibrous Sorbent for Rapid Enrichment of Phosphopeptides. ACS Appl. Mater. Interfaces 2015, 7 (31), 17356–17362. [DOI] [PubMed] [Google Scholar]
- (46).He X-M; Chen X; Yuan B-F; Feng Y-Q Graft Modification of Cotton with Phosphate Group and Its Application to the Enrichment of Phosphopeptides. J. Chromatogr. A 2017, 1484, 49–57. [DOI] [PubMed] [Google Scholar]
- (47).Zhao M; Fujisawa S; Saito T Distribution and Quantification of Diverse Functional Groups on Phosphorylated Nanocellulose Surfaces. Biomacromolecules 2021, 22 (12), 5214–5222. [DOI] [PubMed] [Google Scholar]
- (48).Rol F; Sillard C; Bardet M; Yarava JR; Emsley L; Gablin C; Léonard D; Belgacem N; Bras J Cellulose Phosphorylation Optimization and Analysis of Phosphorus Position on Cellulose Fibers. Carbohyd. Polym 2019, 229, 115294. [DOI] [PubMed] [Google Scholar]
- (49).Wang D; Huang J; Zhang H; Ma M; Xu M; Cui Y; Shi X; Li L ATP-Coated Dual-Functionalized Titanium(IV) IMAC Material for Simultaneous Enrichment and Separation of Glycopeptides and Phosphopeptides. J. Proteome Res 2023, 22 (6), 2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Tyanova S; Temu T; Cox J The MaxQuant Computational Platform for Mass Spectrometry-Based Shotgun Proteomics. Nat. Protoc 2016, 11 (12), 2301–2319. [DOI] [PubMed] [Google Scholar]
- (51).Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Pérez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yılmaz Ş; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaíno JA The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47 (Database issue), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Huang DW; Sherman BT; Lempicki RA Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc 2009, 4 (1), 44–57. [DOI] [PubMed] [Google Scholar]
- (53).Jastrzębski W; Sitarz M; Rokita M; Bułat K Infrared Spectroscopy of Different Phosphates Structures. Spectrochim. Acta - A: Mol. Biomol 2011, 79 (4), 722–727. [DOI] [PubMed] [Google Scholar]
- (54).Hong Y; Yao Y; Zhao H; Sheng Q; Ye M; Yu C; Lan M Dendritic Mesoporous Silica Nanoparticles with Abundant Ti 4+ for Phosphopeptide Enrichment from Cancer Cells with 96% Specificity. Anal. Chem 2018, 90 (12), 7617–7625. [DOI] [PubMed] [Google Scholar]
- (55).Zheng H; Wang Z; Jia Q Simultaneous Profiling of Palmitoylomics and Glycomics with Photo/PH Dual-Responsive Magnetic Nanocomposites. Small Methods 2023, e2300254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


