Summary
Many promising vaccine candidates and licensed vaccines lead to variable immune responses within humans. Studies suggest that environmental exposures in the gastrointestinal tract could contribute to a reduction in vaccine efficacy via immune tolerance at this site; this is partly achieved by a high abundance of regulatory T cells (Tregs). It is unclear if Treg subsets regulate systemic vaccine responses following oral antigen pre-exposure. Here, we implemented a conditional knock-out mouse model of RORγt+ Tregs to examine the role of these cells in mediating this process. Following oral exposure to the model antigen ovalbumin (OVA) prior to immunization, we found similar induction of vaccine-induced antibody responses in mice lacking RORγt expression in Tregs compared to sufficient controls. Use of various adjuvants led to distinct findings. Our data suggest that expression of RORγt+ within Tregs is not required to regulate tolerance to systemic vaccination following oral antigen exposure.
Subject areas: Immunology, Immune response, Cell biology
Graphical abstract
Highlights
-
•
Mice lacking RORγt expression in Tregs (cKO) have skewed immunophenotype
-
•
RORγt expression in Tregs is dispensable for oral tolerance to a systemic vaccine
-
•
Adjuvants lead to increased RORγt+ Tregs in the small intestine lamina propria
-
•
Oral tolerance is maintained if immunostimulatory adjuvant is used in vaccination
Immunology; Immune response; Cell biology
Introduction
Vaccine development is critical to effectively control existing and emerging pathogens that pose a threat to humans. Upon immunization, it is imperative that the immunogen generates an effective immune response to protect from future infection.1 Heterogeneous responses to vaccines have been reported, and numerous studies have attempted to explain why populations respond differentially to a particular vaccine. Recently, evidence from clinical vaccine trials suggest that effective immune responses may be diverted due to previous exposure to antigens, specifically components of gut microbiota, which are structurally similar to those within the vaccine and lead to a non-protective immune response or immunological tolerance.2,3 Analysis of clinical trials of human immunodeficiency virus (HIV) vaccine candidates suggest a shared amino acid sequence between the envelope protein gp41 and proteins derived from Escherichia coli resident to the gastrointestinal (GI) tract.2,4,5,6,7 Similarly, evidence suggests that the BCG vaccine used to prevent tuberculosis is less effective after exposure to environmental mycobacterium, suggesting diversion from building effective immunity against the pathogen due to pre-exposure of homologous antigens.3 Here, we explore how pre-existing immunity in the gut could impact subsequent systemic vaccination.8 Understanding the mechanisms by which vaccine responses are inhibited by previous oral antigen exposure is a significant objective that will inform efforts to maximize vaccine immunogenicity.
The gut-associated lymphoid tissue (GALT) is a site where regulatory mechanisms are critical to prevent inappropriate inflammation in response to innocuous environmental exposures to dietary antigens or commensal microbes.9 Upon systemic exposure to antigens previously introduced in the gut, a reduction in antibody titers and antigen-specific cellular responses develop compared to antigens that are naive to this site10; this is known as oral tolerance. The underlying mechanism of oral tolerance has been studied in depth and involves the passage of antigens through goblet-cell associated antigen passages11 and processing by tolerogenic dendritic cells in the lamina propria for presentation to naive T cells in the mesenteric lymph nodes (MLNs).12,13 These T cells, which can differentiate into regulatory T cells (Tregs), can then home back to the lamina propria where they are critical for inhibiting potent immune responses against the antigen upon subsequent exposure.14 Tregs have been described as critical for oral tolerance, and further, oral tolerance is maintained in the absence of thymically-derived Tregs,15 suggesting that peripherally derived Tregs may be necessary.
Inflammation is controlled by various regulatory mechanisms at homeostasis, including through Tregs that express FoxP3.9,16 Tregs are positioned to reduce effector responses of other CD4+ T-helper subsets and are especially pertinent at mucosal barrier surfaces like the gut, where tolerance to commensals is essential.17 Transcription factors that are typically associated with CD4+ T-helper subsets can also be expressed in Tregs. FoxP3+ Tregs can express additional transcription factors including T-bet, GATA3, and RORγt; each subset has been studied for their effects on local and systemic responses.18,19,20,21 Subsets of Tregs have been shown to specifically suppress the function of certain types of effector responses.22,23,24,25,26 RORγt+ Tregs are found in high numbers in the GALT and are typically peripherally derived with unique TCRs enriched for specificity to microbial antigens.27,28,29,30 Germ-free (GF) mice display markedly reduced RORγt+ Tregs in the small intestine lamina propria (siLP) and colon compared to conventionally raised specific-pathogen-free (SPF) mice.23 Notably, specific commensals have been associated with RORγt+ Treg frequencies in the colon.23 This subset has also been reported to suppress Type 2 responses, as mice lacking RORγt expression in Tregs have elevated total IgE titers and increased frequencies of GATA3+ conventional CD4+ T cells (Tconv) and Tregs.22,31 Recent work also suggests that RORγt+ Tregs may inhibit Th1 and Th17 responses in some models with the same specificity.32 RORγt+ Tregs have also been implicated in suppressing oral vaccine responses33; however, the role of this subset in affecting systemic vaccine responses is uncharacterized.
Our studies have aimed to define if RORγt expression in Tregs influence systemic vaccine responses following oral antigen exposure to the same antigen. GF mice that have few RORγt+ Tregs display altered tolerogenic responses; GF mice exposed to the model antigen ovalbumin (OVA) prior to OVA vaccination displayed increased OVA-IgG titers compared to SPF mice, indicating a break in tolerance.10,34 Because tolerance is maintained in the absence of thymically-derived Tregs,15 we hypothesized that RORγt expression within peripherally derived Tregs is involved in regulating responses upon oral exposure prior to immunization. Our study addresses key questions on the cellular mechanisms of oral tolerance and systemic vaccination and whether this is impacted by the adjuvant used during vaccination. We have utilized a conditional knock-out mouse model of Rorcfl/fl FoxP3cre to characterize the role of RORγt expression within Tregs in altering systemic vaccine responses with multiple adjuvants following oral pre-exposure to OVA. The selection of adjuvants used in a vaccine is critical for eliciting an appropriate response. Use of strong adjuvants in mice have been reported to overcome peripheral anergy,35 and have also been shown to boost frequencies of RORγt+ Tregs in draining lymph nodes above baseline.32 Our study utilizes a variety of adjuvants in a model of oral tolerance to a systemic immunization to characterize how different adjuvants may alter vaccine responses. We propose that suppression of vaccine responses following oral exposure is limited to Type 2 immunity regardless of adjuvant, and that oral tolerance remains intact in the absence of RORγt expression within regulatory T cells.
Results
Oral administration of antigen prior to vaccination with alum suppresses type 2 immune responses
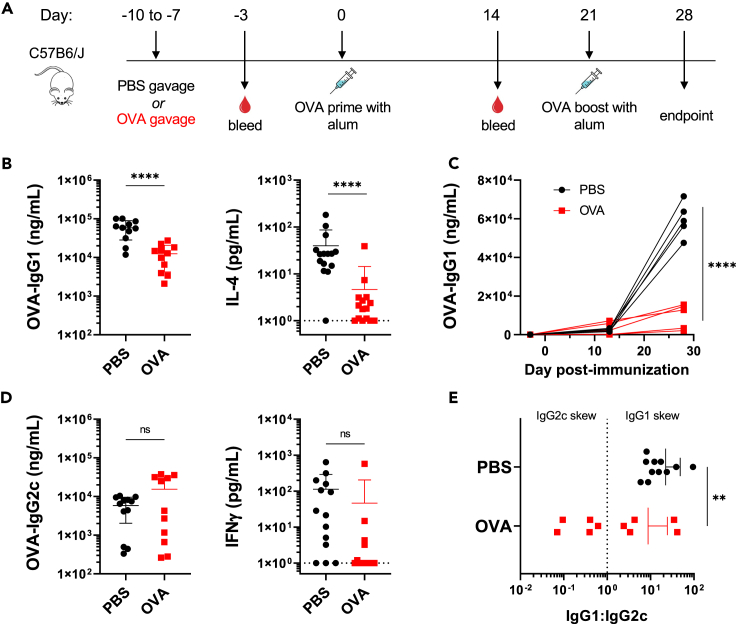
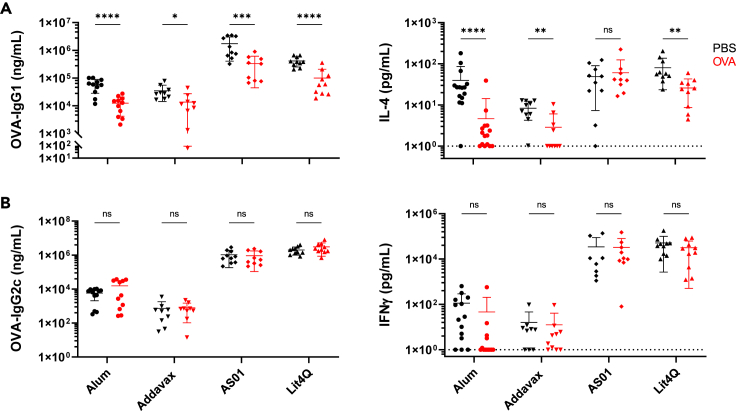
To interrogate the effects of enteric antigen pre-exposure on a systemic vaccine response, C57BL/6 mice were exposed to 1 mg of OVA by oral gavage for four consecutive days, ending one week prior to intraperitoneal (i.p.) vaccination with 10 μg OVA formulated with alum (Figure 1A). Control groups received PBS by oral gavage prior to immunization with the same vaccine. Both groups were boosted on Day 21 and assessed one week later for systemic and local immune responses. Our findings support existing work15 that OVA exposure prior to OVA vaccination suppresses total serum OVA-IgG titers (Figure S1A). This was not observed at Day 14 following prime alone (Figure S1B), but groups differed significantly following boost by Day 28. Using alum as an adjuvant in our oral tolerance model, we observed specific suppression of IgG1 over time (Figures 1B and 1C). Upon splenocyte re-stimulation for 72 h with OVA protein, secreted cytokine production from culture supernatant was assessed by ELISA. We observed that mice that received oral OVA prior to immunization had a reduction of IL-4 secretion compared to PBS pre-treated groups (Figure 1B). Though oral administration of vaccine antigen prior to immunization has been previously described, we noticed this effect was specific for Type 2-associated immune responses. Interestingly, no suppression of OVA-IgG2c or IFNγ upon OVA re-stimulation of splenocytes was observed for OVA gavage groups (Figure 1D).
Figure 1.
Oral administration of antigen prior to vaccination with alum suppresses Type 2 immune responses
Analysis of endpoint (Day 28) of mice gavaged with PBS (black) or OVA (red) prior to immunization with alum.
(A) Experimental timeline (B) OVA-IgG1 titers and secreted IL-4 upon OVA-re-stimulation of splenocytes after 72 h.
(C) OVA-IgG1 from an individual experiment over time, representative of 3 independent trials.
(D) OVA-IgG2c titers and secreted IFNγ upon OVA-re-stimulation of splenocytes after 72 h.
(E) Ratio of IgG1:IgG2c titers for individual mice. Each data point represents an individual mouse.
Data are pooled from 3 independent experiments and represented as mean ± SD. Unpaired t tests were used for statistical analysis. ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
The ratio of IgG1 to IgG2c is commonly used as an indicator of skew induced immunity upon immunization. Here, we observe a higher IgG1:IgG2c ratio in mice treated with PBS prior to systemic vaccination compared to mice pre-exposed to OVA (Figure 1E). This indicates that oral exposure to vaccine antigen specifically suppresses Type 2 induced immunity in our model. Additionally, we examined T-independent subclass OVA-IgG2b. Here, groups exposed to OVA prior to immunization had a significant increase in OVA-IgG2b compared to PBS treated groups. In both OVA and PBS groups, OVA-IgG2b titers were lower than IgG1 and IgG2c titers, suggesting a greater T cell-dependent response (Figure S1C). OVA-IgE titers were also suppressed following oral OVA exposure prior to immunization, in line with specific suppression of Type 2-mediated immune responses, though titers were overall relatively low (Figure S1D). There were also no differences in T follicular helper (TFH) subsets (CXCR5+ PD-1+ CD4+) in splenocytes from both groups (Figure S1E).
To explore mechanisms of Type 2-specific suppression of vaccine induced immunity following oral antigen exposure, we turned to regulatory T cell subsets. Previous work suggests that Tregs are critical in the maintenance of oral tolerance,14,15 and data indicate that Tregs expressing the transcription factor RORγt+ could specifically suppress Type 2 immunity.22 Given that these cells arise from bacterial exposure and are highly prevalent in the siLP,36 the likely site of oral tolerance induction,11,13 we hypothesized that mice orally exposed to OVA prior to systemic immunization could directly alter Treg phenotypes. Though RORγt+ Tregs are typically found in low frequencies systemically, an increased frequency of these cells was observed in the spleens of groups that received OVA prior to immunization compared to PBS treated groups (Figures S2 and S3). However, OVA gavaged mice exhibited no change in FoxP3+ Treg frequencies in the MLN or siLP (Figure S3). We examined CD4+ GATA3+ FoxP3- T conventional Th2 cells and found that in agreement with our serum antibody and IL-4 data (Figure 1B), Th2 responses in the spleen were suppressed in OVA gavaged groups (Figure S3). Surprisingly, in the MLN and siLP, no differences in Th2 cells, RORγt+ Tregs (Figure S3). There were no differences in other Treg subsets examined, including GATA3+ Tregs and IL-33R + Tregs in all tissues examined (data not shown). This indicates that pre-exposure to vaccine antigen prior to systemic immunization using alum contributes to modulation of systemic immune responses, but not detectable changes in T cell subsets in the GALT.
Compensatory increase in GATA3+ and IL-33R + subsets in mice lacking RORγt expression in Tregs does not impact vaccination
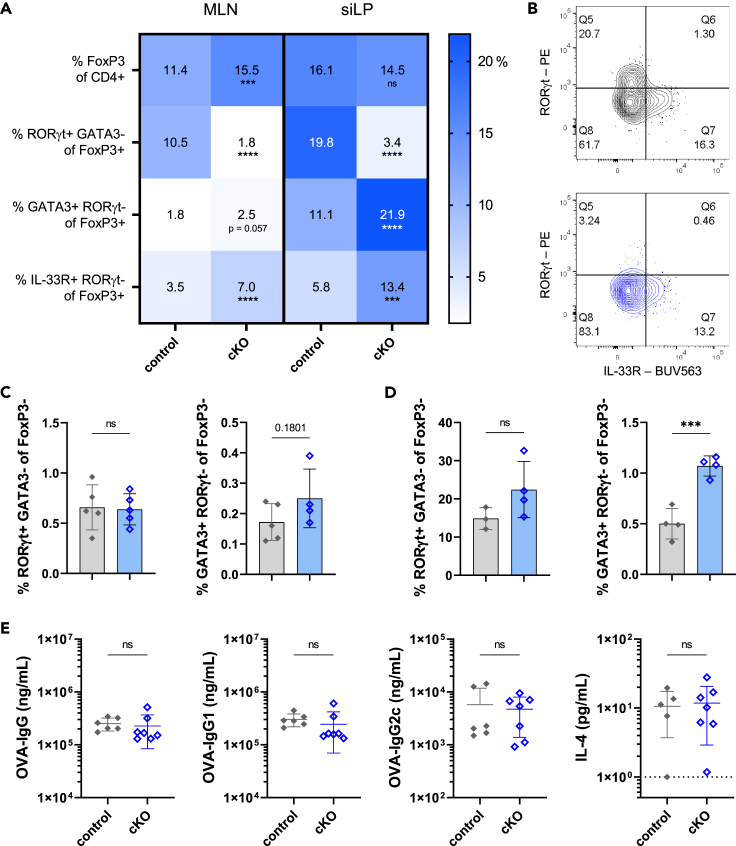
At steady-state in the siLP, peripheral Tregs (pTregs), including those that express RORγt+, maintain gut barrier integrity and mediate effector responses. These cells also contribute to tolerance against gut commensal microbes and controlling homeostatic interactions against environmentally acquired antigens. To examine the role of RORγt Tregs in mediating tolerance to a systemic vaccine following oral exposure, we developed a Rorcfl/fl FoxP3cre conditional knock-out (cKO) line of C57BL/6 mice housed in specific pathogen-free conditions. As previously described,22 these mice that lack RORγt+ expression specifically in FoxP3+ Tregs and have imbalanced immune cell populations in the GALT where these cells are normally present at increased frequencies compared to the spleen. To further investigate these findings in the context of vaccination, we compared immune cell populations and baseline immunoglobulins across various tissues of Rorcfl/fl FoxP3cre cKO mice and Rorc+/+ FoxP3cre controls. As expected, we found a significant reduction in the frequencies of RORγt+ Tregs in Rorcfl/fl FoxP3cre cKO mice compared to controls (Figures 2A and 2B). No significant differences were observed in FoxP3+ frequencies among all CD4+ in the siLP, though frequencies of Tregs were increased in the MLN in cKO mice. Frequencies of GATA3+ RORγt- FoxP3+ Tregs and IL-33R + RORγt- FoxP3+ cells were increased in Rorcfl/fl FoxP3cre cKO mice compared to Rorc+/+ FoxP3cre control mice in both the MLN and siLP (Figure 2A). Effects of the cre-lox system were specific to FoxP3+ Tregs, as there were no differences in frequencies of RORγt+ FoxP3- Th17 cells (Figures 2C and 2D). cKO mice also had increased proportions of Th2 (GATA3+ RORγt- FoxP3-) cells compared to controls in local tissues, both MLN and siLP (Figures 2C and 2D). Similar trends were observed in total cell numbers; there was a significant increase in GATA3+ RORγt- FoxP3- Th2 cells in the siLP of cKO mice, no differences in Treg cell numbers, and greater numbers of GATA3+ RORγt- FoxP3+ Tregs and IL-33R + RORγt- FoxP3+ in the MLN and siLP (Figure S4). In the spleen, we observed a slight increase in FoxP3+ frequencies overall, but similar patterns in Th2, GATA3+ RORγt- FoxP3+ and IL-33R + RORγt- FoxP3+ subsets as observed in the siLP (Figure S5). These trends were not observed in total cell counts in the spleen (Figure S4). We examined serum IgE levels in cKO lines and control mice and did not detect any differences between groups, and though in contrast to previously reported findings,22 this is likely due to a high baseline antibody titer in the Rorc+/+ FoxP3cre control line compared to wild-type (WT) B6 mice (Figure S6A). At baseline, there were no differences in the TFH compartment between cKO and controls in the spleen, MLN, or siLP (Figures S6B and S6C). To assess if the Type 2 skew of the cKO mice affected vaccine responses, both cKO and control mice received an OVA + alum immunization without oral antigen pre-exposure (Figure 2E). Both groups were able to produce anti-OVA-IgG and subclasses IgG1 and IgG2c at comparable levels, as well as IL-4 production upon OVA re-stimulation, indicating that lack of expression of RORγt within Tregs alone does not contribute to differences in vaccine response. Taken together, these data support the role of RORγt+ Tregs in controlling Type 2-associated immunity in gut tissue, though expression of RORγt within Tregs does not contribute to altered antibody titers upon systemic immunization.
Figure 2.
Compensatory increase in GATA3+ and IL-33R + subsets in mice lacking RORγt expression in Tregs does not impact vaccination
(A) Frequencies indicated Treg or Treg subset from Rorc+/+ FoxP3cre (control) or Rorcfl/fl FoxP3cre (cKO) mice in MLN or siLP at steady-state.
(B) Representative gating of RORγt+ Treg population from siLP of control (gray) or cKO (blue) mice.
(C and D) (C) Frequencies of indicated Tconv subsets in MLN or (D) siLP from control (gray) or cKO (blue) mice. Phenotyping data representative of 3 independent experiments. Data represents mean ± SD.
(E) Control or cKO mice were immunized with OVA+ alum at Day 0, boosted at Day 21, and assessed at Day 28 for serum antibody titers (OVA-IgG, IgG1 and IgG2c) and cytokine secretion of IL-4 after 72-h stimulation of splenocytes.
Data are pooled from 2 independent experiments and represented as mean ± SD. Unpaired t tests were used for statistical analysis. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Tolerance remains intact in mice lacking RORγt expression in Tregs upon immunization with alum
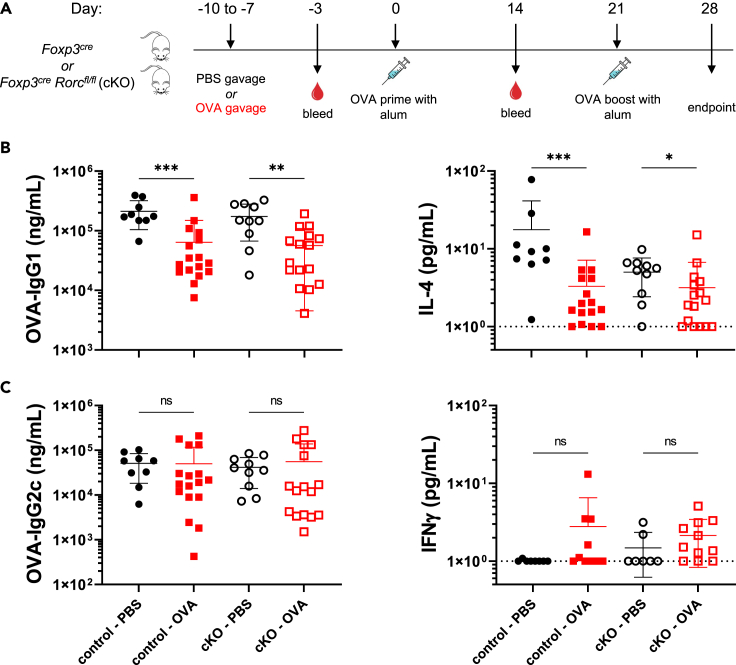
To examine the role of RORγt+ Tregs in oral tolerance to systemic vaccine responses, Rorcfl/fl FoxP3cre cKO mice and Rorc+/+ FoxP3cre controls were used in our tolerance model with alum adjuvant. Both cKO and control mice received OVA or PBS by oral gavage for 4 consecutive days ending one week prior to i.p. immunization with OVA formulated with alum (Figure 3A). We hypothesized that if RORγt expression in Tregs were necessary in regulating oral tolerance to a systemic vaccine, then cKO mice pre-exposed to OVA would have comparable antibody titers, specifically OVA-IgG1, to cKO mice that received PBS. To our surprise, we observed that Rorcfl/fl FoxP3cre cKO mice exposed to OVA maintained tolerance as measured by suppression of OVA-IgG1 and had antibody titers comparable to controls given OVA prior to immunization (Figure 3B). Upon OVA re-stimulation of splenocytes and analysis of secreted cytokines, there was suppression of IL-4 production in both Rorcfl/fl FoxP3cre cKO mice exposed to OVA prior to immunization compared to PBS treated controls (Figure 3B). All Rorc+/+ FoxP3cre controls also maintained tolerance as measured by suppression of IgG1 and IL-4 production in groups that received OVA gavage prior to immunization (Figure 3B). OVA-IgG2c titers and IFNγ production after OVA re-stimulation were comparable among all groups (Figure 3C), consistent with previous findings in WT mice (Figure 1D). In sum, we find that RORγt expression in Tregs is not necessary for the maintenance of oral tolerance to systemic vaccination using alum adjuvant.
Figure 3.
Tolerance remains intact in mice lacking RORγt expression in Tregs upon immunization with alum
Analysis of endpoint (Day 28) of mice gavaged with PBS (black) or OVA (red) from control mice (closed symbols) or cKO mice (open symbols).
(A) Experimental timeline.
(B) OVA-IgG1 titers and secreted IL-4 upon OVA-re-stimulation of splenocytes after 72 h.
(C) OVA-IgG2c titers and secreted IFNγ upon OVA-re-stimulation of splenocytes after 72 h.
Data are pooled from 3-4 independent experiments and represented as mean ± SD. Unpaired t tests were used for statistical analysis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
RORγt+ tregs frequencies increase upon exposure to the gut microbiota and upon immunization with various adjuvants
RORγt+ Tregs have been observed to be largely absent in GF mice, indicating the necessity of commensal stimulation for the development of this subset.23 After observing no differences in systemic immunization in mice lacking RORγt expression in Tregs, we set out to define the optimal strategy for oral exposure prior to immunization in GF and SPF mice. To this end, we characterized frequencies of Tregs and various subsets early in life. We assessed the baseline frequencies of SPF and GF mice over time before (day of life (DOL) 12–15), during (DOL 20–22), and after mice were weaned (DOL 26–29), as well as in adulthood (DOL 75–85). We observed that though there were minor differences in abundance of total FoxP3+ cells between groups at each timepoint, there were no consistently significant differences between Treg frequencies in spleen, MLN, siLP or colonic lamina propria (cLP) (Figure S7A). Despite similarities of Treg frequencies, we saw a significant increase of RORγt+ FoxP3+ Treg frequencies in the spleen, MLN, siLP, and cLP in adult SPF mice compared to GF counterparts of the same age (Figure S7B). Similar trends were observed with cell numbers (Figure S7C). Recent work suggests that development of RORγt+ Tregs is dependent on MHCII+ RORγt+ antigen-presenting cells, including ILC3s.29,36,37,38 Early in life, prior to diverse exposures to commensal microbes, the RORγt+ Treg compartment is not developed in SPF mice. We observed no differences in this subset between SPF and GF mice early in life prior to weaning. Our data confirm that development of RORγt+ Tregs occurs within the first few weeks of life and leads to establishment of this subset in adult mice. We did not observe differences in other Treg subsets including T-bet+ RORγt- FoxP3+ or GATA3+ RORγt- FoxP3+ cells between adult SPF and GF mice (data not shown). Together, these data indicate that RORγt+ Treg frequencies increase during the post-weaning period into adulthood in SPF mice compared to GF settings.
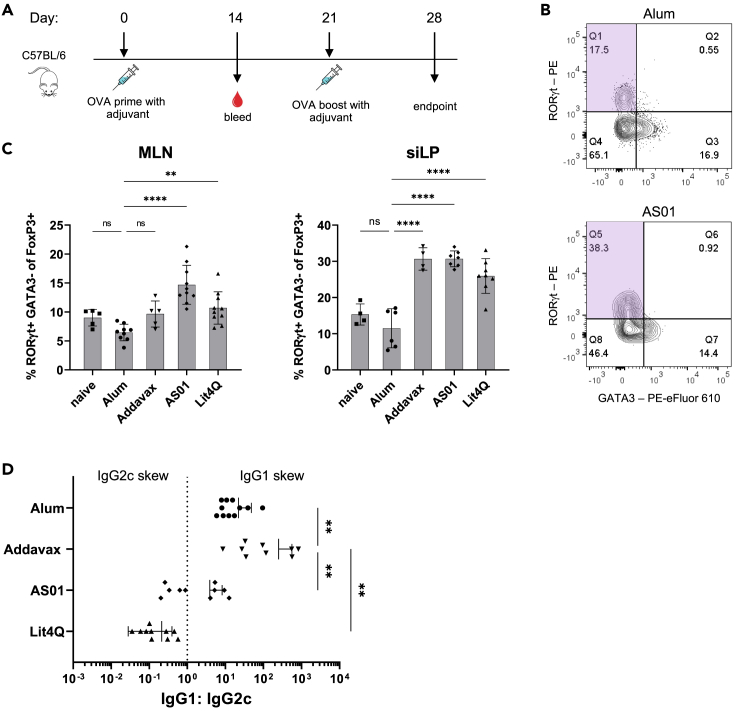
Next, we turned to other factors that could alter RORγt+ Treg population dynamics. To determine how immunization alters frequencies of these cells, we assessed whether this subset is altered by utilizing various adjuvants (Figure 4A). Though alum, the first licensed adjuvant used clinically, was used for many original studies of tolerance, stronger immune stimuli are now used in the development of vaccine candidates.39 Despite their use in licensed vaccines for seasonal influenzas and shingles, the mechanisms of action of adjuvants like AddaVax, similar to MF59, and AS01 are incompletely understood. Characterization of how antigen pre-exposure affects immune suppression upon vaccination with these adjuvants has not been examined prior to this study.
Figure 4.
RORγt+ Tregs frequencies increase upon immunization with various adjuvants
(A) Experimental timeline for remaining figure panels.
(B) Representative gating of RORγt+ Tregs upon immunization with Alum (top) or AS01 (bottom).
(C) Frequencies of RORγt+ Tregs in mice immunized with indicated adjuvant.
(D) Ratio of IgG1:IgG2c titers for individual mice.
Data are pooled from 9 independent experiments and represented as mean ± SD; each adjuvant was tested in 2–3 independent experiments apart from the naive mouse group. Unpaired t tests were used for statistical analysis. ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
Alum induces Th2 cells and facilitates antigen-specific antibody production via NLRP3 activation.40,41 AddaVax is associated with increased TFH formation and acts through NLRP3 independent pathways by recruiting CD11b+ DCs that stimulate both Th1 and Th2 responses.42,43 AS01, comprised of 3D-MPL and QS-21, signals through TLR-4 and induces caspase-1 activation that elicits a balanced Th1 and Th2 response when used in a vaccine.39,44 The novel compound Lit4Q also contains a TLR-4 agonist in association with QS-21 and is expected to lead to comparable responses to AS01 in vivo when given at the same dose (personal communication, PAI Life Sciences). One report using a Complete Freund’s Adjuvant (CFA) vaccine suggests that use of a more potent adjuvant may lead to expansion of RORγt+ Treg in gut-associated tissues.32 In naive mice, frequencies of RORγt+ Tregs locally in MLN and siLP were 9% and 15% respectively, consistent with previous reports23 (Figure 4C). Upon immunization with alum, frequencies of these cells remained relatively unaffected in each tissue. When AddaVax, AS01 and Lit4Q were used to adjuvant OVA immunization, we observed increased frequencies of RORγt+ Tregs in the MLN and siLP (Figures 4B and 4C).
To assess the skew of each of these adjuvants varied responses, we compared IgG1:IgG2c titers in mice after OVA immunization in the absence of oral OVA pre-exposure. While immunization with alum, AddaVax, and AS01 led to a skew toward OVA-specific IgG1, Lit4Q lead to a skew toward IgG2c production, indicating differences in adjuvant mechanism of action (Figure 4D). Therefore, proportions of RORγt+ Tregs in gut tissue are increased in C57BL/6 mice immunized with strong immunostimulatory adjuvants.
Suppression of Type 2 responses maintained upon oral exposure prior to vaccination with various adjuvants
When using alum, Type 2 immune responses were suppressed upon oral exposure to the vaccine antigen (Figure 1). The Type 2 skewing of alum and subsequent suppression of Type 2 immune response upon oral exposure led us to question whether this effect remains when stronger adjuvants or those that elicit a balanced Type 1 and 2 response were used. To this end, we used AddaVax, AS01, and Lit4Q in our model of oral tolerance. Using each of these adjuvants with OVA in our tolerance model in C57BL/6 mice (Figure 1A), we observed suppression of OVA-IgG1, but not OVA-IgG2c, in mice that received OVA gavage prior to immunization compared to PBS gavaged controls regardless of adjuvant strength or skew (Figures 5A and 5B). Overall, both AS01 and Lit4Q adjuvanted vaccines led to greater production of IgG1 and IgG2c antibody titers compared to other adjuvants tested. Similarly, we observed that mice gavaged with OVA prior to immunization with AddaVax and Lit4Q led to suppression of IL-4 upon OVA re-stimulation of splenocytes (Figure 5A). Mice immunized with AS01 had higher overall IL-4 production and no differences between groups. Because AS01 elicited the greatest IgG1 response among those examined, the OVA re-stimulation of splenocytes may have led to higher overall IL-4 production that was not able to be suppressed by pre-exposure to vaccine antigen. No differences were observed in IFNγ production upon OVA re-stimulation, indicating that all the adjuvants suppressed only Type 2, and not Type 1 mediating immune responses upon immunization following oral antigen exposure (Figures 5A and 5B).
Figure 5.
Suppression of Type 2 responses maintained upon oral exposure prior to vaccination with various adjuvants
Experimental timeline from Figure 1A was used with various adjuvants.
(A) OVA-IgG1 titers and secreted IL-4 upon OVA-re-stimulation of splenocytes after 72 h.
(B) OVA-IgG2c titers and secreted IFNγ upon OVA-re-stimulation of splenocytes after 72 h.
For each adjuvant, data are pooled from 2 to 3 independent experiments and represented as mean ± SD. Unpaired t tests were used for statistical analysis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Consistent with our findings using alum, there were no observable impact of oral OVA exposure on Treg or RORγt+ Treg frequencies in the MLN and siLP compared to mice given PBS prior to immunization (Figures S8A and S8B). There were no differences between OVA gavage and PBS gavage on the TFH compartments of spleen, MLN, and siLP, regardless of adjuvant (Figure S9). These data indicate that suppression of Type 2 vaccine-induced immunity is not specific to adjuvants such as alum that skew toward Type 2 but is consistent for a variety of adjuvants examined in our model.
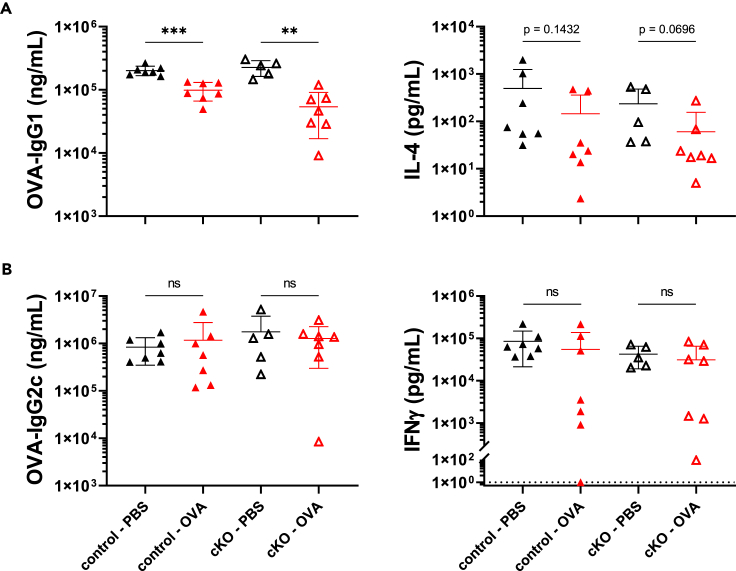
Tolerance maintained in mice lacking RORγt expression in Tregs with Lit4Q adjuvanted immunization
After testing multiple adjuvants in our tolerance model, Lit4Q was chosen for use in the conditional knock-out model of RORγt+ Tregs to examine whether use of different adjuvants would be sufficient for breaking tolerance in a systemic vaccine. Our previous data indicated that use of Lit4Q led to an increased frequency of RORγt+ Tregs compared to immunization with alum in both the MLN and siLP (Figure 4C) and favored IgG2c production over IgG1 (Figures 5A and 5B). Therefore, we decided to investigate if immunization with Lit4Q be sufficient for recovery of IgG1 upon oral OVA exposure in mice lacking RORγt expression in Tregs. We hypothesized that tolerance could be broken in mice lacking RORγt+ expression in Tregs when orally exposed to the vaccine antigen prior to vaccination with Lit4Q. To test this, a similar model to Figure 3A was utilized, but using a Lit4Q adjuvanted OVA vaccine. Similar to alum-adjuvanted immunization, we observed that upon use of Lit4Q, OVA-IgG1 was suppressed in both Rorcfl/fl FoxP3cre cKO and Rorc+/+ FoxP3cre control mice gavaged with OVA prior to immunization (Figure 6A). Upon OVA re-stimulation of splenocytes, OVA gavaged groups had a trend toward suppression of IL-4 production, though these did not reach the threshold for statistical significance (Figure 6A). In line with our findings using alum (Figure 3C) and in WT C57BL/6 mice (Figure 5B), there were no differences in OVA-IgG2c production or IFNγ upon OVA re-stimulation between mice of different genotypes or groups pre-exposed to oral OVA compared to PBS (Figure 6B). This indicates that regardless of adjuvant strength of skew, RORγt expression within Tregs is dispensable for tolerance to a systemic vaccine following oral antigen exposure.
Figure 6.
Tolerance maintained in mice lacking RORγt expression in Tregs with Lit4Q adjuvanted immunization
Experimental timeline from Figure 4A was used with Lit4Q as adjuvant in immunization. Control mice (closed symbols) or cKO mice (open symbols) were gavaged with PBS (black) or OVA (red) prior to immunization.
(A) OVA-IgG1 titers and secreted IL-4 upon OVA-re-stimulation of splenocytes after 72 h.
(B) OVA-IgG2c titers and secreted IFNγ upon OVA-re-stimulation of splenocytes after 72 h.
Data are pooled from 2 independent experiments and represented as mean ± SD. Unpaired t tests were used for statistical analysis. ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
Our data indicate that RORγt expression in Tregs is dispensable for regulating oral tolerance to systemic vaccination. Mice that were exposed to OVA prior to immunization displayed a reduction in OVA-IgG1 and IL-4 production upon re-stimulation, however, we did not observe suppression of Type 1 immunity, including OVA-IgG2c or IFNγ production. To dissect the role of RORγt+ Tregs, we used a conditional knock-out model (Rorcfl/fl FoxP3cre) and observed a local increase of Th2 cells, along with GATA3+ and IL-33R + Treg subsets. In these cKO mice, tolerance to alum immunization was maintained following oral OVA exposure, indicating that expression of the transcription factor RORγt was dispensable within Tregs for mediating the function of suppressing systemic antibody titers. We then characterized RORγt+ Treg frequencies in multiple settings; in early-life, no differences in this population were observed between GF and SPF mice, but as this subset develops upon commensal exposure in SPF mice, there was a significant increase in adult mice. To test whether this subset contribute to tolerance to other systemic vaccines following oral antigen exposure in other settings, we expanded our system to utilize various adjuvants. Upon use of TLR4 agonists AS01 and Lit4Q, frequencies of RORγt+ Tregs were increased compared to naive groups and alum immunized mice. Lit4Q adjuvant which skews toward OVA-IgG2c production and leads to development of an increased antibody titers compared to immunization with alum. Oral OVA exposure prior to OVA + Lit4Q immunization led to similar suppression of Type 2 immunity. In mice lacking RORγt expression within Tregs, immunization with Lit4Q lead to similar maintenance of tolerance, indicating that the type of adjuvant used likely does not determine how systemic immunization is controlled upon prior oral antigen exposure. Here, we found that tolerance to systemic vaccination following oral antigen pre-exposure remains intact in the absence of RORγt expression within Tregs, regardless of adjuvant used in the immunization.
In our model, we disrupt RORγt expression within Tregs, though these pTregs, albeit lacking RORγt, are still present. It is possible that genes controlled by RORγt are non-essential for the maintenance of tolerance to systemic vaccination, though the presence of the cell is still required. While various genes are upregulated in colonic Tregs,23 which consist of ∼40% RORγt+ Tregs, the distinct role this transcription factor and downstream pathways play in the phenotype of the cell has not been well characterized. Sequencing experiments in various settings will be critical in understanding how these cells function. Previous work has also characterized the role of RORγt+ Tregs in suppressing oral vaccine response in environmental enteric dysfunction.33 These studies found vaccine-specific T cells and IgA were altered in the absence of RORγt+ Tregs, though the consequences on systemic vaccination and in the context of non-dysregulated systems remains unknown. Understanding how RORγt+ Tregs may function differently in oral vs. systemic vaccines will be essential for improving vaccine outcomes.
By utilizing a conditional knock-out model, our cKO mice lacking RORγt expression in Tregs exhibit dysregulated local immunity, though reduction of this subset did not contribute to differences in Treg frequencies between groups in the siLP (Figure 2A). Immunization with adjuvants like Lit4Q boosts RORγt+ Treg frequencies above baseline, similar to findings that have been reported for CFA.32 Whether targeting cells in the local gut immune system could affect systemic vaccine responses is not known. By exposing mice to certain commensals, cells in the siLP can be skewed; segmented filamentous bacteria contributes to local Th17 induction,45 while Helicobacter hepaticus elicits RORγt+ Treg expansion at steady-state.46 Addition of commensals from wild mice leads to a more accurate model of human environmental encounters.47 Our model also introduces OVA as an innocuous, dietary exposure, while expression of OVA in a commensal could contribute to innate signaling that is more representative of commensal exposures. Further investigation of how the addition of such perturbations to the system could impact systemic immunization is warranted.
We observed that lack of expression of RORγt in Tregs led to upregulation of other Treg subsets, specifically of GATA3+ Tregs in the MLN and siLP. GATA3+ Tregs are largely thymically-derived, and cKO of GATA3+ Tregs leads to development of inflammatory disorders in mice, indicating these cells control Type 2 immunity.23,26 The increased frequency of these cells may compensate for the lack of RORγt+ expression, as we did observe an increased Treg frequency in the spleen and MLN of cKO mice compared to controls (Figures 2A and 2E). Specific deletion of GATA3+ within Tregs leads to inflammatory disorders and increased susceptibility to allergic reactions.26,34 Studies suggest that dietary antigens induce pTregs in the siLP, but this population is distinct from pTregs induced by microbial antigens.9,34 Whether these distinct subsets play a role in controlling tolerance to systemic vaccination is unclear. It also remains a possibility that Tregs that only express FoxP3 and no transcription factors associated with other T-helper subsets are critical for this response. Recent work has highlighted the necessity of MHCII+ RORγt+ APC subsets for formation of pTregs,36,37,38 though the precise mechanism by which this occurs is not well understood. Further work in characterizing the role of various APC populations in mediating oral tolerance to a systemic vaccine would provide critical insight to this system.
Our model utilizes primarily adult mice in the context of health, though further studies could focus on models of enteric dysfunction, such as was utilized to study of RORγt+ Tregs during oral vaccination.33 Utilizing a model of vaccination against a specific pathogen followed by challenge studies would provide further understanding of the mechanisms involved in r tolerance to systemic vaccines. Examining various timepoints early in life and how RORγt+ Tregs develop during weaning would also provide further insights. Encounters with gut commensals early in life are critical for proper development of pTreg subsets and the local immune system necessary to tolerize innocuous interactions.31,48,49 Variation in laboratory mouse models have also been reported to display altered RORγt+ Tregs frequencies at baseline.50 By disrupting the state of health, altering the timing of exposure to OVA, or testing other strains, a greater understanding of tolerance upon systemic vaccination would be obtained.
In clinical settings, heterogeneous responses to vaccination between individuals results in difficulties predicting success.51 Environmental exposures, including pre-existing immunity (prior vaccinations or infections), microbiota composition, genetics, and epigenetic factors each contribute to how an individual responds following immunization.52,53,54 It is also apparent that components of the vaccine, including adjuvant, immunogen, and route of administration ultimately contribute to effectiveness. These factors must be understood to ensure increased vaccine efficacy and reduce the spread of harmful infections. Specifically, can the system be altered prior to vaccination to better position the immune system to respond? Ongoing studies from many groups aim to address these outstanding questions.55,56 The scope of this work may extend to advance our understanding of the mechanisms involved in autoimmunity and allergy, both of which will be important for improving human health. We intend for this work to elucidate mechanisms of how signals from antigens encountered in the small intestine influence local and systemic immune responses that could be leveraged to improved vaccine efficacy.
Limitations of the study
While our study examines the role of the transcription factor RORγt in Tregs, it does not directly address the role of RORγt+ Tregs or the role of the peripherally derived Treg subset in altering systemic vaccination following oral antigen exposure.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD3e (BUV 395) | BD | Cat# 565992; RRID: AB_2739443 |
| Anti-CD4 (BUV 496) | BD | Cat# 612952; RRID: AB_2813886 |
| Anti-CD4 (PerCP-eFluor 710) | Thermo Fisher Scientific | Cat# 46-0042-82; RRID: AB_1834431 |
| Anti-IL-33R (BUV 563) | BD | Cat# 749324; RRID: AB_2873698 |
| Anti-CD8a (BUV 737) | BD | Cat# 612759; RRID: AB_2870090 |
| Anti-CD45 (BUV 805) | BD | Cat# 748370; RRID: AB_2872789 |
| Anti-CXCR5 (BV 421) | BD | Cat# 562889; RRID: AB_2737868 |
| Anti-CXCR5 (BV 421) | Biolegend | Cat# 145512; RRID: AB_2562128 |
| Anti-CD62L (BV 711) | Biolegend | Cat# 104445; RRID: AB_256421 |
| Anti-CD44 (Alexa Fluor 700) | Biolegend | Cat# 103026; RRID: AB_493713 |
| Anti-CD25 (APC-eFluor 780) | Thermo Fisher Scientific | Cat# 47-0251-82; RRID: AB_1272179 |
| Anti-PD-1 (PE/Cyanine7) | Biolegend | Cat# 135216; RRID: AB_10689635 |
| Anti-GATA3 (PE-eFluor 610) | Thermo Fisher Scientific | Cat# 61-9966-42; RRID: AB_2574686 |
| Anti-FoxP3 (APC) | Thermo Fisher Scientific | Cat# 17-5773-82; RRID: AB_469457 |
| Anti-FoxP3 (FITC) | Thermo Fisher Scientific | Cat# 11-5773-82; RRID: AB_465243 |
| Anti-RORγt (PE) | Thermo Fisher Scientific | Cat# 12-6981-82; RRID: AB_10807092 |
| Anti-RORγt (BV650) | BD | Cat# 564722; RRID: AB_2738915 |
| Anti-CD19 (BV510) | Biolegend | Cat# 115546; RRID: AB_2562137 |
| Anti-F4/80 (BV510) | Biolegend | Cat# 123135; RRID: AB_2562622 |
| Anti-CD11b (BV510) | Biolegend | Cat# 101245; RRID: AB_2561390 |
| Anti-IgG-HRP | SouthernBiotech | Cat# 1033-05; RRID: AB_2737432 |
| Anti-IgG-HRP | BioLegend | Cat# 405306; RRID: AB_31500 |
| Anti-IgG1-HRP | Thermo Fisher Scientific | Cat# A10551; RRID: AB_2534048 |
| Anti-IgG2c-HRP | Thermo Fisher Scientific | Cat# PA1-29288; RRID: AB_10983148 |
| Anti-IgG2b-HRP | Thermo Fisher Scientific | Cat# M32407; RRID: AB_2536647 |
| Anti-CD3e | Thermo Fisher Scientific | Cat# 16-0031-82; RRID: AB_468847 |
| Anti-CD28 | Thermo Fisher Scientific | Cat# 16-0281-82; RRID: AB_468921 |
| Chemicals, peptides, and recombinant proteins | ||
| Endofit Ovalbumin | Invivogen | vac-pova-100 |
| Alum - Alhydrogel adjuvant 2% | Invivogen | vac-alu-250 |
| Addavax | Invivogen | vac-adx-10 |
| Critical commercial assays | ||
| Th1/Th2 mouse uncoated ELISA kits | Invitrogen | 88-7711-44 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | Jackson Laboratory | 000664 |
| Mouse: C57BL/6 FoxP3cre | Jackson Laboratory | 016959 |
| Mouse: C57BL/6 Rorctm3Litt | Jackson Laboratory | 008771 |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad | Version 9 |
| FlowJo version 10.6.0 | FlowJo | 10.6.0 |
| Other | ||
| AccuraCeck count beads | Life Technologies | PCB100 |
| ACK lysis buffer | Invitrogen | 501129751 |
| Live/Dead viability dye | Thermo Scientific | L34962 |
| Live/Dead viability dye | Thermo Scientific | L34966 |
| TMB | Invitrogen | 00-4201-56 |
| FoxP3/transcription factor fixation/permeabilization reagent | Fisher Scientific | 00-5523-00 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, James Kublin (jkublin@fredhutch.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze reported data will be shared by the lead contact upon request.
Experimental model details
Mice
C57BL/6 mice were obtained from The Jackson Laboratory (strain 000664) and maintained in specific-pathogen free conditions at Fred Hutchinson Cancer Center. C57BL/6 GF mice were bred in-house at the University of Washington Gnotobiotic Animal Core and housed in germ-free conditions. FoxP3cre mice on the C57BL/6 background contain a YFP at this locus57 and were received from the Lund lab (Jackson Laboratory, strain 016959). Rorcfl/fl mice, also known as Rorctm3Litt/J were ordered from Jackson Laboratory58 (strain 008771). These strains were crossed to generate the Rorcfl/fl FoxP3cre line used in this study. All mice used in were males and were between 6 and 16 weeks old at the beginning of each experiment (except for early life experiments, Figure S7). Similar results were generated in female mice (data not shown) and data from males was reported for consistency. Experiments were approved by the Institutional Animal Care and Use Committees (IACUC), and all mice were euthanized following AVMA guidelines for CO2 overdose.
Method details
Mouse tissue collection and processing
Single-cell suspensions were prepared from mouse tissues. Spleens were homogenized and treated with ACK lysis buffer (Invitrogen; 501129751) to remove red blood cells. Lymphocytes were then washed and resuspended in PBS +2% FBS. MLNs were homogenized, washed, and resuspended in phosphate buffered saline (PBS) + 2% fetal bovine serum (FBS). To obtain lymphocytes from the small intestine lamina propria, intestines were removed from mice, then cleaned of remaining fat. Peyer’s patches were removed and discarded. The tissue was then sectioned into 6 sections and cut lengthwise to remove intestinal contents. Segments were rinsed in PBS +2% FBS and mucus was removed with tweezers, then rinsed again in clean PBS +2% FBS and placed in a 50 mL conical containing no media. Following dissection, samples were resuspended with 10 mL of Solution A (HBSS without calcium and magnesium +4.2 mM sodium bicarbonate +2% FBS, pH 7.4). Samples were vortexed for 20 s between washes and washed a total of three times with Solution A. Samples were then resuspended in Solution B (Solution A + 5 mM EDTA +1 mM DTT) and incubated at 37°C while shaking for 15 min. Samples were then vortexed for 30 s and resuspended with 25 mL of Solution C (Solution A + 5 mM EDTA) and incubated at 37°C while shaking for 30 min. Intestinal fragments were vortexed for 30 s, washed in Solution A to remove EDTA, then resuspended in 5 mL of collagenase digestion media (HBSS containing Calcium and Magnesium +4.2 mM sodium bicarbonate +10 mM HEPES +0.25 mg/mL Type-II collagenase +1% FBS +1000 Kunitz/mL of DNAse) for 30 to 45 min at 37°C while shaking. Following digestion, 5 mM EDTA was added to halt collagenase activity. Samples were passed through a 18g needle to mechanically disrupt tissue and passed through 100 mm filter, rinsed with cRPMI and passed through a 70 mm filter. Each tissue was then counted on a hemocytometer and normalized for flow cytometry staining.
Ovalbumin oral gavage
Endofit Ovalbumin (Invivogen, vac-pova-100) was reconstituted to 10 mg/mL per vendor instructions and stored at −20°C. Mice were treated with either 1 mg OVA or PBS by oral gavage for four consecutive days. Both groups received the final treatment with OVA or PBS one week prior to OVA immunization.
Ovalbumin immunization
Mice were immunized with 10 μg of Endofit Ovalbumin diluted in PBS with adjuvant. Adjuvants doses & routes are as follows: 100 μg alum (Invivogen, Alhydrogel adjuvant 2%), 25 μL Addavax (Invivogen), both administered intraperitonially, or 4.5 μg AS01, 4.5 μg Lit4Q, both administered subcutaneously at base of tail.
Cell staining for flow cytometry
Upon generating single cell suspension, cells were incubated on ice for 20 min with CD16/32 and live dead blue or aqua viability dye (Thermo Scientific L34962, L34966). The flow panels used include surface and intracellular antibodies: anti-CD3e (BD, clone 145-2C11, Brilliant Ultraviolet 395), anti-CD4 (BD, clone GK1.5, Brilliant Ultraviolet 496 or Invitrogen, clone RM4-5, PerCP-eFluor 710) anti-IL-33R (BD, clone U29-93, Brilliant Ultraviolet 563), anti-CD8a (BD, clone 53-6.7, Brilliant Ultraviolet 737), anti-CD45 (BD, clone 30-F11, Brilliant Ultraviolet 805), anti-CXCR5 (Biolegend or BD, clone L138D7 or 2G8, Brilliant Violet 421), anti-CD62L (Biolegend, clone MEL-14, Brilliant Violet 711), anti-CD44 (Biolegend, clone IM7, Alexa Fluor 700), anti-CD25 (Invitrogen, clone PC61.5, APC-eFluor 780), anti-PD-1 (Biolegend, 29F.1A12, PE/Cyanine7), anti-GATA3 (Invitrogen, clone TWAJ, PE-eFluor 610), anti-FoxP3 (Invitrogen, clone FJK-16s, APC or FITC), anti-RORγt (BD or Invitrogen, clone Q31–378 or B2D, Brilliant Violet 650 or PE), anti-CD19 (Biolegend, clone 6D5, Brilliant Violet 510), anti-F4/80 (Biolegend, clone BM8, Brilliant Violet 510), anti-CD11b (Biolegend, clone M1/70, Brilliant Violet 510). Intracellular proteins were detected using a FoxP3/transcription factor fixation/permeabilization reagent (Fisher Scientific 00-5523-00). Each sample was collected using the BD FACSymphany instrument from the HIV Vaccine Trails Network flow core. Cell counts were analyzed using AccuraCheck count beads (Life Technologies, PCB100) added to each sample prior to acquisition.
ELISA for OVA-specific immunoglobulins
Mouse serum was heat inactivated at 56°C for 30 min prior to storage at 4°C. Plates were coated with 4 μg/mL OVA overnight in 0.1 M NaHCO3. Serum was diluted in milk buffer and incubated at 37°C for 1 h on plates. Secondary antibodies specific for relevant Ig-subclass linked to HRP (SouthernBiotech 1033-05, BioLegend 405306, Thermo Fisher Scientific PA129288, M32407, A10551) were then incubated on plates for 1 h. TMB (Invitrogen 00-4201-56) was used to develop plates and reaction was stopped with 2N H2SO4. Absorbance at 450 nm was read on a SpectraMax i3x plate reader. Samples were run as technical duplicates.
Re-stimulation of splenocytes with OVA
Upon single cell suspension of splenocytes and normalizing counts, cells were plated on a 12-well flat bottom plate at 107 cells/mL and incubated for 72 h at 37°C with cRPMI containing stimulation. Stimulation conditions include unstimulated media only control (used to subtract background signal), Endofit OVA (10 μg/mL), or positive control anti-CD3e (0.5 μg/mL, Invitrogen, clone 145-2C11) and anti-CD28 (0.25 μg/mL Invitrogen, clone 37.51). After incubation, supernatant was collected and frozen for analysis of cytokine secretion by ELISA. Th1/Th2 mouse uncoated ELISA kits from Invitrogen were used and kit instructions were followed to analyze secreted IL-4, IFNγ and IL-10. Samples were run as technical duplicates. For analysis of cytokine production, 1 pg/mL was added to all data points to assess on log scale.
Software
Relevant programs were used for analysis and include GraphPad Prism 9 and FlowJo (version 10.6.0). BioRender was used for creation of graphical abstract.
Quantification and statistical analysis
Data was log transformed for statistical testing on graphs that use a log scale. Each data point represents an individual animal and data is represented as mean ± standard deviation (error bars), unless otherwise noted. Statistical tests were performed using GraphPad Prism 9 software analysis tool using a Gaussian distribution and assuming the same SDs between groups. Details of statistical tests provided in respective figure legends. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 for all figures.
Acknowledgments
We thank Dr. Sean Gray (PAI Life Sciences) for providing adjuvant Lit4Q and Dr. Marguerite Koutsoukos and Dr. Clarisse Lorin (GSK) for providing adjuvant AS01. We thank the Lund lab for providing FoxP3cre mice, as well as providing crucial support for these studies. In addition, we thank Dr. Martin Prlic and lab, as well as our collaborators, Dr. Sam Minot, Dr. Andrew Fiore-Gartland, and Dr. Koshlan Mayer-Blackwell for thoughtful discussions and their contributes to this research. We also thank Fred Hutchinson Cancer Center (FHCC) Comparative Medicine and the HIV Vaccine Trials Network flow cytometry core for support on this work. This work was funded by 1R01AI127100 (to JGK). The University of Washington ITHS TL1 training grant UL1TR002319 was used to support this work (to NBP).
Author contributions
N.B.P., A.M.F.J., J.G., K.H., K.H.L-W., P.V., L.W., and I.C.T. helped with dissection, tissue collection and processing. Data was analyzed by N.B.P. Conception of the work by N.B.P., J.M.L., and J.G.K. N.B.P. wrote the first draft of the manuscript. All authors received the manuscript and provided comments as necessary prior to submission.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 22, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108504.
Supplemental information
References
- 1.Cram J.A., Fiore-Gartland A.J., Srinivasan S., Karuna S., Pantaleo G., Tomaras G.D., Fredricks D.N., Kublin J.G. Human gut microbiota is associated with HIV-reactive immunoglobulin at baseline and following HIV vaccination. PLoS One. 2019;14 doi: 10.1371/journal.pone.0225622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams W.B., Liao H.X., Moody M.A., Kepler T.B., Alam S.M., Gao F., Wiehe K., Trama A.M., Jones K., Zhang R., et al. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015;349:aab1253. doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price D.N., Kusewitt D.F., Lino C.A., McBride A.A., Muttil P. Oral Tolerance to Environmental Mycobacteria Interferes with Intradermal, but Not Pulmonary, Immunization against Tuberculosis. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trama A.M., Moody M.A., Alam S.M., Jaeger F.H., Lockwood B., Parks R., Lloyd K.E., Stolarchuk C., Scearce R., Foulger A., et al. HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe. 2014;16:215–226. doi: 10.1016/j.chom.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao H.X., Chen X., Munshaw S., Zhang R., Marshall D.J., Vandergrift N., Whitesides J.F., Lu X., Yu J.S., Hwang K.K., et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J. Exp. Med. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cram J.A., Hager K.W., Kublin J.G. Utilizing gnotobiotic models to inform the role of the microbiome in vaccine response heterogeneity. Curr. Opin. HIV AIDS. 2018;13:1–8. doi: 10.1097/COH.0000000000000422. [DOI] [PubMed] [Google Scholar]
- 7.Mayer-Blackwell K., Johnson A.M., Potchen N., Minot S.S., Heptinstall J., Seaton K., Sawant S., Shen X., Tomaras G.D., Fiore-Gartland A., Kublin J.G. Multi-trial analysis of HIV-1 envelope gp41-reactive antibodies among global recipients of candidate HIV-1 vaccines. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.983313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jong S.E., Olin A., Pulendran B. The Impact of the Microbiome on Immunity to Vaccination in Humans. Cell Host Microbe. 2020;28:169–179. doi: 10.1016/j.chom.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda K., Littman D.R. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 10.Moreau M.C., Gaboriau-Routhiau V. The absence of gut flora, the doses of antigen ingested and aging affect the long-term peripheral tolerance induced by ovalbumin feeding in mice. Res. Immunol. 1996;147:49–59. doi: 10.1016/0923-2494(96)81548-3. [DOI] [PubMed] [Google Scholar]
- 11.Kulkarni D.H., Gustafsson J.K., Knoop K.A., McDonald K.G., Bidani S.S., Davis J.E., Floyd A.N., Hogan S.P., Hsieh C.S., Newberry R.D. Goblet cell associated antigen passages support the induction and maintenance of oral tolerance. Mucosal Immunol. 2020;13:271–282. doi: 10.1038/s41385-019-0240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esterházy D., Loschko J., London M., Jove V., Oliveira T.Y., Mucida D. Classical dendritic cells are required for dietary antigen-mediated induction of peripheral T(reg) cells and tolerance. Nat. Immunol. 2016;17:545–555. doi: 10.1038/ni.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esterházy D., Canesso M.C.C., Mesin L., Muller P.A., de Castro T.B.R., Lockhart A., ElJalby M., Faria A.M.C., Mucida D. Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature. 2019;569:126–130. doi: 10.1038/s41586-019-1125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadis U., Wahl B., Schulz O., Hardtke-Wolenski M., Schippers A., Wagner N., Müller W., Sparwasser T., Förster R., Pabst O. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Mucida D., Kutchukhidze N., Erazo A., Russo M., Lafaille J.J., Curotto de Lafaille M.A. Oral tolerance in the absence of naturally occurring Tregs. J. Clin. Invest. 2005;115:1923–1933. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vignali D.A.A., Collison L.W., Workman C.J. How regulatory T cells work. Nat. Rev. Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cebula A., Seweryn M., Rempala G.A., Pabla S.S., McIndoe R.A., Denning T.L., Bry L., Kraj P., Kisielow P., Ignatowicz L. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lochner M., Peduto L., Cherrier M., Sawa S., Langa F., Varona R., Riethmacher D., Si-Tahar M., Di Santo J.P., Eberl G. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J. Exp. Med. 2008;205:1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang B.H., Hagemann S., Mamareli P., Lauer U., Hoffmann U., Beckstette M., Föhse L., Prinz I., Pezoldt J., Suerbaum S., et al. Foxp3(+) T cells expressing RORgammat represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2016;9:444–457. doi: 10.1038/mi.2015.74. [DOI] [PubMed] [Google Scholar]
- 20.Park J.H., Eberl G. Type 3 regulatory T cells at the interface of symbiosis. J. Microbiol. 2018;56:163–171. doi: 10.1007/s12275-018-7565-x. [DOI] [PubMed] [Google Scholar]
- 21.Pratama A., Schnell A., Mathis D., Benoist C. Developmental and cellular age direct conversion of CD4+ T cells into RORgamma+ or Helios+ colon Treg cells. J. Exp. Med. 2020;217 doi: 10.1084/jem.20190428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohnmacht C., Park J.H., Cording S., Wing J.B., Atarashi K., Obata Y., Gaboriau-Routhiau V., Marques R., Dulauroy S., Fedoseeva M., et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 23.Sefik E., Geva-Zatorsky N., Oh S., Konnikova L., Zemmour D., McGuire A.M., Burzyn D., Ortiz-Lopez A., Lobera M., Yang J., et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faustino L.D., Griffith J.W., Rahimi R.A., Nepal K., Hamilos D.L., Cho J.L., Medoff B.D., Moon J.J., Vignali D.A.A., Luster A.D. Interleukin-33 activates regulatory T cells to suppress innate gammadelta T cell responses in the lung. Nat. Immunol. 2020;21:1371–1383. doi: 10.1038/s41590-020-0785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch M.A., Tucker-Heard G., Perdue N.R., Killebrew J.R., Urdahl K.B., Campbell D.J. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Su M.A., Wan Y.Y. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanoue T., Atarashi K., Honda K. Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 2016;16:295–309. doi: 10.1038/nri.2016.36. [DOI] [PubMed] [Google Scholar]
- 28.Lathrop S.K., Bloom S.M., Rao S.M., Nutsch K., Lio C.W., Santacruz N., Peterson D.A., Stappenbeck T.S., Hsieh C.S. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephen-Victor E., Chatila T.A. An embarrassment of riches: RORgammat(+) antigen-presenting cells in peripheral tolerance. Immunity. 2022;55:1978–1980. doi: 10.1016/j.immuni.2022.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdel-Gadir A., Stephen-Victor E., Gerber G.K., Noval Rivas M., Wang S., Harb H., Wang L., Li N., Crestani E., Spielman S., et al. Microbiota therapy acts via a regulatory T cell MyD88/RORgammat pathway to suppress food allergy. Nat. Med. 2019;25:1164–1174. doi: 10.1038/s41591-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knoop K.A., McDonald K.G., Hsieh C.S., Tarr P.I., Newberry R.D. Regulatory T Cells Developing Peri-Weaning Are Continually Required to Restrain Th2 Systemic Responses Later in Life. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.603059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim B.S., Lu H., Ichiyama K., Chen X., Zhang Y.B., Mistry N.A., Tanaka K., Lee Y.H., Nurieva R., Zhang L., et al. Generation of RORgammat(+) Antigen-Specific T Regulatory 17 Cells from Foxp3(+) Precursors in Autoimmunity. Cell Rep. 2017;21:195–207. doi: 10.1016/j.celrep.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharjee A., Burr A.H.P., Overacre-Delgoffe A.E., Tometich J.T., Yang D., Huckestein B.R., Linehan J.L., Spencer S.P., Hall J.A., Harrison O.J., et al. Environmental enteric dysfunction induces regulatory T cells that inhibit local CD4+ T cell responses and impair oral vaccine efficacy. Immunity. 2021;54:1745–1757.e7. doi: 10.1016/j.immuni.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim K.S., Hong S.W., Han D., Yi J., Jung J., Yang B.G., Lee J.Y., Lee M., Surh C.D. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science. 2016;351:858–863. doi: 10.1126/science.aac5560. [DOI] [PubMed] [Google Scholar]
- 35.Verkoczy L., Chen Y., Zhang J., Bouton-Verville H., Newman A., Lockwood B., Scearce R.M., Montefiori D.C., Dennison S.M., Xia S.M., et al. Induction of HIV-1 broad neutralizing antibodies in 2F5 knock-in mice: selection against membrane proximal external region-associated autoreactivity limits T-dependent responses. J. Immunol. 2013;191:2538–2550. doi: 10.4049/jimmunol.1300971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyu M., Suzuki H., Kang L., Gaspal F., Zhou W., Goc J., Zhou L., Zhou J., Zhang W., et al. JRI Live Cell Bank ILC3s select microbiota-specific regulatory T cells to establish tolerance in the gut. Nature. 2022;610:744–751. doi: 10.1038/s41586-022-05141-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akagbosu B., Tayyebi Z., Shibu G., Paucar Iza Y.A., Deep D., Parisotto Y.F., Fisher L., Pasolli H.A., Thevin V., Elmentaite R., et al. Novel antigen-presenting cell imparts T(reg)-dependent tolerance to gut microbiota. Nature. 2022;610:752–760. doi: 10.1038/s41586-022-05309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kedmi R., Najar T.A., Mesa K.R., Grayson A., Kroehling L., Hao Y., Hao S., Pokrovskii M., Xu M., Talbot J., et al. A RORgammat(+) cell instructs gut microbiota-specific T(reg) cell differentiation. Nature. 2022;610:737–743. doi: 10.1038/s41586-022-05089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulendran B., S Arunachalam P., O'Hagan D.T. Emerging concepts in the science of vaccine adjuvants. Nat. Rev. Drug Discov. 2021;20:454–475. doi: 10.1038/s41573-021-00163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marrack P., McKee A.S., Munks M.W. Towards an understanding of the adjuvant action of aluminium. Nat. Rev. Immunol. 2009;9:287–293. doi: 10.1038/nri2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisenbarth S.C., Colegio O.R., O'Connor W., Sutterwala F.S., Flavell R.A. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim E.H., Woodruff M.C., Grigoryan L., Maier B., Lee S.H., Mandal P., Cortese M., Natrajan M.S., Ravindran R., Ma H., et al. Squalene emulsion-based vaccine adjuvants stimulate CD8 T cell, but not antibody responses, through a RIPK3-dependent pathway. Elife. 2020;9 doi: 10.7554/eLife.52687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosca F., Tritto E., Muzzi A., Monaci E., Bagnoli F., Iavarone C., O'Hagan D., Rappuoli R., De Gregorio E. Molecular and cellular signatures of human vaccine adjuvants. Proc. Natl. Acad. Sci. USA. 2008;105:10501–10506. doi: 10.1073/pnas.0804699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coccia M., Collignon C., Hervé C., Chalon A., Welsby I., Detienne S., van Helden M.J., Dutta S., Genito C.J., Waters N.C., et al. Cellular and molecular synergy in AS01-adjuvanted vaccines results in an early IFNgamma response promoting vaccine immunogenicity. NPJ Vaccines. 2017;2:25. doi: 10.1038/s41541-017-0027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu M., Pokrovskii M., Ding Y., Yi R., Au C., Harrison O.J., Galan C., Belkaid Y., Bonneau R., Littman D.R. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 2018;554:373–377. doi: 10.1038/nature25500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosshart S.P., Herz J., Vassallo B.G., Hunter A., Wall M.K., Badger J.H., McCulloch J.A., Anastasakis D.G., Sarshad A.A., Leonardi I., et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. 2019;365 doi: 10.1126/science.aaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knoop K.A., Gustafsson J.K., McDonald K.G., Kulkarni D.H., Coughlin P.E., McCrate S., Kim D., Hsieh C.S., Hogan S.P., Elson C.O., et al. Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aao1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al Nabhani Z., Dulauroy S., Marques R., Cousu C., Al Bounny S., Déjardin F., Sparwasser T., Bérard M., Cerf-Bensussan N., Eberl G. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity. 2019;50:1276–1288.e5. doi: 10.1016/j.immuni.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 50.Ramanan D., Sefik E., Galvan-Pena S., Wu M., Yang L., Yang Z., Kostic A., Golovkina T.V., Kasper D.L., Mathis D., Benoist C. An Immunologic Mode of Multigenerational Transmission Governs a. Gut Treg Setpoint. Cell. 2020;181:1276–1290.e1213. doi: 10.1016/j.cell.2020.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimmermann P., Curtis N. The influence of the intestinal microbiome on vaccine responses. Vaccine. 2018;36:4433–4439. doi: 10.1016/j.vaccine.2018.04.066. [DOI] [PubMed] [Google Scholar]
- 52.Ciabattini A., Olivieri R., Lazzeri E., Medaglini D. Role of the Microbiota in the Modulation of Vaccine Immune Responses. Front. Microbiol. 2019;10:1305. doi: 10.3389/fmicb.2019.01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Littman D.R. Do the Microbiota Influence Vaccines and Protective Immunity to Pathogens? If So, Is There Potential for Efficacious Microbiota-Based Vaccines? Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a029355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lynn M.A., Tumes D.J., Choo J.M., Sribnaia A., Blake S.J., Leong L.E.X., Young G.P., Marshall H.S., Wesselingh S.L., Rogers G.B., Lynn D.J. Early-Life Antibiotic-Driven Dysbiosis Leads to Dysregulated Vaccine Immune Responses in Mice. Cell Host Microbe. 2018;23:653–660.e5. doi: 10.1016/j.chom.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Cable J., Graham B.S., Koup R.A., Seder R.A., Karikó K., Pardi N., Barouch D.H., Sharma B., Rauch S., Nachbagauer R., et al. Progress in vaccine development for infectious diseases-a Keystone Symposia report. Ann. N. Y. Acad. Sci. 2023;1524:65–86. doi: 10.1111/nyas.14975. [DOI] [PubMed] [Google Scholar]
- 56.Lynn D.J., Benson S.C., Lynn M.A., Pulendran B. Modulation of immune responses to vaccination by the microbiota: implications and potential mechanisms. Nat. Rev. Immunol. 2022;22:33–46. doi: 10.1038/s41577-021-00554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rubtsov Y.P., Rasmussen J.P., Chi E.Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W.R., Jr., et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 58.Choi G.B., Yim Y.S., Wong H., Kim S., Kim H., Kim S.V., Hoeffer C.A., Littman D.R., Huh J.R. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze reported data will be shared by the lead contact upon request.