Abstract
Background
Acquiring well-focused digital images of cytology slides with scanners can be challenging due to the 3-dimensional nature of the slides. This study evaluates performances of whole-slide images (WSIs) obtained from 2 different cytopreparations by 2 distinct scanners with 3 focus modes.
Methods
Fourteen urine specimens were collected from patients with urothelial carcinoma. Each specimen was equally divided into 2 portions, prepared with Cytospin and ThinPrep methods and scanned for WSIs using Leica (Aperio AT2) and Hamamatsu (NanoZoomer S360) scanners, respectively. The scan settings included 3 focus modes (default, semi-auto, and manual) for single-layer scanning, along with a manual focus mode for 21 Z-layers scanning. Performance metrics were evaluated including scanning success rate, artificial intelligence (AI) algorithm-inferred atypical cell numbers and coverage rate (atypical cell numbers in single or multiple Z-layers divided by the total atypical cell numbers in 21 Z-layers), scanning time, and image file size.
Results
The default mode had scanning success rates of 85.7% or 92.9%, depending on the scanner used. The semi-auto mode increased success to 92.9% or 100%, and manual even further to 100%. However, these changes did not affect the standardized median atypical cell numbers and coverage rates. The selection of scanners, cytopreparations, and Z-stacking influenced standardized median atypical cell numbers and coverage rates, scanning times, and image file sizes.
Discussion
Both scanners showed satisfactory scanning. We recommend using semi-auto or manual focus modes to achieve a scanning success rate of up to 100%. Additionally, a minimum of 9-layer Z-stacking at 1 μm intervals is required to cover 80% of atypical cells. These advanced focus methods do not impact the number of atypical cells or their coverage rate. While Z-stacking enhances the AI algorithm's inferred quantity and coverage rates of atypical cells, it simultaneously results in longer scanning times and larger image file sizes.
Keywords: Urine cytology, Whole-slide image, Digital cytopathology, Z-stacking, Artificial intelligence
Highlights
-
•
Study evaluates performance of slide digitization from 2 cytopreparations with 2 scanners.
-
•
Urine cytology slides from bladder cancer patients scanned with 3 focus modes and Z-stacking.
-
•
Advanced focus modes improved scan success rates but didn't affect median atypical cell numbers.
-
•
Choice of scanner, cytopreparation, and Z-stacking impacts performance of slide digitization.
-
•
Z-stacking enhances AI-inferred atypical cells but increases scanning times and file sizes.
Introduction
A high-quality whole-slide image (WSI) plays an important role in digital pathology, with slide scanning being the initial and the most crucial step in the digital process. Excluding the importance of the specimen procurement, processing, staining, and overall quality of scanned glass slides, quality whole slide imaging is the critical step to the overlay of artificial intelligence (AI) to analyze and interpret an image. Acquiring well-focused WSI in cytology, however, is more challenging due to its 3-dimensional (3D) nature. For example, the thickness of a liquid-based cytology slide can reach up to 30 μm, whereas typical histology slides have a thickness of only 4–6 μm.1 Identifying optimal focus points for cytology slides is difficult and often requiring multilayer stacks.1 In addition, various cytopreparations, e.g., conventional smears, Cytospin, and liquid-based methods, may contribute to diverse slide thicknesses and increase the variability during slide scanning.2
Urine cytology serves as a primary diagnostic method for identifying bladder cancer.3 Recent studies highlight the potential benefits of AI algorithms in digital urine cytology.4, 5, 6, 7, 8 In our prior study, we developed an AI algorithm capable of detecting and quantifying suspicious urothelial cells within single-layer WSIs of urine cytology slides.9 Moreover, we corroborated that the AI-assisted method can effectively aid the cytopathologists in diagnosing urine cytology compared to direct microscopic examination. Nevertheless, further investigation of various focus modes and Z-stack scan settings is necessary to optimize digital urine cytology.
In this study, we attempted to address the major challenge of scanning cytology slides by testing various focus modes and Z-stacking across 2 different cytopreparations and digital scanners. To evaluate performance, we calculated scanning success rate for the total number of slides scanned using different focus modes and scanners. In addition, we utilized AI algorithm-inferred atypical cell numbers and coverage rate, scanning time, and image file size for each sample slide as evaluation metrics.
Material and methods
Specimens and cytopreparations
The study protocol received approval from the Institutional Review Board (IRB) of Tung's Taichung MetroHarbor Hospital (IRB no. 111028). We collected urine specimens from patients diagnosed with high-grade urothelial carcinoma (HGUC) of the urinary bladder at Tung's Taichung MetroHarbor Hospital, Taiwan. We applied a washing procedure to increase cell yield when collecting specimens. Each specimen was evenly divided into 2 portions and prepared as cytology slides using Cytospin (Thermo Fisher Scientific, Waltham, MA, USA) and ThinPrep (Hologic, Marlborough, MA, USA) methods, following the manufacturer's protocols. All slides were stained with Papanicolaou stain and reviewed by a senior cytopathologist, who confirmed the diagnosis of HGUC for all specimens.
Focus modes and Z-stack scan settings
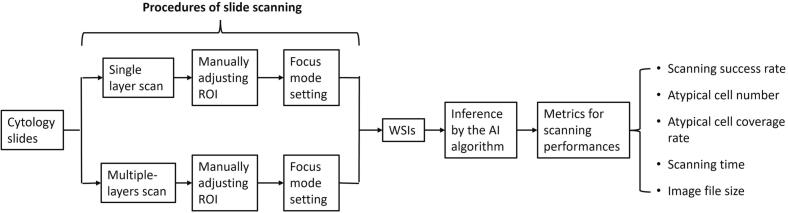
The study flowchart is depicted in Fig. 1. We scanned all cytology slides using Aperio AT2 (Leica Biosystems, Wetzlar, Germany) and NanoZoomer S360 (Hamamatsu Photonics, Shizuoka, Japan) whole-slide imagers, respectively. We started by scanning slides with a single layer. A cytotechnologist manually adjusted the scanning region of interest (ROI) to match the actual sample areas on the slides. Subsequently, we applied 3 focus modes from the scanners to determine the optimal focal plane for each slide. These modes included default (the scanner automatically placed optimal focus points within the ROI), semi-auto (the number or density of optimal focus points placed by the scanner was set to automatically increased based on the scanner console software algorithm), and manual modes (the user manually added focus points directly to the slide based on semi-auto mode). For the Z-stack scan, we tested 21 Z-layer scanning by manually adjusting the ROI and utilizing manual focus mode. The scanner first determined and scanned the optimal focal plane (Z=0) followed by scanning 10 Z-layers above and below the focus plane (Z=0±10) with a 1 μm interval between each layer, resulting in a total of 21 Z-layers WSI. All slides were scanned at 40× magnification and WSIs were acquired in proprietary image file formats, such as SVS (for Leica Aperio AT2) or NPDI (for Hamamatsu NanoZoomer S360). A senior cytotechnologist evaluated each resulting WSI for image quality including focus and color display. The percentage of successfully scanned WSIs relative to the total number of scanned slides was then determined for each scanner. To ensure comparability of scanning results across the same pairs of Cytospin and ThinPrep slides, if any WSI failed the quality check, the corresponding paired specimens were excluded from this study.
Fig. 1.
Flowchart of the study examining various focus modes and Z-stack scan settings using cytology slides. The slides were scanned with Leica and Hamamatsu digital scanners to produce WSI using different settings: (1) selecting between the single or 21 Z-layer scan mode; (2) manually adjusting the region of interest (ROI); (3) selecting a focus mode (default, semi-auto, or manual) and scanning the slide to generate WSI; (4) applying the artificial intelligence (AI) algorithm to determine the number and location of atypical cells in each WSI, whether in single or multiple layers; (5) analyzing metrics for scanning performance, such as scanning success rate, atypical cell numbers and coverage rate, scanning time, and image file size.
The AI algorithm-inferred atypical cells in WSI and scanning performance analysis
We used a deep-learning-based AI algorithm for inference on the single-layer (Z=0) and 21 Z-layers (Z=0±10), aiming to detect and quantify the total number of atypical cells in WSI, following the protocol and procedures from our previous study.9 We utilized several metrics to evaluate scanning performances, including the scanning success rate (the percentage of WSI files that passed the satisfactory image quality check in the total number of scanned slides), the number of atypical cells identified by the AI algorithm and the coverage rate (the ratio of atypical cell numbers in single or multiple layers to the total atypical cell numbers in 21 Z-layers), the scanning time (in seconds), and the image file size (in gigabytes, GB) of each WSI. To assess the performance of multiple-layer scans, we used proprietary software to analyze 21 Z-layers of the AI algorithm-inferred results, including atypical cell numbers and coverage rates. The data was then adjusted by software to align with single-layer and multi-layer results, ranging from 3 layers (Z=0±1), 5 layers (Z=0±2), 7 layers (Z=0±3), 9 layers (Z=0±4), 11 layers (Z=0±5), 13 layers (Z=0±6), 15 layers (Z=0±7), 17 layers (Z=0±8), 19 layers (Z=0±9), and 21 layers (Z=0±10). To ensure comparability of atypical cell numbers and coverage rates between 2 cytopreparations, we standardized these metrics using the sample area of Cytospin (28.27 mm2) or ThinPrep (314.16 mm2) slides, following the manufacturer’s manuals.
Statistical analysis
Median and interquartile range (IQR) values were used to summarize the standardized atypical cell numbers and coverage rates from a total of 20 cytology slides. The Kruskal–Wallis test was utilized to identify differences in these standardized values across different focus modes, cytopreparations, and scanners. For Z-stacking results, the mean response curve was used to display the relationship between the number of Z-layers (n=1–21) and either the standardized number of atypical cells or the coverage rate. Considering the heterogeneity of subjects, a random intercept model was used to evaluate the trend in the coverage rate of atypical cells across various Z-layers. We further explored if this trend varied between 4 different combinations of preparations and scanners by using a quadratic parametric model for data analysis. The statistical significance was set to equal 0.05, and all tests were conducted using a 2-sided approach. All statistical analyses were performed using SAS software (version 9.4; SAS Institute, Cary, NC, USA).
Results
Performance of different focus modes, cytopreparations, and digital scanners in single layer scanning
A total of 14 urine specimens with HGUC diagnosis were selected, prepared with Cytospin and ThinPrep methods, and scanned by Leica and Hamamatsu scanners, respectively. The default focus mode achieved scanning success rates of 85.7%–92.9% across both scanners and cytopreparations (Table 1). Semi-auto and manual focus modes attained scanning success rates of 92.9%–100% and 100%, respectively. WSIs of 4 pairs of specimens that failed the quality check were excluded from this study. A total of 20 paired WSIs (comprising 10 each of Cytospin and ThinPrep slides) were analyzed using the AI algorithm for this study.
Table 1.
Comparative analysis of scanning success rates across different scanners, focus modes, and cytopreparations.
| Scanner | Preparation | Focus mode |
||
|---|---|---|---|---|
| Default | Semi-auto | Manual | ||
| Leica | Cytospin | 12/14 (85.7%) | 13/14 (92.9%) | 14/14 (100.0%) |
| ThinPrep | 13/14 (92.9%) | 14/14 (100.0%) | 14/14 (100.0%) | |
| Hamamatsu | Cytospin | 13/14 (92.9%) | 13/14 (92.9%) | 14/14 (100.0%) |
| ThinPrep | 13/14 (92.9%) | 13/14 (92.9%) | 14/14 (100.0%) | |
We evaluated the AI algorithm-identified atypical cell numbers and coverage rates among the 20 paired WSIs with different focus modes (Table 2). For both Cytospin and ThinPrep slides, there were no significant differences among default, semi-auto, and manual modes for standardized median atypical cell numbers with different scanners (Table 3). For both Hamamatsu and Leica scanners, there were no significant differences in standardized median atypical cell numbers between Cytospin and ThinPrep using 3 different focus modes. However, the atypical cells coverage rates for Cytospin were significantly higher than ThinPrep for most focus modes (Table 3). When comparing the results between Leica and Hamamatsu scanners, only median coverage rates of the semi-auto focus mode in ThinPrep WSIs showed a statistically significant difference (17.55% in Leica versus 26.45% in Hamamatsu, P-value=.041) (Table 4). Those results suggest that utilizing focus modes for advanced users improved the scanning success rate but did not affect the number of atypical cells and coverage rate. The significantly higher coverage rates of atypical cells in Cytospin, compared to ThinPrep, indicate that a greater proportion of these cells were captured in a single layer scanned WSI when using Cytospin slides rather than ThinPrep slides.
Table 2.
Comparison of standardized median AI-inferred atypical cell numbers and coverage rates in single-layer scanning WSIs across 3 focus modes.
| Focus mode |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Default |
Semi-auto |
Manual |
|||||||
| Variable | Scanner | Preparation | Median | IQRa | Median | IQRa | Median | IQRa | P-value |
| Standardized atypical cell numbers | Hamamatsu | Cytospin | 5.96 | 8.14 | 5.62 | 7.85 | 5.55 | 7.78 | .982 |
| ThinPrep | 3.79 | 7.02 | 4.67 | 7.68 | 4.46 | 7.54 | .836 | ||
| Leica | Cytospin | 4.09 | 5.27 | 4.24 | 5.06 | 2.97 | 4.49 | .916 | |
| ThinPrep | 2.44 | 2.01 | 2.47 | 2.05 | 2.72 | 2.77 | .954 | ||
| Atypical cell coverage rate (%) | Hamamatsu | Cytospin | 31.10 | 5.60 | 32.90 | 11.00 | 32.60 | 12.20 | .931 |
| ThinPrep | 23.85 | 11.40 | 26.45 | 10.10 | 24.55 | 10.80 | .656 | ||
| Leica | Cytospin | 28.80 | 13.30 | 30.30 | 17.40 | 25.80 | 19.20 | .791 | |
| ThinPrep | 18.35 | 12.50 | 17.55 | 11.90 | 16.10 | 13.10 | .993 | ||
IQR: interquartile range.
Table 3.
Comparison of standardized median AI-inferred atypical cell numbers and coverage rates in single-layer scanning WSIs between 2 cytopreparations.
| Preparation |
|||||||
|---|---|---|---|---|---|---|---|
| Cytospin |
ThinPrep |
||||||
| Variable | Scanner | Focus mode | Median | IQRa | Median | IQRa | P-value |
| Standardized atypical cell numbers | Hamamatsu | Default | 5.96 | 8.14 | 3.79 | 7.02 | .199 |
| Semi-auto | 5.62 | 7.85 | 4.67 | 7.68 | .326 | ||
| Manual | 5.55 | 7.78 | 4.46 | 7.54 | .290 | ||
| Leica | Default | 4.09 | 5.27 | 2.44 | 2.01 | .326 | |
| Semi-auto | 4.24 | 5.06 | 2.47 | 2.05 | .406 | ||
| Manual | 2.97 | 4.49 | 2.72 | 2.77 | .597 | ||
| Atypical cell coverage rate (%) | Hamamatsu | Default | 31.10 | 5.60 | 23.85 | 11.40 | .028 |
| Semi-auto | 32.90 | 11.00 | 26.45 | 10.10 | .028 | ||
| Manual | 32.60 | 12.20 | 24.55 | 10.80 | .019 | ||
| Leica | Default | 28.80 | 13.30 | 18.35 | 12.50 | .004 | |
| Semi-auto | 30.30 | 17.40 | 17.55 | 11.90 | .006 | ||
| Manual | 25.80 | 19.20 | 16.10 | 13.10 | .070 | ||
IQR: interquartile range.
Table 4.
Comparison of standardized median AI-inferred atypical cell numbers and coverage rates in single-layer scanning WSIs between 2 scanners.
| Scanner |
|||||||
|---|---|---|---|---|---|---|---|
| Hamamatsu |
Leica |
||||||
| Variable | Preparation | Focus mode | Median | IQRa | Median | IQRa | P-value |
| Standardized atypical cell numbers | Cytospin | Default | 5.96 | 8.14 | 4.09 | 5.27 | .406 |
| Semi-auto | 5.62 | 7.85 | 4.24 | 5.06 | .226 | ||
| Manual | 5.55 | 7.78 | 2.97 | 4.49 | .199 | ||
| ThinPrep | Default | 3.79 | 7.02 | 2.44 | 2.01 | .597 | |
| Semi-auto | 4.67 | 7.68 | 2.47 | 2.05 | .545 | ||
| Manual | 4.46 | 7.54 | 2.72 | 2.77 | .496 | ||
| Atypical cell coverage rate (%) | Cytospin | Default | 31.10 | 5.60 | 28.80 | 13.30 | .571 |
| Semi-auto | 32.90 | 11.00 | 30.30 | 17.40 | .762 | ||
| Manual | 32.60 | 12.20 | 25.80 | 19.20 | .364 | ||
| ThinPrep | Default | 23.85 | 11.40 | 18.35 | 12.50 | .199 | |
| Semi-auto | 26.45 | 10.10 | 17.55 | 11.90 | .041 | ||
| Manual | 24.55 | 10.80 | 16.10 | 13.10 | .112 | ||
IQR: interquartile range.
Performance of different cytopreparations and digital scanners in manual focus mode with Z-stack scanning
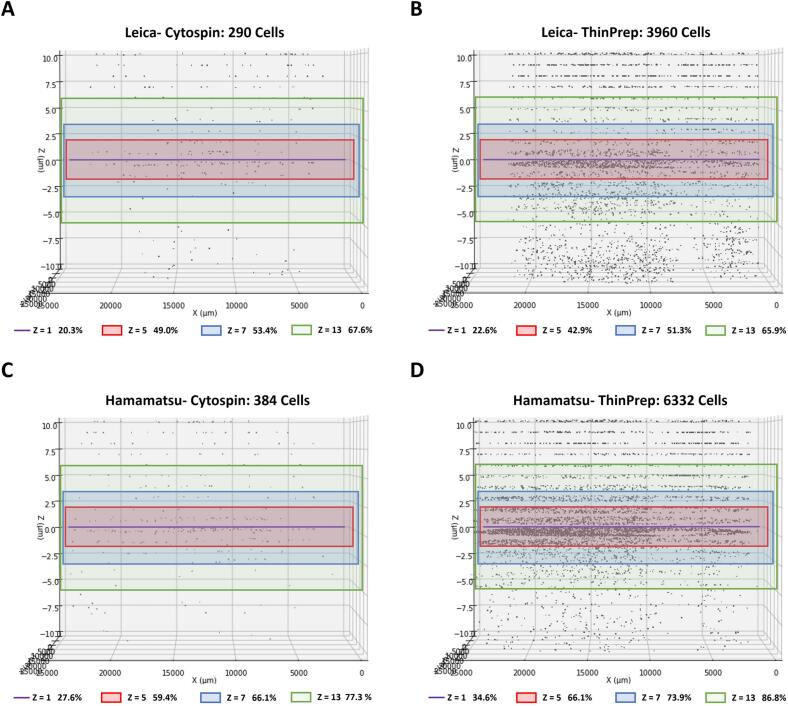
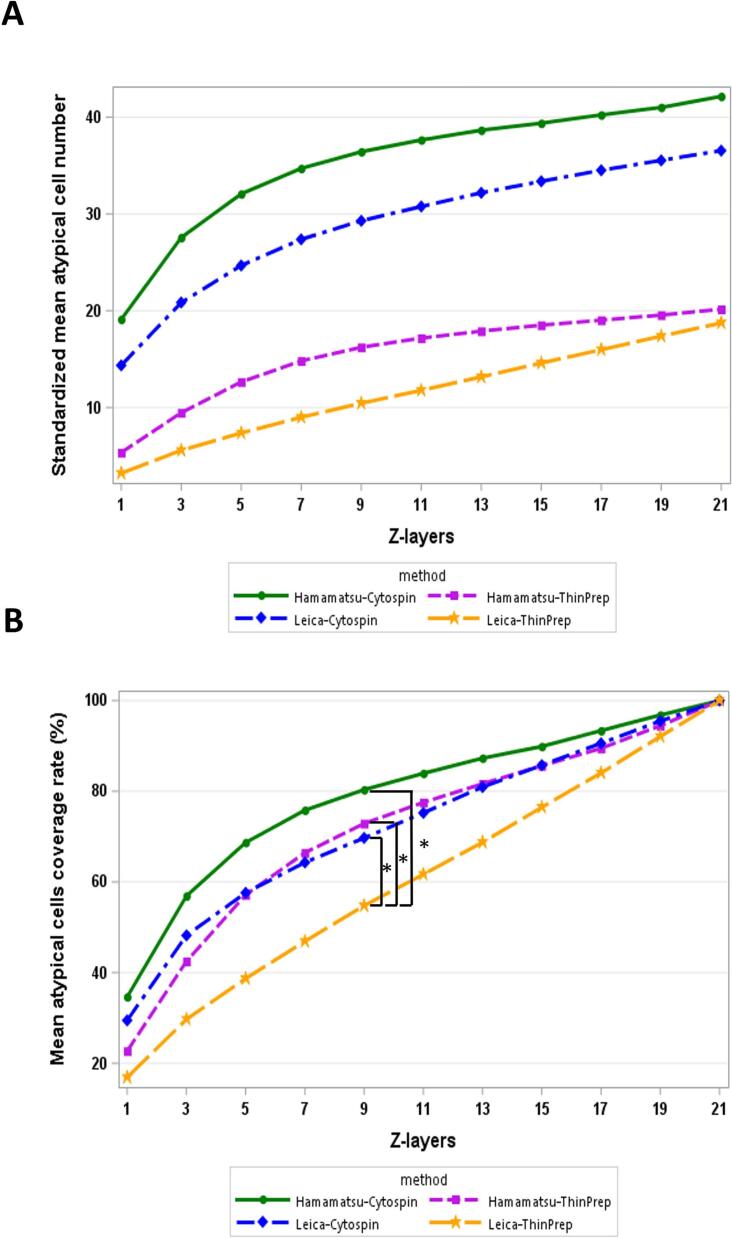
We constructed 3D plots using the total atypical cell numbers from 21 Z-layers, providing a visual representation of the atypical cell distribution within the 3D sample space of paired slides scanned by 2 digital scanners (Fig. 2). The data from both Leica and Hamamatsu scanners highlighted a higher number of total atypical cells in the ThinPrep results compared to the Cytospin results. Next, we analyzed the tendency between atypical cell numbers and coverage rates in relation to the number of Z-layers scanned. The graphs in Fig. 3 depict the standardized mean atypical cell numbers and coverage rates across a single layer and various multiple Z-layers in the 4 test groups: Hamamatsu-Cytospin, Leica-Cytospin, Hamamatsu-ThinPrep, and Leica-ThinPrep. A consistent trend was observed across all groups, where both the standardized mean atypical cell numbers and coverage rates increased proportionately with the number of Z-layers. However, a remarkable observation from Fig. 3B reveals that only the Leica-ThinPrep group showed a significant upward trend in the number of Z-layer scans compared to the other three groups (P-value=.001–.048). The estimated quadratic coefficient for the Leica-ThinPrep group was not significantly different from zero (−0.031, P-value=.220), while that for the other 3 groups were significantly smaller than zero (from −0.103 to −0.164, P-value <.001). This suggests that, compared to the Leica-ThinPrep group, the other 3 groups captured a greater number of atypical cells with fewer Z-layer scans. For example, to achieve a 50% coverage rate for atypical cells, scanning with the Leica scanner would need 5 Z-layers for Cytospin slides and 9 Z-layers for ThinPrep slides. On the other hand, with the Hamamatsu scanner, only 3 Z-layers for Cytospin slides and 5 Z-layers for ThinPrep slides would be required. However, as the number of scanning Z-layers increases, the marginal efficiency in capturing atypical cells noticeably decreases.
Fig. 2.
3D plots to visualize the distribution of AI-inferred atypical cells in representative paired sample slides scanned using 2 digital scanners. The upper panel shows a Cytospin slide (A) and a paired ThinPrep slide (B) scanned by the Leica scanner. The lower panel displays a Cytospin slide (C) and a paired ThinPrep slide (D) scanned by the Hamamatsu scanner. In the 3D slide plots, each black dot represents the location of an atypical cell. The horizontal axis designates the slide's relative distance (μm), and the vertical axis represents the Z-axis distance (μm). Line or rectangle area colors indicate scan ranges of single (Z=1, purple line) and multiple Z-layers (Z=5, red rectangle area; Z=7, blue rectangle area; Z=13, green rectangle area). Each Z-layer indicates the coverage rate of atypical cells defined as the ratio of atypical cell numbers in single or multiple Z-layers to the total number of atypical cells across all 21 Z-layers.
Fig. 3.
The graphs of mean response curve were used to illustrate the relationship between the number of Z-layers and either standardized mean atypical cell numbers (A) or coverage rate (B) across the 4 groups: Hamamatsu-Cytospin (represented by a solid green line), Leica-Cytospin (represented by a blue dashed line), Hamamatsu-ThinPrep (represented by a purple dashed line), and Leica-ThinPrep (represented by an orange dashed line). (A) A consistent pattern across all groups was observed where standardized mean atypical cell numbers increased with the number of Z-layers. (B) Upon utilizing a random intercept model for analysis, only the Leica-ThinPrep group exhibited a significant increase in the trend with the rising number of Z-layers, compared to the other 3 groups. *Statistically significant difference (P<.05)
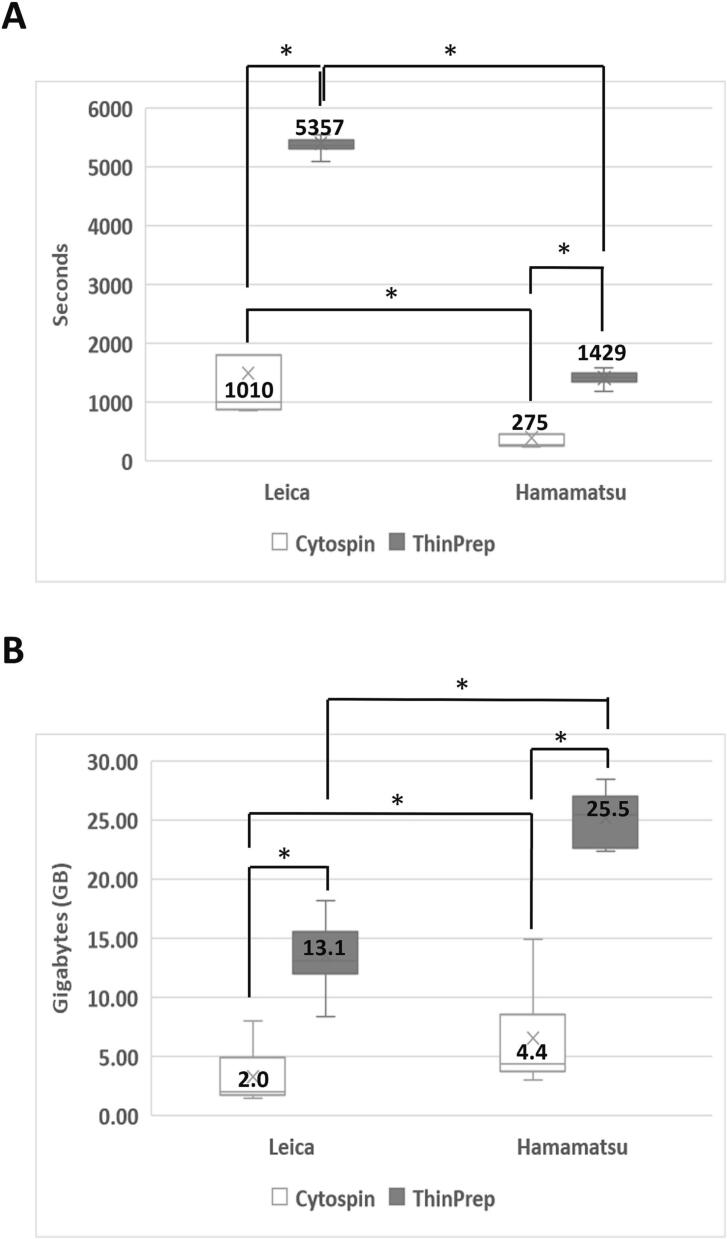
To consider the cost of Z-stacking, we compared the median scanning time and file size of 21 Z-layer WSIs across 2 different cytopreparations and digital scanners (Fig. 4). The results showed that compared to Cytospin slides, ThinPrep slides required a significantly longer median scanning time (1010 versus 5357 s for Leica, P-value <.001; 275 versus 1429 s for Hamamatsu, P-value <.001) and larger median image file size (2.0 versus 13.1 GB for Leica, P-value <.001; 4.4 versus 25.5 GB for Hamamatsu, P-value <.001). When compared to the Hamamatsu scanner, the Leica scanner required a substantially longer median scanning time (275 versus 1010 s for Cytospin, P-value <.001; 1429 versus 5357 s for ThinPrep, P-value <.001) but smaller median image file size (4.4 versus 2.0 GB for Cytospin, P-value=.339; 25.5 versus 13.1 GB for ThinPrep, P-value <.001). Those findings indicate that Z-stack scanning increased atypical cell numbers and coverage rates, albeit at the cost of increased scanning time and image file size.
Fig. 4.
Comparison of scanning time and image file size of different cytopreparation WSIs using 2 distinct scanners with manual focus mode and 21 Z-layer scan settings. The median scanning time (A) and image file size (B) were obtained from 10 paired Cytospin (represented by the white color bar) and ThinPrep (represented by the gray color bar) slides, scanned respectively by Leica and Hamamatsu digital scanners. *Statistically significant difference (P<.05)
Discussion
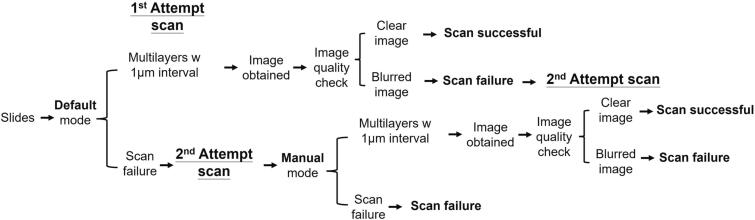
In this study, we demonstrated how 3 focus modes and 21 Z-layers scanning in 2 digital scanners affected the digitization of urine cytology. We recommend a urine cytology slide scanning workflow designed for roughly 80% atypical cell coverage, compatible with Cytospin and ThinPrep slides and operational with Leica Aperio AT2 and Hamamatsu NanoZoomer S360 scanners. The workflow comprises two main scanning attempts as outlined in Fig. 5. The first attempt uses the “default” mode, utilizing at least 9 scanning layers with a 1 μm interval and is followed by an image assessment to determine its success. If failed, a second scan is initiated in “manual” mode, remaining to the same interval policy. The scan quality is reassessed, determining the final status of the slide image. This procedure ensures that the images obtained are suitable for clinical interpretation.
Fig. 5.
Recommended urine cytology slide scanning workflow. In the first attempt, users should use the “default” mode to scan the urine cytology slides, utilizing at least 9 scanning layers (depending on the specific scanner and cytopreparation slides) with a 1 μm interval between each layer. If a slide image is successfully obtained on the first attempt, users must conduct an image quality assessment, determining the scan as either “clear” (indicating no artifacts or distortions in the image) and hence “successful,” or “blur” (highlighting any noticeable distortion or lack of clarity), designating the scan as a “failure” and requiring a second scan using the “manual” mode. If the initial scan does not meet the quality standards, a second attempt should be undertaken using the “manual” mode, maintaining a 1 μm interval between multilayer scans. Following this attempt, users should once again assess the image quality to ascertain whether the scan can be deemed “successful” with a clear image or “failure” with a blurry output, denoting the slide as unsuitable for further scanning. This 2-step approach is designed to enhance the quality of slide images making them fit for clinical interpretation.
In digital urine cytology, qualities that would improve the technology for diagnosis, research, and AI evaluation include rapid slide scanning times (preferably less than 5 min per slide), detection and selection of only atypical urothelial cells, sharp focus of target cells, ideal color balance, and a low image file size. A pivotal factor for the “quality” of WSI is the inclusion of well-focused candidate cells, to facilitate accurate examination by cytologists or for the AI algorithm inference. In our study, we leveraged the number of atypical cells inferred by an AI algorithm as a benchmark to evaluate the “quality” of WSI. This algorithm has been trained on a dataset of well-focused atypical cells annotated by a senior cytopathologist. It is essential to note that a current limitation of our AI algorithm is its inability to identify atypical cells that are out of focus. Thus, we advocate for the utilization of varied focus modes (semi-auto or manual) and/or multiple Z-layer scans (9-layers minimum) to secure well-focused candidate cells, a strategy critical to enhancing digital urine cytology for diagnostic, research, and AI-based evaluations.
Both Leica and Hamamatsu scanners are equipped with multiple focus modes for advanced requirements of slide scanning. Beyond the default mode, which automatically determines the number and location of focus points, both semi-auto and manual modes allow users to manually add additional focus points within the sample area of the slide to obtain the optimal focal plane and then scan well-focused cells in WSI. To the best of our knowledge, no studies have evaluated these focus modes on digital scanners while scanning urine cytology slides. Our data reveals that there is no significant difference in standardized atypical cell numbers and coverage rates among the 3 focus modes on either Leica or Hamamatsu scanners (Table 2). However, a large-scale study is required to verify our initial findings.
Cytospin and ThinPrep are the 2 primary urine cytology preparation methods used in the United States and globally.10,11 Our study represents the first attempt to evaluate the scanning of cytology slides from the same urine specimen prepared using both methods. The results exhibited no statistically significant difference in the standardized atypical cell numbers between 2 cytopreparations, suggesting considering different sample areas, cytopreparations are independent to the AI algorithm-inferred atypical cell numbers in WSIs. However, ThinPrep demonstrated a significantly lower coverage rate compared to Cytospin (Table 3). These results might be attributed to a more dispersed cell distribution in 3D sample space of ThinPrep than commonly expected. Despite being known as a thin-layer cytology slide compared to Cytospin, ThinPrep appears to have a broader cell distribution when visualized in the 3D plots of cell distribution in this study (Fig. 2).
Our study represents the initiative, in-depth assessment of urine cytology WSIs, utilizing Z-stack scanning with 2 distinct digital scanners for the examination of 2 varied cytopreparations. This was achieved by evaluating metrics such as the quantity of atypical cells, coverage rate, scanning time, and image file size. The results indicate that an analysis of 21 Z-layer scans shows an ascending pattern between the number of Z-layers and both the quantity and coverage rates of atypical cells, across 4 different combinations of preparations and scanners (Fig. 3). Our findings imply that the selection of scanner and cytopreparation influences the capture efficiency of atypical cells during Z-stack scanning. When considering the cost of Z-stacking, ThinPrep slides need longer scanning time and larger image file size compared to Cytospin slides for a 21 Z-layer scan, irrespective of the scanner used. Moreover, compared to the Hamamatsu scanner, the Leica scanner required a longer scanning time but results in a smaller image file size (Fig. 4). Those results suggest that different cytopreparations and scanners impact the capture efficiency of atypical cells and cost of digital cytology, such as scanning time and image file size. Aside from accounting for the cost implications of Z-stacking, the significant increase in workload for cytopathologists makes the review of WSIs from multiple Z-layers a complex task, currently posing considerable challenges for digital cytology in clinical practice. This highlights an urgent need for technological innovations in slide scanning, to overcome the key obstacles faced in the field of digital cytology.
In this study, we demonstrated that the AI algorithm's inference of atypical cell numbers in WSIs is not influenced by specific cytopreparations or digital scanners (Table 3, Table 4). This suggests that our AI algorithm maintains impartiality when inferring atypical cells in WSIs, regardless of the source of the WSIs, whether they originate from different cytopreparations or digital scanners. In addition, quantitative analysis is a key strength of AI technology, particularly valuable for localizing and quantifying atypical cells in Z-stacked WSIs of urine cytology.4, 5, 6, 7, 8 In a previous study, we demonstrated capability of the AI algorithm to quantify high and low-risk atypical cells in single-layer WSIs of urine cytology.9 In this study, we utilized 3D plots to visualize the distribution of AI-inferred atypical cells in WSIs, displaying a potential application for cytopathologists and cytotechnologists (Fig. 2). This enables them to quickly locate atypical cells across any Z-layer if they need to review multiple layers within a WSI, making it a significant development for digital cytology in future clinical practice.
In conclusion, our study examined various focus modes and Z-stacking for scanning urine cytology slides using 2 different commercial digital scanners, while also evaluating the cost-effectiveness of digitization. Both semi-auto and manual focus modes yielded a higher scanning success rate, but they did not affect the numbers and coverage rates of atypical cells on the identical cytology slide. Therefore, Z-stacking still provides a primary solution for increasing the detection of atypical cells and coverage rate in WSIs. However, it requires longer scanning time and results in larger image file size. Furthermore, compared to ThinPrep slides, Cytospin slides facilitated faster scanning times and generated smaller image files. Moreover, the Hamamatsu scanner required less time to scan and generated smaller image files compared to the Leica scanner. Our study highlights the challenges associated with optimizing digital urine cytology for AI-assisted applications. Additionally, our recommended urine cytology slide scanning workflow equips digital scanner users and manufacturers with the guidance to prepare high-quality WSI for digital cytology.
Funding support
This study was financially supported by AIxMed, Inc.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Tien-Jen Liu reports financial support was provided by AIxMed, Inc. Tien-Jen Liu reports a relationship with AIxMed, Inc. that includes: employment.
Wei-Lei Yang, Cheng-Hung Yeh, Chi-Bin Li, En-Yu Hsu and Po-Yen Hung reports financial support was provided by AIxMed, Inc. Wei-Lei Yang, Cheng-Hung Yeh, Chi-Bin Li, En-Yu Hsu and Po-Yen Hung reports a relationship with AIxMed, Inc. that includes: employment.
Contributor Information
Tien-Jen Liu, Email: tienjen.liu@aixmed.com.
Min-Che Tung, Email: tungminche@gmail.com.
References
- 1.Lee R.E., McClintock D.S., Laver N.M., Yagi Y. Evaluation and optimization for liquid-based preparation cytology in whole slide imaging. J Pathol Inform. 2011;2:46. doi: 10.4103/2153-3539.86285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.da Cunha Santos G., Saieg M.A. Preanalytic specimen triage: smears, cell blocks, cytospin preparations, transport media, and cytobanking. Cancer Cytopathol. Jun 2017;125(S6):455–464. doi: 10.1002/cncy.21850. [DOI] [PubMed] [Google Scholar]
- 3.Wojcik E.M., Kurtycz D.F.I., Rosenthal D.L. We’ll always have Paris The Paris System for Reporting Urinary Cytology 2022. J Am Soc Cytopathol. Mar-Apr 2022;11(2):62–66. doi: 10.1016/j.jasc.2021.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Nojima S., Terayama K., Shimoura S., et al. A deep learning system to diagnose the malignant potential of urothelial carcinoma cells in cytology specimens. Cancer Cytopathol. May 12 2021;129(12):984–995. doi: 10.1002/cncy.22443. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko M., Tsuji K., Masuda K., et al. Urine cell image recognition using a deep-learning model for an automated slide evaluation system. BJU Int. Jun 18 2021;130(2):235–243. doi: 10.1111/bju.15518. [DOI] [PubMed] [Google Scholar]
- 6.Awan R., Benes K., Azam A., et al. Deep learning based digital cell profiles for risk stratification of urine cytology images. Cytometry A. Jul 2021;99(7):732–742. doi: 10.1002/cyto.a.24313. [DOI] [PubMed] [Google Scholar]
- 7.Vaickus L.J., Suriawinata A.A., Wei J.W., Liu X. Automating the Paris System for urine cytopathology-a hybrid deep-learning and morphometric approach. Cancer Cytopathol. Feb 2019;127(2):98–115. doi: 10.1002/cncy.22099. [DOI] [PubMed] [Google Scholar]
- 8.Sanghvi A.B., Allen E.Z., Callenberg K.M., Pantanowitz L. Performance of an artificial intelligence algorithm for reporting urine cytopathology. Cancer Cytopathol. Oct 2019;127(10):658–666. doi: 10.1002/cncy.22176. [DOI] [PubMed] [Google Scholar]
- 9.Ou Y.C., Tsao T.Y., Chang M.C., et al. Evaluation of an artificial intelligence algorithm for assisting the Paris System in reporting urinary cytology: a pilot study. Cancer Cytopathol. Jun 21 2022;130(11):872–880. doi: 10.1002/cncy.22615. [DOI] [PubMed] [Google Scholar]
- 10.Barkan G.A., Tabatabai Z.L., Kurtycz D.F.I., et al. Practice patterns in urinary cytopathology prior to the Paris System for Reporting Urinary Cytology. Arch Pathol Lab Med. Feb 2020;144(2):172–176. doi: 10.5858/arpa.2019-0045-CP. [DOI] [PubMed] [Google Scholar]
- 11.Kurtycz D., Brimo F., Rosenthal D.L., et al. Perceptions of Paris: an international survey in preparation for The Paris System for Reporting Urinary Cytology 2.0 (TPS 2.0) J Am Soc Cytopathol. 2021;10(5):S4. doi: 10.1016/j.jasc.2022.09.002. [DOI] [PubMed] [Google Scholar]