Abstract
In Escherichia coli an autoregulatory mechanism of programmed ribosomal frameshifting governs the level of polypeptide chain release factor 2. From an analysis of 20 sequences of genes encoding release factor 2, we infer that this frameshift mechanism was present in a common ancestor of a large group of bacteria and has subsequently been lost in three independent lineages.
The advent of complete genome sequences provides the opportunity to assess the conservation of programmed ribosomal frameshifting in the expression of particular genes. The sequences also permit deductions about single or multiple origins of frameshift cassettes and the degree of conservation of the signals involved. A second reason to examine published sequences for a particular frameshift cassette is to highlight the need for caution in assigning genes by comparative methods. Gene assignments in some genome-sequencing papers have been based exclusively on homology to a single open reading frame (ORF), while other investigators have been mindful of the fact that the synthesis of some proteins involves a programmed ribosomal frameshift event to link the information from two ORFs.
Expression of the Escherichia coli release factor 2 (RF2) gene, prfB, requires ribosomes at codon 25, CUU, to shift to the +1 frame, which encodes the main part of the protein (7, 8, 18) (Fig. 1). Codon 26 in the initiating frame is a UGA stop codon. RF2 mediates release at UGA, and in the presence of excess RF2, a high proportion of ribosomes terminate at codon 26 and only a small proportion shift to the +1 frame. The released 25-amino-acid peptide is degraded, and little full-length active RF2 is synthesized. However, when there is a deficit of RF2, the UGA, and pertinently its 1st base, U, is temporarily free. This U forms the 3rd base of a +1-frame UUU codon with which peptidyl tRNALeu pairs following disengagement from the 0-frame CUU (40). This re-pairing involves first-position wobble pairing.
FIG. 1.
Current model for RF2 frameshifting. See text for details.
The RF2 frameshift site is conserved in a large number of distantly related bacteria.
The nucleotide sequence of the region that signals programmed frameshifting in the RF2 gene in Bacillus subtilis is strikingly similar to that of its E. coli counterpart (26), but in Streptomyces coelicolor frameshifting does not seem to be involved (25). With the recent increase in available genome sequence information, we collected 20 RF2 sequences from different bacteria. This was achieved with the help of the Entrez Browser (12a), the Blast (2) server at the Institute for Genome Research (15a), the Gonococcal and Streptococcal Genome Projects (31, 32), the Pseudomonas Genome Project (30), and the Chlamydia Genome Project (37). In addition, we obtained the Aquifex aeolicus RF2 sequence from R. Swanson at Diversa Corp. All sequences were aligned with the PILEUP program of the Genetics Computer Group package and manually searched for possible frameshift sites. Particular caution was taken in the alignment of the first part of the RF2 amino acid sequences (Fig. 2), and homology before the potential frameshift sites was examined carefully to determine whether a frameshift was likely to take place or not. All sequences allowed unambiguous detection of the absence or presence of a frameshift site. Nonframeshifters lacked an in-frame stop codon and had continuous sequence similarity to the coding frames of other RF2 sequences both before and after their frameshift sites. Sequences with the frameshift site all had possible start sites only in the 0 frame upstream of the shift site and had a UGA stop codon in the 0 frame at a position corresponding to the beginning of the +1-frame homologous sequences. Their products were also homologous RF2s. In no case did we detect any homologs of RF2 other than RF1 and occasionally RF-H (release factor homolog); therefore, these organisms are likely to harbor only one prfB gene.
FIG. 2.
Alignment of the N-terminal end of RF2. The last preshift and first postshift amino acids are boldfaced. For the sources of the sequences, see the reference for each organism, as follows: A. aeolicus (38), Bacillus firmus (4), B. subtilis (33), Borrelia burgdorferi (14), Chlamydia trachomatis (37), C. acetobutylicum (15), Deinococcus radiodurans (15a), E. coli (8), Haemophilus influenzae (13), Helicobacter pylori (39), Neisseria gonorrhoeae (31), Mycobacterium tuberculosis (27), Mycobacterium leprae (12), Pseudomonas aeruginosa (30), Salmonella typhimurium (19), S. coelicolor (25), Streptococcus pneumoniae (15a), Streptococcus pyogenes (32), Synechocystis sp. strain PCC6803 (17), and Treponema pallidum (38).
All RF2 frameshift sites identified in the DNA sequences have a conserved CTTTGAC motif (Fig. 3). The position of the frameshift site is the same in all organisms, and even in some organisms that do not shift, the leucine-aspartate codons are conserved, which suggests that these amino acids are structurally important in the RF2 molecule. Maybe this provides additional selection pressure, together with the autoregulatory mechanism, to keep the sequence in the organisms that do frameshift. The stop codon is always UGA, which allows RF2 autoregulation. The codon preceding the shift is always CUU. In E. coli, replacement of CUU by other codons which permit their decoding tRNA to re-pair with the overlapping +1-frame codon allows frameshifting (9, 40), although CUU itself causes the most efficient frameshifting (9). As in other systems, rather weak preshift pairing and relatively strong postshift pairing is important for RF2 frameshifting (9). The identities of the two carboxy-terminal amino acids of the nascent chain influence termination in E. coli (3, 5). Although there seems to be a preference for tyrosine, valine, or serine as the amino acid preceding leucine, this is likely to reflect demands on the RF2 structure rather than effects on termination. In test constructs, when the UGA stop codon is replaced with a sense codon with U as its 1st base, the potential for pairing with the overlapping +1-frame UUU is retained. However, the level of frameshifting is substantially reduced. The flanking stop codon is important but not essential for the frameshifting (11, 36, 40, 42), although it is crucial for autoregulation.
FIG. 3.
Alignment of RF2 frameshifting sites and the nonfunctional similar site in E. coli prfH. Sequences were obtained from the sources cited in the legend to Fig. 2.
The 1st codon in the new frame is in all cases a GAC aspartate codon. The alternative aspartate codon, GAT, is not found, presumably because the UGA stop codon is most efficient when followed by a C. Following several early studies (see references 6 and 34), numerous reports have shown that the identity of the base following a triplet stop codon substantially influences the efficiency of termination. The termination codon may effectively be a quadruplet (28, 29). In E. coli, UGAC is a comparatively poor terminator, and it is probably not coincidental that the UGA at codon 26 in the gene for RF2 is followed by C (23, 28). Since there is competition between frameshifting and termination, as well as in-frame readthrough (1, 10), having a poor terminator permits more efficient frameshifting.
In all cases the shift site is preceded by a G-rich sequence at a variable distance from the shift site. This element is important for a Shine-Dalgarno-like interaction, which involves translocating, rather than initiating, ribosomes (10, 35, 40–42). Pairing between 16S rRNA of ribosomes and a Shine-Dalgarno sequence 3 bases 5′ of the shift site directly stimulates +1 frameshifting. Mutagenesis experiments have shown that precise positioning of the Shine-Dalgarno sequence is required (40) and that spacing between the Shine-Dalgarno sequence and the shift site influences the directionality of shifting (20, 21). In E. coli this spacing has to be 3 nucleotides. This spacing is conserved in most of the organisms analyzed, although, interestingly, Clostridium acetobutylicum and Synechocystis sp. strain PCC6803 seem to be exceptions to this rule.
Several bacterial lineages have independently lost the RF2 frameshift site.
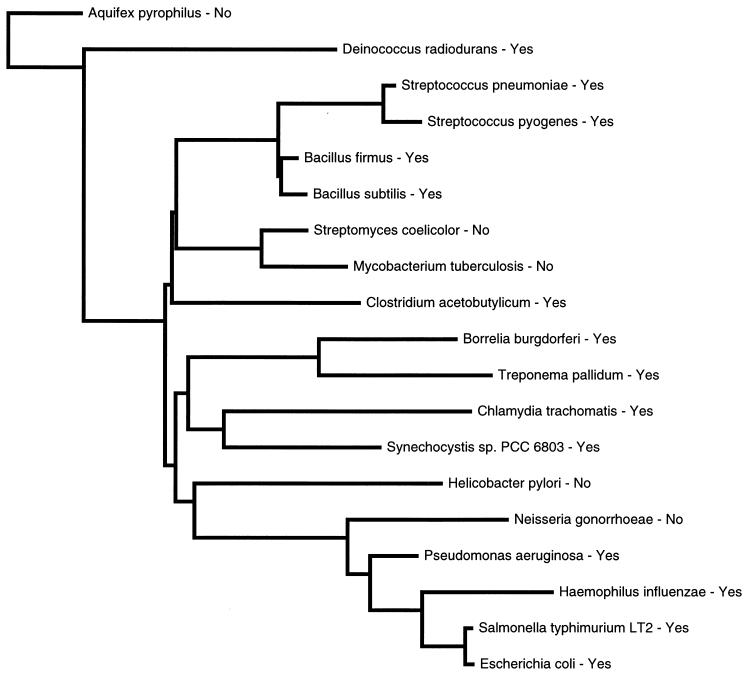
With the help of the Ribosomal Database Project web site (22), a phylogenetic tree based on the 16S rRNA of these organisms was constructed (Fig. 4). The phylogenetic tree of bacteria with the RF2 frameshift site suggests that this autoregulatory element was acquired by an early ancestor of a large group of present-day bacteria ranging from green nonsulfur bacteria and cyanobacteria to purple and gram-positive bacteria. Then the frameshift mechanism seems to have been independently lost in at least three branches of the bacterial phylogenetic tree, leading to its absence in mycobacteria, Streptomyces, Neisseria, and Helicobacter. It will be interesting to see how the RF2 levels are regulated in these organisms.
FIG. 4.
Phylogenetic tree based on 16S rRNA obtained from the Ribosomal Database Project. Because the 16S rRNA sequence from A. aeolicus was unavailable, the sequence of Aquifex pyrophilus was used. Yes, RF2 frameshift site present; No, RF2 frameshift site absent.
We searched for the sequence GGGGGNNNCTTTGAC at other locations in the genome of E. coli. Several sequences with resemblance to this motif were found, but none was found in the productive reading frame within a coding region. There is one gene in E. coli, prfH, that encodes a protein homologous to RF2. In the beginning of this gene there is a sequence with some similarity to the frameshift site of prfB (Fig. 3). We tested this sequence for its frameshifting ability in vivo by inserting the sequence between gst and lacZ, with gst being fused in the 0 frame and lacZ being fused in the +1 frame. No frameshifting activity could be detected by assaying for β-galactosidase (data not shown).
Many cases of programmed frameshifting are known, or suspected, in the decoding of viral genes and transposable elements, and a small number are known for cellular gene decoding. Very little is yet known about the phylogeny of frameshifting cassettes, but dnaX frameshifting in widely divergent eubacteria (21, 24, 43) is being compared, as is antizyme frameshifting in Drosophila and humans (16).
Acknowledgments
We acknowledge the following people and organizations for making unpublished sequence data available to us: The Institute for Genomic Research, the Genome Therapeutics Corporation, R. Swanson at Diversa Corp., the Chlamydia Genome Project, R. S. Stephens, S. Kalman, C. Fenner, R. Davis, the Cystic Fibrosis Foundation, the University of Washington Genome Center, the PathoGenesis Corporation, the Gonococcal and Streptococcal Genome Sequencing Projects, B. A. Roe, S. P. Lin, L. Song, X. Yuan, S. Clifton, D. W. Dyer, M. McShan, and J. Ferretti. We thank Ray Gesteland for characteristically generous support and comments.
This work was supported by the Howard Hughes Medical Institute (R. F. Gesteland is an Investigator). This work was also supported by a grant (to J.F.A.) from the National Institutes of Health (RO1-GM48152-05).
REFERENCES
- 1.Adamski F M, Donly B C, Tate W P. Competition between frameshifting, termination and suppression at the frameshift site in the Escherichia coli release factor-2 mRNA. Nucleic Acids Res. 1993;21:5074–5078. doi: 10.1093/nar/21.22.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arkov A L, Korolev S V, Kisselev L L. Termination of translation in bacteria may be modulated via specific interaction between peptide chain release factor 2 and the last peptidyl-tRNASer/Phe. Nucleic Acids Res. 1993;21:2891–2897. doi: 10.1093/nar/21.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, M., and E. Baeuerlein. GenBank accession no. P96314.
- 5.Björnsson A, Mottagui-Tabar S, Isaksson L A. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J. 1996;15:1696–1704. [PMC free article] [PubMed] [Google Scholar]
- 6.Bossi L, Roth J R. The influence of codon context on genetic code translation. Nature. 1980;286:123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- 7.Craigen W J, Caskey C T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 8.Craigen W J, Cook R G, Tate W P, Caskey C T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci USA. 1985;82:3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curran J F. Analysis of effects of tRNA:message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucleic Acids Res. 1993;21:1837–1843. doi: 10.1093/nar/21.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curran J F, Yarus M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J Mol Biol. 1988;203:75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- 11.Curran J F, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- 12.Eiglmeier K, Honore N, Woods S A, Caudron B, Cole S T. Use of an ordered cosmid library to deduce the genomic organization of Mycobacterium leprae. Mol Microbiol. 1993;7:197–206. doi: 10.1111/j.1365-2958.1993.tb01111.x. [DOI] [PubMed] [Google Scholar]
- 12a.Entrez Browser.http://www3.ncbi.nlm.nih.gov/Entrez/index.html.
- 13.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, Fitzhugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 14.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Whattey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 15.Genome Therapeutics Corporation.http://www.genomecorp.com/htdocs/sequences/clostridium/.
- 15a.Institute for Genome Research (TIGR). BLAST search web site. http://www.ncbi.nlm.nih.gov/BLAST/tigrbl.html.
- 16.Ivanov I P, Simin K, Letsou A, Atkins J F, Gesteland R F. The Drosophila gene for antizyme requires ribosomal frameshifting for expression and contains an intronic gene for snRNP Sm D3 on the opposite strand. Mol Cell Biol. 1998;18:1553–1561. doi: 10.1128/mcb.18.3.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 18.Kawakami K, Jönsson Y H, Björk G R, Ikeda H, Nakamura Y. Chromosomal location and structure of the operon encoding peptide-chain-release factor 2 of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:5620–5624. doi: 10.1073/pnas.85.15.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawakami K, Nakamura Y. Autogenous suppression of an opal mutation in the gene encoding peptide chain release factor 2. Proc Natl Acad Sci USA. 1990;87:8432–8436. doi: 10.1073/pnas.87.21.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen B, Wills N M, Gesteland R F, Atkins J F. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen B, Gesteland R F, Atkins J F. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50% J Mol Biol. 1997;271:47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maidak B I, Olsen G A, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Major L L, Poole E S, Dalphin M E, Mannering S A, Tate W P. Is the in-frame termination signal of the Escherichia coli release factor-2 frameshift site weakened by a particularly poor context? Nucleic Acids Res. 1996;24:2673–2678. doi: 10.1093/nar/24.14.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McHenry C S, Seville M, Cull M G. A DNA polymerase III holoenzyme-like subassembly from an extreme thermophilic eubacterium. J Mol Biol. 1997;272:178–189. doi: 10.1006/jmbi.1997.1238. [DOI] [PubMed] [Google Scholar]
- 25.Ogawara H, Urabe H, Ohtaki R, Nakamura Y. Properties of peptide chain release factor 2 from Streptomyces coelicolor A3(2): conserved primary structure but no frameshift regulation. J Bacteriol. 1995;177:5342–5345. doi: 10.1128/jb.177.18.5342-5345.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pel H J, Rep M, Grivell L A. Sequence comparison of new prokaryotic and mitochondrial members of the polypeptide chain release factor family predicts a five-domain model for release factor structure. Nucleic Acids Res. 1992;20:4423–4428. doi: 10.1093/nar/20.17.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Jr, Cole S T. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole E S, Brown C M, Tate W P. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole E S, Brimacombe R, Tate W P. Decoding the translational termination signal: the polypeptide chain release factor in Escherichia coli crosslinks to the base following the stop codon. RNA. 1997;3:974–982. [PMC free article] [PubMed] [Google Scholar]
- 30.Pseudomonas Genome Project.http://www.pseudomonas.com/.
- 31.Roe, B. A., S. P. Lin, L. Song, X. Yuan, S. Clifton, and D. W. Dyer. Gonococcal Genome Sequencing Project. http://dna1.chem.uoknor.edu/gono.html.
- 32.Roe, B. A., S. P. Linn, L. Song, X. Yuan, S. Clifton, M. McShan, and J. Ferretti. Streptococcal Genome Sequencing Project. http://dna1.chem.uoknor.edu/strep.html.
- 33.Sadaie Y, Takamatsu H, Nakamura K, Yamane K. Sequencing reveals similarity of the wild-type div+ gene of Bacillus subtilis to the Escherichia coli secA gene. Gene. 1991;98:101–105. doi: 10.1016/0378-1119(91)90110-w. [DOI] [PubMed] [Google Scholar]
- 34.Salser W. The influence of the reading context upon the suppression of nonsense codons. Mol Gen Genet. 1969;105:125–130. doi: 10.1007/BF00445682. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz R, Curran J F. Analyses of frameshifting at UUU-pyrimidine sites. Nucleic Acids Res. 1997;25:2005–2011. doi: 10.1093/nar/25.10.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sipley J, Goldman E. Increased ribosomal accuracy increases a programmed translational frameshift in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:2315–2319. doi: 10.1073/pnas.90.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens, R. S., S. Kalman, C. Fenner, and R. Davis. Chlamydia Genome Project. http://chlamydia-www.berkeley.edu:4231/index.html.
- 38.Swanson, R. (Diversa Corp.). Personal communication.
- 39.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann D R, Ketchum K A, Klenk H-P, Gills S, Dougherty B A, Nelson K, Quackenbusch J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodeka A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Whattey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 40.Weiss R B, Dunn D M, Atkins J F, Gesteland R F. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5 and +6 ribosomal frameshifting. Cold Spring Harbor Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- 41.Weiss R B, Dunn D M, Dahlberg A E, Atkins J F, Gesteland R F. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss R B, Dunn D M, Atkins J F, Gesteland R F. Ribosomal frameshifting from −2 to +50 nucleotides. Prog Nucleic Acid Res Mol Biol. 1990;39:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]
- 43.Yurieva O, Skangalis M, Kuriyan J, O’Donnell M. Thermus thermophilus dnaX homolog encoding γ and τ-like proteins of the chromosomal replicase. J Biol Chem. 1997;272:27131–27139. doi: 10.1074/jbc.272.43.27131. [DOI] [PubMed] [Google Scholar]