Abstract
Objectives:
We aimed to investigate the role of genetics in the respiratory response of asthmatic children to air pollution, with a genome-wide level analysis of gene by nitrogen dioxide (NO2) and carbon monoxide (CO) interaction on lung function and to identify biological pathways involved.
Methods:
We used a two-step method for fast linear mixed model computations for genome-wide association studies, exploring whether variants modify the longitudinal relationship between 4-month average pollution and post-bronchodilator FEV1 in 522 Caucasian and 88 African-American asthmatic children. Top hits were confirmed with classic linear mixed-effect models. We used the improved gene-set enrichment analysis for GWAS (i-GSEA4GWAS) to identify plausible pathways.
Results:
Two SNPs near the EPHA3 (rs13090972, rs958144) and one in TXNDC8 (rs7041938) showed significant interactions with NO2 in Caucasians but we did not replicate this locus in African-Americans. SNP-CO interactions did not reach genome-wide significance. The i-GSEA4GWAS showed a pathway linked to the HO-1/CO system to be associated with CO-related FEV1 changes. For NO2-related FEV1 responses, we identified pathways involved in cellular adhesion, oxidative stress, inflammation, and metabolic responses.
Conclusion:
The host lung function response to long-term exposure to pollution is linked to genes involved in cellular adhesion, oxidative stress, inflammatory and metabolic pathways.
Keywords: air pollution, asthma, genome-wide, gene-environment interaction, lung function, pathways
Introduction
Epidemiological studies have demonstrated a strong association between exposure to ambient air pollution and adverse effects on childhood respiratory health1–3, with asthmatic children being more susceptible to the negative effects of air pollution4–6. Lower lung function levels in asthmatic and non-asthmatic children have been associated with short-term exposure to air pollution3, 7, 8, but the long-term effects of pollution on lung function are less well studied in asthmatic children9–12.
Known biological mechanisms by which air pollution can impair health include autonomic dysfunction, oxidative stress, and systemic inflammatory responses13–16. Respiratory response to air pollution varies between individuals suggesting that genetic susceptibility likely plays a role17. Recent genome-wide interaction analyses of chronic air pollution exposure indicated that gene-environment interactions are important for asthma development18 and for lung function decline in non-asthmatic adults19.
In asthma, also genes play a role in determining the susceptibility to the harmful effects of air pollution20 but the underlying biological mechanisms of air pollution-mediated health effects are not fully understood warranting further examination of the genes and pathways that might be involved.
We previously investigated the longitudinal relationship between the 4-month average exposure to air pollution and post bronchodilator (BD) forced expiratory volume in 1 second (FEV1) and showed that among the measured air pollutants, long-term exposures to carbon monoxide (CO) and nitrogen dioxide (NO2) are associated with reduced levels of FEV1 in children with asthma21. In the current study we use a hypothesis-free, genome-wide analysis to investigate whether genetic variants modify the long-term effects of CO and NO2 on lung function in children with asthma, and with a pathway analysis we explore further plausible underlying biological pathways of CO and NO2 mediated effects on lung function in asthmatic children.
Materials and Methods
The Childhood Asthma Management Program (CAMP; ClinicalTrials.gov Identifier: NCT00000575) study design and methods have been described elsewhere22. Additional details on all methods used in the present report are provided in an online data supplement. In summary, children enrolled in CAMP were 5–12 years of age and had airway hyper-responsiveness to methacholine at study entry. 1,041 children entered the randomization phase and 311, 312, 418 children received budesonide, nedocromil, and placebo, respectively. All subjects were treated and followed for four years with visits at two and four months after randomization and at four-month intervals thereafter. Each parent or guardian signed a consent form and participants of 7 years of age and older signed an assent form approved by each clinical center’s institutional review board.
Spirometry before and after the administration of two puffs of albuterol (bronchodilator) was conducted at randomization (RZ) and at follow up visits (n=13) according to the American Thoracic Society Standards23. Twenty-four hour average concentrations of CO and NO2 were estimated for each metropolitan area using data from the United States Environmental Protection Agency’s Atmospheric Integrated Research Monitoring Network. The ZIP or postal code centroid coordinates were used to link participants to daily concentrations from the nearest monitor within 50 km that did not have missing data on that day (December 1993 through June 1999). Averaging the daily pollution concentrations for the 4-month intervals between the clinic visits for lung function measurement created the moving averages.
Genome-wide single nucleotide polymorphisms (SNP) genotyping for CAMP subjects (their families and iControlDB controls) was performed on Illumina’s HumanHap550 Genotyping BeadChip (Illumina, Inc., San Diego, CA).
Statistical Analysis
Genome-wide interaction study
In a genome-wide interaction analysis the computational effort needed to evaluate the effects of hundreds of thousands SNPs on the longitudinally measured trait is prohibitively large with a classic linear mixed model (LMM) approach. We followed the Sikorska et al. conditional two-step approach for fast linear mixed model computations for genome-wide association studies (GWAS)24, a method to explore whether the longitudinal relationship between 4-month averaged pollution (CO and NO2) and post-BD FEV1 %predicted is modified by SNPs in the human genome. The practical application of this approach is to be used as a surrogate of classic LMM, hence we performed the genome-wide scan for hundreds of thousands SNPs in a fast manner.
In summary, in the first step we fitted a LMM with subject-specific (random) intercept and slope for pollution exposure with all SNP terms omitted (main effect and interaction with pollutant) from the model. LMM tests were performed in the R programming language (version 3.5.0 (2018–04-23)), and code availability can be requested by the corresponding author.
At the second step, simple linear regression tests of SNPs genome-wide with an individual’s FEV1 response to CO and NO2 (subject-specific-random slopes of pollution as given by LMM in step 1), respectively were performed in PLINK25, using an additive allelic model. SNPs included in the genome-wide analysis had a minor allele frequency > 5% (n=474,792).
For estimating the exact effect size of the interactions and confirm statistical significance, top signals for SNP-pollution interaction as given by 2-step approach (P-value <10−5) were tested with the classic LMM including terms of pollution, SNP and SNP-pollution interaction, (e.g., Bonferroni corrected minimally significant P-value being 0.05/474,792=1.05E-07). Non-Hispanic white (Caucasian) CAMP subjects (n=522) were used as the primary study population and African-American CAMP subjects (n=88) served as the replication study population.
Pathway-level analysis for the genome-wide SNP by pollutant interaction analysis
To analyze pathway-level SNP- pollutant interactions we used the improved gene-set enrichment analysis for GWAS (i-GSEA4GWAS; http://gsea4gwas.psych.ac.cn/inputPage.jsp)26, GSEA evaluates whether the distribution of genes sharing a biochemical or cellular function is different from the distribution of a ranked genome-wide gene list26, 27. Details on the i-GSEA4GWAS method are given in the supplementary material.
Input data to perform the pathway-level analysis of the SNP-pollution interaction analysis were P-values of the two-step genome-wide SNP-pollution interaction analysis in Caucasian CAMP subjects. We changed the default settings and selected specific parameters for the gene-set enrichment analysis; to avoid overrepresentation of SNPs in more than one gene we restricted mapping SNPs to +/−20kb around a gene. We selected additional filtering for gene set size, set to at least 5 genes, so any narrow functional categories would not be missed. Next, the default canonical pathway method of gene-sets was used for further analysis. These canonical pathways were extracted and curated from Molecular Signatures Database from a variety of online resources (MSigDB v2.5; http://www.broadinstitute.org/gsea/msigdb/). The genome-wide P-values were transformed to –log (P-values), represented genes were mapped based on SNPs P-values, and the enrichment score was calculated.
Significant genes in a pathway are defined as the genes mapped with at least one of the top 5% P-values of all SNPs (0.05* 474,792 = 23,740 SNPs). Each significant gene was represented by the SNP in that gene with the lowest genome-wide SNP-pollutant interaction P-value (top SNP per significant gene). We selected the top SNPs of all given pathways and with classic LMM we estimated the interaction effect size for the gene-sets most significant SNP-pollutant interactions in Caucasians.
Results
All subjects in CAMP considered in this analysis were randomized and followed up during the trial period. A total of 1,003 of the 1,041 randomized children (96.3%) had pollution data available of which 610 were studied in the genetic analysis. At study entry the mean (SD) age was 9 (2.1) and geometric mean (min-max) PC20 1.1 (0.02–2.5) mg/ml. Table 1 shows the main characteristics of the participants. 82.5% of the children attended all visits during the 4-year trial (median number of completed visits=14 (range: 1–14)). Each participant had a median of 10 (range: 1–10) post-BD lung function measurements. Repeated FEV1 measurements increase the power of our statistical analysis to detect significant differences between means (8200 and 8600 observations for NO2 and CO analysis, respectively). Tables S1-S2 summarizes the 4-month moving averages pollutant concentrations during December’93-June’99, with number of observations, percentiles and interquartile range (IQR). CO and NO2 were weakly correlated (spearman rho=0.30).
Table 1.
Population characteristics
| N= 1003 | |
|---|---|
|
| |
| City; n (%) | |
| Albuquerque | 121 (12.1) |
| Baltimore | 126 (12.6) |
| Boston | 123 (12.3) |
| Denver | 141 (14.1) |
| San Diego | 122 (12.2) |
| Seattle | 136 (13.6) |
| Saint Louis | 133 (13.3) |
| Toronto | 101 (10.1) |
|
| |
| Sex; n (%) | |
| Males Females |
602 (60) 401 (40) |
|
| |
| Treatment Group; n (%) | |
| Placebo | 407 (40.6) |
| Budesonide | 298 (29.7) |
| Nedocromil | 298 (29.7) |
|
| |
| Ethnicity; n (%) | |
|
| |
| Caucasians | 677 (67.5) |
| African-Americans | 137 (13.7) |
| Hispanics | 97 (9.7) |
| Other | 92 (9.2) |
|
| |
| Annual Income ≥30K USD; n (%) | |
|
| |
|
Yes
No |
728 (76) 235 (24) |
|
| |
| In utero smoking exposure; n (%) | |
|
| |
|
Yes
No |
114 (14) 854 (86) |
| Pre bronchodilator lung function at randomization; mean (SD) | |
| FEV1 %predicted | 93.8 (14.3) |
| FVC %predicted | 104.0 (13.1) |
| FEV 1 /FVC% | 79.7 (8.3) |
| Post bronchodilator lung function at randomization; mean (SD) | |
| FEV1 %predicted | 103.0 (12.8) |
| FVC %predicted | 106.5 (12.8) |
| FEV 1 /FVC% | 85.5 (6.5) |
FEV1 : forced expiratory volume in 1 second; FVC: forced vital capacity; SD: standard deviation; =>30K USD: equal or more than 30,000 United State Dollars
Two-step genome-wide SNP by pollutant interaction analysis
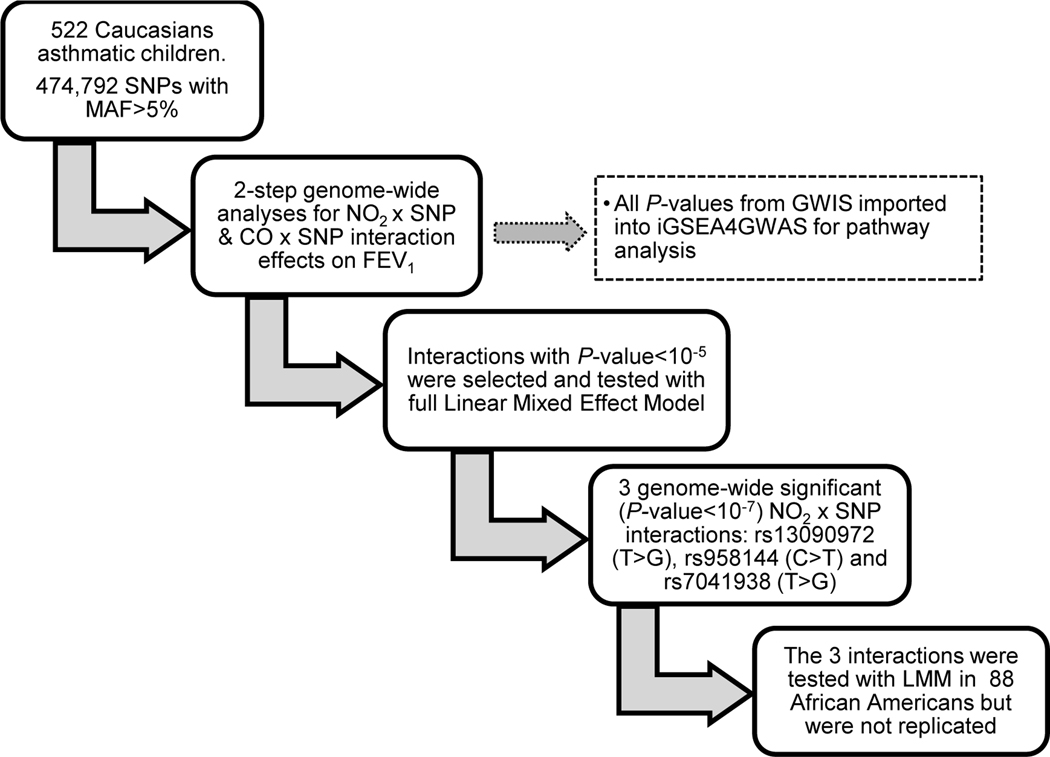
Figure 1 presents an overview of our study design and results of the GWIS. After MAF pruning, 474,792 SNPs were included in the primary analysis, and the smallest P-values for SNP-NO2 and SNP-CO interactions with the 2-step approach were 1.37E-06 and 2.04E-06 respectively, showing only suggestive evidence for genome-wide SNP-pollutant interactions (Table 2 and tables S3A and S4). The quantile-quantile (QQ) plots of the two-step GWIS are presented in Figures S1-S2, showing that the distribution of association P-values was similar to that expected for a null distribution and that no P-values met the conventional genome-wide statistically significant levels (e.g., Bonferroni corrected minimally significant P-value being 0.05/474,792=1.05E-07; see Figures S1 and S2).
Figure 1.
presents the flow chart with the analytic steps and summary of results of the genome-wide gene by pollutant(s) interaction study. Top hit SNPs (P<10−5) interacting with pollutants in Caucasians were selected and with LMM we assessed the interaction effect size and p-values. Genome-wide significant interactions (P<10−7) were tested for replication in African-Americans.
CO: carbon monoxide; NO2: nitrogen dioxide; LMM: linear mixed model; SNP: single nucleotide polymorphism, MAF: minor allele frequency; GWAS: genome wide interaction study; iGSEA4GWAS: improved gene-set enrichment analyses for GWAS; FEV1: forces expiratory volume in 1 second
Table 2:
Top genome-wide gene by nitrogen dioxide interaction loci and suggested functions
| Top genome-wide interaction locus | SNP | 2-Step approach P-values | Classic LMM P value | LMM Change per IQR | Function(s) related to genesa | Identified pathwaysb linked to those gene functions |
|---|---|---|---|---|---|---|
|
| ||||||
|
Near EPHA3;
chr3:88994672 and 89076896 |
rs13090972 (T>G) rs958144 (C>T) |
1.37E-06 4.81E-06 |
1.33E-08
*
2.94E-08 * |
-1.33 −1.28 |
Receptor tyrosine kinase of Eph family
Cell adhesion; Immune surveillance; Tissue remodeling |
Cell adhesion molecules; Calcium regulation; Glycosphingolipids metabolism, Glycosaminoglycan (chondroitin) biosynthesis |
|
| ||||||
|
TXNDC8; chr9:113091523
|
rs7041938 (T>G) |
7.35E-06 | 1.04E-07 * | 1.14 |
Thioredoxin reductase family
Cell redox homeostasis |
HSP27, iNOS, IL10, Heme biosynthesis–Heme oxyganase-1/CO, Calcium regulation |
|
in LD=0.8 with SEVP1; chr9:113133588 |
rs12684188 (C>T) |
> 1E-05 | 3.88E-07 | 1.20 | Cell adhesion; Immune surveillance |
Cell adhesion molecules, Glycosphingolipids metabolism, Calcium regulation |
SNP: single nucleotide polymormpism, LMM: linear mixed model, IQR: interquartile range of 4-month average nitrogen dioxide concentration (4 parts per billion).
genome-wide significance (P< 1.05E-07)
based on coding protein’s function(s). Details for each gene are given in the text .
based on pathway analysis
Confirmation by classic linear mixed model testing
We selected the six top signals (P-value <10−5) SNP-NO2 interactions given by the two-step approach and with the classic LMM model we assessed the effect size of these interactions and compared their P-value as given by the two approaches. In Caucasians, change in post-BD FEV1 %predicted per IQR increase in NO2 level ranged from −1.3 to 1.1 for the 6 SNP-NO2 interactions. With the classic LMM model the P-values decreased for 5 out of 6 SNP-NO2 interactions with values ranging from 1.3E-08 to 8.5E-06 (table S3A). Three SNP-NO2 interactions reached genome-wide significance with the classic LMM: rs13090972 (80kb 5’ of EPHA3) and rs958144 (162kb 5’ of EPHA3) near EPHA3 (LD between 2 SNPs r2=0.55) and rs7041938 in TXNDC8 – the latter in high linkage disequilibrium (r2=0.8) with rs12684188 in SVEP1 (Table 2). Similarly, in African Americans the P-values of associations were lower with LMM, but none reached genome-wide statistical significance (all P-values > 0.05; see table S3B). Table S4 shows that the seven top signals (P-value <10–5) SNP-CO interactions as given by the two-step approach did not reach genome-wide statistical significance with LMM. The change in post-BD FEV1 %predicted per IQR increase in CO level ranged from −0.98 to 0.83 and P-values range from 9.69E-07 to 1.26E-05.
Pathway-level analysis for the two-step genome-wide SNP by pollutant interaction analysis on FEV1 %predicted
For the i-GSEA4GWAS in Caucasian CAMP subjects, -log (P-values) of 474,792 gene variants were imported and 265,485 variants were mapped on genes +/−20kb (total number of genes: 16,854). We identified one pathway interacting with CO (P-value=0.001) and 23 pathways interacting with NO2 (P-values: 0.0001–0.01). Table S5 presents the i-GSEA4GWAS suggested pathways for the two pollutants. Details for each individual pathway (SNPs, mapped genes, gene sets, FDR, P-value, description) of NO2 and CO mediated effects can be found http://gsea4gwas.psych.ac.cn/getResult.do?result=13F3A972887892430E6A5C369D76FEAD_1372283303807 and http://gsea4gwas.psych.ac.cn/getResult.do?result=13F3A972887892430E6A5C369D76FEAD_1372284527739, respectively. All the pathways we present in our findings had FDR<0.25. In summary, the i-GSEA4GWAS showed a pathway (PAC1R; receptor of pituitary adenylate cyclase-activating polypeptide (PACAP)) to be associated with CO-related FEV1 changes. For NO2-related FEV1 responses, we identified several pathways involved in inflammation, oxidative stress, the HO-1/CO system, calcium homeostasis, cellular adhesion and metabolic responses.
Within each gene-set/pathway there were significant genes (genes mapped with at least one of the top 5% of all SNPs-pollutant interactions in the 2-step genome-wide analysis). Each significant gene is represented by the SNP in that gene with the lowest genome-wide P-value of SNP by pollutant interaction (the top SNP per significant gene). Effect sizes of interaction of those SNPs with pollutants as given by LMM are shown in the supplementary material (see tables S6 and S7).
Discussion
Most gene–air pollution studies have focused on a few candidate genetic variations and investigated short-term exposures to pollution17. Although these small hypothesis-driven studies can contribute to our understanding of specific gene–pollution effects, they often fail to uncover novel disease-causing mechanisms and in some cases have not been replicated by subsequent studies28, 29. To the best of our knowledge, this is the first longitudinal GWIS on lung function response to ambient air pollution in asthmatic children. We used the 2-step approach as a screening tool to identify genes that may interact with air pollution while gaining computational time, and we confirmed the top hits of the 2-step approach with the classic LMM; we used the genome-wide output for a iGSEA4GWAS. Below we discuss the putative genes involved in air pollution effects on lung function in childhood asthma and the identified pathways.
At SNP-level, two loci, the EPHA3 (receptor tyrosine kinase of Eph family; location 3p11.2) and TXNDC8 (thioredoxin domain containing 8 (spermatozoa) or Spermatocyte/Spermatid-Specific Thioredoxin-3; location 9q31.3) genes showed genome-wide statistical evidence for interaction with NO2 (with the classic LMM). The best-documented function of the Eph-receptor/ephrin-A signaling is the regulation of cell adhesion and migration processes critical for a wide variety of including tissue remodeling and immune surveillance30, 31. Recent findings suggest that Eph-signaling is involved in pathological conditions such as lung cancer, yet its role in asthma is unknown32, 33. The fact that receptor tyrosine kinase pathways contribute to aspects of airway inflammation, airway hyper-responsiveness and remodeling of asthma34, suggests that we may have identified a novel receptor tyrosine kinases (EPHA3) important for the pathogenesis of asthma in response to NO2 in Caucasian children.
The second top signal locus, TXNDC8, belongs to the thioredoxin reductase enzymes, a well-characterized subfamily of selenoproteins that perform an essential redox role in immune cells35. Recent studies indicated that Thioredoxin system may contribute to the pathogenesis of COPD, asthma and lung injury and suggest that this pathway may be used in future therapeutic applications36. The genome-wide top hit SNP (rs7041938) in TXNDC8 found to modify the NO2 effects on FEV1 in Caucasian subjects is in high linkage disequilibrium (r2>0.8) with rs12684188 in SVEP1 (sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1; location 9q32). In a recent GWAS, a locus containing the SVEP1 gene showed signals of association with FEV1 decline in non-asthmatic adults37. In our asthmatic children the interaction P-value of the SVEP1 variant did not reach significance. SVEP1 codes for a protein called polydom, which is recognized as a cell adhesion molecule with a biological role in cellular adhesion and/or in the immune system38, 39; but its role in asthma has not been investigated so far. We were unable to replicate these loci in African-Americans and it would be important to replicate our finding in other populations in the future.
The pathway analysis helps to clarify biological plausible connections for our GWAS hits with one another. Some of the pathways identified from our iGSEA4GWAS analysis have been previously found to play a role in asthma and be related to cellular adhesion and immune response, as do so our GWAS top loci. The first genome-wide gene by interaction study on asthma development identified genes involved in glycosphingolipids biosynthesis, G-protein coupled receptor signalling and adhesion18. Similarly, our iGSEA4GWAS identified sphingolipid (glycosphingolipid metabolism pathway), G-coupled receptor (gs-pathway, agpcr-pathway, plce-pathway) and epithelial adhesion (HSA04514 cell adhesion molecules pathway) pathways in lung function response to NO2, pointing to the same direction. Sphingolipids and altered sphingolipid metabolism have emerged as potential key contributors to the pathogenesis of asthma40. Orosomucoid-like 3 gene (ORMDL3) and the asthma susceptibility locus 17q21 have been strongly and reproducibly linked to childhood asthma41.
The role of airway epithelial barrier function (HSA04514 cell adhesion molecules pathway) in the susceptibility to develop allergic asthma has been extensively studied and polymorphisms in adhesion molecules genes have been associated with asthma and asthma severity42–44. It is plausible that exposure to NO2 induces oxidative stress with cellular barrier damage and inflammatory responses. In the supplementary material we describe in more detail how pathways involved in inflammation and oxidative stress (NOS1, HSP27, IL10, Heme biosynthase) may be linked to NO2 exposure and how they are inter-related.
Metabolic pathways (feeder of glycolysis and obesity pathways) are activated to compensate the cellular demands to stress and the HO-1/CO system may protect against oxidative stress and inflammation. In line with our findings, a GWIS study of non-asthmatic adults, identified a mechanistic link between adiponectin (a metabolic biomarker with modulating action on inflammatory processes systemically and locally in the lung) and cadherin 13 as a biologically plausible pathway for modifying the air pollution exposure effect on lung decline19.
Oxidative stress has been associated with calcium influx regulation, two responses observed in our pathway analysis as well45, 46. Interestingly, a proteomic-based study has shown that allergen-induced early asthma response in rats is associated with glycolysis, calcium binding and mitochondrial activity47, supporting our identified underlying molecular mechanisms for response to environmental toxicants in asthma.
The iGSEA4GWAS of CO interactions suggested the neuropeptide pituitary adenylate cyclase-activating peptide receptor (PAC1R) pathway in CO-related response. The ligand of PAC1R (PACAP) can induce bronchodilation and endogenous regulation of airway tone by means of a CO-dependent mechanism with local HO-1/CO release in the airway smooth muscle, and it also has pro-inflammatory functions that require calcium regulation48–50. Furthermore, PACAP, acting through type 1 PACAP receptor, exerts a potent protective effect against oxidative stress-induced apoptosis51.
Our childhood asthma study had the advantage of having a long follow-up period with high attendance of the subjects and repeated lung function measurements, air pollution levels during that period and genomic data. The two-step approached used for longitudinal data24 provided shorter processing time and we confirmed its accuracy, i.e., at a second stage the genome-wide top signals found by the two-step approach were confirmed by LMM testing.
We could not find a second study of asthmatic children with similar design, repeated lung function measurements, population characteristics, genome-wide genotyping and air pollution data. Although population stratification is less likely to bias estimates of gene-environment interaction effects52, we used as our primary study only Caucasian CAMP subjects and found no evidence of stratification in our Q/Q plots. For replication studies, definition and measurement of the exposure and/or outcome is critical to the success of gene–environment investigations, therefore we decided to use the second largest ethnic subgroup of the CAMP as our replication population (although of relative small size), to ensure that the genotyping, outcome and exposure were measured reliably and consistently. This reduced power and potential different linkage disequilibrium patterns in the replication population represent limitations of this study.
After testing for pollution effect modification at the SNP-level, we performed the pathway approach to assess the overall evidence of interaction of pollution with a group of functionally related genes, thus incorporating prior biological knowledge. Our pathway-level analysis of SNP-pollution interactions identified biological plausible mechanisms for pollution-mediated asthma progression in children that are generally consistent with the SNP-level analysis.
Our findings highlight the promise of pursuing genome-wide gene-environment interaction studies in smaller populations with high quality longitudinal exposure information by showing that they can identify biologically relevant effects of these exposures. We conclude that genetic susceptibility to traffic-related air pollutants such as with CO and NO2 are linked to oxidative stress and inflammation pathways, while metabolic pathways including calcium homeostasis and the HO-1/CO pathway may play a cytoprotective role against oxidative stress and inflammation. Our findings may represent the first step for functional research and pharmacological developments for protection against the detrimental effects of air pollution on asthma severity and progression.
Supplementary Material
Acknowledgment
We would like to thank Steve Melly for his contribution on the air pollution database preparation and our colleagues Paul V. Williams, Teal S. Hallstrand and Anne N. Fuhlbrigge for the Childhood Management Asthma (CAMP) Program Group. We dedicate this manuscript to the memory of our friend and colleague Dr. Gail G. Shapiro who passed away unexpectedly during this study. Dr. Shapiro dedicated her life to understanding the causes of childhood asthma and determining the best treatments for asthma. She is deeply missed by her colleagues, patients, and the asthma community. A special thank you to all participants of the CAMP study and their families.
Financial support:
The Childhood Asthma Management Program trial and CAMP Continuation Study were supported by contracts NO1-HR-16044, 16045, 16046, 16047, 16048, 16049, 16050, 16051, and 16052 with the National Heart, Lung, and Blood Institute and General Clinical Research Center grants M01RR00051, M01RR0099718-24, M01RR02719-14, and RR00036 from the National Center for Research Resources. The CAMP Continuation Study/Phases 2 and 3 were supported by grants U01HL075232, U01HL075407, U01HL075408, U01HL075409, U01HL075415, U01HL075416, U01HL075417, U01HL075419 and U01HL075420 from the National Heart, Lung, and Blood Institute. The National Jewish Health site was also supported in part by Colorado CTSA grant UL1RR025780 from NCRR/NIH and UL1TR000154 from NCATS/NIH. C:\lm_camp\misc\campdua.wpd June 1, 2013 (2:07pm). In addition, all work on data collected from the CAMP Genetic Ancillary Study was conducted at the Channing Laboratory of the Brigham and Women’s Hospital under appropriate CAMP policies and human subject’s protections. The CAMP Genetics Ancillary Study is supported by U01 HL075419, U01 HL65899, P01 HL083069, R01 HL086601, and RC2 HL101543 from the National Heart, Lung and Blood Institute, National Institutes of Health.
This study was also funded by: the National Institutes of Health (NHLBI P01 HL083069, U01 HL075419, U01 HL65899, R01 HL 086601; NIEHS P01 ES09825, R21 ES020194, P30 ES000002); the U.S. Environmental Protection Agency (RD 83241601, RD 83479801), and the International Initiative for Environment and Public Health Cyprus Program of HSPH. The contents of this publication are solely the responsibility of the grantee and do not necessarily represent the official views of the US EPA. Further, US EPA does not endorse the purchase of any commercial products or services mentioned in the publication.
Footnotes
The authors declare no conflict of interest.
Supplementary information is available at Journal of Exposure Science and Environmental Epidemiology’s website.
References
- 1.Searing DA, Rabinovitch N. Environmental pollution and lung effects in children. Curr Opin Pediatr. 2011. Jun;23(3):314–8. [DOI] [PubMed] [Google Scholar]
- 2.Schwela D. Air pollution and health in urban areas. Rev Environ Health. 2000. Jan-Jun;15(1–2):13–42. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. Effects of air pollution on children’ s health and developement. A review of the evidence. Bonn. http://www.euro.who.int/__data/assets/pdf_file/0010/74728/E86575.pdf. 2005. Date last accessed: April 2013.
- 4.Schildcrout JS, Sheppard L, Lumley T, Slaughter JC, Koenig JQ, Shapiro GG. Ambient air pollution and asthma exacerbations in children: An eight-city analysis. Am J Epidemiol. 2006. Sep 15;164(6):505–17. [DOI] [PubMed] [Google Scholar]
- 5.Sunyer J, Spix C, Quenel P, Ponce-de-Leon A, Ponka A, Barumandzadeh T, et al. Urban air pollution and emergency admissions for asthma in four European cities: The APHEA project. Thorax. 1997. Sep;52(9):760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romieu I, Meneses F, Ruiz S, Sienra JJ, Huerta J, White MC, et al. Effects of air pollution on the respiratory health of asthmatic children living in Mexico city. Am J Respir Crit Care Med. 1996. Aug;154(2 Pt 1):300–7. [DOI] [PubMed] [Google Scholar]
- 7.HEI panel on the Health Effects of Traffic-Related Air Pollution. Traffic-related air pollution: A critical review of the literature on emissions, exposure, and health effects. Boston, MA: 2010. Report No.: HEI Special Report 17. [Google Scholar]
- 8.Li S, Williams G, Jalaludin B, Baker P. Panel studies of air pollution on children’s lung function and respiratory symptoms: A literature review. J Asthma. 2012. Nov;49(9):895–910. [DOI] [PubMed] [Google Scholar]
- 9.Gao Y, Chan EY, Li LP, He QQ, Wong TW. Chronic effects of ambient air pollution on lung function among Chinese children. Arch Dis Child. 2013. Feb;98(2):128–35. [DOI] [PubMed] [Google Scholar]
- 10.Rosenlund M, Forastiere F, Porta D, De Sario M, Badaloni C, Perucci CA. Traffic-related air pollution in relation to respiratory symptoms, allergic sensitisation and lung function in schoolchildren. Thorax. 2009. Jul;64(7):573–80. [DOI] [PubMed] [Google Scholar]
- 11.Gauderman WJ, Vora H, McConnell R, Berhane K, Gilliland F, Thomas D, et al. Effect of exposure to traffic on lung development from 10 to 18 years of age: A cohort study. Lancet. 2007. Feb 17;369(9561):571–7. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz J. Lung function and chronic exposure to air pollution: A cross-sectional analysis of NHANES II. Environ Res. 1989. Dec;50(2):309–21. [DOI] [PubMed] [Google Scholar]
- 13.Kelly FJ. Oxidative stress: Its role in air pollution and adverse health effects. Occup Environ Med. 2003. Aug;60(8):612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel MM, Chillrud SN, Deepti KC, Ross JM, Kinney PL. Traffic-related air pollutants and exhaled markers of airway inflammation and oxidative stress in new york city adolescents. Environ Res. 2013. Feb;121:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emmerechts J, Hoylaerts MF. The effect of air pollution on haemostasis. Hamostaseologie. 2012;32(1):5–13. [DOI] [PubMed] [Google Scholar]
- 16.Auerbach A, Hernandez ML. The effect of environmental oxidative stress on airway inflammation. Curr Opin Allergy Clin Immunol. 2012. Apr;12(2):133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang IA, Fong KM, Zimmerman PV, Holgate ST, Holloway JW. Genetic susceptibility to the respiratory effects of air pollution. Postgrad Med J. 2009. Aug;85(1006):428–36. [DOI] [PubMed] [Google Scholar]
- 18.Gref A, Kebede Merid S, Gruzieva O, Ballereau S, Becker A, Bellander T, et al. Genome-wide interaction analysis of air pollution exposure and childhood asthma with functional follow-up. Am J Respir Crit Care Med. 2016. Nov 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imboden M, Kumar A, Curjuric I, Adam M, Thun GA, Haun M, et al. Modification of the association between PM10 and lung function decline by cadherin 13 polymorphisms in the SAPALDIA cohort: A genome-wide interaction analysis. Environ Health Perspect. 2015. Jan;123(1):72–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno-Macias H, Dockery DW, Schwartz J, Gold DR, Laird NM, Sienra-Monge JJ, et al. Ozone exposure, vitamin C intake, and genetic susceptibility of asthmatic children in Mexico city: A cohort study. Respir Res. 2013. Feb 4;14:14,9921–14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ierodiakonou D, Zanobetti A, Coull BA, Melly S, Postma DS, Boezen HM, et al. Ambient air pollution, lung function, and airway responsiveness in asthmatic children. J Allergy Clin Immunol. 2016. Feb;137(2):390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Childhood Asthma Management Program Research Group. The childhood asthma management program (CAMP): Design, rationale, and methods. childhood asthma management program research group. Control Clin Trials. 1999. Feb;20(1):91–120. [PubMed] [Google Scholar]
- 23.American Thoracic Society. Standardization of spirometry, 1994 update. Am J Respir Crit Care Med. 1995. Sep;152(3):1107–36. [DOI] [PubMed] [Google Scholar]
- 24.Sikorska K, Rivadeneira F, Groenen PJ, Hofman A, Uitterlinden AG, Eilers PH, et al. Fast linear mixed model computations for genome-wide association studies with longitudinal data. Stat Med. 2013. Jan 15;32(1):165–80. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007. Sep;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Cui S, Chang S, Zhang L, Wang J. i-GSEA4GWAS: A web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res. 2010. Jul;38(Web Server issue):W90–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005. Oct 25;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romieu I, Moreno-Macias H, London SJ. Gene by environment interaction and ambient air pollution. Proc Am Thorac Soc. 2010. May;7(2):116–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowatte G, Lodge CJ, Perret JL, Matheson MC, Dharmage SC. Interactions of GST polymorphisms in air pollution exposure and respiratory diseases and allergies. Curr Allergy Asthma Rep. 2016. Nov;16(12):85. [DOI] [PubMed] [Google Scholar]
- 30.Miao H, Wang B. EphA receptor signaling--complexity and emerging themes. Semin Cell Dev Biol. 2012. Feb;23(1):16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arvanitis D, Davy A. Eph/ephrin signaling: Networks. Genes Dev. 2008. Feb 15;22(4):416–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008. Apr 4;133(1):38–52. [DOI] [PubMed] [Google Scholar]
- 33.Zhuang G, Song W, Amato K, Hwang Y, Lee K, Boothby M, et al. Effects of cancer-associated EPHA3 mutations on lung cancer. J Natl Cancer Inst. 2012. Aug 8;104(15):1182–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guntur VP, Reinero CR. The potential use of tyrosine kinase inhibitors in severe asthma. Curr Opin Allergy Clin Immunol. 2012. Feb;12(1):68–75. [DOI] [PubMed] [Google Scholar]
- 35.Huang Z, Rose AH, Hoffmann PR. The role of selenium in inflammation and immunity: From molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2012. Apr 01;16(7):705–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Li T, Wu H, Xu T. Role of thioredoxin in lung disease. Pulm Pharmacol Ther. 2012. Apr;25(2):154–62. [DOI] [PubMed] [Google Scholar]
- 37.Imboden M, Bouzigon E, Curjuric I, Ramasamy A, Kumar A, Hancock DB, et al. Genome-wide association study of lung function decline in adults with and without asthma. J Allergy Clin Immunol. 2012. May;129(5):1218–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwanzer-Pfeiffer D, Rossmanith E, Schildberger A, Falkenhagen D. Characterization of SVEP1, KIAA, and SRPX2 in an in vitro cell culture model of endotoxemia. Cell Immunol. 2010;263(1):65–70. [DOI] [PubMed] [Google Scholar]
- 39.Gilges D, Vinit MA, Callebaut I, Coulombel L, Cacheux V, Romeo PH, et al. Polydom: A secreted protein with pentraxin, complement control protein, epidermal growth factor and von willebrand factor A domains. Biochem J. 2000. Nov 15;352 Pt 1:49–59. [PMC free article] [PubMed] [Google Scholar]
- 40.Ono JG, Worgall TS, Worgall S. Airway reactivity and sphingolipids-implications for childhood asthma. Mol Cell Pediatr. 2015. Dec;2(1):13,015–0025-3. Epub 2015 Dec 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao CN, Fan Y, Huang JJ, Zhang HX, Gao T, Wang C, et al. The association of GSDMB and ORMDL3 gene polymorphisms with asthma: A meta-analysis. Allergy Asthma Immunol Res. 2015. Mar;7(2):175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heijink IH, Nawijn MC, Hackett TL. Airway epithelial barrier function regulates the pathogenesis of allergic asthma. Clin Exp Allergy. 2014;44(5):620–30. [DOI] [PubMed] [Google Scholar]
- 43.Faura Tellez G, Willemse BW, Brouwer U, Nijboer-Brinksma S, Vandepoele K, Noordhoek JA, et al. Protocadherin-1 localization and cell-adhesion function in airway epithelial cells in asthma. PLoS One. 2016. Oct 4;11(10):e0163967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ierodiakonou D, Postma DS, Koppelman GH, Boezen HM, Gerritsen J, Ten Hacken N, et al. E-cadherin gene polymorphisms in asthma patients using inhaled corticosteroids. Eur Respir J. 2011. May 3. [DOI] [PubMed] [Google Scholar]
- 45.Jiang LH, Yang W, Zou J, Beech DJ. TRPM2 channel properties, functions and therapeutic potentials. Expert Opin Ther Targets. 2010. Sep;14(9):973–88. [DOI] [PubMed] [Google Scholar]
- 46.Xu R, Li Q, Zhou XD, Perelman JM, Kolosov VP. Oxidative stress mediates the disruption of airway epithelial tight junctions through a TRPM2-PLCgamma1-PKCalpha signaling pathway. Int J Mol Sci. 2013. Apr 29;14(5):9475–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu YD, Cui JM, Wang Y, Yin LM, Gao CK, Liu YY, et al. The early asthmatic response is associated with glycolysis, calcium binding and mitochondria activity as revealed by proteomic analysis in rats. Respir Res. 2010. Aug 6;11:107,9921–11-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinhult J, Andersson JA, Uddman R, Stjarne P, Cardell LO. Pituitary adenylate cyclase-activating peptide 38 a potent endogenously produced dilator of human airways. Eur Respir J. 2000. Feb;15(2):243–7. [DOI] [PubMed] [Google Scholar]
- 49.Kinhult J, Uddman R, Cardell LO. The induction of carbon monoxide-mediated airway relaxation by PACAP 38 in isolated guinea pig airways. Lung. 2001;179(1):1–8. [DOI] [PubMed] [Google Scholar]
- 50.Linden A, Cardell LO, Yoshihara S, Nadel JA. Bronchodilation by pituitary adenylate cyclase-activating peptide and related peptides. Eur Respir J. 1999. Aug;14(2):443–51. [DOI] [PubMed] [Google Scholar]
- 51.Douiri S, Bahdoudi S, Hamdi Y, Cubi R, Basille M, Fournier A, et al. Involvement of endogenous antioxidant systems in the protective activity of pituitary adenylate cyclase-activating polypeptide against hydrogen peroxide-induced oxidative damages in cultured rat astrocytes. J Neurochem. 2016. Jun;137(6):913–30. [DOI] [PubMed] [Google Scholar]
- 52.Thomas D. Gene--environment-wide association studies: Emerging approaches. Nat Rev Genet. 2010. Apr;11(4):259–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.