Abstract
“Epilepsy is a chronic brain disorder that affects people of all ages. The cause of epilepsy is often unknown and its effect in different age groups is not yet investigated. The main objective of this study is to introduce a novel approach that successfully detects epilepsy even from noisy EEG signals. In addition, this study also investigates population specific epilepsy detection for providing novel insights. Correspondingly, we utilized the TUH EEG corpus database, publicly available challenging multi-channel EEG database containing detailed patient information. We applied a band-pass filter and manual noise rejection to remove noise and artifacts from EEG signals. We then utilized statistical features and correlation to select channels, and applied different transform analysis methods such as continuous wavelet transform, spectrogram, and Wigner-Ville distribution, with and without ensemble averaging, to construct an image dataset. Afterwards, we used various deep-learning models for general analysis. Our findings suggest that different models such as DenseNet201, DenseNet169, DenseNet121, VGG16, VGG19, Xception, InceptionV3, and MobileNetV2 performed better while using images generated from different approaches in general analysis. Furthermore, we split the dataset into two sections according to age for population analysis. All the models that performed well in the general analysis were used for population analysis, which provided novel insights in epilepsy detection from EEG. Our proposed framework for epilepsy detection achieved 100% accuracy, which outperforms other concurrent methods."
Keywords: Convolutional neural network, EEG signal, Epilepsy, Population analysis, Transform analysis
1. Introduction
Recurrent, sudden, and transitory paroxysmal electrical activity in the brain is an indication of epilepsy, the fourth most common neurological condition [[1], [2], [3]]. The cerebral cortex is principally affected by the “epileptogenesis” process, that entails the rapid conversion of the regular neural network into a hyper-excitable network [4]. With seizures affecting over 1–2% of individuals and negatively impacting productivity, accurate and efficient diagnosis is pivotal. The symptoms of seizure intensify with age and therefore it's prevalence rate increases in aged people [5].
Among the diagnostic procedures for epilepsy, electroencephalogram (EEG) monitoring, complemented by alternative magnetoencephalogram signals as well as brain scans like computed tomography, positron emission tomography, and magnetic resonance imaging, is commonly employed. EEG stands out among them as the most effective, non-invasive, and economical technique for analyzing brain electrical activity, offering insightful data. Using the 10–20 electrode placement technique, each channel in a multi-channel scalp EEG functions as a sensory source, providing detailed information in five frequency bands (alpha, beta, delta, theta, and gamma) [6].
Although professional neurologists use visual interpretation of recorded data to diagnose epilepsy, the manual scoring approach is time-consuming, expensive, and prone to errors. Numerous hours of continuous EEG recording are necessary for epilepsy diagnosis, which is expensive, time-consuming, and difficult. Additionally, because there is no common reference standard and each doctor's judgment is based on their own personal experience, the diagnosis results of the same record may vary considerably [7]. The requirement for creating automatic methods to assess and diagnose epileptic seizure EEG signals is ever-increasing in order to avoid the danger of the analyst losing information. Due to their tremendous capacity for learning high-level patterns from natural data, deep learning algorithms have successfully demonstrated promising results in the identification and classification of epilepsy using EEG. Diverse techniques for the classification and detection of epilepsy from EEG based on CNN have been the subject of research for several years [7,8].
Despite the encouraging outcomes reported by earlier deep EEG seizure detection methods, there are still a number of issues that need to be resolved. Firstly, the objective functions of the majority of deep learning-based approaches do not explicitly take into account the innate correlations between EEG channels. On the one hand, there are some channels that are ineffective since the region beneath them might not detect seizure activity. Although the influence of redundant and irrelevant features can be somewhat reduced by the typical deep learning structure, it might not be sufficient to fully harness the power of multi-channel information. Owing to the low number of seizure occurrences as well as the significant costs associated with labeling the data, imbalanced along with unlabeled data issues could possibly hinder the efficacy of deep learning algorithms in real-world applications. Moreover, the prevalence rate gets higher with an aging population and the elevated incidence of epilepsy in belated life [5].
This study introduces a novel method that merges statistical features and transform analysis techniques to create a specialized image dataset form the Temple University Hospital (TUH) dataset [9] for epilepsy detection through deep learning models. We employed various transform analysis techniques, including the continuous wavelet transform, the spectrogram analysis, and the Winger-Ville distribution, as well as ensemble approaches like the ensemble continuous wavelet transform, ensemble spectrogram, and ensemble Winger-Ville distribution. In addition, it will also investigate age specific population analysis for epilepsy detection to provide new insights. Considering the prevalence rate of epilepsy, the data has also been dissected into two age groups [10]. To the best of our knowledge, this specific combination type is both unique and distinct in this field.
The remainder of the paper is presented as follows. In Section 2, we provide a brief review of related literature. Section 3 presents a detailed description of the proposed methodology, including the transform analysis methods used in this study and the epilepsy detection techniques. We also provide a brief overview of the benchmark EEG data and experimental design. Section 4, 5 respectively addresses the experimental results and the performance evaluation of the multiple CNN models, as well as the findings and insights gained from this study along with the limitations subsequently. Finally, we provide our concluding remarks in Section 6.
2. Literature review
Several researchers have investigated methods for detecting epilepsy. One study utilized wavelet transform on a dataset described by Andrzejak et al. [11] with K-means clustering [12] has achieved 96.67% accuracy. In a different study, a wavelet scalogram was utilized in the dataset described by Andrzejak et al. [11] with AlexNet and achieved 100% accuracy [13]. Wavelet entropy was also used on the dataset described by Andrzejak et al. [11], achieving the highest accuracy (100%) when combined with support vector machine [8]. In another study, 5 s epileptic segment was utilized with sample entropy and distributed entropy, using GA-SVM (genetic algorithm for feature selection and parameter optimization of support vector machine) on the dataset described by Andrzejak et al. [11] and achieved a maximum AUC of 96.67% [4]. Another study used optimum allocation technique on the dataset described by Andrzejak et al. [11] with the logistic model tree achieving 96.67% accuracy [14]. A study using time and frequency domain features from the ARMOR project dataset with a Bayesian Net achieved maximum accuracy of 95% [15]. Additionally, wavelet transform-based features were extracted from the dataset described by Andrzejak et al. [11] and utilizing the Random forest classifier achieved 95% accuracy [16]. The Teager energy feature was extracted by using data obtained from (the Institute of Neuroscience, Ramaiah Memorial Hospital, Bengaluru, India) in a supervised backpropagation neural network and achieved 95% accuracy [17]. The sub-frequency-based feature was extracted from a Kaggle dataset (www.kaggle.com) which was put on for use in an open seizure prediction competition conducted on the website achieved 91.6% accuracy in a generalized regression neural network [7]. Fuzzy distribution entropy and wavelet packet decomposition were extracted from the dataset described by Andrzejak et al. [11] and were utilized in K-nearest neighbor. This method achieved 98.33% accuracy [18]. Discrete wavelet transform coupled with a feature set from the dataset described by Andrzejak et al. [11] was utilized in artificial neural networks to achieve 95% accuracy [19]. Hurst exponent was calculated from the dataset described by Andrzejak et al. [11] then the KNN classifier is utilized for classification and achieved 100% accuracy [20]. Symlet wavelet processing and grid search optimizer feature extracted from dataset described by Andrzejak et al. [11] and were used in gradient boosting machine. This method achieved 96.1% accuracy [2]. Self-organized map (SOM) method used in radial basis function (RBF) neural network approaches tested on the Bonn University epilepsy dataset achieved 97.47% accuracy [21]. The spectral, spatial, and temporal features were extracted from the CHB-MIT dataset along with Seoul national university hospital utilized in multi-scale 3D CNN with deep neural network and achieved 99.4% accuracy [22]. Short-time Fourier transform was used in the Bern Barcelona dataset then constructed images were utilized in a convolutional neural network. This method achieved 91.8% accuracy [23]. Fourier-based synchro squeezing transform (SST) and convolutional neural network were applied in the publicly available CHB-MIT dataset and achieved 99.63% accuracy [24]. Multi-view feature learning was utilized with convolutional deep learning applied in the CHB-MIT dataset achieving 94.37% accuracy [25]. Waveform level classification was done by the convolutional neural network to extract features then these features were utilized in SVM applied in clinical dataset recorded at Massachusetts general hospital, Boston achieving 83.86% accuracy [26]. Spectrogram images of EEG signals were utilized in a pre-trained convolutional neural network applied in the TUH dataset, which achieved the highest 88.3% accuracy [27]. A one-dimensional deep convolutional neural network was used for automated identification of abnormal EEG signals without any feature extraction and applied in the TUH dataset, which achieved 79.34% accuracy [1].
3. Methodology
3.1. Dataset
In this study, we utilized the world's largest publicly available EEG database which is called the TUH EEG corpus database [9]. This dataset has some unique feature such as each EEG signal of this dataset has a report provided by the clinician. This report apprises us with multifarious information about the patient including sex, age, heart rate, medications, clinical history, and so on. This database comprises a multi-channel EEG signal. The number of channels utilized for the recording of EEG signal is 32. This dataset carries annotation files by dint of it we can acquire knowledge about the event of each channel.
From the TUH EEG corpus dataset, we employ a fragment of data for our study. Here, we utilized data from 5 disparate patients for epileptic EEG signal. Additionally, we utilized data from 5 disparate patients for Normal Epilepsy data. We utilized 1h 34min data for our study which is significantly larger than dataset utilized for most other pervious studies. The percentage of dataset split for training, validation and testing are (Training- 90%, Test- 5%, Validation- 5%). Delineate information regarding patients is given in Table 1.
Table 1.
Patients information.
| Types | Patient Id | Medications | Sex | Age | Heart Rate |
|---|---|---|---|---|---|
| Epilepsy | 258 | Dilantin, Phenobarbital, and Baclofen | M | 41 | 84 |
| 675 | Albuterol | F | 4 | 126 | |
| 1027 | Dilantin and Phenobarbital | M | 50 | 72 | |
| 1770 | Trileptal, Valproate, and Klonopin | F | 26 | 126 | |
| 1984 | Trileptal | M | 6 | 72 | |
| Normal | 258 | Dilantin, Phenobarbital, and Baclofen | M | 41 | 84 |
| 629 | Tegretol and Topamax | F | 22 | 78 | |
| 1027 | Dilantin and Phenobarbital | M | 50 | 72 | |
| 1278 | Dilantin, Topamax, Remeron, and Reglan | F | 33 | 96 | |
| 1981 | Tegretol | M | 56 | 72 |
3.2. Data pre-processing
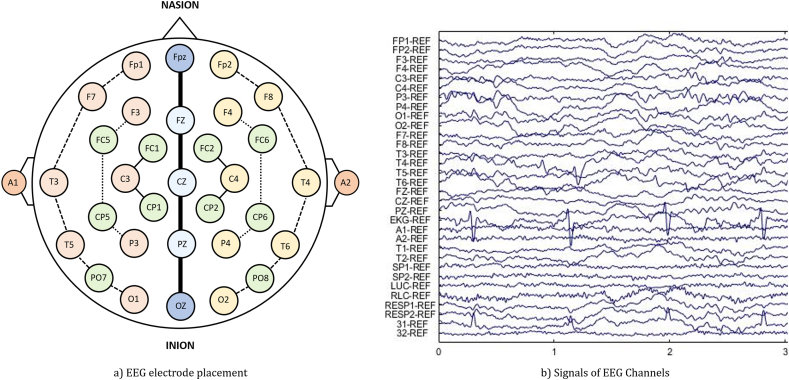
While recording the EEG signal noise and artifacts also get recorded with the signal. These noises as well as artifacts are obviated from the signal by employing data preprocessing. We employed a filtering technique along with manual noise and artifact rejection owing to data preprocessing (see Fig. 1).
Fig. 1.
Typical graphical representation of (a) EEG electrode placement and its corresponding (b) signals of EEG channels recorded from the brain.
3.2.1. Filtering
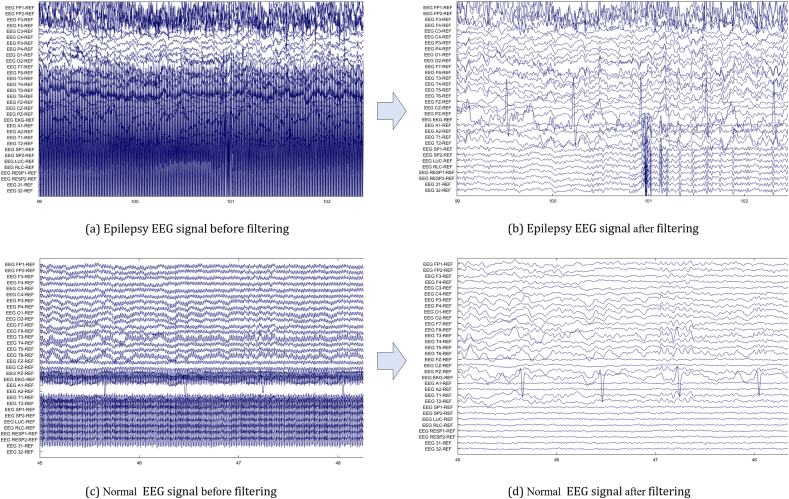
A bandpass filter is the most effective way to remove noise from the EEG signal. In our study, EEG signals were passed across a bandpass filter with a cut-off frequency between 0.1 and 44 Hz, resulting in high-frequency noise along with low-frequency superfluous signal being detached. MATLAB EEGLAB is utilized for the visualization as well as filtering of the signal. EEG signals before as well as after filtering are depicted in Fig. 2.
Fig. 2.
EEG signal (Epilepsy and Normal) before and after using bandpass filter for removing noise and artifact. (a) Epilepsy EEG signal before filtering, (b) Epilepsy EEG signal after filtering, (c) Normal EEG before filtering and (d) Normal EEG signal after filtering.
3.2.2. Manual noise rejection
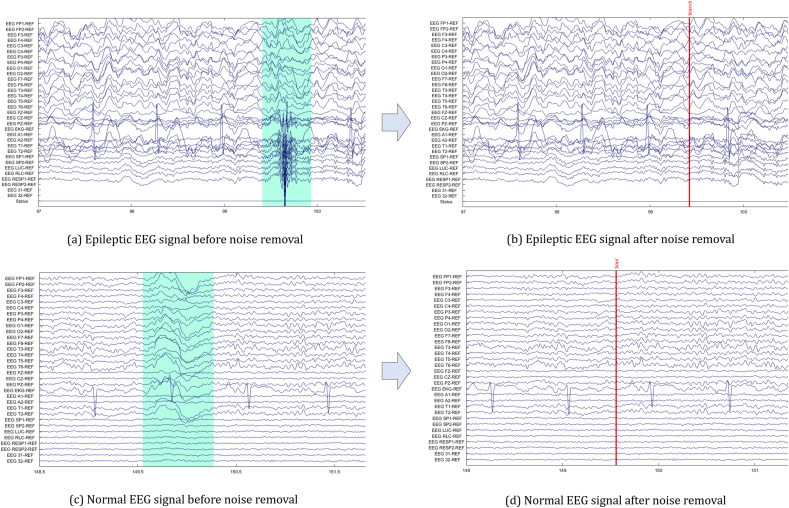
While recording EEG signals, noise and artifacts such as multiple eye blink, single eye blink, EMG artifacts, electrode movement, electrostatic, jerking, and lead artifacts also got recorded. TUH EEG corpus dataset contains annotation files where these noise and artifacts images are exhibited. After filtering the signal, noise, and artifacts are obviated from the signal manually by taking advantage of the annotation files. To discern the noise, we employ the image exhibited in the annotation files. EEG signals before as well as after removing the noise along with the artifact are depicted in Fig. 3.
Fig. 3.
EEG signal (Epilepsy and Normal) before and after manual noise rejection. (a) Epileptic EEG signal before noise removal, Epileptic EEG signal after noise removal, (c) Normal EEG signal before noise removal and (d) Normal EEG signal after noise removal. Blue highlighted regions indicate noise and artifact which are removed after manual noise rejection.
3.3. Channel selection
As the TUH EEG corpus dataset comprises a multi-channel EEG signal, it is necessary to select the appropriate channels. We used statistical features such as mean, median, and standard deviation for channel selection. Initially, we calculated statistical features for each channel of the EEG signal. Subsequently, we constructed co-relation as well as reckoned the p-value of the mean, median and standard deviation for each channel of the EEG signal. Afterward, we selected two channels that have upmost co-relation and p-value. These selected two channels were utilized for the analysis.
3.4. Ensemble averaging
The ensemble average is defined as the mean of a quantity. In our analysis, we have done two sorts of ensemble averaging.
-
I.
Ensemble averaging of selected two channels- Initially, selected two channels' signals were added together. Then, the added signal was divided by 2.
-
II.
Ensemble averaging of all the channels- Here, signals from all the channels (32) were added together. Then, the added signal was divided by 32.
3.5. Data segmentation
In this study, we segmented the Normal EEG signal and Epileptic EEG signal into 5s fragments. Then, this 5s EEG signal was used for transform analysis to create an image. Different signal fragments have been used in other research studies such as 0.1s [28], 2s [29,30], 4s [31], 5s [4,32], and 60s [1].
3.6. Transform analysis
Transform analysis is defined as altering the representations of the signal from one form to a disparate form by employing some mathematical transformation [13]. Transform analysis such as continuous wavelet transform (CWT), Spectrogram using short-time Fourier transform, and Winger-Ville distribution (WVD) was utilized during our analysis.
3.6.1. Continuous wavelet transform (CWT)
Continuous wavelet transform (CWT) is a formal tool that renders an overcomplete representation of a signal by letting the translation as well as scale parameters of the wavelets differ continuously. EEG signals were converted into a 2-dimensional image by utilizing CWT. We have utilized the MATLAB function for accomplishing the continuous wavelet transform. MATLAB function which is utilized for CWT is [wt, f, coi] = cwt(X, fs).
Where X represents the EEG signal and fs represents the sampling frequency. X and fs are utilized as input of the function which returns wt, f, and coi. Here wt represents the wavelet transform of the EEG signal X, f represents scale-to-frequency conversions, and coi represents the cone of influence.
CWT is a formal (i.e., non-numerical) tool that can render an over-complete representation of a signal by letting the translation, as well as the scale parameter vary continuously.
The CWT of a function x(t) at a scale (a>0) along with translational value b ∈ R is expressed by following integral,
| (1) |
in equation (1), Ψ(t) is a continuous function twain time domain along with frequency domain called the mother wavelet as well as the over line representation operation of complex conjugate. The prime intention of the mother wavelet is to render a source function to engender the daughter wavelets which are simply translated along with scaled versions of the mother wavelet.
3.6.2. Short-time Fourier transform (STFT)
Short-time Fourier transform is used to determine the sinusoidal frequency as well as phase content of local sections as it alters over time. EEG signals were converted into a 2-dimensional image by utilizing a spectrogram Short-time Fourier transform (STFT). We have utilized the MATLAB function for accomplishing the Short-time Fourier transform. MATLAB function which is utilized for STFT is S = spectrogram (X, window, noverlap, f, fs).
Where X represents the EEG signal. Function utilizes window to dissect the signal into segments along with performing windowing. Function utilizes noverlap samples of overlap betwixt adjoining segments, f represents sampling points which is used to calculate the discrete Fourier transform, and fs represents the sampling frequency. The function returns S as output which represents the spectrogram of the EEG signal X.
In practice, the strategy for reckoning STFT is to dissect a prolonged time signal into shorter segments of identical length as well as then compute the Fourier transform independently on each shorter segment. This discloses the Fourier spectrum of one and all segments.
In the discrete-time case, the data to be transformed could be broken up into chunks or frames (which customarily overlap each other, to mitigate artifacts at the boundary). Each chunk is Fourier transformed, as well as the complex result is connected to a matrix, which records magnitude along with phase for one and all points in time along with frequency as depicted in equation (2).
| (2) |
Likewise, with signal x[n] along with window w[n]. In this respect, m is discrete and ω is continuous, nevertheless in the majority of archetypical; applications the STFT is performed on a computer utilizing the fast Fourier transform, so the twain variables are discrete as well as quantized.
The magnitude squared of the STFT yields the spectrogram representation of the power spectral density of the function is depicted in equation (3).
| (3) |
3.6.3. Winger-Ville distribution (WVD)
The Winger-Ville distribution (WVD) function is utilized in signal processing as a transform in time-frequency analysis. EEG signals were converted into a 2-dimensional image by utilizing WVD. We have utilized the MATLAB function for accomplishing WVD. MATLAB function which is utilized for WVD is D = wvd(X, fs).
Where X represents the EEG signal and fs represents the sampling frequency. X and fs are utilized as input of the function which returns D. Here, D represents the Winger-Ville distribution of the EEG signal X.
In mathematics, there are disparate definitions for the Winger distribution function. Given the time series x[t], its non-stationary autocorrelation function is given by equation (4) as
| (4) |
Where ⟨ … ⟩ denotes the average over all possible realizations of the process and μ(t) is the mean, which may or may not be a function of time. The Winger function is given by first expressing the autocorrelation function in terms of the average time )/2 and time lag and then Fourier transforming the lag. This is expressed in equation (5),
| (5) |
for a single (mean-zero) time series, the Winger function is
| (6) |
The motivation for the Winger function is that it mitigates to the spectral density function at all times t for stationary processes, yet it is thoroughly equivalent to the non-stationary autocorrelation function as shown in In equation (6). Therefore, the Winger function informs us (roughly) how spectral density alters.
3.7. Image crop and resize
Succeeding transform analysis of the EEG signal, we obtained the images of that stipulated signal. In the wake of the analysis, the images we perceived that albeit these images contain information utterly on the x-axis nevertheless it does not contain a great deal of information on the y-axis. Therefore, to eliminate unnecessary space from the image, we cropped 40 % of the image from the y-axis. We did not perform any cropping on the x-axis. Afterward, we resized the cropped image to the original size of the image.
3.8. Sample data
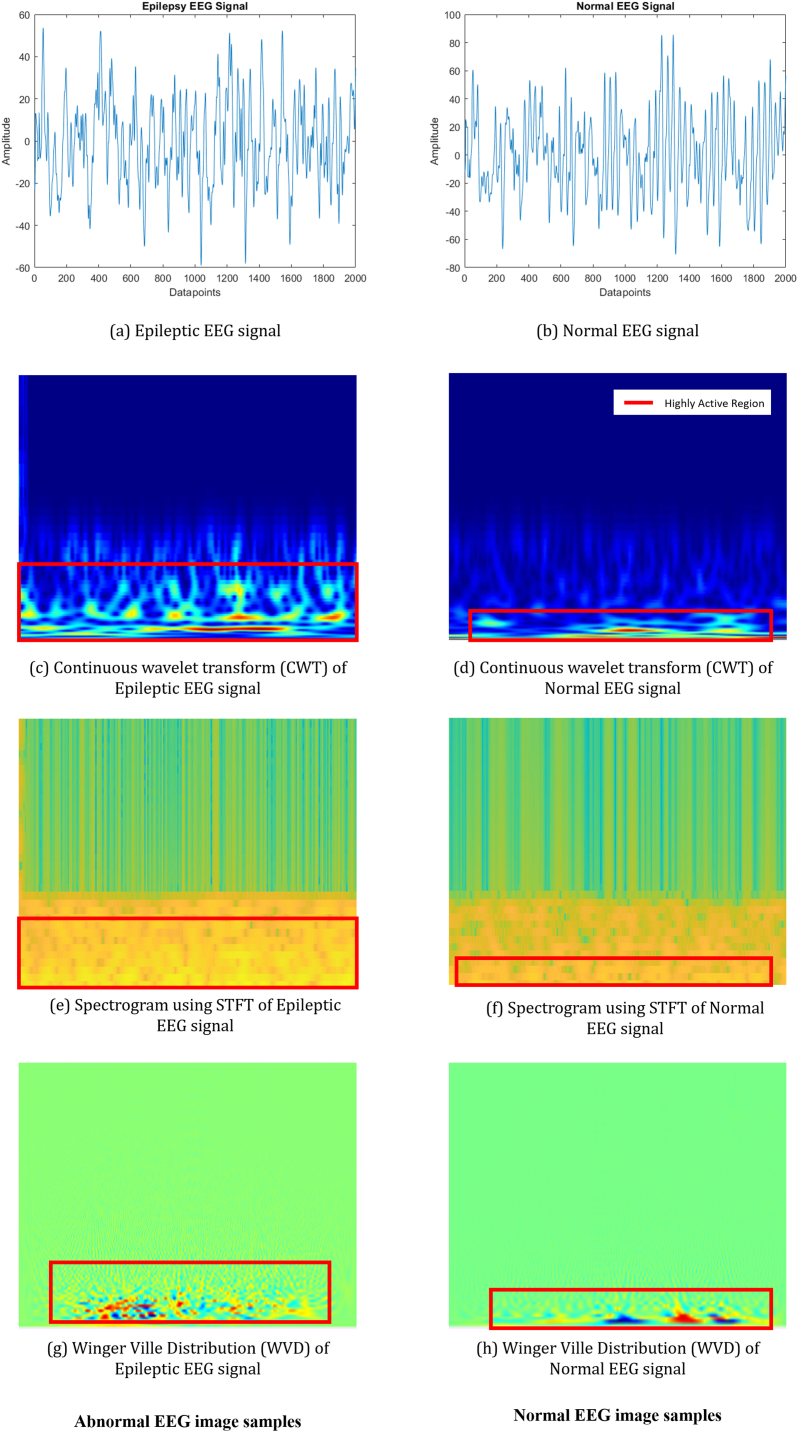
Epileptic EEG signals along with Normal EEG signals with their corresponding image generated by transform analysis (CWT, Spectrogram, WVD) are depicted in Fig. 4.
Fig. 4.
Sample EEG data along with different transform analysis. (a) Epileptic EEG signal, (b) Normal EEG signal, (c) Continuous wavelet transform (CWT) of Epileptic EEG signal, (d) Continuous wavelet transform (CWT) of Normal EEG signal, (e) Spectrogram using STFT of Epileptic EEG signal, (f) Spectrogram using STFT of Normal EEG signal (g) Winger-Ville Distribution (WVD) of Epileptic EEG signal and (h) Winger-Ville Distribution (WVD) of Normal EEG signal. The red boxes indicate the region that has significant activity.
In Fig. 4, the red boxes indicate the region that has significant activity. By examining all the images, we observed that the images generated from the epilepsy signal exhibit the most activity, while the images generated from the normal signal exhibit substantially less activity. This unique pattern can be found in all the transform analysis that we performed in this study.
3.9. Convolutional neural network
Convolutional neural network in other words CNN is a deep learning network fabricated in order to analyze structured arrays of data such as images. CNNs exhibit outstanding results for picking up patterns in the input image [33]. CNN architecture is a multifarious-layered feed-forward neural network, which is constructed by stacking many hidden layers’ peaks of each other in sequence. It is this sequential arrangement that permits CNNs to comprehend hierarchical features. The hidden layers are customarily convolutional layers that comply with activation layers, some of them followed by polling layers. Initially in our study, we strived all the convolutional neural networks. On that occasion some CNN models namely DenseNet201, DenseNet169, DenseNet121, VGG16, VGG19, Xception, InceptionV3, MobileNetV2 exhibited better result.
3.10. Input image resize
Pre-trained CNN model has a default image size. While inputting the image in the CNN model for classification we must resize the image according to the default image size of that stipulated CNN model. We have utilized all the pre-trained CNN architectures in our study. A few of them exhibited sublime results. The default input size of some of these CNN architectures are given in Table 2.
Table 2.
Input image size of disparate CNN model.
| Model | Input Size |
|---|---|
| DenseNet201 | 224*224*3 |
| DenseNet169 | 224*224*3 |
| DenseNet121 | 224*224*3 |
| VGG16 | 224*224*3 |
| VGG19 | 224*224*3 |
| Xception | 299*299*3 |
| InceptionV3 | 256*256*3 |
| MobileNetV2 | 224*224*3 |
| Resnet50 | 227*227*3 |
3.11. Hyperparameter tuning
A hyperparameter is a parameter whose value is used to control the learning process. Hyperparameter optimization is an imperative task to get the utmost result in any deep learning approach. In this study, we optimized various hyperparameters, such as the optimizer, learning rate, batch size, and number of epochs, to achieve the best results in our deep learning process [33]. We have utilized different learning rates for our study to get the upmost result. However, for most of the model, the range of learning rate was 0.001 to 0.00001 for the best result. We utilized disparate optimizer to get the upmost result. However, for most of the model we observed that SGD, RMSprop and adam gave the best results.
3.12. Evaluation metrics
In our analysis, we reckoned accuracy ( to quantify the performance of the CNN models [33],
| (7) |
in equation (7), , , , and respectively stands for the true positive, true negative, false positive, and false negative of a 2 × 2 confusion matrix.
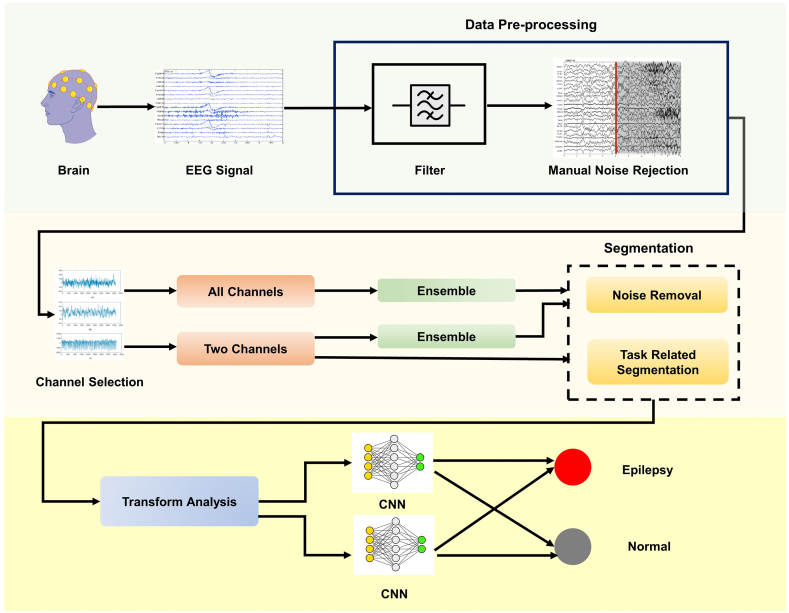
Fig. 5 illustrates the complete methodological framework of this study. This includes data pre-processing (filtering and manual noise rejection), image dataset generation by transform analysis (CWT, Spectrogram, WVD) with and without ensemble averaging and classification using CNN architecture to distinguish between epileptic and normal EEG.
Fig. 5.
Proposed methodology.
4. Result
4.1. General analysis
In this study, we utilized deep learning models for the automatic classification of normal and epilepsy EEG signals. The dataset was divided into three parts: training, testing, and validation. Data were selected randomly. A few models exhibited preferable performance. However, it is not feasible to list the results of all deep learning models. Therefore, we adopted a threshold accuracy value of 80%. All the models which demonstrated an accuracy of more than 80% for training, validation as well as test that is put down in Table 3.
Table 3.
Results of general analysis of epilepsy.
| Transform analysis | Convolutional neural network model | Train Accuracy |
Test Accuracy |
Validation Accuracy |
|---|---|---|---|---|
| CWT | VGG16 | 100% | 98.33% | 96.67% |
| VGG19 | 99.6% | 96.67% | 96.67% | |
| Xception | 97.2% | 91.67% | 93.33% | |
| InceptionV3 | 100% | 96.67% | 100% | |
| MobileNetV2 | 100% | 98.33% | 100% | |
| DenseNet201 | 100% | 96.67% | 98.33% | |
| DenseNet121 | 100% | 96.67% | 98.33% | |
| Ensemble CWT |
Xception | 82.7% | 88.33% | 86.44% |
| DenseNet201 | 86.5% | 90% | 93.22% | |
| DenseNet121 | 83.7% | 93.33% | 93.22% | |
| DenseNet169 | 81.3% | 95% | 94.92% | |
| Spectrogram | VGG16 | 92.2% | 86.67% | 96.67% |
| Xception | 100% | 90% | 96.67% | |
| InceptionV3 | 86.5% | 83.33% | 90% | |
| DenseNet201 | 87.7% | 88.33% | 93.3% | |
| DenseNet121 | 83.6% | 88.3333% | 83.33% | |
| DenseNet169 | 86.8% | 81.67% | 86.67% | |
| Ensemble Spectrogram | Xception | 89.4% | 81.67% | 93.33% |
| DenseNet169 | 89.8% | 80% | 81.67% | |
| WVD | VGG16 | 90.2% | 93.33% | 93.33% |
| VGG19 | 90.6% | 85% | 86.67% | |
| Xception | 100% | 80.1% | 83.33% | |
| MobileNetV2 | 100% | 91.67% | 91.67% | |
| DenseNet201 | 100% | 93.33% | 90% | |
| DenseNet121 | 100% | 86.67% | 91.67% | |
| DenseNet169 | 100% | 91.67% | 91.67% | |
| Ensemble WVD | Xception | 91.1% | 98.33% | 100% |
| InceptionV3 | 87.6% | 93.33% | 91.67% | |
| MobileNetV2 | 98.5% | 100% | 100% | |
| DenseNet201 | 95.7% | 95% | 95% | |
| DenseNet121 | 94.3% | 93.33% | 96.67% |
We generated images using six different transform analyses, including three with ensemble averaging and three without ensemble averaging. These included CWT, Spectrogram, and WVD without ensemble averaging, and ensemble CWT, ensemble Spectrogram, and ensemble WVD with ensemble averaging. We conducted our analysis using these different images independently.
For CWT images, few deep learning models exhibited excellent training accuracy. 100% training accuracy was achieved for VGG16, InceptionV3, MobileNetV2, DenseNet201, and DenseNet121. VGG19 achieved 99.6% training accuracy and the least training accuracy was 97.2% for Xception. Test accuracy was more than 96% for all the models except Xception which exhibited 91.67% test accuracy. The upmost test accuracy was achieved by VGG16 and MobileNetV2 which is 98.33%. While in the validation Inception as well as MobileNetV2 have exhibited 100% accuracy. Others model also achieved more than 96% except Xception which exhibited 93.33% validation accuracy.
In the case of ECWT images, we can sight that accuracy has dropped. Only Xception, DenseNet201, DenseNet121, and DenseNet169 have exhibited an accuracy of more than 80%. The utmost training accuracy was 86.5% for DenseNet201 and the training accuracy was 81.3% which is for DenseNet169. For test accuracy, DenseNet169 has exhibited the utmost 95% accuracy and the least 88.33% accuracy was achieved by Xception. For validation, DenseNet169 has achieved the utmost 94.92% accuracy and the least 86.44% accuracy was achieved by Xception.
For spectrogram images, Xception exhibited better results than all other models and achieved sublime 100% training accuracy. DenseNet121 has achieved the lowest training accuracy which was 83.6%. Xception also achieved the utmost test accuracy which was 90% and the least test accuracy was 81.67% which was for DenseNet169. Upmost 96.67% validation accuracy was achieved by Xception. 83.33% was the lowest validation accuracy which was achieved by DenseNet201.
For ensemble spectrogram images, only Xception and DenseNet169 exhibited accuracy greater than 80%. For Xception training, test and validation accuracy was 89.4%, 81.67%, and 93.33% respectively. For DenseNet169 training, test and validation accuracy was 89.8%, 80%, and 81.67% respectively. From the table, we can sight that the utmost test and validation accuracy was achieved by Xception, and the utmost training accuracy was achieved by DenseNet169.
For WVD images, Xception, MobileNetV2, DenseNet201, DenseNet121, and DenseNet169 have achieved 100% training accuracy. 90.2% was the lowest training accuracy achieved by VGG16. Test and validation accuracy was up to the mark like training accuracy. The utmost test accuracy was 93.33% which was achieved by DenseNet201 as well as VGG16. 91.67% test accuracy was achieved by both MobileNetV2 and DenseNet169. 80.1% was the lowest training accuracy which was achieved by Xception. 93.33% was the highest validation accuracy and it was achieved by VGG16. DenseNet169, MobileNetV2, and DenseNet201 exhibited 91.67% validation accuracy. The lowest validation accuracy was 83.33% which was achieved by Xception.
For EWVD, Xception, InceptionV3, MobileNetV2, and DenseNet201 exhibited an accuracy of more than 80%. 98.5% was the highest training accuracy achieved by MobileNetV2. 87.6% was the lowest training accuracy achieved by InceptionV3. 100% test accuracy was achieved by MobileNetV2. 93.33% was the lowest test accuracy achieved by InceptionV3 and DenseNet121. For validation, MobileNetV2 and Xception both achieved 100% accuracy. 91.67% was the lowest validation accuracy achieved by InceptionV3. We can sight from the result that ensemble WVD images performed better than WVD images.
4.2. Population analysis
We divided the dataset into two parts based on the age of the patients. This is an unprecedented approach in epilepsy detection research and was done to gain new insights. We utilized a threshold age of 40 for the dissociation since it segregates the dataset into two more or less identical numbers. Additionally, we utilized 40 years as a threshold value since in some other studies such as endometrial cancer, arterial disease along with heart disease 40 years was used as a threshold value [[34], [35], [36]]. The two groups were labeled as “40-Minus” and “40-Plus,” allowing us to compare the results of the classification of older and younger individuals.
To first with, in general, we employed all the deep learning models for detection and exhibited the result of some stipulate models which gave an accuracy of more than 80%. Following, we utilized the models (VGG16, VGG19, Xception, InceptionV3, MobileNetV2, DenseNet201, DenseNet121, and DenseNet169) for population analysis that exhibited better accuracy in general analysis. We employed these models for all six types of images. The results of the population analysis are presented in Table 4.
Table 4.
Results of population analysis of epilepsy.
| Transform analysis | Convolutional neural network model | 40-Minus |
40-Plus |
||||
|---|---|---|---|---|---|---|---|
| Train Accuracy |
Test Accuracy |
Validation Accuracy |
Train Accuracy |
Test Accuracy |
Validation Accuracy |
||
| CWT | VGG16 | 100% | 100% | 100% | 99.86% | 95% | 94.29% |
| VGG19 | 100% | 100% | 100% | 99.86% | 89.2% | 91.43% | |
| Xception | 100% | 100% | 100% | 100% | 90% | 88.57% | |
| InceptionV3 | 100% | 100% | 100% | 100% | 87.2% | 91.43% | |
| MobileNetV2 | 100% | 100% | 100% | 100% | 100% | 90% | |
| DenseNet201 | 100% | 100% | 100% | 100% | 92.86% | 94.29% | |
| DenseNet121 | 100% | 100% | 100% | 100% | 91.4% | 92.86% | |
| DenseNet169 | 100% | 100% | 100% | 100% | 92.86% | 92.86% | |
| Ensemble CWT |
VGG16 | 98.33% | 100% | 100% | 87.67% | 87.5% | 90% |
| VGG19 | 98% | 100% | 100% | 82% | 85% | 87.5% | |
| Xception | 99.17% | 100% | 100% | 83% | 87% | 92% | |
| InceptionV3 | 99.17% | 100% | 100% | 88.33% | 82.5% | 85% | |
| MobileNetV2 | 99.67% | 100% | 100% | 97% | 90% | 85% | |
| DenseNet201 | 99.5% | 100% | 100% | 87.17% | 92.5% | 90% | |
| DenseNet121 | 99.17% | 100% | 100% | 88.17% | 87.5% | 87.5% | |
| DenseNet169 | 99.83% | 100% | 100% | 91% | 87.5% | 90% | |
| Spectrogram | VGG16 | 96.5% | 100% | 100% | 89.07% | 87.37% | 72% |
| VGG19 | 96.7% | 97% | 100% | 91.47% | 78% | 76% | |
| Xception | 98.6% | 100% | 100% | 100% | 74.51% | 64% | |
| InceptionV3 | 98.9% | 100% | 100% | 100% | 85% | 74% | |
| MobileNetV2 | 99.8% | 98% | 100% | 100% | 95% | 86% | |
| DenseNet201 | 99.3% | 100% | 100% | 100% | 90% | 80% | |
| DenseNet121 | 99.6% | 100% | 100% | 100% | 95% | 80% | |
| DenseNet169 | 99.4% | 100% | 100% | 100% | 90% | 74% | |
| Ensemble Spectrogram | VGG16 | 95.33% | 95% | 97.5% | 55.5% | 57.5% | 60% |
| VGG19 | 94.33% | 95% | 95% | 59.5% | 70% | 65% | |
| Xception | 99% | 100% | 95% | 84.83% | 82.5% | 72.5% | |
| InceptionV3 | 99% | 100% | 100% | 77.67% | 85% | 87.5% | |
| MobileNetV2 | 99.67% | 97.5% | 97.5% | 84.67% | 87.5% | 87.5% | |
| DenseNet201 | 99.67% | 100% | 100% | 79.33% | 77.5% | 85% | |
| DenseNet121 | 99% | 98% | 97.5% | 82.67% | 72.5% | 70% | |
| DenseNet169 | 99.67% | 100% | 100% | 76.83% | 77.5% | 77.5% | |
| WVD | VGG16 | 71.2% | 71% | 68% | 92.8% | 85% | 87.5% |
| VGG19 | 75.3% | 76% | 70% | 94% | 85% | 95% | |
| Xception | 88.6% | 94% | 90% | 100% | 83.15% | 82.5% | |
| InceptionV3 | 91.5% | 94% | 87% | 100% | 95% | 90% | |
| MobileNetV2 | 92.9% | 91% | 86% | 100% | 95% | 92.5% | |
| DenseNet201 | 88.8% | 97% | 96% | 100% | 85% | 95% | |
| DenseNet121 | 88.7% | 96% | 96% | 100% | 90% | 92.5% | |
| DenseNet169 | 93.2% | 97% | 95% | 100% | 90% | 95% | |
| Ensemble WVD |
VGG16 | 100% | 100% | 97.5% | 50% | 70% | 72.5% |
| VGG19 | 100% | 97.5% | 97.5% | 63.67% | 50% | 50% | |
| Xception | 100% | 95% | 90% | 89.17% | 97.5% | 92.5% | |
| InceptionV3 | 100% | 95% | 100% | 91% | 87.5% | 87.5% | |
| MobileNetV2 | 100% | 97.5% | 97.5% | 97.33% | 97.5% | 82.5% | |
| DenseNet201 | 100% | 97.5% | 100% | 92.5% | 95% | 92.5% | |
| DenseNet121 | 100% | 95% | 100% | 93.17% | 97.5% | 92.5% | |
| DenseNet169 | 100% | 97.5% | 100% | 93.83% | 95% | 87.5% | |
For CWT images, the 40-Minus group exhibited exceptional results (100% accuracy). As seen from the table, all models achieved 100% training, test, and validation accuracy. On the other hand, in the case of 40-Plus albeit the models have shown 100% training accuracy except for VGG16 and VGG19 nevertheless only MobileNetV2 exhibited 100% test accuracy as well as no model gave 100% validation accuracy. The utmost validation accuracy was 94.29% for VGG16 and DenseNet201.
For ensemble CWT images, the 40-Minus group gave the highest training accuracy of 99.83% for DenseNet169. Others models also exhibited good training accuracy. For test and validation, all the models have exhibited 100% accuracy which is pleasing. In contrast, in the event of 40-Plus, we can sight that the highest training, test as well as validation accuracy are lower than in the case of 40-Minus. Here, the highest training accuracy was 97%, the highest test accuracy was 92.5% and the highest validation accuracy was 92%. We can observe the downfall of accuracy in all the models for all the aspects of training, testing, and validation.
For spectrogram images, the 40-Minus group exhibited the highest training accuracy of 99.8% for MobileNetV2. Others models also exhibited pretty good accuracy in Table 4. All the models have exhibited 100% test accuracy except VGG19 along with MobileNetV2. All the models have shown exquisite 100% validation accuracy. In opposition, in the case of 40-Plus, we can sight that all the models have shown 100% training accuracy except VGG16 and VGG19. However, our test and validation accuracy plummeted. The utmost test accuracy of 95% was achieved by MobileNetV2 along with DenseNet121 models. Consequently, the utmost validation accuracy (85%) was achieved by MobileNetV2. Validation accuracy has descended at a greater range than the test accuracy.
For ensemble spectrogram images, in the 40-Minus group, the highest training accuracy was 99.67% which was exhibited by MobileNetV2, DenseNet201 along with DenseNet169. Utmost test accuracy was 100% which was achieved by Xception, InceptionV3, DenseNet201 as well as DenseNet169. Here, the utmost validation accuracy was also 100% achieved by InceptionV3, DenseNet201 as well as DenseNet169. In contrast, for 40-Plus group, we can sight the downfall of the accuracy for training, validation as well as test. In the case of 40-Plus, the highest training accuracy was 84.83% achieved by Xception. The highest test accuracy was 87.5% which was achieved by MobileNetV2. It also gained 87.5% validation accuracy by InceptionV3 and MobileNetV2 models.
For WVD images, the 40-Minus group, has achieved the 93.2% highest accuracy by DenseNet169 model. 97% was the utmost test accuracy achieved by DenseNet169 as well as DenseNet201. 96% is the utmost validation accuracy achieved by DenseNet201 along with DenseNet121. On the flip side, in the 40-Plus group, we can sight the escalation of training accuracy greatly. 100% training accuracy was achieved by all the models except VGG16 along with VGG19. 95% was the utmost test accuracy achieved by InceptionV3 as well as MobileNetV2. It has also achieved the utmost 95% validation accuracy by VGG19, DenseNet201 as well as DenseNet169.
For ensemble WVD images, in the 40-Minus group, all the models have achieved 100% training accuracy. The highest test as well as validation accuracy was 100% too. Here, VGG16 has achieved 100% test accuracy only. 100% validation accuracy was achieved by InceptionV3, DenseNet201, DenseNet121 as well as DenseNet169. In opposition, for the 40-Plus group, 97.33% was the highest training accuracy which is slightly lower than the 40-Minus group. 97.5% was the utmost test accuracy achieved by Xception, MobileNetV2 as well as DenseNet121. 92.5% was the utmost validation accuracy achieved by Xception, DenseNet201 along with DenseNet121.
5. Discussion
The key finding of this study was the ability to work with a challenging dataset and achieve high accuracy through manual noise rejection and innovative channel selection methods. Additionally, the ensemble averaging approach escalates the accuracy to a greater extent in WVD. Moreover, achieving utmost accuracy even after using 5s epochs of signals. Last but not the least, supplementary scrutiny such as population analysis renders additional novel findings such as 40-Minus epileptic signal was detected better than 40-Plus.
5.1. Addressing challenging dataset
In practice, EEG signals are often contaminated by noise and artifacts such as eye blinks, EMG artifacts, electrode movement, electrostatic, jerking, and lead artifacts. Albeit high-frequency noise along with low-frequency superfluous signal can be obviated by utilizing filtering the EEG signal, nevertheless, there still may be remnant noise and artifacts. In Table 5, we can sight that, in some previous studies [1,27] where the TUH EEG corpus dataset was utilized, we observed that they have achieved significantly low accuracy. The profound reason that we detected this lessened accuracy was remnant noise and artifacts after filtering. Thusly, we manually observed all the data and obviated the remnant noise and artifacts. We manually detected the noise and artifacts by employing the annotation files rendered with the TUH EEG corpus dataset where disparate noise and artifacts were visualized. We utilized an unhackneyed channel selection procedure. To perceive the most significant channels, we reckoned statistical values (mean, median, and standard deviation) and then calculated co-relation and p-value. Thusly, it was feasible for us to detect the most significant channel. In addition, we have also done hyperparameter tuning. Afterward, we described a significant escalation in accuracy.
Table 5.
Comparison results of proposed method with previous works.
| Feature | Classifier | Dataset | Result | Reference |
|---|---|---|---|---|
| Wavelet transform | K-means clustering | Bonn | Acc. 96.67% | [12] |
| Wavelet scalogram | AlexNet | Bonn | Acc. 100% | [13] |
| Wavelet entropy | SVM | Bonn | Acc. 100% | [8] |
| Sample entropy, distributed entropy | GA-SVM | Bonn | AUC.96.67% | [4] |
| Optimum allocation technique | Logistic model tree | Bonn | Acc. 95.33% | [14] |
| Wavelet transform | Random forest | Bonn | Acc. 95.00% | [16] |
| Fuzzy distribution entropy and wavelet packet decomposition | KNN | Bonn | Acc. 98.33% | [18] |
| Discrete wavelet transform and wavelet energy distribution | ANN | Bonn | Acc. 95% | [19] |
| Hurst exponent | KNN | Bonn | Acc. 100% | [20] |
| Symlet wavelet and grid search optimizer feature | Gradient boosting machine | Bonn | Acc. 96.1% | [2] |
| Self-organized map | RBF neural network | Bonn | Acc. 97.47% | [21] |
| Spectral, spatial and temporal feature | 3D CNN | CHB-MIT | Acc. 99.4% | [22] |
| Short time Fourier transform | CNN | Bern Barcelona | Acc. 91.8% | [23] |
| Fourier-based SST | CNN | CHB-MIT | Acc. 99.63% | [24] |
| Multi view feature learning | Convolutional deep learning | CHB-MIT | Acc. 94.37% | [25] |
| Waveform level classification by CNN | SVM | Clinical dataset | Acc. 83.86% | [26] |
| Time domain and frequency domain feature | Bayesian Net | ARMOR project dataset | Acc. 95% | [15] |
| Teager energy | Supervised backpropagation neural network | (Institute of Neuroscience, India) | Sensitivity 96.66% | [17] |
| Spectrogram | CNN | TUH | Acc. 88.3% | [27] |
| Automated identification without feature extraction | One dimensional deep convolutional neural network | TUH | Acc. 79.34% | [1] |
| Automatic feature extraction | CNN | TUH | Acc. 100% | This study |
5.2. Significance of ensemble averaging
We employed an ensemble averaging approach to enhance the accuracy of our results. Our findings indicate that the ensemble Winger-Ville method resulted in improved accuracy compared to the traditional Winger-Ville approach. This suggests that ensemble averaging can be an effective technique for increasing accuracy when combined with other methods.
5.3. Age specific population analysis
Additionally, further analysis uncovered some significant new insights. This is a novel finding as it has not been previously studied. In our study, we divided the dataset into two classes: 40-Minus and 40-Plus. The results showed that the 40-Minus class produced better detection of epileptic signals and higher accuracy compared to the 40-Plus class. Moreover, the accuracy in the case of 40-Minus was better than general analysis (without age-specific dataset splitting). Hence, our supplemental study suggests that splitting the dataset based on age can lead to improved results in detecting epileptic signals.
5.4. Comparing results
In Table 5, a comparison of the results of our study and others is depicted. We perceived that in the case of the TUH EEG corpus dataset [1] and clinical dataset [26] the accuracy was significantly low (79.34% [1], 88.3% [27], and 83.86% [26]). We were unable to use the clinical dataset as it was not publicly accessible. However, other studies using different datasets achieved significantly higher accuracy, with some reaching more than 90% and even 100%.
While some studies employed CNN and filtering, the TUH EEG corpus dataset still showed low accuracy compared to others, particularly the Bonn University dataset which showed extremely high accuracy. Our analysis revealed that this was due to the presence of additional noise in the TUH EEG corpus dataset, while the Bonn University dataset was nearly noise-free.
In practice, data often contains significant amounts of noise and artifacts. As such, the TUH EEG corpus dataset is more representative of real-world data. Although other studies have achieved satisfactory accuracy, our study is more relevant to practical applications as we used a TUH EEG corpus dataset with a similar level of noise.
Table 6 compares our study to others based on epoch size. Some studies segmented the data, while in our study we divided the EEG signal into 5s epochs. As shown in Table 6, our approach outperforms other studies that utilized data segmentation. Our study achieved an error rate of 0%, which is an outstanding result.
Table 6.
Performance comparison of the proposed method with other method using three parameters (Epoch Size, Results and Classifier).
| Classifier | Epoch Size | Results | Reference |
|---|---|---|---|
| One dimensional deep convolutional neural network | 60s | Error rate 20.6 % | [1] |
| KNN | 0.1s | Error rate 41.8 % | [28] |
| RF | 0.1s | Error rate 31.7 % | [28] |
| GA-SVM | 5s | AUC 96.67 % | [4] |
| Supervised backpropagation neural network | 1s | Sensitivity 96.66 %, Specificity 99.15 % |
[17] |
| 1-D CNN | 2s | Error rate 2.75 % | [29] |
| KNN | 2s | Error rate 1.39 % | [30] |
| KNN | 5s | ROC 91 % | [32] |
| Bi-LSTM | 4s | Sensitivity 93.61 %, Specificity 91.85 % |
[31] |
| CNN | 5s | Error rate 0 % | This study |
It is important to note that there is a tradeoff between epoch size and signal information. Decreasing the epoch size also reduces the amount of information. Therefore, it is not practical to use an excessively small epoch size. Despite this, our approach still achieved an error rate of 0% after segmenting the data.
In spite of that, our approach has significant utility. We used 5s epochs of data, which is sufficient for detecting epileptic signals. In contrast, other methods that do not use data segmentation require a longer signal for epilepsy detection.
5.5. Limitation and future work
Although this study represents a robust approach for detecting epileptic EEG signals, there are opportunities for advancing a capable detection tool for diagnosing and treating epilepsy in the future. The sample of only 10 subjects is small as TUH EEG is a large dataset and therefore, further research incorporating more patient might provide a better overall scenario. Moreover, progressing beyond binary classification to seizure subtypes could better characterize differences in epilepsy. Considering other demographic characteristics besides age group like sex, medications, and history may reveal additional insights tied to seizures. While this method detects seizures, nevertheless localizing the epileptic region from the EEG channels could provide additional insights in diagnosis and treatment. Integrating simultaneous EEG, clinical, imaging, and genetic data within a benchmark framework may provide insights into biological markers and concurrent knowledge in epilepsy research.
6. Conclusion
In this study, epilepsy detection was performed using various deep learning models on the TUH EEG corpus dataset. Preprocessing was performed to significantly improve the quality of the dataset. Three different transformations were applied prior to classification (CWT, Spectrogram, and WVD both with and without ensemble averaging). These transformations emphasized unique features in the signals, leading to more precise detection. The dataset was divided into two groups, 40-Plus and 40-Minus, for population analysis. Results showed that age has an impact on epilepsy detection. This may assist healthcare professionals in detecting epilepsy. Future research plans include exploring multi-classification and hybrid machine learning and deep learning models, as well as determining epilepsy severity to aid in patient diagnosis.
Data availability statement
The TUH EEG Epilepsy Corpus dataset was collected from the weblink- https://isip.piconepress.com/projects/tuh_eeg/html/downloads.shtml.
CRediT authorship contribution statement
Torikul Islam: Conceptualization, Investigation, Methodology, Software, Validation, Visualization, Writing - original draft, Writing - review & editing. Redwanul Islam: Investigation, Methodology, Software, Validation, Visualization. Monisha Basak: Investigation, Methodology, Software, Validation, Visualization. Amit Dutta Roy: Conceptualization, Data curation, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the facilities provided by the Department of Biomedical Engineering, Khulna University of Engineering & Technology, Khulna, Bangladesh. Additionally, the authors would like to thank the Temple University Hospital, Pennsylvania, USA, for permitting to use their database for research.
Contributor Information
Torikul Islam, Email: torikulislam142@gmail.com.
Monisha Basak, Email: monishabasak.kuet.bme@gmail.com.
Redwanul Islam, Email: redwanul10@gmail.com.
Amit Dutta Roy, Email: amit@bme.kuet.ac.bd, amitbme.bd@gmail.com.
References
- 1.Yildirim Ö., Baloglu U.B., Acharya U.R. A deep convolutional neural network model for automated identification of abnormal EEG signals. Neural Comput. Appl. 2020;32(20) doi: 10.1007/s00521-018-3889-z. [DOI] [Google Scholar]
- 2.Wang X., Gong G., Li N. Automated recognition of epileptic EEG states using a combination of symlet wavelet processing, gradient boosting machine, and grid search optimizer. Sensors. 2019;19(no. 2) doi: 10.3390/s19020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Islam R., Debnath S., Raen R., Islam N., Palash T.I., Ali R. Epileptic seizure detection from EEG signal using. ANN-LSTM Model. 2023 doi: 10.1007/978-981-99-1916-1_10. [DOI] [Google Scholar]
- 4.Li P., Karmakar C., Yan C., Palaniswami M., Liu C. Classification of 5-S epileptic EEG recordings using distribution entropy and sample entropy. Front. Physiol. 2016;7(APR) doi: 10.3389/fphys.2016.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Council J.E. Jt. Epilepsy Counc; Leeds, no. December: 2005. Epilepsy Prevalence, Incidence and Other Statistics. [Google Scholar]
- 6.Yuan Y., Jia K. Fusionatt: deep fusional attention networks for multi-channel biomedical signals. Sensors. 2019;19(11) doi: 10.3390/s19112429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudalaimani C., Sivakumaran N., Elizabeth T.T., Rominus V.S. Automated detection of the preseizure state in EEG signal using neural networks. Biocybern. Biomed. Eng. 2019;39(1):160–175. doi: 10.1016/j.bbe.2018.11.007. [DOI] [Google Scholar]
- 8.Kumar Y., Dewal M.L., Anand R.S. Epileptic seizure detection using DWT based fuzzy approximate entropy and support vector machine. Neurocomputing. 2014;133 doi: 10.1016/j.neucom.2013.11.009. [DOI] [Google Scholar]
- 9.Obeid I., Picone J. The temple university hospital EEG data corpus. Front. Neurosci. 2016;10(MAY) doi: 10.3389/fnins.2016.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shackleton D.P., Kasteleijn-Nolst Trenité D.G.A., De Craen A.J.M., Vandenbroucke J.P., Westendorp R.G.J. Living with epilepsy: long-term prognosis and psychosocial outcomes. Neurology. 2003;61(1):64–70. doi: 10.1212/01.WNL.0000073543.63457.0A. [DOI] [PubMed] [Google Scholar]
- 11.Andrzejak R.G., Lehnertz K., Mormann F., Rieke C., David P., Elger C.E. Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Phys. Rev. E - Stat. Physics, Plasmas, Fluids, Relat. Interdiscip. Top. 2001;64(6) doi: 10.1103/PhysRevE.64.061907. [DOI] [PubMed] [Google Scholar]
- 12.Orhan U., Hekim M., Ozer M. EEG signals classification using the K-means clustering and a multilayer perceptron neural network model. Expert Syst. Appl. 2011;38(10) doi: 10.1016/j.eswa.2011.04.149. [DOI] [Google Scholar]
- 13.Roy A.D., Islam M.M. 2020. Detection of Epileptic Seizures from Wavelet Scalogram of EEG Signal Using Transfer Learning with AlexNet Convolutional Neural Network. [DOI] [Google Scholar]
- 14.Kabir E., Siuly, Zhang Y. Epileptic seizure detection from EEG signals using logistic model trees. Brain Informatics. 2016;3(2) doi: 10.1007/s40708-015-0030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pippa E., et al. Improving classification of epileptic and non-epileptic EEG events by feature selection. Neurocomputing. 2016;171 doi: 10.1016/j.neucom.2015.06.071. [DOI] [Google Scholar]
- 16.Tzimourta K.D., et al. A robust methodology for classification of epileptic seizures in EEG signals. Health Technol. 2019;9(2) doi: 10.1007/s12553-018-0265-z. [DOI] [Google Scholar]
- 17.Sriraam N., et al. Multichannel EEG based inter-ictal seizures detection using Teager energy with backpropagation neural network classifier. Australas. Phys. Eng. Sci. Med. 2018;41(4) doi: 10.1007/s13246-018-0694-z. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T., Chen W., Li M. Fuzzy distribution entropy and its application in automated seizure detection technique. Biomed. Signal Process Control. 2018;39 doi: 10.1016/j.bspc.2017.08.013. [DOI] [Google Scholar]
- 19.Wani S.M., Sabut S., Nalbalwar S.L. vol. 810. 2018. Detection of epileptic seizure using wavelet transform and neural network classifier. (Advances in Intelligent Systems and Computing). [DOI] [Google Scholar]
- 20.Lahmiri S., Shmuel A. Accurate classification of seizure and seizure-free intervals of intracranial EEG signals from epileptic patients. IEEE Trans. Instrum. Meas. 2019;68(3) doi: 10.1109/TIM.2018.2855518. [DOI] [Google Scholar]
- 21.Osman A.H., Alzahrani A.A. New approach for automated epileptic disease diagnosis using an integrated self-organization map and radial basis function neural network algorithm. IEEE Access. 2019;7 doi: 10.1109/ACCESS.2018.2886608. [DOI] [Google Scholar]
- 22.Choi G., et al. 2019. A Novel Multi-Scale 3D CNN with Deep Neural Network for Epileptic Seizure Detection. [DOI] [Google Scholar]
- 23.Sui L., Zhao X., Zhao Q., Tanaka T., Cao J. vol. 559. 2019. Localization of epileptic foci by using convolutional neural network based on iEEG. (IFIP Advances in Information and Communication Technology). [DOI] [Google Scholar]
- 24.Ozdemir M.A., Cura O.K., Akan A. Epileptic EEG classification by using time-frequency images for deep learning. Int. J. Neural Syst. 2021;31(8) doi: 10.1142/S012906572150026X. [DOI] [PubMed] [Google Scholar]
- 25.Yuan Y., Xun G., Jia K., Zhang A. A multi-view deep learning framework for EEG seizure detection. IEEE J. Biomed. Heal. Informatics. 2019;23(1) doi: 10.1109/JBHI.2018.2871678. [DOI] [PubMed] [Google Scholar]
- 26.Thomas J., Comoretto L., Jin J., Dauwels J., Cash S.S., Westover M.B. vol. 2018. EMBS; 2018. EEG CLassification via convolutional neural network-based interictal epileptiform event detection. (Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raghu S., Sriraam N., Temel Y., Rao S.V., Kubben P.L. EEG based multi-class seizure type classification using convolutional neural network and transfer learning. Neural Network. 2020;124 doi: 10.1016/j.neunet.2020.01.017. [DOI] [PubMed] [Google Scholar]
- 28.Lopez S., Suarez G., Jungreis D., Obeid I., Picone J. 2016. Automated Identification of Abnormal Adult EEGs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharan R.V., Berkovsky S. vol. 2020. EMBS; 2020. Epileptic seizure detection using multi-channelchannel EEG wavelet power spectra and 1-D convolutional neural networks. (Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society). July. [DOI] [PubMed] [Google Scholar]
- 30.Li M., Sun X., Chen W. Patient-specific seizure detection method using nonlinear mode decomposition for long-term EEG signals. Med. Biol. Eng. Comput. 2020;58(12) doi: 10.1007/s11517-020-02279-6. [DOI] [PubMed] [Google Scholar]
- 31.Hu X., Yuan S., Xu F., Leng Y., Yuan K., Yuan Q. Scalp EEG classification using deep Bi-LSTM network for seizure detection. Comput. Biol. Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103919. [DOI] [PubMed] [Google Scholar]
- 32.Ryu S., et al. Pilot study of a single-channel EEG seizure detection algorithm using machine learning. Child’s Nerv. Syst. 2021;37(7) doi: 10.1007/s00381-020-05011-9. [DOI] [PubMed] [Google Scholar]
- 33.Palash T.I., Islam R., Basak M., Dutta Roy A. 2021. Automatic Classification of COVID-19 from Chest X-Ray Image Using Convolutional Neural Network. [DOI] [Google Scholar]
- 34.Duska L.R., Garrett A., Rueda B.R., Haas J., Chang Y., Fuller A.F. Endometrial cancer in women 40 years old or younger. Gynecol. Oncol. 2001;83(2) doi: 10.1006/gyno.2001.6434. [DOI] [PubMed] [Google Scholar]
- 35.Bertomeu V., et al. Prevalence and prognostic influence of peripheral arterial disease in patients ≥40 Years old admitted into hospital following an acute coronary event. Eur. J. Vasc. Endovasc. Surg. 2008;36(2) doi: 10.1016/j.ejvs.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Yildirin N., Arat N., Doǧan M.S., Sökmen Y., Özcan F. Comparison of traditional risk factors, natural history and angiographic findings between coronary heart disease patients with age <40 and ≥40 years old. Anadolu Kardiyol. Derg. 2007;7(2) [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The TUH EEG Epilepsy Corpus dataset was collected from the weblink- https://isip.piconepress.com/projects/tuh_eeg/html/downloads.shtml.