Abstract
Introduction
The aim of the study was to determine the genetic diversity of Echinococcus multilocularis in pigs in highly endemic areas in Poland, as well as to attempt to confirm the occurrence and geographical distribution of haplotypes characteristic for these areas, which were previously described on the basis of examination of adult tapeworms isolated from foxes.
Material and Methods
Twenty samples of E. multilocularis larval forms were obtained from pigs’ livers in four provinces of Poland. Genetic analyses were conducted on sequences of two mitochondrial genes: cox1 and nad2.
Results
Seven haplotypes were found for the cox1 gene (OQ874673–OQ874679) and four haplotypes for nad2 (OQ884981–OQ884984). They corresponded to the haplotypes described earlier in foxes in Poland (some of them differing only in one nucleotide). The analysis showed the presence of the Asian-like haplotype in both the cox1 and nad2 genes. The remaining haplotypes were grouped in the European clade. The geographical distribution of haplotypes identified in the pig samples was noticed to bear a similarity to the distribution of haplotypes previously isolated from foxes in the same regions.
Conclusion
The characteristic geographical distribution of E. multilocularis haplotypes in Central Europe (including the presence of the Asian-like haplotype) previously described in the population of definitive hosts (foxes) has now been confirmed by the analysis of samples from non-specific intermediate hosts (pigs).
Keywords: Echinococcus multilocularis, pigs, haplotypes, genetic diversity
Introduction
Alveolar echinococcosis is a parasitic zoonosis dangerous to human health and life, which is caused by larval forms of the E. multilocularis tapeworm. A typical definitive host, in the intestines of which adult forms of this tapeworm develop, is the red fox. Other rare definitive hosts are raccoon dogs, arctic foxes, golden jackals, wolves and also dogs and cats. Adult tapeworms produce eggs that are excreted into the environment and are a source of infection for intermediate hosts, which are typically rodents (31). However, eggs can also infect humans and other animal species (e.g. pigs and horses) acting as non-specific (aberrant) intermediate hosts. In the tissues of these hosts, the larva develops in an unusual way and most often it does not produce protoscolices or degenerate (31).
In recent years, much attention has been devoted to the genetic diversity of E. multilocularis. The studies have been carried out in two main ways, namely by analysing the sequence of selected genes (usually mitochondrial genes) (6, 11, 27) and by an innovative method of analysing tandemly repeated microsatellites (1, 20, 34). They revealed the divisions between the main clades grouping the most common genotypes according to the geographical distribution of parasites.
Thus the genotypes typical of Europe, Asia and North America were described (20, 27). This genetic diversity research also made it possible to trace the migration of parasites with their hosts and to determine the primary foci of occurrence and peripheral areas colonised by parasites later (on the pattern of the mainland-island model). Knowledge of genetic diversity provided the means to observe the mixing of genotypes characteristic for one continent with those characteristic for others. Example of this are the finding of Asian-like haplotypes in Europe (18, 37) or European-like examples in North America (12, 32).
Poland’s geographical location and relatively high prevalence of E. multilocularis in red foxes (15, 17) make it an interesting site for the analysis of the genetic diversity of this tapeworm. There is a specific mixing of western (European) and eastern (probably Asian) genetic pools. Recent studies in Poland conducted on adult worms isolated from red foxes showed the presence of dominant haplotypes characteristic for this area of Europe. What is most interesting is that the presence of the Asian-like haplotype was also found (18, 37).
The good availability of genetic material of tapeworms isolated from the intestines of definitive hosts dictates that a lot of data on genetic diversity come from studies of such – most often from red fox tapeworms (8, 18, 35, 36). However, samples from other animal species are also investigated, including some from pigs, which may act as non-specific intermediate hosts in the life cycle of E. multilocularis. There is small number of studies on the prevalence of E. multilocularis in pigs. The only ones that refer to a wider population come from Switzerland, where the prevalence was estimated at 0.009% in 2017–2018 (26), and from Japan, where E. multilocularis larvae were identified in 0.0002% of slaughtered pigs in 2005–2008 (19). Despite the relatively low prevalence, this animal species can be a specific indicator of environmental contamination with E. multilocularis eggs, which is an important element in assessing the risk of human infection in a given region. Recently, extensive studies were carried out in a pig population in France using analysis of the tandemly repeated microsatellite EmsB, confirming the possibility of monitoring this species as an additional indicator of the risk of human infection, as well as one by which to observe the genetic diversity of these tapeworms. In Poland, the first detections of larval forms in pigs have already been described (16), but no wide investigation has been conducted on the genetic diversity of E. multilocularis based on material obtained from this host.
The aim of the study was to determine the genetic diversity of E. multilocularis in pigs in highly endemic areas of Poland, as well as to attempt to confirm the occurrence and geographical distribution of haplotypes characteristic for this region, which were previously described on the basis of examination of adult tapeworms isolated from red foxes.
Material and Methods
Samples of Echinococcus multilocularis
Echinococcus multilocularis larval forms were obtained in 2011–2016 from pigs’ livers in four provinces of Poland: Warmińsko-Mazurskie and Podlaskie (northeast) and Podkarpackie and Małopolskie (south-east). Fragments of livers with lesions were collected individually by vets in slaughterhouses, and after freezing were sent to the laboratory of the National Veterinary Research Institute in Puławy, Poland. Samples were first investigated macroscopically to ascertain the content of lesions and microscopically using a stereo microscope at 50× magnification to detect the protoscolices. Extraction of DNA was carried out using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. That DNA matched the E. multilocularis species was confirmed molecularly by a nested PCR (10) with modifications (16). Overall, 20 DNA samples of E. multilocularis were used for genetic analysis.
PCR and sequencing
Two mitochondrial genes, nad2 and cox1, were amplified by PCR according to Nakao et al. (27). Sequencing was performed at a commercial company (Genomed, Warsaw, Poland) using Sanger dideoxy sequencing.
Data analysis
Phylogenetic analyses were conducted separately for each molecular marker (nad2 and cox1). The forward and reverse sequences were analysed, aligned and trimmed using the Geneious Alignment algorithm in the Geneious Prime bioinformatics software platform (Biomatters, Auckland, New Zealand). The obtained consensus sequences were aligned with sequences from GenBank using the BLAST nucleotide algorithm. Phylogenetic analysis was also conducted using sequences available in GenBank as outgroups. A phenogram was created by applying the Tamura–Nei genetic distance model and the neighbour-joining building method with 1,000 bootstrap replications in Geneious Prime. The nucleotide sequences obtained in this study were submitted to the GenBank database under the accession numbers OQ874673–OQ874679 (cox1) and OQ884981–OQ884984 (nad2). To estimate the phylogenetic position of the Polish isolates, homologous mitochondrial DNA sequences obtained in previous research (12, 13, 18, 22–25, 27, 30, 38) were retrieved from GenBank and used in analyses.
Results
The macroscopic examination demonstrated the following types of lesions: in 15 samples there were white, thick-walled cysts 1–10 mm in diameter deeply recessed in the liver tissue with whitish degenerated mass or thick fluid, and in 5 samples white nodular forms 2–4 mm in diameter were found on the surface of liver tissue (Fig. 1). Protoscolices were not found microscopically in any of the examined samples.
Fig. 1.
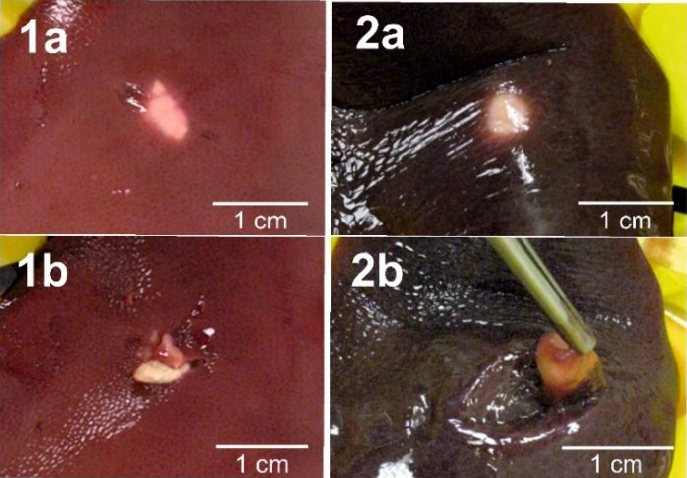
Two examples of Echinococcus multilocularis lesions found in pigs’ livers 1a and 2a – before cutting; 1b and 2b – after cutting
All 20 samples gave specific products in PCRs for cox1 and nad2. Good-quality sequences were obtained for 19 cox1 and 19 nad2 products, but one sample only had a cox1 sequence and another sample only had a nad2 sequence. One cox1 sequence (EmPL_cox_E) was incomplete and lacked its first 207 nucleotides.
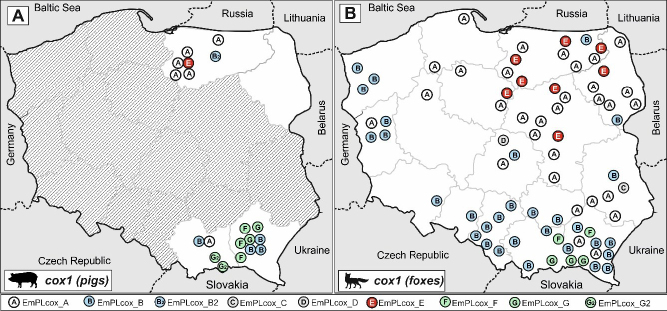
In the analysis of the cox1 gene, seven haplotypes were found (GenBank accession numbers OQ874673– OQ874679) (Fig. 2). Five of them, EmPL_cox_A, EmPL_cox_B, EmPL_cox_E, EmPL_cox_F and EmPL_cox_G, corresponded exactly to the haplotypes described previously in foxes in Poland (KY205683, KY205691, KY205685, KY205689 and KY205690) (18). The other two, EmPL_cox_B2 and EmPL_cox_G2, differed by only one nucleotide from the previously described ones.
Fig. 2. The phylogenetic tree of Echinococcus multilocularis based on the cox1 gene.
EmPL_cox_A–EmPL_cox_G – Polish haplotypes (* – sequences of this study, numbers of isolates in each haplotype are shown in curly brackets); Aus – Austria; Can_SK1 – Canada; CHM – China (Inner Mongolia); CHQ – China (Quinhai); CHS – China (Sichuan); Est2 – Estonia; Fra – France; Jap – Japan, Kaz – Kazakhstan; Kyr – Kyrgyzstan; RUS5, RUS14, RUS17 – Russia; Slo – Slovakia. US-A – USA (Alaska - St. Lawrence Island); US-I – USA (Indiana). Values on the tree nodes are bootstrap proportions (%)
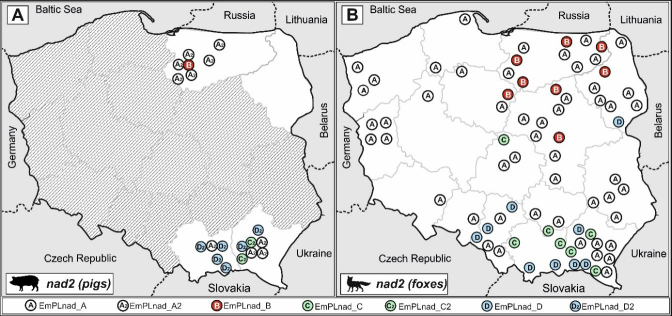
The nad2 gene analysis revealed four haplotypes (GenBank accession numbers OQ884981–OQ884984) (Fig. 3). They were EmPL_nad_A2, EmPL_nad_B2, EmPL_nad_C2 and EmPL_nad_D2, and all of them corresponded to similar haplotypes described previously in foxes in Poland (18), EmPL_nad_A, EmPL_nad_B, EmPL_nad_C and EmPL_nad_D (GenBank accession numbers KY205706, KY205700, KY205704 and KY205705), with a difference of only one nucleotide.
Fig. 3. The phylogenetic tree of Echinococcus multilocularis based on the nad2 gene.

EmPL_nad_A–EmPL_nad_D – Polish haplotypes (* – sequences of this study, numbers of isolates in each haplotype are shown in curly brackets); Aus – Austria; Can – Canada; CHM – China (Inner Mongolia); CHS – China (Sichuan); Est2 – Estonia; Fra – France; Jap – Japan, Kaz – Kazakhstan; Kyr – Kyrgyzstan; Slo – Slovakia. US-A – USA (Alaska - St. Lawrence Island); US-I – USA (Indiana); US-M – USA (Missouri). Values on the tree nodes are bootstrap proportions (%)
The analysis showed the presence of the Asian-like haplotype, to which one sample’s genetic material affiliated, in both the cox1 (EmPL_cox_E) (OQ874676) and nad2 (EmPL_nad_B) (OQ884982) genes. These genes’ sequences were similar to those previously identified in foxes in Poland (KY205670 and KY205700) (18). The remaining haplotypes were grouped in the European clade.
Comparing the geographical distribution of haplotypes identified in the pig samples, a similarity could also be noticed with the distribution of haplotypes previously isolated from foxes. This is particularly evident in the more diverse cox1 gene and the EmPL_cox_A and E haplotypes. From both foxes and pigs these were present, or were in significant predominance, only in the north of the country. In contrast, the EmPL_cox_F, G and B haplotypes from both pig and fox samples were found only in the south (Fig. 4). A resemblance could also be observed in the distributions of pig and fox nad2 haplotypes – similar groups of haplotypes dominated in the same regions in pigs and foxes (Fig. 5).
Fig. 4. Geographic distribution of Echinococcus multilocularis cox1 gene haplotypes in isolates.
A – in pigs in this study; B – in foxes according to Karamon et al. (18)
Fig. 5. Geographic distribution of Echinococcus multilocularis nad2 gene haplotypes in isolates.
A – in pigs in this study; B – in foxes according to Karamon et al. (18)
Discussion
In pigs, the lesions caused by E. multilocularis are often characteristic of the early phase of larval development. In our studies, these were usually small nodules/cysts embedded in the liver parenchyma, often filled with caseous matter indicating degeneration of the larva. This is consistent with the description of lesions observed in the first case of E. multilocularis in pigs in Poland (16). These lesions are also comparable to those caused by E. multilocularis larvae found in commercially reared pigs and described in the few published reports on the subject. Kimura et al. (19) identified six lesions positive for E. multilocularis in Japan among 109 slaughtered pigs with whitish nodules in the liver. In Switzerland, sharply demarcated dense white nodules 5–15mm in diameter were observed (33).
Larvae of E. multilocularis as nodular foci (from 3 to 22 mm in diameter) consisting of white capsules surrounding whitish to light yellow material with paste consistency were noted in Germany (3). Necrotic and calcified E. multilocularis lesions were found in Lithuania (7). Similar lesions were also obtained during experimental infections of pigs with E. multilocularis eggs (9, 28), and the same forms were also observed in infected wild boar livers (4, 29). Characteristic for all the described studies was the lack of protoscolices. As a contrasting finding, the infectivity of larval forms in pigs or wild boars was confirmed by inoculation of gerbils with parasitic tissue material isolated from the livers of infected animals, as a result of which characteristic cysts containing protoscolices were obtained in gerbils immediately after the first inoculation (14) or after the second passage (29).
The genetic diversity of E. multilocularis is highly dependent on geographical location. In the early 1990s, two different genotypes of E. multilocularis were characterised: M1 among isolates from China, Alaska and North America and M2 as an isolate from Europe (5, 6). With more isolates from different parts of the world becoming available, more extensive investigations were carried out using mitochondrial genes, and in them genotypes characteristic for Asia, Europe and North America, and specific to selected areas of China were distinguished (27). The study of genetic diversity also involved the analysis of microsatellite tandem repetitions (1), which facilitated the identification of genetic profiles characteristic for individual regions of Europe and the tracking of the routes of infection spread from originally endemic areas (the “core region”) to peripheral areas (20, 36). The type of spread of this infection has been theorised to be the mainland-island model of parasite transmission. It assumes greater genetic diversity of E. multilocularis in the “core region” or “mainland”, which in Europe is southern Germany, Switzerland and south-eastern France, from which the parasites gradually spread to create areas with less genetic diversity (“islands”) (20). Therefore, according to this theory, it is believed that the genetic groups of tapeworms detected in central-eastern Europe reached there via hosts from the historical “core region” referred to above. On the other hand, the results obtained first in Russia (100 km from Moscow) (22) and later in central-eastern Europe (Poland) (18) suggested that the genetic diversity of E. multilocularis on the continent is also significantly influenced by parasites migrating with hosts from Asia. This was implied by these studies’ confirmation of the presence of a haplotype belonging to the Asian clade. The probable Asian origin of this haplotype in Poland was further confirmed by EmsB analysis (37). In studies in Poland, all Asian-like isolates contained one haplotype from the Asian clade, which suggests a relatively early arrival of these parasites from Asia (18). Poland is an interesting area where parasite genetic diversity manifests the mixed influences of western and southern Europe and Asia. This is also observed in the case of other parasites, e.g. Trichinella: in western Poland T. spiralis was significantly dominant in wild boars (70–85% of infected animals), while in the eastern part of the country approximately half of the infected wild boars showed infection with T. britovi, a species more characteristic of areas located east of Poland (2).
The research carried out in pigs described in this article confirmed the presence of an Asian-like haplotype in Poland similar to that previously described in red foxes, and did so by analysing the cox1 (EmPL_cox_E) and nad2 (Em_PL_nad_B) genes. Other genetic diversity studies conducted with swine-derived material in Switzerland using EmsB only showed the presence of different genetic profiles typical of Europe (21).
The geographical distribution of haplotypes found in pigs in Poland corresponds to the characteristic distribution of them in the red fox population in the country, which was particularly visible in the analysis of the more diverse cox1 gene. The E. multilocularis isolates from both red foxes and pigs in the south of the country were dominated by EmPL_cox_F, G and B haplotypes (similar to the haplotypes found in Slovakia, bordering Poland to the south). Similarly, the EmPL_cox_A haplotype (also found in Estonia (24)) was in the majority in the isolates from the north-east of Poland in both host species. The Asian-like haplotype was also detected in pigs in similar areas to where it was identified in red foxes.
Unusual geographical locations of E. multilocularis genotypes in terms of the continent have also recently been observed in North America, where the presence of genotypes characteristic for Europe was noted in adult tapeworms detected in coyotes (11) and in larvae derived from a dog (13). This was also confirmed by recent studies conducted in red foxes and coyotes in Canada, where various European-like haplotypes were found among North American haplotypes (32). Phylogenetic analysis showed some differences among European-like haplotypes isolated in North America, which suggests different scenarios with multiple introductions of European strains of E. multilocularis to Canada, probably occurring over several hundred years (e.g. through dogs originally imported from Europe) (32).
Summarising, the original division into continental clades was previously decided on the results given by a limited number of samples (27). Subsequent studies using more samples revealed further interesting facts probably related to the intercontinental migration of parasites with their hosts and their actual characteristic location.
In conclusion, studies on the genetic diversity of E. multilocularis deepen the knowledge of the ways in which this parasite spreads across continents. The results obtained in our present investigation conducted on samples of larval forms complement and confirm our previous studies conducted on material isolated from mature parasites. This is important due to the complex life cycle of tapeworms. The characteristic geographical distribution of haplotypes in this part of Europe, and most interestingly the presence of the haplotype from the Asian clade, which had previously been described in the population of definitive hosts (red foxes), have now been confirmed by the analysis of samples from accidental intermediate hosts (pigs).
Footnotes
Conflict of Interests Statement:
The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement:
The study was supported by a project implemented as part of the “Protection of Animal and Public Health” Multiannual Programme funded by the Polish Ministry of Agriculture.
Animal Rights Statement:
According to Polish law, the conducted study did not require the consent of the local Ethical Committee.
References
- 1.Bart J.M., Knapp J., Gottstein B., El-Garch F., Giraudoux P., Glowatzki M.L., Berthoud H., Maillard S., Piarroux R. EmsB, a tandem repeated multi-loci microsatellite, new tool to investigate the genetic diversity of Echinococcus multilocularis Infect Genet Evol. 2006;6:390. doi: 10.1016/j.meegid.2006.01.006. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 2.Bilska-Zając E., Różycki M., Grądziel-Krukowska K., Bełcik A., Mizak I., Karamon J., Sroka J., Zdybel J., Cencek T. Diversity of Trichinella species in relation to the host species and geographical location Vet Parasitol. 2020;279 doi: 10.1016/j.vetpar.2020.109052. : . , , doi: . [DOI] [PubMed] [Google Scholar]
- 3.Boettcher D., Bangoura B., Schmaeschke R., Mueller K., Fischer S., Vobis V., Meiler H., Wolf G., Koller A., Kramer S., Overhoff M., Gawlowska S., Schoon H.-A. Diagnostics and epidemiology of alveolar echinococcosis in slaughtered pigs from large-scale husbandries in Germany Parasitol Res. 2013;112:629. doi: 10.1007/s00436-012-3177-2. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 4.Boucher J.M., Hanosset R., Augot D., Bart J.M., Morand M., Piarroux R., Pozet-Bouhier F., Losson B., Cliquet F. Detection of Echinococcus multilocularis in wild boars in France using PCR techniques against larval form Vet Parasitol. 2005;129:259. doi: 10.1016/j.vetpar.2004.09.021. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 5.Bowles J., Blair D., McManus D.P. Genetic variants within the genus Echinococcus identified by mitochondrial-DNA sequencing Mol Biochem Parasitol. 1992;54:165. doi: 10.1016/0166-6851(92)90109-w. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 6.Bowles J., McManus D.P. NADH dehydrogenase-1 gene-sequences compared for species and strains of the genus Echinococcus Int J Parasit. 1993;23:969. doi: 10.1016/0020-7519(93)90065-7. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 7.Bruzinskaite R., Sarkunas M., Torgerson P.R., Mathis A., Deplazes P. Echinococcosis in pigs and intestinal infection with Echinococcus spp. in dogs in southwestern Lithuania Vet Parasitol. 2009;160:237. doi: 10.1016/j.vetpar.2008.11.011. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 8.Casulli A., Bart J.M., Knapp J., La Rosa G., Duscher G., Gottstein B., Di Cerbo A., Manfredi M.T., Genchi C., Piarroux R., Pozio E. Multi-locus microsatellite analysis supports the hypothesis of an autochthonous focus of Echinococcus multilocularis in northern Italy Int J Parasit. 2009;39:837. doi: 10.1016/j.ijpara.2008.12.001. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 9.Deplazes P., Grimm F., Syder T., Tanner I., Kapel C.M.O. Experimental alveolar echinococcosis in pigs, lesion development and serological follow up Vet Parasitol. 2005;130:213. doi: 10.1016/j.vetpar.2005.03.034. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 10.Dinkel A., von Nickisch-Rosenegk M., Bilger B., Merli M., Lucius R., Romig T. Detection of Echinococcus multilocularis in the definitive host: Coprodiagnosis by PCR as an alternative to necropsy J Clin Microbiol. 1998;36:1871. doi: 10.1128/jcm.36.7.1871-1876.1998. : . , , –. , doi: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gesy K.M., Hill J.E., Schwantje H., Liccioli S., Jenkins E.J. Establishment of a European-type strain of Echinococcus multilocularis in Canadian wildlife Parasitology. 2013;140:1133. doi: 10.1017/s0031182013000607. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 12.Gesy K.M., Jenkins E.J. Introduced and Native Haplotypes of Echinococcus multilocularis in Wildlife in Saskatchewan, Canada J Wildl Dis. 2015;51:743. doi: 10.7589/2014-08-214. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 13.Jenkins E.J., Peregrine A.S., Hill J.E., Somers C., Gesy K.M., Barnes B., Gottstein B., Polley L. Detection of European Strain of Echinococcus multilocularis in North America Emerg Infect Dis. 2012;18:1010. doi: 10.3201/eid1806.111420. : . , , –. , doi: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamiya M., Ooi H.K., Oku Y., Okamoto M., Ohbayashi M., Seki N. Isolation of Echinococcus multilocularis from the liver of swine in Hokkaido, Japan Jpn J Vet Res. 1987;35:99. : . , , –. . [PubMed] [Google Scholar]
- 15.Karamon J., Kochanowski M., Sroka J., Cencek T., Różycki M., Chmurzyńska E., Bilska–Zając E. The prevalence of Echinococcus multilocularis in red foxes in Poland –current results (2009–2013) Parasitol Res. 2014;113:317. doi: 10.1007/s00436-013-3657-z. : . , , –. , doi: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karamon J., Sroka J., Cencek T. The first detection of Echinococcus multilocularis in slaughtered pigs in Poland Vet Parasitol. 2012;185:327. doi: 10.1016/j.vetpar.2011.09.022. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 17.Karamon J., Sroka J., Cencek T., Michalski M.M., Zięba P., Karwacki J. Prevalence of Echinococcus multilocularis in red foxes in two eastern provinces of Poland Bull Vet Inst Pulawy. 2011;55:429. : . , , –. , [Google Scholar]
- 18.Karamon J., Stojecki K., Samorek-Pieróg M., Bilska-Zając E., Różycki M., Chmurzyńska E., Sroka J., Zdybel J., Cencek T. Genetic diversity of Echinococcus multilocularis in red foxes in Poland: the first report of a haplotype of probable Asian origin Folia Parasit. 2017;64 doi: 10.14411/fp.2017.007. : . , , doi: . [DOI] [PubMed] [Google Scholar]
- 19.Kimura M., Toukairin A., Tatezaki H., Tanaka S., Harada K., Araiyama J., Yamasaki H., Sugiyama H., Morishima Y., Kawanaka M. Echinococcus multilocularis detected in slaughtered pigs in Aomori, the northernmost prefecture of mainland Japan Jpn J Infect Dis. 2010;63:80. : . , , –. . [PubMed] [Google Scholar]
- 20.Knapp J., Bart J.-M., Giraudoux P., Glowatzki M.-L., Breyer I., Raoul F., Deplazes P., Duscher G., Martinek K., Dubinsky P., Guislain M.-H., Cliquet F., Romig T., Malczewski A., Gottstein B., Piarroux R. Genetic diversity of the cestode Echinococcus multilocularis in red foxes at a continental scale in Europe PLoS Neglect Trop Dis. 2009;3 doi: 10.1371/journal.pntd.0000452. : . , , e452, doi: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knapp J., Meyer A., Courquet S., Millon L., Raoul F., Gottstein B., Frey C.F. Echinococcus multilocularis genetic diversity in Swiss domestic pigs assessed by EmsB microsatellite analyzes Vet Parasitol. 2021;293 doi: 10.1016/j.vetpar.2021.109429. : . , , doi: . [DOI] [PubMed] [Google Scholar]
- 22.Konyaev S.V., Yanagida T., Nakao M., Ingovatova G.M., Shoykhet Y.N., Bondarev A.Y., Odnokurtsev V.A., Loskutova K.S., Lukmanova G.I., Dokuchaev N.E., Spiridonov S., Alshinecky M.V., Sivkova T.N., Andreyanov O.N., Abramov S.A., Krivopalov A.V., Karpenko S.V., Lopatina N.V., Dupal T.A., Sako Y., Ito A. Genetic diversity of Echinococcus spp. in Russia Parasitology. 2013;140:1637. doi: 10.1017/s0031182013001340. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 23.Kuroki K., Morishima Y., Neil J., Beerntsen B.T., Matsumoto J., Stich R.W. Intestinal echinococcosis in a dog from Missouri J Am Vet Med Assoc. 2020;256:1041. doi: 10.2460/javma.256.9.1041. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 24.Laurimaa L., Sueld K., Moks E., Valdmann H., Umhang G., Knapp J., Saarma U. First report of the zoonotic tapeworm Echinococcus multilocularis in raccoon dogs in Estonia, and comparisons with other countries in Europe Vet Parasitol. 2015;212:200. doi: 10.1016/j.vetpar.2015.06.004. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 25.Li J.Q., Li L., Fan Y.L., Fu B.Q., Zhu X.Q., Yan H.B., Jia W.Z. Genetic Diversity in Echinococcus multilocularis From the Plateau Vole and Plateau Pika in Jiuzhi County, Qinghai Province, China Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02632. : . , , doi: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer A., Olias P., Schupbach G., Henzi M., Barmettler T., Hentrich B., Gottstein B., Frey C.F. Combined cross-sectional and case-control study on Echinococcus multilocularis infection in pigs in Switzerland Vet Parasitol. 2020;277 doi: 10.1016/j.vpoa.2020.100031. : . , , doi: . [DOI] [PubMed] [Google Scholar]
- 27.Nakao M., Xiao N., Okamoto M., Yanagida T., Sako Y., Ito A. Geographic pattern of genetic variation in the fox tapeworm Echinococcus multilocularis Parasitol Int. 2009;58:384. doi: 10.1016/j.parint.2009.07.010. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 28.Pfister K., Franck W. Experimentelle Untersuchungen zur Empfänglichkeit des Schweines für Echinococcus multilocularis (Experimental studies on the susceptibility of pigs to Echinococcus multiocularis – in German) Mitt Österr Ges Tropenmed Parasitol. 1988;10:103. : . , , –. . [Google Scholar]
- 29.Pfister T., Schad V., Schelling U., Lucius R., Frank W. Incomplete development of larval Echinococcus multilocularis (Cestoda, Taeniidae) in spontaneously infected wild boars Parasitol Res. 1993;79:617. doi: 10.1007/bf00932250. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 30.Rojas C.A.A., Kronenberg P.A., Aitbaev S., Omorov R.A., Abdykerimov K.K., Paternoster G., Mullhaupt B., Torgerson P., Deplazes P. Genetic diversity of Echinococcus multilocularis and Echinococcus granulosus sensu lato in Kyrgyzstan: The A2 haplotype of E. multilocularis is the predominant variant infecting humans PLoS Neglect Trop Dis. 2020;14 doi: 10.1371/journal.pntd.0008242. : . , , e0008242, doi: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romig T., Deplazes P., Jenkins D., Giraudoux P., Massolo A., Craig P.S., Wassermann M., Takahashi K., de la Rue M. Ecology and Life Cycle Patterns of Echinococcus Species Adv Parasitol. 2017;95:213. doi: 10.1016/bs.apar.2016.11.002. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 32.Santa M.A., Umhang G., Klein C., Grant D.M., Ruckstuhl K.E., Musiani M., Gilleard J.S., Massolo A. It’s a small world for parasites: evidence supporting the North American invasion of European Echinococcus multilocularis Proc R Soc B-Biol Sci. 2023;290 doi: 10.1098/rspb.2023.0128. : . , , doi: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sydler T., Mathis A., Deplazes P. Echinococcus multilocularis lesions in the livers of pigs kept outdoors in Switzerland Eur J Vet Pathol. 1998;4:43. : . , , –. . [Google Scholar]
- 34.Umhang G., Bastid V., Avcioglu H., Bagrade G., Bujanić M., Čabrilo O.B., Casulli A., Dorny P., van der Giessen J., Guven E., Harna J., Karamon J., Kharchenko V., Knapp J., Kolarova L., Konyaev S., Laurimaa L., Losch S., Miljević M., Miterpakova M., Moks E., Romig T., Saarma U., Snabel V., Sreter T., Valdmann H., Boué F. Unravelling the genetic diversity and relatedness of Echinococcus multilocularis isolates in Eurasia using the EmsB microsatellite nuclear marker Infect Genet Evol. 2021;92:104863. doi: 10.1016/j.meegid.2021.104863. : . , , , doi: . [DOI] [PubMed] [Google Scholar]
- 35.Umhang G., Karamon J., Hormaz V., Knapp J., Cencek T., Boué F. A step forward in the understanding of the presence and expansion of Echinococcus multilocularis in Eastern Europe using microsatellite EmsB genotyping in Poland Infect Genet Evol. 2017;54:176. doi: 10.1016/j.meegid.2017.07.004. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 36.Umhang G., Knapp J., Hormaz V., Raoul F., Boué F. Using the genetics of Echinococcus multilocularis to trace the history of expansion from an endemic area Infect Genet Evol. 2014;22:142. doi: 10.1016/j.meegid.2014.01.018. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]
- 37.Umhang G., Knapp J., Wassermann M., Bastid V., de Garam C.P., Boué F., Cencek T., Romig T., Karamon J. Asian Admixture in European Echinococcus multilocularis Populations: New Data From Poland Comparing EmsB Microsatellite Analyses and Mitochondrial Sequencing Front Vet Sci. 2021;7:620722. doi: 10.3389/fvets.2020.620722. : . , , , doi: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao N., Qiu J.M., Nakao M., Li T.Y., Yang W., Chen X.W., Schantz P.M., Craig P.S., Ito A. Echinococcus shiquicus n. sp., a taeniid cestode from Tibetan fox and plateau pika in China Int J Parasitol. 2005;35:693. doi: 10.1016/j.ijpara.2005.01.003. : . , , –. , doi: . [DOI] [PubMed] [Google Scholar]