Abstract
A Bacillus subtilis strain with the polynucleotide phosphorylase gene deleted was sensitive for growth in the presence of tetracycline. This strain was used to select for tetracycline-resistant mutants. A point mutation in the tetA(L) promoter and a spontaneously occurring tetA(L) gene copy number mutant were characterized.
The tetA(L) gene of Bacillus subtilis Marburg 168 strains confers low-level resistance to tetracycline (TET). This gene was identified by Williams and Smith (17), who isolated mutants of B. subtilis that were able to grow on 50 μg of TET/ml. Sakaguchi and Shishido (15) cloned this gene and showed that its presence on a plasmid conferred inducible TET resistance on B. subtilis. The tetA(L) gene normally confers resistance to concentrations of TET up to 2 μg/ml. Upon protoplast regeneration (10, 18) or transformation with a tetA(L)-containing plasmid (9), amplification of the chromosomal tetA(L) locus can occur, resulting in significantly elevated levels of TET resistance. Krulwich and colleagues demonstrated that the tetA(L) gene product functions as a TET-metal/H+ antiporter and as a physiologically significant Na+/H+ antiporter (3, 5). By using tetA(L)-lacZ fusion strains, it was demonstrated that expression of tetA(L) was induced 25-fold in the presence of 0.25 μg of TET/ml (4). The tetA(L) leader region, which contains a ribosome binding site and a short open reading frame (Fig. 1B), is thought to be involved in the regulation of tetA(L) expression. Northern blot analysis showed a two- to threefold increase in tetA(L) mRNA in response to inducing concentrations of TET. Reverse transcriptase analysis of tetA(L) mRNA indicated two transcriptional start sites 5 nucleotides apart (Fig. 1C).
FIG. 1.
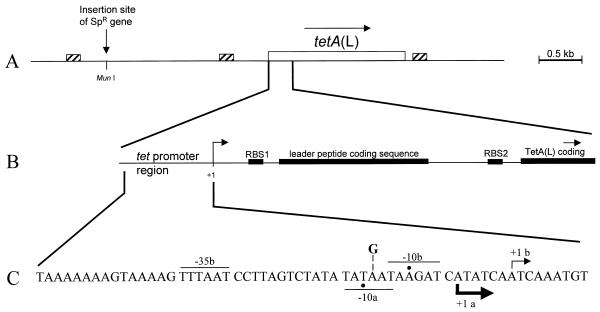
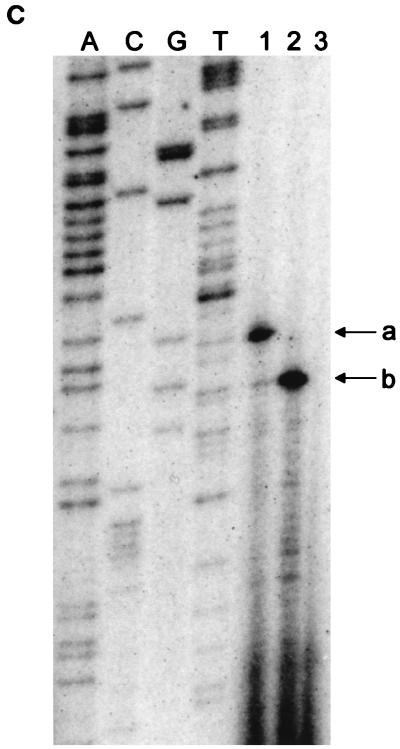
Map of the tetA(L) chromosomal locus. (A) Schematic diagram of the 4.5-kb HindIII fragment cloned into pJBC1 (4). The location of the MunI site, into which the Spr gene was inserted, is shown. Small boxes with diagonal stripes mark the locations of homologous 170-bp direct repeats (1). Open box, tetA(L) gene. (B) Expanded view of the tetA(L) promoter and leader region. Filled boxes, translational signals and coding sequences. The site of transcription initiation is marked +1. RBS, ribosome binding site. (C) Nucleotide sequence of the tetA(L) promoter region. Transcriptional start sites (+1) a and b are indicated, along with the respective −10 and −35 sequences. Actual −10 positions are marked by dots. The thicknesses of the arrows showing the direction of transcription indicate the relative amounts of transcription initiation from the two start sites in the wild type. The location of the A→G change at position −14 (relative to start site b) is shown.
Polynucleotide phosphorylase (PNPase), encoded by the pnpA gene (11, 12), is a 3′-to-5′ exoribonuclease thought to be involved in mRNA decay. We constructed a pnpA deletion strain, designated BG119, and found that this strain was more sensitive to TET than the wild type (16). In this study, BG119 was tested quantitatively for sensitivity to TET. The growth of BG119 was severely compromised in 0.8 μg of TET/ml and was completely inhibited in 1.0 μg of TET/ml (data not shown). The wild-type B. subtilis strain, on the other hand, grew well in concentrations of TET up to 2 μg/ml (see Fig. 2). We took advantage of the TET sensitivity of the pnpA deletion strain to select for TET-resistant mutants. (All strains used in this study are listed in Table 1.)
FIG. 2.
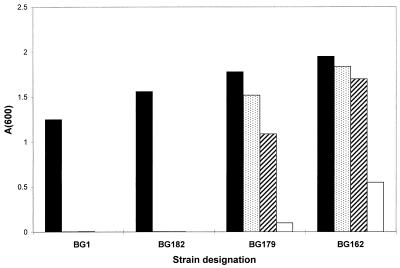
Growth of wild-type and Tcr mutant strains in increasing concentrations of TET. A 2.5-μl portion of an overnight culture was used to inoculate 5 ml of Luria-Bertani broth containing the indicated concentration of TET. Cultures were grown for 16 h with shaking at 37°C, after which the A600 was measured. TET (a 4-mg/ml solution in 50% ethanol) was stored for no more than a few weeks at −10°C. TET concentrations, in micrograms per milliliter, were as follows: 2 (filled bars), 4 (stippled bars), 6 (diagonally striped bars), and 8 (open bars).
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant characteristic(s) | Description |
|---|---|---|
| BG1 | thr-5 trpC2 | Wild type |
| BG119 | BG1 pnpA::Kmr | Replacement of pnpA coding sequence with Kmr gene |
| BG149 | BD99 amyE::tetA(L)-lacZ | Wild-type tetA(L)-lacZ gene fusion integrated at amyE locus |
| BG161 | BG119 Spr | Insertion of Spr gene upstream of tetA(L) |
| BG162 | BG161 Tcr | Mutant selected for Tcr; contains A→G change in tetA(L) promoter |
| BG178 | BG161 Tcr | Mutant selected for Tcr; contains amplified tetA(L) |
| BG179 | BG161 Tcr | Mutant selected for Tcr; contains amplified tetA(L) |
| BG182 | BG161 Tcr | Mutant selected for Tcr; mutation unlinked to tetA(L) |
| BG194 | BD99 amyE::tetA(L)-lacZ | tetA(L)-lacZ fusion with A→G change in tetA(L) promoter |
| BG226 | BG1 Tcr | A→G change in promoter of tetA(L) gene in wild-type strain |
To map mutations in BG119 relative to the tetA(L) locus, a spectinomycin resistance gene, cloned as an EcoRI fragment, was inserted into the chromosome at the MunI site, located 1.65 kb upstream of the tetA(L) gene (Fig. 1A), producing strain BG161. Cultures of BG161 were grown overnight, and aliquots were plated on solid medium containing 3 μg of TET/ml. After overnight growth at 37°C, TET-resistant colonies arose at a frequency of about 10−8.
The Tcr colonies were mostly small (1 mm in diameter or smaller), but several larger colonies were obtained. The growth of one large colony and two smaller colonies, in the presence of TET, is shown in Fig. 2. The least-resistant strains (represented by BG182) could grow in the presence of 2 μg of TET/ml, which was similar to the level of TET resistance of the wild type (BG1). The mutations in these strains were unlinked to the tetA(L) locus, and they were not characterized further. A second class of mutants (represented by BG179) was able to grow well in 6 μg of TET/ml but grew poorly at 8 μg/ml. In two of these, designated BG178 and BG179, the mutations that conferred TET resistance were found to be 60 to 70% linked to the Spr marker. Sequence analysis showed that these strains contained only the wild-type tetA(L) gene. By Southern blot analysis, it was determined that these strains had a fourfold amplification of the tetA(L) locus (data not shown).
The single large colony that was picked (BG162) represented the third class, for which reasonable growth was observed in 8 μg of TET/ml (Fig. 2). The mutation in BG162 was more than 90% linked to the Spr marker. Sequencing of the tetA(L) promoter in BG162 revealed an A→G change in the −10 sequence for the upstream start site (−10a) (Fig. 1C). The same A→G change was found in a large Tcr colony from a second plating experiment.
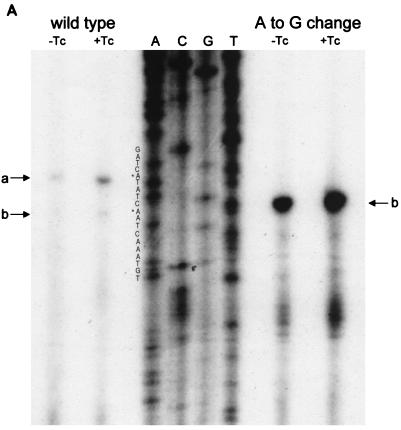
The transcriptional start site for tetA(L) mRNA was examined by reverse transcriptase analysis (Fig. 3A). The amount of tetA(L) mRNA from the mutant strain (BG162) was approximately 10-fold greater than that from the wild-type strain (BG1). In both the wild-type and mutant strains, the level of transcriptional induction by TET was about twofold. Strikingly, the minor downstream transcriptional start site of the wild-type tetA(L) (+1b) was the sole transcriptional start site in BG162. Examination of the promoter sequence (Fig. 1C) shows that this change created a TG dinucleotide at the −15 and −14 positions relative to the downstream start site. This −15,−14 sequence is highly conserved among B. subtilis promoter sequences (7) and has been found to exert a strong positive effect on transcription by B. subtilis RNA polymerase (8).
FIG. 3.
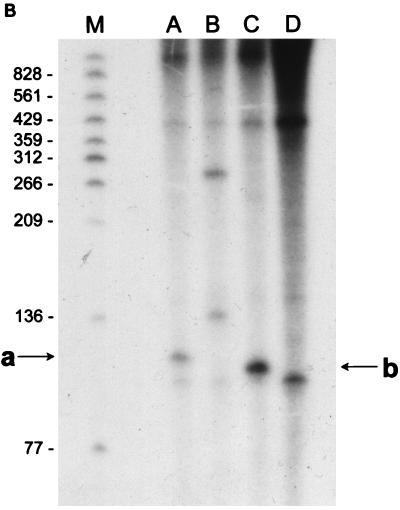
(A) Reverse transcriptase analysis of in vivo tetA(L) mRNA. RNA was isolated from the wild-type strain and from the strain containing the A→G change in the tetA(L) promoter. Strains were grown in the absence or presence of 0.25 μg of TET/ml. Transcriptional start sites are marked a and b, as in Fig. 1. Lanes A, C, G, and T are a sequence ladder obtained with the primer used for the reverse transcriptase reactions. The complement of the ladder sequence, which is the same as the sequence shown in Fig. 1C, is shown on the left of the sequencing ladder. Transcriptional start sites are marked by asterisks. The amount of radioactivity in the bands was quantitated with a PhosphorImager (Molecular Dynamics). (B) In vitro transcription. Plasmid pAN583 contained either a wild-type tetA(L) promoter-leader DNA fragment (lanes A and B), a promoter-leader DNA fragment with the A→G change (lane C), or no tetA(L) DNA (lane D). Plasmid DNA was linearized with XbaI (lanes A, C, and D) or with HindIII (lane B). Digestion with HindIII linearized the plasmid at a site 25 bp downstream of the XbaI site. Comparison of lanes A and B demonstrates that the band indicated by “a” at the left represents transcription from transcriptional start site a. In lane C, the transcription product migrates faster and represents transcription from transcriptional start site b. A nonspecific transcription product runs below band b and is present in all lanes, including lane D [no tetA(L) DNA]. Lane M contained end-labeled DNA fragments from a TaqI restriction endonuclease digest of pSE420 (2). Sizes (in base pairs) of the labeled DNA fragments are indicated on the left. (C) Reverse transcriptase analysis of in vitro-transcribed tetA(L) RNA. Lane 1, wild-type tetA(L) RNA; lane 2, mutant (A→G change) tetA(L) mRNA; lane 3, no RNA (labeled primer only). Transcriptional start sites are marked a and b, as in Fig. 1. Lanes A, C, G, and T are a sequence ladder obtained with the primer used for the reverse transcriptase reactions.
To demonstrate that the A→G change was the only mutation necessary for the increase in tetA(L) transcription, and to quantitate the effect of the mutation on tetA(L) expression, tetA(L)-lacZ fusions containing either wild-type or mutant promoter regions were constructed at the amyE locus of BD99. Uninduced and induced β-galactosidase levels were seven- to eightfold higher in the mutant than in the wild type. However, the levels of induction of tetA(L) expression in the presence of TET were similar for wild-type and mutant tetA(L)-lacZ strains: 16- and 21-fold, respectively.
To demonstrate that the changes in tetA(L) transcription were due solely to the cis-acting effect of the A→G point mutation and were not dependent on a trans-acting factor, a 260-bp EcoRI-XbaI fragment containing the tetA(L) promoter and leader region was cloned from wild-type and mutant tetA(L) genes into the transcription vector plasmid pAN583 (14). This plasmid contains a transcriptional terminator sequence downstream of a multicloning site. Transcription was performed on supercoiled DNA or on DNA that had been linearized with XbaI or with HindIII; the HindIII site in the vector is located 25 bp downstream of the XbaI site. Transcription reaction mixtures contained 40 mM Tris (pH 7.9), 50 mM NaCl, 10 mM MgCl2, 1 mM each of the four nucleoside triphosphates, 8 μM β-mercaptoethanol, 10 μCi of [α-32P]UTP, and 0.5 μg of linearized or supercoiled plasmid DNA. The reaction was started by the addition of 5 μg of B. subtilis RNA polymerase holoenzyme. After incubation at 37°C for 25 min, 1 μl of a 6-mg/ml solution of heparin was added, and the incubation was continued at 37°C for 5 min. After DNase treatment for 15 min, the reaction product was extracted with phenol-chloroform (1:1), and nucleic acid was precipitated. Reaction products were separated on a 6% denaturing polyacrylamide gel. Transcription using the mutant template gave a fourfold-higher amount of RNA than transcription using the wild-type template (Fig. 3B, lanes A and C). It was further evident from the results in Fig. 3B that the transcription start sites differed for the mutant and wild-type templates. This was confirmed by reverse transcriptase analysis of in vitro-transcribed tetA(L) RNA (Fig. 3C). The +1a nucleotide was the major start site for transcription in the wild type, while the +1b nucleotide was the exclusive start site for the mutant. We conclude that the change in the level of transcription and the start site choice in BG162 are determined solely by the point mutation.
BG162 was able to grow well in 8 μg of TET/ml, which was four times the highest level of TET in which the wild-type BG1 strain could grow. However, the BG162 strain also contains the pnpA deletion, which we have shown causes an increase in TET accumulation (16). To test the level of resistance conferred by the A→G change on wild-type B. subtilis, the mutation was transferred to BG1 to produce strain BG226. BG226 was able to grow well at TET concentrations up to 20 μg/ml, which was the challenge concentration used in previous studies (10, 18).
Unlike the findings of the previous work, in which a protoplast regeneration regimen or transformation with a plasmid carrying the tetA(L) gene was required to obtain highly amplified tetA(L) regions, we isolated strains with a fourfold amplification of the tetA(L) locus without prior manipulations. Although the tetA(L) gene is flanked by three highly homologous copies of a small (170-bp) direct repeat sequence (Fig. 1A) (1), it is unlikely that these elements are involved in the duplication. From additional Southern blot analyses, we found that the amplified units in BG178 and BG179 were larger than the segments between these repeat sequences.
We note that Hashiguchi et al. (6) have reported endogenous chromosomal amplification in B. subtilis, in cells treated with a chemical mutagen, which resulted in tunicamycin resistance. Also, Neyfakh et al. (13) have characterized a B. subtilis strain with an amplified bmr gene, but this was isolated after growth for several weeks in the presence of increasing drug concentrations.
The shift in transcription start site that resulted from the A→G mutation did not appear to affect the expression of tetA(L). In other studies (15a), we have found that tetA(L) expression is regulated at the transcriptional, posttranscriptional, and translational levels. The existence of two transcriptional start sites adds another level of complexity. Multiple controls on tetA(L) gene expression imply that the level of TetA(L) protein must be precisely regulated, which supports earlier evidence that tetA(L) plays a major role in B. subtilis pH homeostasis and monovalent cation efflux (3, 4).
Acknowledgments
This work was supported by Public Health Service grant GM-52837.
We thank I. Smith for providing B. subtilis RNA polymerase holoenzyme and K. Hue-Roye for technical assistance.
REFERENCES
- 1.Amano H, Shishido K. Bacillus subtilis strains carry highly homologous direct repeat sequences on their chromosomes. Biosci Biotechnol Biochem. 1995;59:2149–2150. doi: 10.1271/bbb.59.2149. [DOI] [PubMed] [Google Scholar]
- 2.Brosius J. Compilation of superlinker vectors. Methods Enzymol. 1992;216:469–483. doi: 10.1016/0076-6879(92)16043-j. [DOI] [PubMed] [Google Scholar]
- 3.Cheng J, Guffanti A A, Krulwich T A. The chromosomal tetracycline-resistance locus of Bacillus subtilis encodes a Na+/H+ antiporter that is physiologically important at elevated pH. J Biol Chem. 1994;269:27365–27371. [PubMed] [Google Scholar]
- 4.Cheng J, Guffanti A A, Wang W, Krulwich T A, Bechhofer D H. Chromosomal tetA(L) gene of Bacillus subtilis: regulation of expression and physiology of a tetA(L) deletion strain. J Bacteriol. 1996;178:2853–2860. doi: 10.1128/jb.178.10.2853-2860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guffanti A A, Krulwich T A. Tetracycline/H+ antiport and Na+/H+ antiport catalyzed by the Bacillus subtilis TetA(L) transporter expressed in Escherichia coli. J Bacteriol. 1995;177:4557–4561. doi: 10.1128/jb.177.15.4557-4561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashiguchi K, Tanimoto A, Nomura S, Yamane K, Yoda K, Harada S, Mori M, Furusato T, Takatsuki A, Yamasaki M, Tamura G. Amplification of the amyE-tmrB region on the chromosome in tunicamycin-resistant cells of Bacillus subtilis. Mol Gen Genet. 1986;204:36–43. doi: 10.1007/BF00330184. [DOI] [PubMed] [Google Scholar]
- 7.Helmann J D. Compilation and analysis of Bacillus subtilis ςA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;13:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henkin T M, Sonenshein A L. Mutations of the Escherichia coli lacUV5 promoter resulting in increased expression in Bacillus subtilis. Mol Gen Genet. 1987;209:467–474. doi: 10.1007/BF00331151. [DOI] [PubMed] [Google Scholar]
- 9.Ives C L, Bott K F. Cloned Bacillus subtilis chromosomal DNA mediates tetracycline resistance when present in multiple copies. J Bacteriol. 1989;171:1801–1810. doi: 10.1128/jb.171.4.1801-1810.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ives C L, Bott K F. Characterization of chromosomal DNA amplification with associated tetracycline resistance in Bacillus subtilis. J Bacteriol. 1990;172:4936–4944. doi: 10.1128/jb.172.9.4936-4944.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luttinger A, Hahn J, Dubnau D. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol Microbiol. 1996;19:343–356. doi: 10.1046/j.1365-2958.1996.380907.x. [DOI] [PubMed] [Google Scholar]
- 12.Mitra S, Bechhofer D H. Polynucleotide phosphorylase activity of B. subtilis. Mol Microbiol. 1996;19:329–342. doi: 10.1046/j.1365-2958.1996.378906.x. [DOI] [PubMed] [Google Scholar]
- 13.Neyfakh A A, Bidnenko V E, Chen L B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci USA. 1991;88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Predich M, Nair G, Smith I. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing ςH. J Bacteriol. 1992;174:2771–2778. doi: 10.1128/jb.174.9.2771-2778.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakaguchi R, Shishido K. Molecular cloning of a tetracycline-resistance determinant from Bacillus subtilis chromosomal DNA and its expression in Escherichia coli and B. subtilis. Biochim Biophys Acta. 1988;949:49–57. doi: 10.1016/0167-4781(88)90053-x. [DOI] [PubMed] [Google Scholar]
- 15a.Stasinopoulos, S. J., and D. H. Bechhofer. Unpublished data.
- 16.Wang W, Bechhofer D H. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J Bacteriol. 1996;178:2375–2382. doi: 10.1128/jb.178.8.2375-2382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams G, Smith I. Chromosomal mutations causing resistance to tetracycline in Bacillus subtilis. Mol Gen Genet. 1979;177:23–29. doi: 10.1007/BF00267249. [DOI] [PubMed] [Google Scholar]
- 18.Wilson C R, Morgan A E. Chromosomal DNA amplification in Bacillus subtilis. J Bacteriol. 1985;163:445–453. doi: 10.1128/jb.163.2.445-453.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]