Abstract
The repressor gene of the Lactobacillus phage A2 has the following properties: it (i) encodes a 224-residue polypeptide with DNA binding and RecA cleavage motifs, (ii) is expressed in lysogenic cultures, and (iii) confers superinfection immunity on the host. Adjacent, but divergently transcribed, lies another open reading frame whose product resembles the λ Cro protein. In the 161-bp intergenic segment, putative promoters and operators have been detected.
Bacteriophages are recognized to be the main source of disruption in industrial food fermentations (5). The temperate phage A2 infects strains of Lactobacillus casei and Lactobacillus paracasei of industrial relevance. The virions present isometric heads and noncontractile tails. The phage genome is a 44.02-kb double-stranded DNA molecule with 3′-protruding cohesive ends (6, 7). A2 can be recovered from lysogens through mitomycin C induction, suggesting that the phage repressor becomes inactivated by proteolytic cleavage during the mitomycin-induced SOS response as it occurs with bacteriophage λ (17). The λ repressor binds to promoters PL and PR, which results in repression of the genes that lead to the lytic development. One consequence of this regulation is that lysogens are immune to superinfection by the same or related viruses. In addition, cI is autogenously regulated through differential binding to three adjacent operator sites.
Characterization of A2 clear plaque deletion mutants.
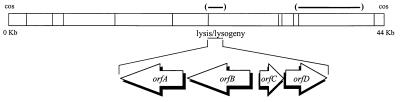
The gene that encodes the viral repressor was localized through the selection of deletion mutants unable to lysogenize L. casei ATCC 393. To get them, phage suspensions were treated with 10 mM sodium pyrophosphate, pH 7.4, at 37°C for 30 min, which resulted in survival of 7 × 10−4 phage. Appropriate dilutions were plated onto MCM (4), and the surviving phage was collected, suspended in SM buffer, and subjected to new rounds of treatment until a plateau was reached at around 10% survival. Phage from isolated plaques, obtained after each round, was repurified, and their DNA restriction patterns were compared with that of the wild type. The deletions ranged from 0.5 to 3.5 kb and mapped in three EcoRI fragments that defined two regions of the genome, comprising up to 7.9 kb dispensable for lytic development (Fig. 1). The phage whose deletion was located in the center of the physical map showed a clear plaque phenotype and was unable to lysogenize its hosts. In contrast, lysogenization was easily obtained with the mutants lacking segments in the right arm of the genome.
FIG. 1.
EcoRI restriction map of A2 DNA with indication of dispensable regions for lytic development (thick lines in parentheses). Below the map is shown the organization of the region that controls the phage cycles. The arrows indicate the relative sizes and directions of transcription of the indicated genes.
Structural characterization of the repressor region of bacteriophage A2.
Since the deletions located at the center of the A2 physical map resulted in impairment of lysogenization, we started the analysis of this region by cloning and sequencing it. In this sequence, four open reading frames (orfA to orfD), which read in opposite directions, were found (Fig. 1). The products of orfB and orfC were hypothesized to be the functional homologs of the λ proteins CI and Cro, respectively. This was based on their sizes (224 and 81 amino acids for ORFB and ORFC, respectively, which correspond to 25,277- and 9,180-Da polypeptides), their transcription in opposite directions, with an intergenic region of 161 nucleotides (Fig. 2), and the similarities shown by ORFB (pI, 4.56) to phage repressors and also to regulatory proteins involved in SOS induction (Fig. 3). The NH2-terminal end of ORFB presents a helix-turn-helix motif, which is possibly involved in binding of a specific DNA target, while its carboxy-terminal part shows a domain for protease RecA recognition. It includes conserved Ser and Lys residues and the Ala-Gly motif (marked with asterisks in Fig. 3), in front of which cleavage has been reported to occur in the λ repressor, as the first step towards lytic development of the prophage (18).
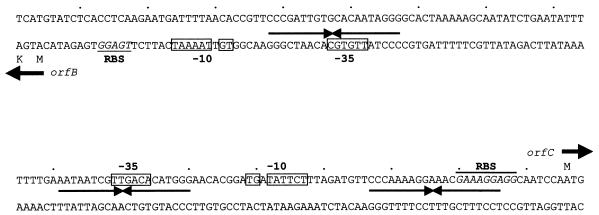
FIG. 2.
Nucleotide sequence of the region between cI and the putative cro. Potential ribosomal binding sites (RBS), consensus promoter sequences (−35 and extended −10 boxes), the starts of translation, and the inverted repeats found are indicated.
FIG. 3.
Amino acid sequence alignment of ORFB with the CI proteins of phages λ (16), φ80 (12), and r1t (11); the putative repressor of Tuc2009 (19); and the SOS response-related proteins LexA (8) and DinR (14). The DNA binding motifs of the amino termini are boxed. Conserved amino acids are shown in boldface, and the RecA cleavage point is marked with asterisks.
As stated above, ORFC is hypothesized to be the functional homolog of Cro, a competitor of CI for binding at the operators of the promoters that regulate the lytic-lysogenic pathways. Nevertheless, ORFC does not show any significant similarity to Cro at the amino acid sequence level, although it does with a putative DNA binding transcription repressor of a Pseudomonas aeruginosa bacteriophage (9), and with a cro topological homolog of φSfi21, which infects Streptococcus thermophilus (3). Additionally, it shares some of the essential amino acids present in Cro homologs and has a basic isoelectric point (pI, 10.80).
In the intergenic region lying between orfB and orfC, two putative divergent promoters were identified. Both presented the dinucleotide TG positioned 1 base upstream of the −10 hexamer, a feature that has been shown to enhance both promoter strength and utilization (20). These are followed by potential ribosomal binding sites, complementary to the Lactobacillus delbrueckii 3′ end of the 16S rRNA (10). In addition, three imperfect palindromic sequences that might act as operators in regulation of the phage developmental cycles were observed (Fig. 2).
The stop triplet of orfC overlaps with the start of orfD in the sequence ATGA, which suggests that both have a coupled translation. The predicted ORFD polypeptide is 160 amino acids long, with a mass of 17,844 Da and a pI of 5.39. It shows some homology to putative proteins encoded by open reading frames located in similar genomic positions of several bacteriophages, such as BK5-T, φSfi21, and r1t, that infect other lactic acid bacteria (2, 3, 11). In turn, the corresponding gene of the temperate S. thermophilus phage φSfi21 has some homology with the gene ant of P1, which encodes an antirepressor (15).
The start codon of orfA is located 58 nucleotides downstream of orfB. In this region an inverted repeat with a ΔG of −17 kcal/mol, followed by a stretch of T’s, was found; it may act as a rho-independent transcription terminator (see below). orfA encodes a 225-amino-acid polypeptide with a mass of 24,447 Da and a pI of 4.4, which is preceded by a canonical ribosome binding site. Comparison of the sequence deduced from orfA with those present in databases did not reveal similarities to relevant proteins.
Expression of orfB confers immunity to A2 superinfection.
If orfB coded for the repressor, it should confer superinfection immunity against A2 when cloned into L. casei cells. To test this possibility, a 0.8-kb DNA segment containing orfB was amplified by using a primer that included its putative promoter sequence and a converging one located just after the stop codon, into which EcoRI restriction sites were introduced. The amplified DNA segment was purified, EcoRI cleaved, and ligated to pEM40 digested with the same enzyme to generate pEM40::orfB. This plasmid is a pUC18 derivative that contains an erythromycin resistance gene for selection in gram-positive bacteria. It does not replicate in L. casei but carries the integrase gene and the attP sequence of A2, which allows its insertion into a tRNA gene of several lactobacilli (1). Challenge of several independently obtained pEM40::orfB transformants with bacteriophage A2 resulted in complete immunity to superinfection (no plaques were produced by a phage suspension with a titer on the untransformed host of 1010 PFU/ml). This is consistent with the suggested function of orfB as the gene that encodes the A2 repressor (cI).
cI-specific transcripts produced during the lytic and lysogenic cycles of A2.
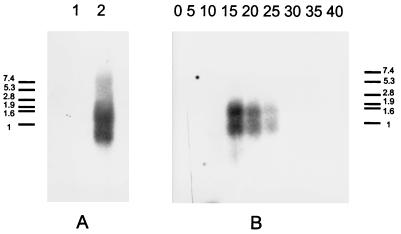
As a final test of cI identity, its transcription pattern was investigated. A Northern blot of total RNA from L. casei ATCC 393 (without infection and at several times postinfection) and from one A2 lysogen derivative was probed with a PCR-generated 32P-labelled DNA fragment that exactly spans cI. Two transcripts, of 0.8 and 1.4 kb, were observed in the lysogen (Fig. 4A). The size of the first fits with the distance between the putative cI promoter and the rho-independent terminator identified 3′ of that gene. The 1.4-kb transcript most probably corresponds to cI plus orfA. In productively infected cultures of L. casei ATCC 393 (Fig. 4B), the same pattern of cI-specific RNAs was found at early times postinfection (from 15 to 25 min) and fading afterwards (the eclipse period of the phage under the propagation conditions used lasts about 120 min).
FIG. 4.
Northern blot analysis of the cI transcripts. (A) Equal amounts of total RNA from uninfected L. casei (lane 1) and from an A2 lysogen (lane 2) were used. (B) Total RNA from L. casei at various times postinfection (in minutes) with bacteriophage A2. The numbers beside the size standards are in kilobases.
Several lines of evidence seem to indicate that the central region of the A2 genome is involved in the genetic switch that directs phage development into the lytic or lysogenic cycles. First is its resemblance to the homologous region of bacteriophage λ (13), with two genes reading in opposite directions, separated by a short intergenic region in which three putative operator sequences could be discerned. In addition, the role of ORFB as the λ CI homolog is suggested by its sequence, in which the relevant DNA binding and RecA protease recognition motifs are present; by its transcription pattern, both in lysogens and at early times postinfection; and, surely most important, through the superinfection immunity phenotype conferred on L. casei upon insertion of cI into its genome. The overall resemblance of the genetic switch regions of phage A2 and λ is a proof of the suitability of this regulation system, which is present in phages not only of gram-negative but also of gram-positive bacteria. However, the distance between the putative cro and cI genes of phage A2 (161 bp) is longer than it is in λ (100 bp), possibly indicating that the DNA-protein and protein-protein interactions that lead to growth cycle regulation may differ between the phages.
Furthermore, lactobacilli are used in many food fermentations and also as probiotics in health promotion. The confirmation that phage repressors, even in single copy but stably integrated into the host’s genome, protect the host from phage infection may suggest ways to avoid one of the main causes of industrial fermentation failure; in fact, we have determined that protection by CI against A2 infection is extended to L. casei growing under fermented milk production conditions (unpublished data). Of course, further refinements would be necessary before it can be used for industrial purposes, mainly to replace the Escherichia coli plasmid-derived sequences of the vector with generally-regarded-as-safe bacterial sequences. Our efforts will be devoted to demonstrating, we hope, the general utility of stable repressor expression in protection against phage attack on valuable bacterial strains.
Nucleotide sequence accession number.
The sequence of the repressor region of bacteriophage A2 has been submitted to EMBL under accession no. Y12813.
Acknowledgments
This work was supported by the BIOTECH Program of the European Communities on Lactic Acid Bacteria (grants BIOT CT94-3055 and BIOT CT96-0402) and by the Comisión Interministerial de Ciencia y Tecnología of Spain (grant BIO94-189). V.L. was the recipient of a CICYT grant connected with the last project.
REFERENCES
- 1.Alvarez, M. A., M. Herrero, and J. E. Suárez. Site-specific integration of bacteriophage A2 and construction of vectors able to integrate both in Gram positive and Gram negative bacteria. Submitted for publication.
- 2.Boyce J D, Davidson B E, Hillier A J. Identification of prophage genes expressed in lysogens of the Lactococcus lactis bacteriophage BK5-T. Appl Environ Microbiol. 1995;61:4099–4104. doi: 10.1128/aem.61.11.4099-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruttin A, Desiere F, Lucchini S, Foley S, Brüssow H. Characterization of the lysogeny DNA module from the temperate Streptococcus thermophilus bacteriophage φSfi21. Virology. 1997;233:136–148. doi: 10.1006/viro.1997.8603. [DOI] [PubMed] [Google Scholar]
- 4.Caso J L, De los Reyes-Gavilán C J, Herrero M, Montilla A, Rodríguez A, Suárez J E. Isolation and characterization of temperate and virulent bacteriophages of Lactobacillus plantarum. J Dairy Sci. 1995;78:741–750. [Google Scholar]
- 5.Daly C, Fitzgerald G F, Davis R. Biotechnology of lactic acid bacteria with special reference to bacteriophage resistance. In: Venema G, Huis in’t Veld J H J, Hugenholtz J, editors. Lactic acid bacteria: genetics, metabolism and applications. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 3–14. [Google Scholar]
- 6.García P, Alonso J C, Suárez J E. Molecular characterization of the cos region of the Lactobacillus casei bacteriophage A2. Gene product 3, gp3, specifically binds to its downstream cos region. Mol Microbiol. 1997;23:505–514. doi: 10.1046/j.1365-2958.1997.d01-1863.x. [DOI] [PubMed] [Google Scholar]
- 7.Herrero M, de los Reyes-Gavilán C G, Caso J L, Suárez J E. Characterization of 393-A2, a bacteriophage that infects Lactobacillus casei. Microbiology. 1994;140:2585–2590. [Google Scholar]
- 8.Horii T, Ogawa T, Ogawa H. Nucleotide sequence of the lexA gene of Escherichia coli. Cell. 1981;23:689–697. doi: 10.1016/0092-8674(81)90432-3. [DOI] [PubMed] [Google Scholar]
- 9.Lee F K N, Dudas K C, Nelson M B, LoVerde P T, Apicella M A. Accession no. L06240. 1992. Unpublished data. [Google Scholar]
- 10.Mikkonen M, Vouristo J, Alatossava T. Ribosome binding site consensus sequence of Lactobacillus delbrueckii subsp. lactis bacteriophage LL-H. FEMS Microbiol Lett. 1994;116:315–320. doi: 10.1111/j.1574-6968.1994.tb06721.x. [DOI] [PubMed] [Google Scholar]
- 11.Nauta A, van Sideren D, Karsens M, Smit E, Venema G, Kok J. Inducible gene expression mediated by a repressor-operator system isolated from Lactococcus lactis bacteriophage r1t. Mol Microbiol. 1996;19:1331–1341. doi: 10.1111/j.1365-2958.1996.tb02477.x. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa T, Ogawa H, Tomizawa J. Organization of the early region of bacteriophage φ 80. Genes and proteins. J Mol Biol. 1988;202:527–550. doi: 10.1016/0022-2836(88)90284-7. [DOI] [PubMed] [Google Scholar]
- 13.Ptashne M. A genetic switch. 2nd ed. Cambridge, Mass: Blackwell Scientific Publications; 1992. [Google Scholar]
- 14.Raymond-Denise A, Guillen N. Identification of dinR, a DNA damage-inducible regulator gene of Bacillus subtilis. J Bacteriol. 1991;173:7084–7091. doi: 10.1128/jb.173.22.7084-7091.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riedel H, Heinrich J, Heising A, Choli T, Schuster H. The antirepressor of phage P1. Isolation and interaction with the C1 repressor of P1 and P7. FEBS Lett. 1993;334:165–169. doi: 10.1016/0014-5793(93)81705-5. [DOI] [PubMed] [Google Scholar]
- 16.Sauer R T, Andreg R. Primary structure of the λ repressor. Biochemistry. 1978;17:1092–1100. doi: 10.1021/bi00599a024. [DOI] [PubMed] [Google Scholar]
- 17.Sauer R T, Ross M J, Ptashne M. Cleavage of the λ and P22 repressors by RecA protein. J Biol Chem. 1982;257:4458–4462. [PubMed] [Google Scholar]
- 18.Slilaty S N, Little J W. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc Natl Acad Sci USA. 1987;84:3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Guchte M, Daly C, Fitzgerald G F, Arendt E K. Identification of the putative repressor-encoding gene cI of the temperate lactococcal bacteriophage Tuc2009. Gene. 1994;144:93–95. doi: 10.1016/0378-1119(94)90209-7. [DOI] [PubMed] [Google Scholar]
- 20.Voskuil M I, Voepel K, Chambliss G H. The −16 region, a vital sequence for the utilization of a promoter in Bacillus subtilis and Escherichia coli. Mol Microbiol. 1995;17:271–279. doi: 10.1111/j.1365-2958.1995.mmi_17020271.x. [DOI] [PubMed] [Google Scholar]