Abstract
Introduction
The HIV-1 prevalence has been steadily increasing in Jiangsu, China. HIV-1 genetic transmission network can be used to explore the transmission kinetics and precision intervention in high-risk populations. Thus, we generated an HIV-1 genetic transmission network, explored key risk populations based on different risk factors and found out the risk factors for HIV-1 prevention and control among the newly-diagnosed HIV-1 cases from 2017 to 2018.
Method
We amplified the HIV-1 pol sequences from the plasma samples of the newly-diagnosed HIV-1 cases from 2017 to 2018 and obtained the infection data from The National HIV/AIDS Surveillance System. HIV-Trace and Cytoscape Software were both used to construct the HIV-1 genetic network with a gene distance of <0.005. The R software was used to analyze the risk factors for inclusion into the network.
Results
We obtained 3362 sequences with the pol gene region, of which 3316 contained detailed individual information. CRF01_AE accounted for 42.3 % of the HIV-1 subtypes in the samples. The median CD4+T lymphocyte count was 329 cells/μL in 2017 and 313 cells/μL in 2018. At the gene distance threshold of 0.005, 481 sequences were incorporated into the HIV-1 gene network, constructing 202 clusters. Age over 60 years old, heterosexual transmission route, subtype (CRF105_0107, CRF55_01 B, and CRF67_01 B) and CD4+T lymphocyte count (>200) were the risk factors influencing inclusion into the HIV-1 gene network. Moreover, south Jiangsu cities had higher inclusion in the network. Thus, key risk populations in the clusters with different transmission routes, new emerging subtypes, drug resistance nodes, and individuals above 60 years of age in the network represented the critical risk populations that should be focused more on for intervention.
Conclusion
The HIV-1 genetic transmission network is adept at discovering undiagnosed HIV-infected cases and linking all diagnosed cases for determination of risk infections. Therefore we should pay more attention to these risk infections with further investigation and intervention, helping to achieve the goal of 95 % use combination prevention from the World Health Organization, and push to end AIDS epidemic.
Keywords: HIV-1, Genetic transmission network, Newly-diagnosed, Risk population
1. Introduction
HIV-1 is still a public health concern worldwide, even though antiretroviral therapy (ART) can inhibit viral replication and prolong life expectancy. The HIV epidemic has been gradually increasing in China, as shown by the number of people (1.14 million) living with HIV and new HIV diagnoses (approximately 133,000) reported in 2021 [1,2]. Jiangsu, an economically developed province in China, had a steady increase in new HIV-1 diagnoses in the last three years, with about 4000 infections annually [3]. However, the actual number of new diagnoses is underestimated, considering the effect of the COVID-19 epidemic. Many prevention strategies have been proved to be effective in controlling the spread of HIV among different high-risk populations. Unfortunately, implementing these interventions to all high-risk populations is often infeasible in various settings [4]. Thus, it is imperative to find out key risk populations from a new perspective for further a new precision intervention [5,6]. An increasing number of studies support that molecular network analyses could contribute to reveal among whom and to where the HIV infection is spreading and to estimate the speed of HIV transmission. It was a break to be widely used in recent years in HIV-1 transmission kinetics and high-risk populations' precision intervention research. Several aspects have been incorporated into the network to illustrate characteristics of the HIV-1 spread at the population level, thus providing critical information to help improve the case-finding strategies and activity control of critical groups in the network [[7], [8], [9]].
Studies on HIV prevalence in Jiangsu have focused more on the male-to-male sexual contact (MMSC) group, which only accounts for half of the HIV infection cases [10,11]. To conduct detailed individual analysis of the new HIV-1 diagnoses and identify more undiagnosed infections and more HIV-negative network members at high risk of infection, we used this molecular network-based strategy. In spite of MMSC group, we also pay attention to the role played by females or people over 60 years old in the genetic transmission network. All these conductions are to achieve the first 95 % target of UNAIDS that could be a part of the global push to end the AIDS epidemic by 2030.
2. Materials and methods
2.1. Laboratory detection
The newly-diagnosed HIV-1 whole blood samples between 2017 and 2018 were collected for sequences amplification. The sequences further were used to construct the Jiangsu HIV genetic transmission network. The CD4+T lymphocyte counts (CD4 counts) were completed within 24 h after the sample collection. The whole blood samples were centrifuged at 3000 r/min for 15 min to collect plasma for HIV-1 gene test. Genotypic drug resistance (DR) was detected via RT-PCR and Nest-PCR [12]. The PCR products were Sanger sequenced by the Sangon Biotech company (Shanghai, China). We obtained partial HIV-1 polymerase (pol) coding region, including the protease gene (1–99 amino acids) and the reverse transcriptase gene (1–270 amino acids). All sequences were submitted to the HIV DR database (https://hivdb.stanford.edu/) for the DR results including mutations and resistance levels to drugs. Sequences with low degree level or above resistance to any drugs against protease and reverse transcriptase regions were defined as DR sequences.
2.2. HIV genetic transmission network inference
The sequences were aligned on MEGA 7.0, with reference sequences downloaded from the HIV Sequence Database (https://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html). Aligned sequences eventually had 1000 bp bases. After the sequences were aligned with the reference sequences, the ML phylogenetic tree was constructed. The subtype of the sequences in the same branch with the reference strains was determined as the reference strain sequence subtype. The Hyphy software was used to compute the distance between the two sequences, and the sequences with a distance of <0.005 were used to generate an HIV-1 genetic transmission network via the HIV-1 Trace and Cytoscape Software following the protocol advised by China CDC.
2.3. Statistical analysis
The function “glm” of the R (version 4.1.3) package “rms” was used for statistical analysis. The univariate logistic analysis determined the associations between the variables and the formed network. The variables with p < 0.2 (from the univariate logistic analysis) were incorporated into the logistic regression model to determine the significance level via binary logistic regression. Variables with p < 0.05 were considered statistically significant.
2.4. Individual information
The individual demographic information were acquired from the National HIV/AIDS Surveillance System, including identification card (ID) number, gender, age, ethnicity, transmission route, education degree, marital status, occupation, the institutions where patients being discovered, the confirmation date of HIV-1 infection, CD4+T cell counts before on ART.
3. Results
3.1. Information of the study population
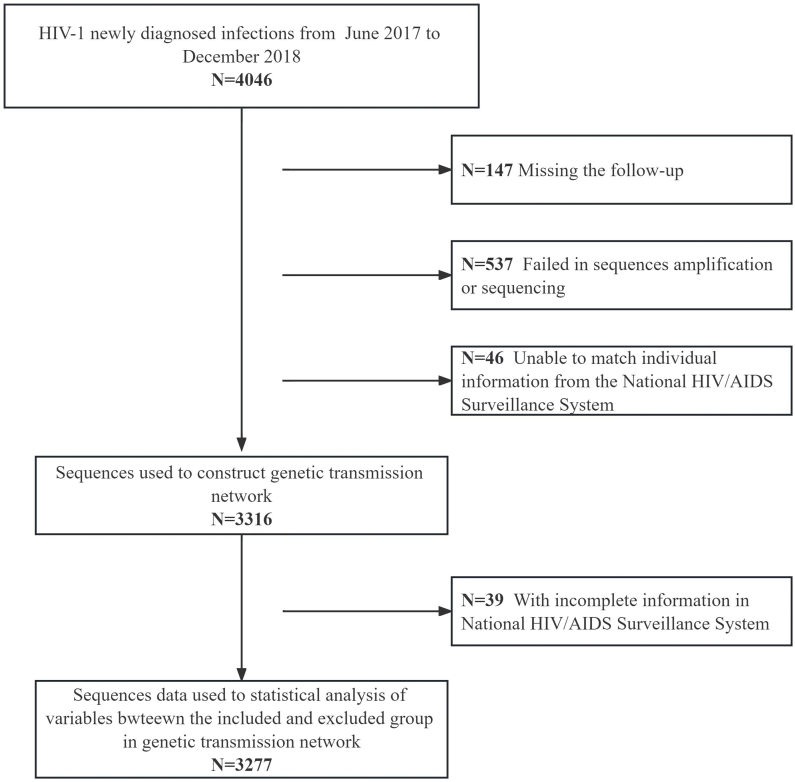
We obtained 3362 sequences, among which 3316 had detailed individual information accessed from the National HIV/AIDS Surveillance System (Fig. 1). Moreover, 87.7 % of the infections were male, with a median age of 36 (interquartile range [IQR], 27–50). Nearly half of the individuals were married, 22.2 % of the population worked as commercial service providers, 19.2 % were farmers, and 17.2 % were housekeeping workers. The population percentage with education level of high school or above was 54.4 %. Furthermore, the transmission route was mostly through sexual transmission, with MMSC accounting for 60.3 %. The median CD4 count was 320 cells/μL (IQR: 204–464). Patients diagnosed at the voluntary counseling and testing (VCT) centers and the comprehensive hospitals accounted for 42.9 % and 38.0 %, respectively. The main circulating subtypes were CRF01_AE and CRF07_BC, accounting for 42.6 % and 31.1 %, respectively. Moreover, 90 % of the HIV-1 sequences were sensitive strains without any drug resistance mutation.
Fig. 1.
The flow chart of individuals included in the study.
3.2. HIV genetic transmission network
HIV-1 pol sequences from 3316 HIV-infected individuals were used to infer an HIV genetic transmission network. Eventually, the network consisted of 399 putative transmission links involving 15 % (481/3316) of individuals overall and 6 % (196/3316) and 9 % (285/3316) of individuals in 2017 and 2018, respectively. There were 202 connected clusters in the network, among which 39 had at least 3 nodes, denoted as big clusters. The largest cluster had 16 nodes, and the following clusters had 9, 7, and 6 nodes. In addition to the big clusters described above, there were 2 clusters each with 5 nodes, 8 clusters each with 4 nodes, and 24 clusters each with 3 nodes.
3.3. Characteristics of the key population in the unique HIV-1 clusters of transmission network
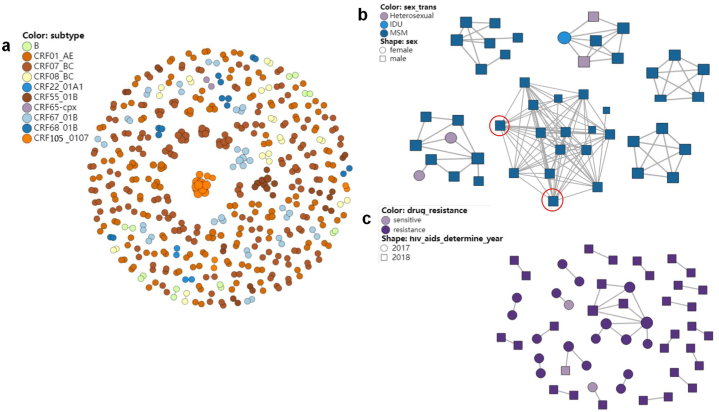
We first present a complete genetic network of the sequences subtype from 2017 to 2018 (Fig. 2a). In this genetic network, the sequences of subtype CRF105_0107 constructed three clusters, one of which was the largest cluster in the network with 16 nodes. The subtype of the second largest cluster with 9 nodes in the network was CRF55_01 B, and the sequences of subtype CRF55_01 B totally constructed 10 clusters. The subtype of the third largest cluster with 7 nodes was CRF67_01 B, and the sequences of subtype CRF67_01 B constructed 15 clusters in this network. HIV-1 subtypes in these three big clusters were all emerging subtypes in Jiangsu. The subtype with the most numerous clusters in the network was CRF07_BC (75 clusters), followed by CRF01_AE (72 clusters). The transmission route presented in the genetic network is an important way to identify individuals at high risk of HIV transmission. In this study, We highlighted the clusters with numbers of nodes in top 6 in the network (Fig. 2b). Among these 6 clusters, 4 clusters exhibited the route of MMSC, and one of them is the largest cluster in the network, with two nodes each containing 14 links (marked in the red circles in Fig. 2b). The number of links was defined as nodal degree. The other two clusters contained heterosexual and injection drug user (IDU) transmissions. In the cluster with 9 nodes composed of heterosexual and homosexual transmission routes, one heterosexual female had one degree, and the other had two degrees. Moreover, in the cluster consisting of 6 nodes, composed of heterosexual, IDU and homosexual transmission routes, the two heterosexual males had six degrees (four for one and two for the other) and both connected to the IDU female. The IDU female had five degrees connected to the heterosexuals and MMSC males. Drug resistance is another element for assessing the cluster risk because of its declining effect on ART. In the network, there were 52 DR nodes. 8 sequences exhibited resistance to nucleotide reverse transcriptase (NRTI) and 45 sequences showed resistance to non-nucleotide reverse transcriptase (NNRTI). The mutations in NRTI region were M41L, S68G, A62V, D67 N and K219E, accounting for 3.85 % (2/52) each. The mutations in NNRTI region were much more. The most numerous mutation was V179E (27/52), followed by V106I (6/52), and the third was V179D (4/52). The other mutations were V179T (3/52), Y181C (3/52), K103 N (3/52), E138G (2/52), H221Y (2/52), respectively. Through the network, we found a cluster with all 9 CRF07_BC resistance nodes from the 2017 and 2018 samples, and the mutation in the cluster was V179E. Another cluster with 3 nodes contained two resistant nodes from the 2017 samples and one sensitive node from the 2018 samples, respectively. Additionally, the other 20 resistant transmission clusters occurred in pairs (Fig. 2c).
Fig. 2.
The HIV-1 genetic transmission network. 2a: The subtype clusters exhibition in HIV-1 genetic transmission network. 2 b: The exhibition of top six clusters in number of nodes through transmission route. The nodes with the most of 14° were red circled. 2c: The exhibition of clusters consisted of resistance mutations.
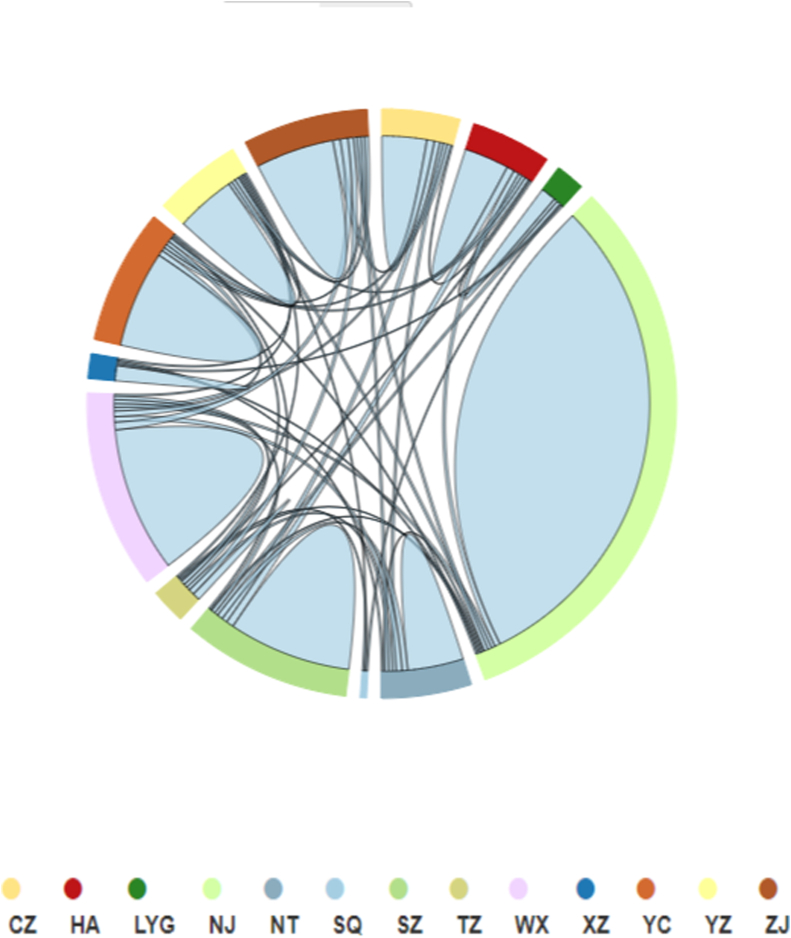
The distributions of infections can also reflect the activity level in HIV-1 transmission. We showed the 13 cities in Jiangsu with their attributions in the network (Fig. 3). The connections between cities had a typical scheme. We found that the infections in cities mainly connected within cities. For example, 95 % (268/282) in Nanjing were mainly connected within Nanjing. 80 % (66/82) of those in Suzhou frequently interacted within Suzhou. Those infected with HIV in Nanjing, Suzhou and Nantong interacted with those in numerous cities because of the convenient transportation by the high-speed railway. Cities in north Jiangsu, like Xuzhou, Taizhou, Lianyungang, and Suqian, were highly associated with those in south Jiangsu. Thus, these results located cities with key risk populations.
Fig. 3.
The distributions and associations of Jiangsu cities in the HIV-1 genetic transmission network.
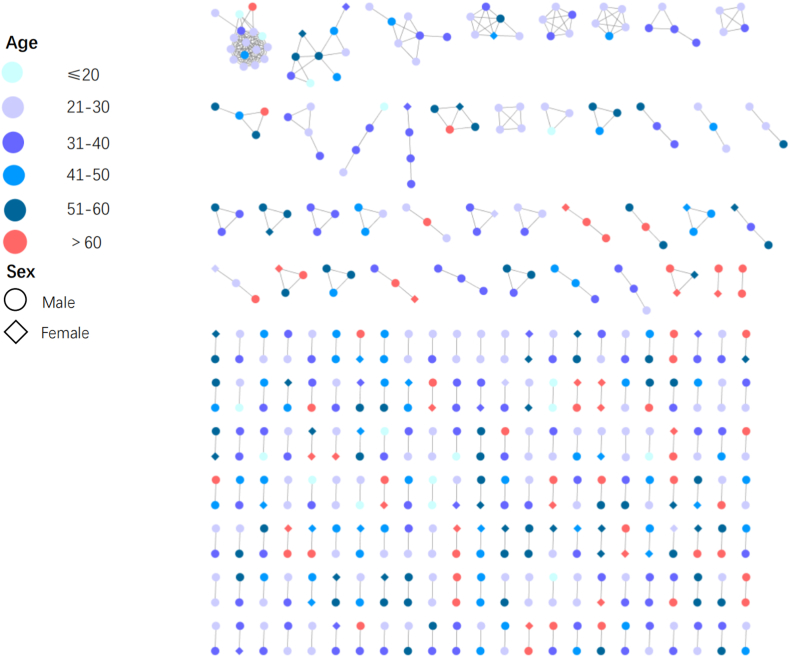
Older infections were becoming more and more in Jiangsu, and we constructed an age interval network using the Cytoscape software to visually present the transmission population based on age (Fig. 4). There were 59 nodes whose age was over 60 years in the genetic transmission network, among which 43 nodes were males, and 16 nodes were females. Of the 39 clusters with at least 3 nodes, only 10 clusters contained people over 60 years old, and the total number of people over 60 years old was only 15. The other nodes of individuals over 60 years old all showed in pairwise clusters. According to individual information, among 59 nodes, 47 nodes were involved in the heterosexual transmission, 11 were engaged in MMSC, and 1 was unknown but showed a pairwise connection with another MMSC. Moreover, in the transmission network, 21 males whose age was over 60 years connected with females, of which 14 were individuals over 60 years old. Two females had odd pairwise clusters in the network.
Fig. 4.
Clustering of different age groups in the HIV-1 genetic transmission network.
3.4. The analysis of sequences included in the genetic transmission network
To find the infection characteristics contributing to inclusion into the network, we compared the risk of variables included in the network with those not included (Table 1). We identified significant differences in age, transmission route, HIV-1 subtype, DR profiles, and CD4+ T cell count. Interestingly, individuals aged 21–40 and 41–60 were less likely to be incorporated into the network compared with individuals over 60 years old. Females were more likely to be included in the network, but had no significant difference. Unlike the sensitive viral hosts, resistant hosts had a higher risk of inclusion into the network. Compared with CRF01_AE, the CRF55_01 B and CRF67_01 B subtypes had a higher risk of evolving in the network. The pairwise comparison analysis showed that the difference in each CD4 count group was significant. We also found that a high CD4 count played a key role in inclusion in the network, as opposite to its protective role in HIV-1 infection immunity.
Table 1.
The risk variables with statistical significance between the included and non-included groups in the HIV-1 transmission network.
| Variables | Total number of cases (n = 3277) | Number of cases in the network | Rate of cases in the network% | One-factor analysis |
Multi-factors logistic analysis |
||
|---|---|---|---|---|---|---|---|
| OR (95%CI) | P Value | OR (95%CI) | P Value | ||||
| Gender | |||||||
| Male | 2876 | 409 | 14.2 | 1 | Ref | Ref | |
| Female | 401 | 67 | 16.7 | 1.21 (0.912–1.605) | 0.186* | 1.279 (0.942–1.716) | 0.108 |
| Age | Ref | Ref | |||||
| ≤20 | 152 | 29 | 19.1 | 1.015 (0.617–1.670) | 0.954 | 0.854 (0.503–1.428) | 0.552 |
| 21–40 | 1736 | 249 | 14.3 | 0.721 (0.523–0.993) | 0.045* | 0.637 (0.457–0.900) | 0.009# |
| 41–60 | 1092 | 142 | 13.0 | 0.643 (0.458–0.904) | 0.011* | 0.594 (0.421–0.846) | 0.003# |
| ≥61 | 297 | 56 | 18.9 | 1 | 1 | ||
| Ethnicity | |||||||
| Han | 3209 | 470 | 14.6 | 1 | Ref | ||
| Others | 68 | 6 | 8.8 | 0.564 (0.243–1.311) | 0.183* | ||
| Educational degree | |||||||
| Junior high school and below | 1491 | 205 | 13.7 | 1 | Ref | ||
| High school or technical secondary school | 790 | 114 | 14.4 | 1.058 (0.826–1.355) | 0.655 | ||
| college and above | 996 | 157 | 15.8 | 1.174 (0.937–1.471) | 0.163* | ||
| Marital status | |||||||
| Unmarried | 1337 | 197 | 14.7 | 1 | Ref | ||
| Married with spouse | 1474 | 215 | 14.6 | 0.988 (0.802–1.218) | 0.912 | ||
| Divorced or widowed | 466 | 64 | 13.7 | 0.921 (0.68–1.249) | 0.597 | ||
| Occupation | |||||||
| Clerk | 232 | 41 | 17.7 | 1 | Ref | ||
| Commercial service | 730 | 99 | 13.6 | 0.731 (0.491–1.089) | 0.123* | ||
| Worker | 545 | 89 | 16.3 | 0.909 (0.605–1.366) | 0.647 | ||
| Housekeeping and houseworker | 562 | 69 | 12.3 | 0.652 (0.428–0.993) | 0.046* | ||
| Farmer | 630 | 95 | 15.1 | 0.827 (0.554–1.236) | 0.355 | ||
| Retired | 113 | 17 | 15.0 | 0.825 (0.445–1.528) | 0.541 | ||
| Student | 172 | 33 | 19.2 | 1.106 (0.666–1.838) | 0.697 | ||
| Others or unknown | 293 | 33 | 11.3 | 0.591 (0.36–0.97) | 0.037* | ||
| Transmission route | |||||||
| Homosexual transmission | 1981 | 311 | 15.7 | 1 | Ref | 1 | Ref |
| Heterosexual transmission | 1263 | 164 | 13.0 | 0.801 (0.654–0.983) | 0.033* | 0.809 (0.654–0.997) | 0.005# |
| IDU and others | 33 | 1 | 3.0 | 0.168 (0.023–1.229) | 0.079* | 0.156 (0.009–0.740) | 0.069 |
| Institutions where patients being discovered | |||||||
| Institution of blood donation and donor | 111 | 15 | 13.5 | 1 | Ref | ||
| Dermatosis and STD clinics | 386 | 62 | 16.1 | 1.225 (0.667–2.25) | 0.514 | ||
| Medical examinations and others | 127 | 15 | 11.8 | 0.857 (0.399–1.844) | 0.693 | ||
| VCT | 1412 | 227 | 16.1 | 1.226 (0.699–2.151) | 0.478 | ||
| Hospitals and medical organizations | 1241 | 157 | 12.7 | 0.927 (0.525–1.638) | 0.794 | ||
| HIV genotype | |||||||
| CRF01_AE | 1399 | 165 | 11.8 | 1 | Ref | 1 | Ref |
| CRF07_BC | 1018 | 170 | 16.7 | 1.499 (1.190–1.890) | 0.001* | 1.365 (1.078–1.730) | 0.009# |
| CRF08_BC | 177 | 20 | 11.3 | 0.953 (0.582–1.560) | 0.847 | 0.914 (0.534–1.494) | 0.731 |
| CRF55_01 B | 100 | 27 | 27.0 | 2.766 (1.728–4.428) | <0.001* | 3.061 (1.852–4.947) | <0.001# |
| B | 127 | 10 | 7.9 | 0.639 (0.329–1.244) | 0.187* | 0.669 (0.321–1.252) | 0.242 |
| CRF67_01 B | 171 | 37 | 21.6 | 2.065 (1.386–3.076) | <0.001* | 2.083 (1.374–3.101) | <0.001# |
| CRF68_01 B | 90 | 15 | 16.7 | 1.496 (0.839–2.665) | 0.172* | 1.559 (0.839–2.727) | 0.137 |
| CRF105_0107 | 54 | 18 | 34.0 | 3.739 (2.076–6.737) | <0.001* | 3.650 (1.966–6.567) | <0.001# |
| Others | 141 | 14 | 9.9 | 0.824 (0.464–1.466) | 0.511 | 0.807 (0.433–1.397) | 0.469 |
| DR or not | |||||||
| Not resistant | 2950 | 443 | 15.0 | 1 | Ref | 1 | Ref |
| Resistance | 327 | 33 | 10.1 | 0.635 (0.437–0.923) | 0.017* | 0.649 (0.432–0.947) | 0.03# |
| CD4+T cell count | |||||||
| ≤200 | 746 | 60 | 8.0 | 1 | Ref | 1 | Ref |
| 201–350 | 1100 | 181 | 16.5 | 2.252 (1.654–3.065) | <0.001* | 2.150 (1.577–2.965) | <0.001# |
| 351–500 | 773 | 142 | 18.4 | 2.573 (1.867–3.545) | <0.001* | 2.382 (1.720–3.333) | <0.001# |
| >501 | 658 | 93 | 14.1 | 1.882 (1.335–2.652) | <0.001* | 1.827 (1.283–2.618) | <0.001# |
Note: Sequences with any missing variable information were excluded from the statistical analysis, and total number was 3277.
1. *: P < 0.2, values acceptable for multi-factors logistic analysis.
2. #: P < 0.05, statistically significant values.
3DR: drug resistance; STD: sexual transmitted disease; VCT: voluntary counseling and testing.
4. Discussion
HIV-1 genetic transmission network is usually used to demonstrate characteristics of a specific colony, like MMSC or heterosexual infections [13,14]. However, to fully and deeply analyze the genetic transmission network, we integrated all sequences of the new diagnoses from our database and identified the key populations in the network. Previous studies on the genetic transmission network mostly prioritized the largest clusters or clusters that recently enlarged to generate more nodes for public health surveillance [15,16]. However, the risk factors considered in our study were more comprehensive. We prioritized the HIV subtype, transmission route, drug resistance, geographical distribution, and age interval. Each factor can identify a specific risk population.
We chose 0.005 gene distance as a threshold in this study to reflect the probable transmission pattern in the last three years [17]. CRF01_AE is the principal circulating strain in Jiangsu province, accounting for 42.3 % [18]. However, the strain did not form the largest cluster; instead, CRF105_0107, CRF55_01 B, and CRF67_01 B formed the most active clusters with attracting nodes from 2017 to 2018. It suggested that the epidemic of new subtypes should be monitored and considered as a key risk factor, regardless of the cluster size or number. CRF105_0107 was previously identified as a new recombination strain emerging among MMSC in Jiangsu [19]. Similarly, CRF55_01 B and CRF67_01 B also emerged from MMSC [20,21]. However, our results indicated that there were also females in these recombinant type clusters. That is means that these females served as the transmission bridge from the MMSC population to other populations. In our study, CRF105_0107, CRF55_01 B, CRF67_01 B, and CRF07_BC were risk variables instead of CRF01_AE, the primary circulating strain in Jiangsu. All these four subtypes are the recombinants of the B subtype [22,23]. Thus, the gene distance was based on the B subtype. However, this raised doubts about whether the gene distance was appropriate for the HIV-1 genetic transmission network study in CRF01_AE or other subtypes except for the B subtype. Thus, further research is needed to explore the gene distance suitable for different subtypes in the genetic transmission network.
The clusters containing multi-transmission routes were considered a risk priority compared to the largest clusters. In most studies focusing on MMSC, a high-risk behavior population, the group had a relatively constant behavior pattern [24,25]. Multi-transmission clusters portrayed complicated sex patterns, implying a higher risk of transmitting the virus to other sexual groups [[26], [27], [28]]. For example, an IDU female had 5 links to other males in the MMSC category, suggesting heterosexual transmission evolving in MMSC relationship. The IDUs may fall under the sexual transmission route if they abuse drugs that stimulate high-frequency sexual behavior without protection. This risk population can only be identified by constructing the HIV-1 genetic transmission network. Compared to MMSC, other sexual populations are not likely to test for HIV-1, and thus have no idea about their HIV status and could be potential viral transmitters. These populations may be the base of the iceberg, with a large number but hard to identify. In our genetic transmission network, the IDU female connected to two males who described themselves infected through heterosexual transmission. It implied that she maybe a sex worker or one of the five men maybe an undisclosed IDU. In any case, an in-depth investigation and intervention on this female would be beneficial in curbing the spread of HIV. If she was a sex worker, the benefits were even more obvious. A review revealed that in low-prevalence epidemics (≤5 % HIV prevalence), the estimated impact of sex-worker interventions included: 6–100 % infections averted; 0.9–6.2 HIV infections averted per 100,000 adults; 11–94 % and 4–47 % relative reduction in prevalence and incidence, respectively [29]. So even though they were not included in the bigger clusters, they could still represent a major risk factor. To sum up, female infections in the MMSC clusters with more than one edge implied that the individuals were probably contributing to HIV-1 transmission and should be precision intervened. However, the transmission effects played by female infections were probably ignored due to not appearing in the results revealed in genetic transmission network.
The UNAIDS established 2025 Global AIDS strategy targets of “95%-95%–95 %" [20], proposed the “U U″ view for HIV prevention, and emphasized the vital role of ART success [21,30]. In our study, we found a cluster with drug resistant sites from 2017 to 2018, indicating a steady transmission and a threat to the success of ART. The other DR nodes had a pairwise connection, and the nodes with resistant sites did not form bigger clusters; however, they still represent the crucial populations determining the success of ART. Therefore, these individuals should be paid more attention in ART.
The network showed that the interconnections among the cities of south Jiangsu formed large clusters. However, cities of north Jiangsu interacted with themselves through one-to-one connection, which could also evolve into big clusters when connected to cities of south Jiangsu. Therefore, the city with higher infection cases can be selected for risk behavior analysis. These results suggest that conducting transmission studies in these risk cities can identify infection rates or new potential infections.
The HIV-1 prevalence among the individuals over 60 years old is rising yearly, but only a few literatures are reported. This is probably due to the less attention paid to the individuals over 60 years old because of the degeneration of their health and low sexual activity [31]. Interestingly, individuals over 60 years old easily formed networks in our study compared with the other age groups. However, all individuals over 60 years old only formed pairwise or triple clusters, indicating their low mobility in HIV-1 transmission. We observed two female-female pairwise cluster among the individuals over 60 years old in the network, indicating that several non-reported male infections could connect to the clusters and may be form a big HIV-1 genetic transmission network because female infections over 60 years old were almost infected through sexual transmission between spouses. As reported, old adults are less likely to use protective measures such as condoms and have never had an HIV test [32,33]. These factors make them easy to infect HIV-1. Among persons living with HIV, it is estimated that more than half will be aged 50 years or older in the near future. In high prevalence areas, HIV infection has been present in up to 5 % of persons who died older than 60 years [34]. Therefore, any individual over 60 years old in the genetic transmission network should not be ignored. Any cluster connection, even one-to-one, could provide a clue for identifying possible infections and further preventing subsequent transmissions. Accordingly, harmful education and enlarging HIV-1 testing based on the genetic transmission network should be implemented among older adults to ensure timely diagnosis.
CD4 count is a key index to determine the immune level of HIV-infected individuals. And so far, CD4 counts have also been used to assess late presentation. HIV-infected individuals with CD4 counts below 350 cells/μL are considered as late presenters. In our study, most infections had blood taken immediately after diagnosis, and the median CD4 count was 320 cells/μL, meaning an unoptimistic immune profile and late HIV presentation status. Combining with the age concentrated between 20 and 60 years old, the newly diagnosed infections were considered as sexually active population, so the risk of transmission by these infections is higher and it is easier for them to enter the transmission network.
4.1. Limitations of the study
This study was an original research of HIV-1 genetic transmission network. All results were only from laboratory sequences and theoretical analysis. The genetic transmission network role in precision intervention on high-risk populations was still deficiency. This precision intervention is carrying out and the effects on stopping the spread of HIV are still waiting to see.
5. Conclusion
Our study elaborated on the characteristics of the HIV-1 genetic transmission network in Jiangsu province. Clusters containing drug resistant nodes and individuals over 60 years old should be intervened, whatever the sizes of clusters nor the numbers of nodes in clusters. CRF01_AE may be ignored in the genetic transmission network based on gene distance <0.005 compared with CRF105_0107, CRF55_01 B, and CRF67_01 B. Besides factors above, we still should pay more attention to high-risk population with active sexual behavior in Jiangsu. Furthermore, identification, testing, and treatment interventions for the key population in the transmission network could help achieve the goal of 95%-95%–95 % from the World Health Organization and push to end AIDS epidemic.
Data availability statement
Data associated with the study has not been deposited into a publicly available repository and data will be made available on request.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics declaration
Informed consent was not required for this study because all personal information were obtained from the National HIV/AIDS Surveillance System.
Additional information
No additional information is available for this paper.
CRediT authorship contribution statement
Ying Zhou: Writing - review & editing, Writing - original draft, Formal analysis, Data curation, Conceptualization. Jing Lu: Writing - original draft, Methodology, Investigation, Formal analysis, Data curation. Zhi Zhang: Writing - original draft, Methodology, Investigation. Qi Sun: Software, Investigation. Xiaoqin Xu: Visualization, Validation, Project administration. Haiyang Hu: Writing - review & editing, Writing - original draft, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Ying Zhou, Email: zhouy@jscdc.cn.
Jing Lu, Email: lujing@jscdc.cn.
Zhi Zhang, Email: zhangzhi@jscdc.cn.
Qi Sun, Email: sq_njmu@163.com.
Xiaoqin Xu, Email: xqxu18@sina.com.
Haiyang Hu, Email: huhaiyang@jscdc.cn.
References
- 1.National Health Commission of the People's Republic of China . 2021. Overview of Notifiable Infectious Diseases Nationwide. [Google Scholar]
- 2.China Central Television, On World AIDS Day . 2022. China Reported 1.14 Million People Living with AIDS. [Google Scholar]
- 3.Jiangsu Commission of Health . 2022. The Work Progress of HIV Prevention and Control in 2022. [Google Scholar]
- 4.Han X., Zhao B., An M., Zhong P., Shang H. Molecular network-based intervention brings us closer to ending the HIV pandemic. Front. Med. 2020;14:136–148. doi: 10.1007/s11684-020-0756-y. [DOI] [PubMed] [Google Scholar]
- 5.Ma N., Chen X.H., Zhao Y., Kang X., Pan S., Yao W.Q. HIV-1 molecular transmission network among sexually transmitted populations in liaoning province, China. Medicine (Baltim.) 2021;100 doi: 10.1097/MD.0000000000026640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ragonnet-Cronin M., Hu Y.W., Morris S.R., Sheng Z., Poortinga K., Wertheim J.O. HIV transmission networks among transgender women in Los Angeles county, Ca, USA: a phylogenetic analysis of surveillance data. Lancet HIV. 2019;6:e164–e172. doi: 10.1016/S2352-3018(18)30359-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikolopoulos G.K., Pavlitina E., Muth S.Q., Schneider J., Psichogiou M., Williams L.D., Paraskevis D., Sypsa V., Magiorkinis G., Smyrnov P., Korobchuk A., Vasylyeva T.I., Skaathun B., Malliori M., Kafetzopoulos E., Hatzakis A., Friedman S.R. A network intervention that locates and intervenes with recently HIV-infected persons: the transmission reduction intervention Project (trip) Sci. Rep. 2016;6 doi: 10.1038/srep38100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasquale D.K., Doherty I.A., Miller W.C., Leone P.A., Sampson L.A., Ledford S.L., Sebastian J., Dennis A.M. Factors associated with human immunodeficiency virus infections linked in genetic clusters but disconnected in partner tracing. Sex. Transm. Dis. 2020;47:80–87. doi: 10.1097/OLQ.0000000000001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gan M.Z., Feng Y., Xing H. [Progress in research on the transmission characteristics of HIV-infected persons based on molecular network method] Chin. J. Epidemiol. 2019;40:1487–1491. doi: 10.3760/cma.j.issn.0254-6450.2019.11.026. [DOI] [PubMed] [Google Scholar]
- 10.Yin Y., Zhou Y., Lu J., Liu X., Shi L., Fu G., Huan X., Fang S., Liu Y., Wang B., Ding J. Molecular epidemiology of HIV-1 in Jiangsu province, southeast China: genotypes and hiv-1 transmission networks among newly diagnosed men having sex with men in 2017. AIDS Res. Hum. Retrovir. 2021;37:62–69. doi: 10.1089/AID.2020.0165. [DOI] [PubMed] [Google Scholar]
- 11.Hu H., Liu X., Zhang Z., Xu X., Shi L., Fu G., Huan X., Zhou Y. Increasing HIV incidence among men who have sex with men in Jiangsu province, China: results from five consecutive surveys, 2011-2015. Int. J. Environ. Res. Publ. Health. 2016;13:795. doi: 10.3390/ijerph13080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y., Lu J., Wang J., Yan H., Li J., Xu X., Zhang Z., Qiu T., Ding P., Fu G., Huan X., Hu H. Prevalence of HIV antiretroviral drug resistance and its impacts on hiv-1 virological failures in Jiangsu, China: a cross-sectional study. BioMed Res. Int. 2016;2016 doi: 10.1155/2016/1752437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pines H.A., Wertheim J.O., Liu L., Garfein R.S., Little S.J., Karris M.Y. Concurrency and HIV transmission network characteristics among MSM with recent HIV infection. AIDS. 2016;30:2875–2883. doi: 10.1097/QAD.0000000000001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mann L.M., Llata E., Flagg E.W., Hong J., Asbel L., Carlos-Henderson J., Kerani R.P., Kohn R., Pathela P., Schumacher C., Torrone E.A. Trends in the prevalence of anogenital warts among patients at sexually transmitted disease clinics-sexually transmitted disease surveillance network, United States, 2010-2016. J. Infect. Dis. 2019;219:1389–1397. doi: 10.1093/infdis/jiy684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wertheim J.O., Murrell B., Mehta S.R., Forgione L.A., Kosakovsky Pond S.L., Smith D.M., Torian L.V. Growth of hiv-1 molecular transmission clusters in New York city. J. Infect. Dis. 2018;218:1943–1953. doi: 10.1093/infdis/jiy431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragonnet-Cronin M., Hayford C., D'Aquila R., Ma F., Ward C., Benbow N., Wertheim J.O. Forecasting hiv-1 genetic cluster growth in Illinois, United States. J. Acquir. Immune Defic. Syndr. 2022;89:49–55. doi: 10.1097/QAI.0000000000002821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Center for AIDS/STD Control and Prevention . 2021. Technical Guide for HIV Transmission Networks Monitoring and Intervention. [Google Scholar]
- 18.Li W., Chua J.J., Wei F., Hea Y., Dong X.X., Gea Y., Jia Y., Musaa T.H., Cao S., Ni Q., Wei P.M., Li X.S. Molecular characteristic of HIV-1 CRF01_AE in nanjing from 2015 to 2017. Infect. Genet. Evol. 2019;75 doi: 10.1016/j.meegid.2019.104038. [DOI] [PubMed] [Google Scholar]
- 19.Yin Y., Zhou Y., Lu J., Guo H., Chen J., Xuan Y., Yuan D., Hu H., Xu X., Fu G., Wang B. First detection of a cluster novel hiv-1 second-generation recombinant (CRF01_AE/CRF07_BC) among men who have sex with men in nanjing, eastern China. Intervirology. 2021;64:81–87. doi: 10.1159/000512135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.UNAIDS, 2025 AIDS TARGETS. Available from:: http://aidstargets2025.unaids.org.
- 21.Smith P., Buttenheim A., Schmucker L., Bekker L.G., Thirumurthy H., Davey D.L.J. Undetectable = untransmittable (U = U) messaging increases uptake of HIV testing among men: results from a pilot cluster randomized trial. AIDS Behav. 2021;25:3128–3136. doi: 10.1007/s10461-021-03284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han X., An M., Zhang W., Cai W., Chen X., Takebe Y., Shang H. Genome sequences of a novel hiv-1 circulating recombinant form, CRF55_01b, identified in China. Genome Announc. 2013;1 doi: 10.1128/genomeA.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosaka M., Fujisaki S., Masakane A., Hattori J., Shiino T., Gatanaga H., Shigemi U., Okazaki R., Hachiya A., Matsuda M., Ibe S., Iwatani Y., Yokomaku Y., Sugiura W. H. I. V. Surveillance network team Japanese drug resistance, HIV-1 CRF01_AE and subtype B transmission networks crossover: a new AE/B recombinant identified in Japan. AIDS Res. Hum. Retrovir. 2016;32:412–419. doi: 10.1089/aid.2015.0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonett S. Networks among racial and ethnic minority men who have sex with men in HIV research in the United States: a concept analysis. J. Assoc. Nurses AIDS Care. 2020;31:379–391. doi: 10.1097/JNC.0000000000000160. [DOI] [PubMed] [Google Scholar]
- 25.Jing F., Zhang Q., Tang W., Wang J.Z., Lau J.T., Li X. Reconstructing the social network of HIV key populations from locally observed information. AIDS Care. 2021:1–8. doi: 10.1080/09540121.2021.1883514. [DOI] [PubMed] [Google Scholar]
- 26.Chow E.P., Wilson D.P., Zhang L. What is the potential for bisexual men in China to act as a bridge of HIV transmission to the female population? Behavioral evidence from a systematic review and meta-analysis. BMC Infect. Dis. 2011;11:242. doi: 10.1186/1471-2334-11-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernando D. Men who have sex with men and women (msmw), biphobia and the CDC: a bridge ignored? Prev. Med. 2017;105:368–369. doi: 10.1016/j.ypmed.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Ito H., Yamamoto T., Morita S. The effect of men who have sex with men (MSM) on the spread of sexually transmitted infections. Theor. Biol. Med. Model. 2021;18:18. doi: 10.1186/s12976-021-00148-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra S., Steen R., Gerbase A., Lo Y.R., Boily M.C. Impact of high-risk sex and focused interventions in heterosexual HIV epidemics: a systematic review of mathematical models. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auid-Orcid Gupta N., Gilleece Y., Orkin C. Implementing U=U in clinical practice: results of a British HIV association members survey. Sex. Transm. Infect. 2021;97:619–620. doi: 10.1136/sextrans-2020-054462. [DOI] [PubMed] [Google Scholar]
- 31.Olalla J., Garcia de Lomas J.M., Marquez E., Gonzalez F.J., Del Arco A., De La Torre J., Prada J.L., Cantudo F., Martin M.D., Nieto M., Perez Stachowski J., Garcia-Alegria J. Experience of using an app in HIV patients older than 60 Years: pilot program. JMIR Mhealth Uhealth. 2019;7 doi: 10.2196/mhealth.9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan D., Yao Y.M., Zheng M., Wang F.L., Sun J.Y., Xiong R., Han M.J., Hu Y.Y. [a survey on sexual needs and factors of HIV risky sexual behaviors among elderly men at different ages in two communities of qiandongnan miao and dong autonomous prefecture] Zhonghua Yufang Yixue Zazhi. 2020;54:1227–1231. doi: 10.3760/cma.j.cn112150-20200113-00018. [DOI] [PubMed] [Google Scholar]
- 33.Lieberman R. HIV in older Americans: an epidemiologic perspective. J. Midwifery Wom. Health. 2000;45:176–182. doi: 10.1016/s1526-9523(00)00002-7. [DOI] [PubMed] [Google Scholar]
- 34.Brooks J.T., Buchacz K., Gebo K.A., Mermin J. HIV infection and older Americans: the public health perspective. Am. J. Publ. Health. 2012;102:1516–1526. doi: 10.2105/AJPH.2012.300844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with the study has not been deposited into a publicly available repository and data will be made available on request.