Abstract
The staphylococcal qacB-encoding multidrug resistance plasmid pSK156, isolated from a clinical strain dating from 1951, was characterized. Comparison of the regions flanking qacB with other qacA- and qacB-encoding plasmids provided insights into the evolution and dissemination of these multidrug efflux genes and led to the detection of the earliest known copy of the insertion sequence IS257.
The antiseptic resistance gene qacA from the opportunistic pathogen Staphylococcus aureus encodes one of the best-characterized bacterial multidrug efflux proteins (10). qacA confers resistance to monovalent and divalent organic cations via a proton motive force-dependent efflux mechanism, while the closely related qacB determinant confers resistance primarily to monovalent organic cations (7). Molecular comparisons of qacA and qacB have indicated that their nucleotide sequences differ by only 7 bp, and mutagenesis studies have determined that a single amino acid substitution is responsible for their different phenotypes (9). qacA and qacB are both found in association with a divergently encoded gene, qacR, whose product has been purified and demonstrated to act as a trans-acting repressor of qacA expression (5).
The qacA and qacB genes have typically been found to be plasmid encoded in S. aureus strains, although the relationships between these plasmids remain unclear. qacB has been characterized on β-lactamase and heavy-metal resistance plasmids, such as pSK23, isolated from strains dating from the early 1980s (7). qacA was first located on plasmids also dating from the early 1980s, such as the β-lactamase and heavy-metal resistance plasmid pSK57 (4), and it has more frequently been found on pSK1 family plasmids in epidemic staphylococcal strains from Australia and the United Kingdom since 1980 (8). Additionally, qacB has been reported to be plasmid encoded in clinical S. aureus strains isolated in the early 1950s (7). This paper describes a qacB-encoding plasmid, pSK156, present in a clinical strain isolated in 1951 from a patient of the Alfred Hospital, Melbourne, Australia, which represents the earliest known multidrug efflux-encoding plasmid.
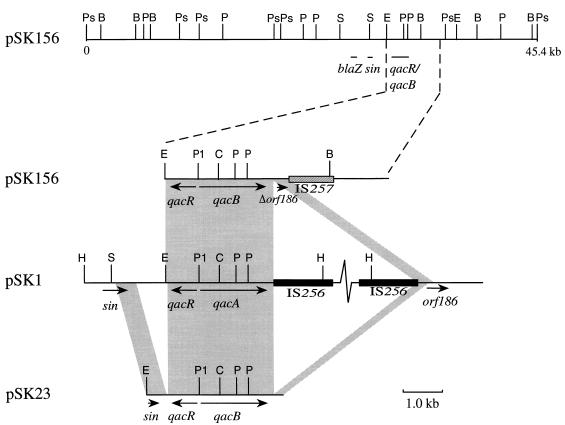
The qacB-encoding plasmid pSK156 is 45.4 kb in size and additionally encodes penicillin resistance (7). A restriction endonuclease map of pSK156 was constructed (Fig. 1), and qacB and the penicillin resistance gene blaZ were localized by Southern hybridization (15), using a 0.33-kb PvuII fragment from pSK1 (13) and a 0.7-kb EcoRV fragment from pSK4 (3) as qacA/B- and blaZ-specific probes, respectively. Comparison of pSK156 with the restriction maps of the plasmids pSK1 and pSK23 (8) revealed no obvious restriction map similarity between these plasmids outside of the qacA- and qacB-encoding regions.
FIG. 1.
Physical and genetic map of the multidrug resistance plasmid pSK156. The locations of genes involved in the following functions are represented: penicillin resistance (blaZ), multidrug resistance (qacR and qacB), and site-specific recombination (sin). Restriction sites are abbreviated as follows: B (BglII), E (EcoRI), Ps (PstI), P (PvuII), and S (SalI); only selected EcoRI sites are displayed. Beneath the pSK156 map is a diagrammatic comparison of the sequenced regions of pSK156 and pSK23 with the homologous regions of pSK1; a kilobase scale is indicated. Using a series of synthetic oligonucleotides, the nucleotide sequences of pSK23 and pSK156 were determined on both strands. The nucleotide sequence of pSK1 was determined previously (3, 15, 17). The shaded regions represent areas with high degrees of sequence similarity (>85% identity). qacA, qacB, qacR, sin, orf186, and Δorf186 (a truncated copy of orf186) are represented by arrows, with the orientation of the arrow indicating the direction of transcription. The insertion sequences IS256 and IS257 are represented by solid and hatched boxes, respectively. Additional restriction sites shown are abbreviated as follows: C (ClaI), H (HindIII), and P1 (PvuI).
The qacB gene from pSK156 was cloned on a 6.3-kb EcoRI fragment (Fig. 1) which was inserted into the corresponding site of the Escherichia coli vector pUC18 (17). MIC analyses and ethidium bromide transport assays were performed as described by Littlejohn et al. (7), and the results indicated that qacB cloned from pSK156 displayed a phenotype identical to that of the previously cloned qacB gene from pSK23 (9).
Nucleotide sequencing of the cloned qacB and qacR genes from pSK156 and of the regions flanking qacB on both pSK23 and pSK156 was undertaken to investigate the evolutionary relationship between these temporally distinct plasmids. The qacB and qacR genes from pSK156 proved to be identical to their counterparts from pSK23, with the exception of a single nucleotide substitution in qacB at codon 184 (GTG [Val] in pSK156→GCG [Ala] in pSK23). As can be seen in Fig. 1, the sequences of the regions flanking qacB on pSK23 and pSK156 are identical to each other and to the sequences flanking qacA on pSK1, with the exceptions detailed below.
pSK1 contains an insertion of the aminoglycoside resistance transposon Tn4001 located 113 bp distal to qacA. Both pSK156 and pSK23 lack this transposon and possess only a single copy of the 8-bp direct repeats flanking Tn4001 on pSK1 (1). pSK156 contains a copy of the staphylococcal insertion sequence IS257, located 488 bp distal to qacB, which is not present in the corresponding region of pSK23 or pSK1. This IS257 element is not flanked by sequence duplications, and comparison with other sequenced IS257 elements indicated that it had four base pair changes relative to the most similar element, IS257L from Tn4003 on pSK1 (14). IS257 characteristically contains an internal BglII site; hence, the other five BglII sites on pSK156 potentially represent additional copies of this element. The sequence of pSK156 further 3′ of the IS257 element did not exhibit similarity to the sequence of the contiguous region on pSK1 (2) (Fig. 1). This region on pSK1 contained an open reading frame, orf186, coding for a 186-amino-acid product of unknown function (2). The copy of orf186 on pSK156 is truncated at codon 97, and the remainder of this open reading frame is not located further 3′ of the IS257 element. Preliminary DNA sequence data suggest that orf186 is also present at an equivalent position on pSK23.
IS257 elements are often associated with various resistance determinants, implicating them in the evolution of multiresistant S. aureus strains (11). In particular, IS257 is often found in association with the chromosomal methicillin resistance (mec) locus in S. aureus (16). The earliest-reported S. aureus isolates carrying mec date from 1960 (6), and it has not been previously determined whether the carriage of IS257 in S. aureus predates the acquisition of the mec region by this organism. The presence of IS257 on the plasmid pSK156, which dates from 1951, represents the earliest reported instance of IS257 in S. aureus.
As for the region left of the qacR gene, pSK1 and pSK156 are identical within the area sequenced (Fig. 1). The sequence of pSK23 diverged from those of pSK1 and pSK156 61 bp downstream of qacR. This region on pSK23 contains a palindromic repeat, of which only one-half is present in pSK1, which instead includes an additional 700-bp region encoding orf112, whose function is unknown. pSK1 contains the putative site-specific recombinase gene sin (12), and partial sequencing of pSK23 indicated that this plasmid also encodes sin (Fig. 1). The sin genes from these two plasmids are distinct, exhibiting 86% identity at the nucleotide level within the regions sequenced. Restriction mapping and PCR analysis using primers internal to and flanking the sin gene indicated that pSK156 also contains a copy of sin in a position equivalent to its location in pSK1.
Despite the apparent lack of similarity among the restriction maps of multidrug resistance plasmids pSK1, pSK23, and pSK156, sequencing of the regions flanking qacA or qacB on these plasmids revealed a high level of sequence similarity, ranging from 85 to 100% identity. There are two possible explanations for this: either the entire plasmids are related and have diverged from a common ancestor, or the region carrying qacA or qacB has acted as a mobile cartridge which has been horizontally transferred between different plasmids, presumably via homologous recombination between regions adjacent to qacA or qacB.
The similarity between the qacA and qacB antiseptic resistance genes, their respective dates of isolation, and the wider substrate specificity of the QacA protein compared with QacB support the conjecture that qacA has evolved from qacB. This is consistent with the proposal that the emergence of the qacA determinant among S. aureus clinical isolates during the 1980s resulted from the extensive use in hospital environments of divalent cations, such as chlorhexidine and pentamidine, to which qacA confers resistance.
Nucleotide sequence accession numbers.
The nucleotide sequences of pSK156 and pSK23 were submitted to GenBank and assigned accession no. AF053771 and AF053772, respectively.
Acknowledgments
We thank Libby Kerr for technical assistance.
This work was supported by a grant from the National Health and Medical Research Council (Australia). I.T.P. was the recipient of a C. J. Martin Fellowship from the National Health and Medical Research Council (Australia).
REFERENCES
- 1.Byrne M E, Rouch D A, Skurray R A. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene. 1989;81:361–367. doi: 10.1016/0378-1119(89)90197-2. [DOI] [PubMed] [Google Scholar]
- 2.Firth, N., S. Apisiridej, and R. A. Skurray. Unpublished data.
- 3.Gillespie M T, Lyon B R, Skurray R A. Structural and evolutionary relationships of β-lactamase transposons from Staphylococcus aureus. J Gen Microbiol. 1988;134:2857–2866. doi: 10.1099/00221287-134-11-2857. [DOI] [PubMed] [Google Scholar]
- 4.Gillespie M T, May J W, Skurray R A. Plasmid-encoded resistance to acriflavine and quaternary ammonium compounds in methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 1986;34:47–51. [Google Scholar]
- 5.Grkovic, S., M. H. Brown, N. J. Roberts, I. T. Paulsen, and R. A. Skurray. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem., in press. [DOI] [PubMed]
- 6.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 7.Littlejohn T G, Paulsen I T, Gillespie M T, Tennent J M, Midgley M, Jones I G, Purewal A S, Skurray R A. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol Lett. 1992;95:259–266. doi: 10.1016/0378-1097(92)90439-u. [DOI] [PubMed] [Google Scholar]
- 8.Lyon B R, Skurray R. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev. 1987;51:88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulsen I T, Brown M H, Littlejohn T G, Mitchell B A, Skurray R A. Molecular characterization of the multidrug resistance proteins QacA and QacB: membrane topology and identification of residues involved in specificity for divalent cations. Proc Natl Acad Sci USA. 1996;93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulsen I T, Firth N, Skurray R A. Resistance to antimicrobial agents other than β-lactams. In: Archer G L, Crossley K B, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 175–212. [Google Scholar]
- 12.Paulsen I T, Gillespie M T, Littlejohn T G, Hanvivatvong O, Rowland S J, Dyke K G, Skurray R A. Characterisation of sin, a potential recombinase-encoding gene from Staphylococcus aureus. Gene. 1994;141:109–114. doi: 10.1016/0378-1119(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 13.Rouch D A, Cram D S, DiBerardino D, Littlejohn T G, Skurray R A. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol. 1990;4:2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 14.Rouch D A, Messerotti L J, Loo L S, Jackson C A, Skurray R A. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol Microbiol. 1989;3:161–175. doi: 10.1111/j.1365-2958.1989.tb01805.x. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 16.Stewart P R, Dubin D T, Chikramane S G, Inglis B, Matthews P R, Poston S M. IS257 and small plasmid insertions in the mec region of the chromosome of Staphylococcus aureus. Plasmid. 1994;31:12–20. doi: 10.1006/plas.1994.1002. [DOI] [PubMed] [Google Scholar]
- 17.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]